Abstract
Odin has been implicated in the downstream signaling pathway of receptor tyrosine kinases, such as the epidermal growth factor and Eph receptors. However, the physiologically relevant function of Odin needs to be further determined. In this study, we used Odin heterozygous mice to analyze the Odin expression pattern; the targeted allele contained a β-geo gene trap vector inserted into the 14th intron of the Odin gene. Interestingly, we found that Odin was exclusively expressed in ependymal cells along the brain ventricles. In particular, Odin was highly expressed in the subcommissural organ, a small ependymal glandular tissue. However, we did not observe any morphological abnormalities in the brain ventricles or ependymal cells of Odin null-mutant mice. We also generated BAC transgenic mice that expressed the PTB-deleted Odin (dPTB) after a floxed GFP-STOP cassette was excised by tissue-specific Cre expression. Strikingly, Odin-dPTB expression played a causative role in the development of the hydrocephalic phenotype, primarily in the midbrain. In addition, Odin-dPTB expression disrupted proper development of the subcommissural organ and interfered with ependymal cell maturation in the cerebral aqueduct. Taken together, our findings strongly suggest that Odin plays a role in the differentiation of ependymal cells during early postnatal brain development.
Keywords: ependymal cells, hydrocephalus, Odin, subcommissural organ
INTRODUCTION
Odin is a member of the Anks1 adaptor protein family, which contains six ankyrin repeats (Ank), two sterile alpha motifs (SAM), and a phosphotyrosine binding domain (PTB) (Uhlik et al., 2005). Odin has been identified as one of the tyrosine phosphorylated proteins induced by activating epidermal growth factor (EGF) or platelet-derived growth factor receptor tyrosine kinases (Pandey et al., 2002). Odin has been implicated in negative regulation of the epidermal growth factor receptor (EGFR) signaling pathway (Kristiansen et al., 2004). Odin also has a role in EGFR recycling. For example, Odin level is correlated with the degree of increased EGF-induced EGFR trafficking to recycle endosomes and recycle back to the cell surface (Tong et al., 2013). In contrast, Odin is a key adaptor protein regulating the EphA receptor signaling pathway (Kim et al., 2010; Shin et al., 2007). In that study, Odin was critical for regulating EphA8-mediated cell migration and neurite outgrowth (Shin et al., 2007). In addition, the Odin SAM domain binds to ubiquitinated EphA receptors and regulates their protein stability (Kim et al., 2010). Interestingly, Odin forms a ternary complex with both EphA8 and RINL, a guanine nucleotide exchange factor for Rab5 subfamily proteins (Kajiho et al., 2012). Although RINL has been implicated in degradation of EphA8 via its interaction with Odin, whether this ternary complex has physiological relevance in vivo needs to be determined.
Ependymal cells are thin epithelial-like cells lining the brain ventricles and are considered a type of neuroglial cell, as they are derived from radial glial cells (Ihrie and Alvarez-Buylla, 2011; Lacar et al., 2010). Ependymal cells are cube-shaped and highly polarized cells with multiple cilia on their apical cell surface, which are a key regulators of normal cerebrospinal fluid (CSF) flow (Breunig et al., 2010). Motile cilia from a single ependymal cell beat coordinately in one direction, and the sum of the beats from multiple cells is thought to produce the stereotypical flow of ventricular CSF (Breunig et al., 2010). FoxJ1 is a key transcription factor involved in ciliogenesis (Yu et al., 2008). The FoxJ1 gene is expressed abundantly in multi-ciliated cells, including the airway lining the lung, the oviducts, and the ependymal cells lining the brain ventricles (Blatt et al., 1999; Whitsett and Tichelaar, 1999). Consistent with its specific expression in ependymal cells, FoxJ1 null-mutant mice display a hydrocephalic brain phenotype caused by a severe defect in the transcription of genes critical for producing motile ependymal cell cilia (Jacquet et al., 2009). Although ependymal cells do not self-renew, it appears that they act as a potential progenitor reservoir in the ventricular zone of the forebrain to produce new neurons (Carlen et al., 2009; Zhao et al., 2009). For example, when ependymal cells are activated by stroke, they are subsequently depleted as they become new neurons (Carlen et al., 2009). Another function of ependymal cells is to secrete hormones and glycoproteins into the brain ventricles (Cottrell and Ferguson, 2004). For example, the subcommissural organ (SCO) is a small ependymal gland located in the dorsocaudal region of the third ventricle and is highly conserved throughout the vertebrate phylum (Rodriguez et al., 1998). It secretes glycoproteins such as spondin, which forms the single fibrous Reissner’s fiber (RF), into the CSF (Rodriguez et al., 1998). RF runs along the aqueduct, the fourth ventricle, and the central canal (Picketts, 2006; Rodriguez et al., 1998). Although the role of the SCO has not been clearly defined, its potential function is to regulate CSF flow and maintain brain homeostasis (Huh et al., 2009; Picketts, 2006). Evidence suggests that abnormal development of the SCO leads to congenital hydrocephalus (Huh et al., 2009; Ortloff et al., 2013; Perez-Figares et al., 2001; Picketts, 2006). Interestingly, the Rfx or Msx gene is expressed in the SCO during postnatal brain development and their null-mutant mice have a hydrocephalic brain phenotype consistent with a size reduction or agenesis of the SCO (Baas et al., 2006; Huh et al., 2009; Ramos et al., 2004; Zhang et al., 2006).
In this study, we found that Odin is primarily expressed in ependymal cells lining the brain ventricles, such as the third ventricle and cerebral aqueduct. Ectopic expression of the PTB-deleted Odin protein in ependymal tissue was responsible for immature development of ependymal cells in the SCO and midbrain. Accordingly, a severe hydrocephalic phenotype developed in the midbrain of transgenic mice expressing Odin-dPTB. Therefore, we propose that Odin is critically involved in the development of ependymal cells during early postnatal brain development.
MATERIALS AND METHODS
Generation of BAC transgenic mice
Odin knockout mice and Odin-dPTB cDNA have been described previously (Kim et al., 2010; Shin et al., 2007). RP24-258K7, which includes homology arms A (824 bp) and B (530 bp) flanking the mouse Odin translation start site (ATG), were synthesized by polymerase chain reaction (PCR) using the following primers sets: 5′-TGCTCTTAACAGGGAACCACCT-3′ (forward primer for A arm), 5′-CCCACCACCGCCACCCCTCGG-3′ (reverse primer for A arm), 5′-GACCGAATTCGCGGAATCCCTCTCAC (forward primer for B arm), 5′-ATGCACTCTGACAGAGCAAT-3′ (reverse primer for B arm) to generate a targeting vector for inserting floxed green fluorescent protein (GFP) plus Odin-dPTB into Odin BAC. Next, homology arms A and B were inserted into a plasmid vector (pGEM11z) containing the frt-Kana-frt cassette. The floxed GFP expression cassette (with three SV40 polyadenylation sites) and the Odin-dPTB expression cassette were inserted into the vector described above. The resulting insert containing the A arm, floxed GFP, Odin-dPTB, frt-Kana-frt, and the B arm was digested out with Sfil/Nsil and then recombined with Odin BAC genomic DNA using a bacterial homologous recombination method, as described previously (Kim et al., 2007). The recombinant Odin BAC was injected into fertilized C57BL/6 mouse eggs as described previously (Kim et al., 2007). Each BAC transgenic line was identified by PCR analysis of DNA extracted from tail biopsy specimens: a 200 bp PCR product was synthesized with primers 5′-GGACCATGGCGGCCTCTGCAGC-3′ and 5′-AG-CGTAATCTGGAACATCGTATGGGTA-3′. Wnt1-Cre mice were purchased from The Jackson Laboratory (USA). All mice were generated and maintained in accordance with the institutional guidelines approved by the Sookmyung Women’s University Animal Care and Use Committee.
Histology, X-gal staining, and immunohistochemistry
Mouse brains at embryonic (E) day 18.5 or postnatal (P) day P5 were collected and fixed overnight in 4% paraformaldehyde (PFA), dehydrated in ethanol, cleared in Histoclear, and embedded in paraffin as described previously (Park et al., 2013). Sections were cut to 10 μm and collected on slides. The slides were rinsed in a Histoclear series in ethanol and then in an ethanol series in PBS to rehydrate the sections. Eosin staining and cresyl violet staining were performed as described previously (Park et al., 2013).
Mouse brains were fixed in 4% PFA and permeated with detergent buffer containing 2% NP-40 and 1% deoxycholate acid sodium salt at room temperature with shaking, stained with X-gal for 3 days at 4°C, and then the X-gal-stained tissues were washed in 1X PBS, dehydrated in ethanol, cleared in Histoclear, and embedded in paraffin. Sections were cut at 10 μm and collected on slides as described previously (Park et al., 2013).
Sections were rinsed in PBS, immersed in antigen unmasking solution (Vector Laboratories, USA), and boiled in a pressure cooker. After washing in cooled-PBS, the sections were blocked in 0.3% Triton X-100/2% horse serum in PBS for 1 h at room temperature (RT) and incubated with primary antibodies overnight at 4°C. The sections were washed and incubated with secondary antibodies for 2 h at RT. Then, the sections were washed in PBS and mounted in VECTASHIELD (Vector Laboratories) with DAPI (Molecular Probes, USA)
Antibodies
A monoclonal mouse anti-acetylated tubulin was purchased from Sigma-Aldrich (USA). A polyclonal rabbit antibody specific for Odin was generated in our lab. Goat anti-mouse IgG conjugated with FITC and goat anti-rabbit with TRITC were purchased from Life Technologies (USA).
RESULTS
Odin is primarily expressed in the brain ventricle-lining cells during early postnatal development
In our previous study, we reported on Odin null-mutant mice carrying a β-geo gene trap inserted into the 14th intron of the Odin gene (Kim et al., 2010). We further used X-gal staining of brain sections from heterozygous mice to analyze Odin gene expression during early postnatal brain development: LacZ activity of the β-geo gene trapped in the intron was expected to recapitulate endogenous Odin expression. Interestingly, we found that Odin was prominently expressed along the cells lining the brain ventricles, such as the third ventricle, cerebral aqueduct, and fourth ventricle (Fig. 1A). We also observed that Odin was expressed from E16.5 to P10, whereas it was barely expressed after P10 (data not shown). Importantly, the SCO, a small ependymal gland of the circumventricular system, was strongly positively stained with X-gal, suggesting that it is the main brain tissue expressing high levels of Odin. The choroid plexus, where CSF is produced, also expressed Odin (Fig. 1B). Taken together, our Odin expression analysis suggests its potential role in the development of brain ependymal tissue. We further investigated whether the SCO was morphologically abnormal in Odin null-mutant mice. Immunohistochemical staining analysis revealed that the SCO of wild-type mice displayed positive staining for anti-Odin antibodies, whereas that of the null-mutant mice did not (Figs. 1C and 1D). However, we did not observe any significant morphological differences in the SCO in between wild-type and null-mutant mice. These results suggest that Odin knockout mice may be incomplete to analyze the biological function of Odin possibly because the Odin-related PTB adaptors provide a redundant function in the SCO and other ependymal tissue.
Fig. 1.
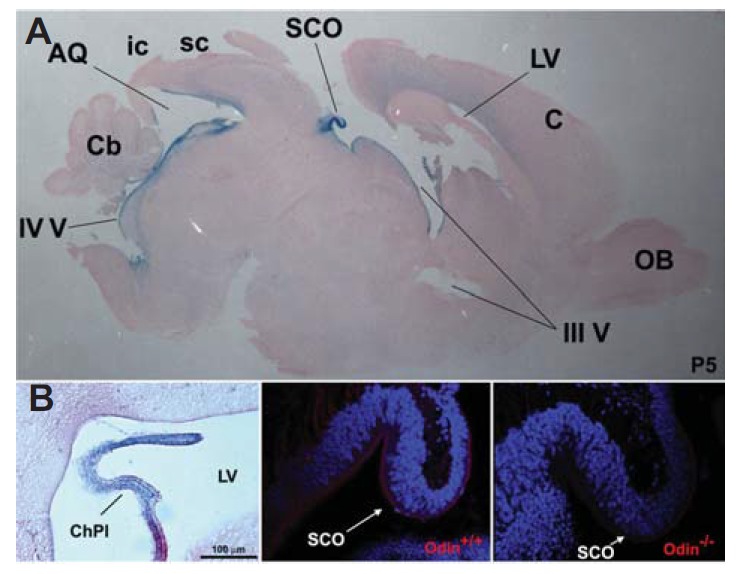
Odin expression analysis during early postnatal brain development. (A) The whole brain was dissected out of Odin heterozygous mice at postnatal (P) day 5 and processed for cryostat sections. The mid-sagittal section was subjected to X-gal staining to reveal LacZ-positive cells lining the brain ventricle. AQ, cerebral aqueduct; C, cerebral cortex; Cb, cerebellum; ic, inferior colliculus; LV, lateral ventricle; OB, olfactory bulb; sc, superior colliculus; SCO, subcommissural organ; III V, third ventricle; IV V, fourth ventricle. (B) LacZ expression analysis of the choroid plexus (ChPl) found in the lateral ventricle (LV). (C and D) The mid-sagittal section from the indicated Odin mice was subjected to immunohistochemical staining using anti-Odin antibodies.
Identification of the BAC clone recapitulating endogenous Odin gene expression
The Odin gene contains 25 exons and is about 153 kb in length (Fig. 2A). A BAC clone, RP24-258K7, contained about 117 kb upstream and 33 kb downstream of the genomic region from the first exon, respectively. To investigate whether this BAC contained most of the regulatory elements and locus control regions for Odin gene expression, the first exon in the BAC was modified to contain a GFP expression cassette flanked by two loxP sites followed by the PTB-deleted Odin mutant (Odin-dPTB) expression cassette. Odin-dPTB was not expressed without Cre-mediated excision of the floxed GFP-STOP cassette in this BAC expression system. We generated three independent BAC transgenic lines, which were used for intensive expression analyses using brain sections. As a result, we found that GFP expression was exclusively restricted to the ependymal tissue lining the brain ventricles in all transgenic lines (Fig. 2B). This result indicates that the Odin BAC clone contains the genetic control elements and promoters sufficient for recapitulating endogenous Odin gene expression. In addition, this result suggests that Odin BAC is a powerful tool to test whether in vivo function of the wild-type Odin protein is disturbed by ectopic expression of Odin-dPTB in ependymal cells.
Fig. 2.
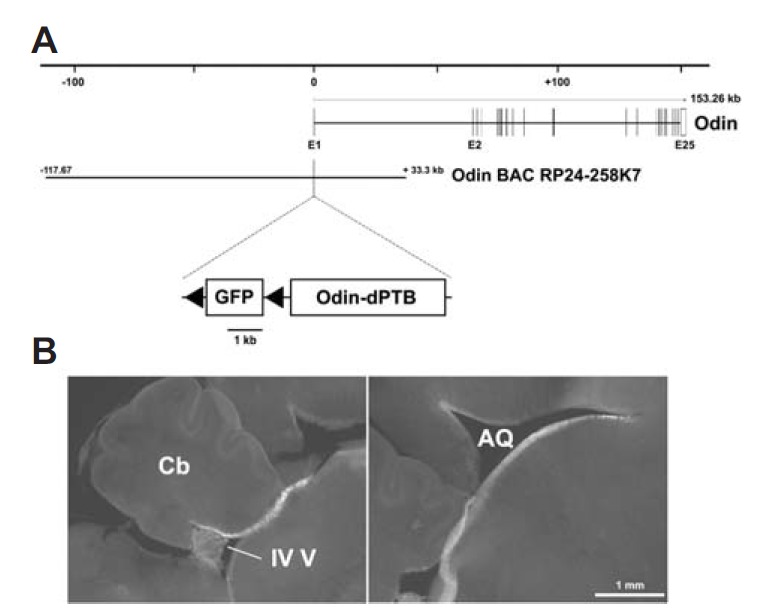
Green fluorescent protein (GFP) expression analysis using Odin BAC transgenic mice. (A) Schematic map of the mouse Odin genomic locus with the Odin BAC clone (RP24-258K7). The recombinant Odin BAC clone contained a floxed GFP-STP cassette followed by the Odin-dPTB expression cassette inserted upstream into the start codon of the first exon. (B) GFP fluorescent image of the mid-sagittal section prepared by vibratome (100 μm-thick). Similar results were reproducibly observed in three different transgenic mice.
Ectopic expression of the PTB-deleted Odin mutant disrupts SCO development
The Odin PTB domain is critically involved in the signaling pathway downstream of EphA8 (Shin et al., 2007). We postulated that the Odin-dPTB mutant may function as a dominant-negative mutant against endogenous Odin because it may compete for signaling proteins. We selected the wnt1-Cre (Dietrich et al., 2009) transgenic line to cross with the Odin-dPTB transgenic line for tissue-specific expression of Odin-dPTB (Fig. 3). We predicted that a floxed GFP expression-STOP cassette would be specifically excised primarily in the dien- and mesencephalon in the wnt1-Cre Odin-dPTB double transgenic line and that Odin-dPTB would be expressed subsequently instead of GFP in the corresponding brain tissues. Strikingly, all double transgenic lines (100% penetrance) displayed enlarged brains and a hydrocephalic phenotype, primarily in the midbrain (Figs. 3A and 3F). Cresyl violet staining analysis using coronal sections revealed that the SCO of double transgenic mice was much smaller than that of the Odin-dPTB transgenic lines and lacked Cre expression (Figs. 3B, 3C, 3G, and 3H). We used Odin-dPTB mice without the Cre transgene as a control throughout all experiments as a comparison with the double transgenic mice. To further investigate whether ependymal cells in the SCO are immature, immunohistochemical staining analysis using anti-acetylated tubulin antibody was performed (Lang et al., 2006; Qin et al., 2011). As a result, ependymal cells in the SCO of the double transgenic lines were mostly negative for acetylated tubulin, a marker for ependymal cell multicilia (Figs. 3D, 3E, 3I, and 3J). Ependymal tissues near the SCO also weakly stained for the acetylated tubulin antibody. Taken together, these results support our hypothesis that Odin-dPTB functions as a dominant-negative mutant against wild-type Odin in vivo. In addition, these findings suggest a potential role for Odin in the proper development of ependymal cells in the SCO.
Fig. 3.
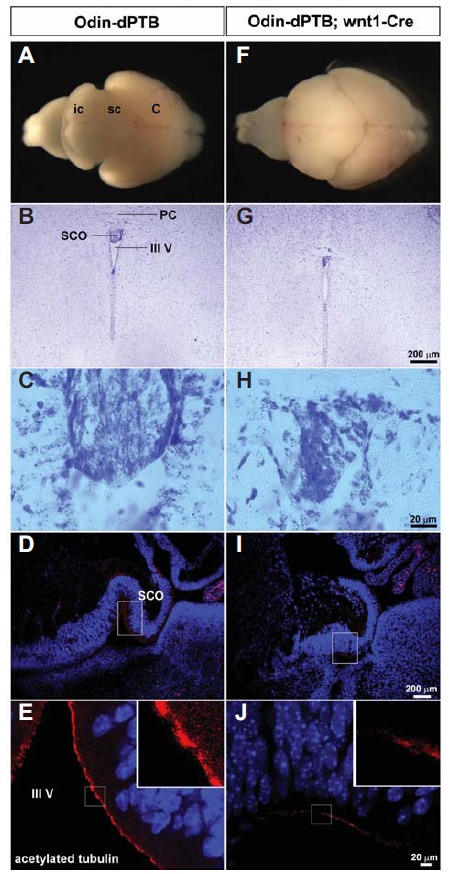
Subcommissural organ (SCO) development is defective in mice ectopically expressing Odin-dPTB. Littermate mice at P5 were obtained from a cross between the Odin-dPTB BAC transgenic line and wnt1-Cre mice to analyze their morphological abnormalities. (A and F) Whole brains were dissected out of the indicated mice for morphological comparison. Note that the brain of the double transgenic mice reveals a severe hydrocephalic phenotype with prominent enlargement in the midbrain. (B and G) Cresyl violet staining analysis of the coronal section. Note that the SCO is much smaller in the brain of the double transgenic line. PC, posterior commissure. (C and H) Enlarged view of the SCO regions in B and G, respectively. (D and I) The mid-sagittal section was subjected to immunohistochemical staining using anti-acetylated tubulin antibody, a marker for multicilia of ependymal cells. (E and J) Enlarged view of the SCO regions in D and I, respectively. Note that the ependymal cells in the SCO display much weaker staining with anti-acetylated tubulin antibody.
PTB-deleted Odin mutant expression interferes with proper development of ependymal cells in the cerebral aqueduct
The SCO is located in a dorsocaudal region of the third ventricle, at the entrance of the cerebral aqueduct (Rodriguez et al., 1998). Therefore, malformation of the SCO may have a causative role in the severe midbrain hydrocephalic phenotype (Picketts, 2006). We further examined the anatomical structure of the midbrain in the double transgenic lines. Consistent with the hydrocephalic morphology of the whole brain, the midbrain and cerebral aqueduct were severely enlarged in double transgenic mice (Figs. 4A, 4B, 4D, and 4E). In contrast, the sizes of the cerebellum and medulla were not significantly altered in double transgenic mice. In addition, neither apoptotic cells nor proliferative (pH3-positive) cells increased significantly in the midbrain region (data not shown). Importantly, ependymal cells of control mice appeared to be well organized with epithelial-like morphology, whereas those of double transgenic mice looked irregular and disorganized (Figs. 4C and 4F). To further investigate whether ependymal cells lining the cerebral aqueduct were properly differentiated in double transgenic mice, we performed acetylated tubulin antibody staining on midbrain sagittal sections. As we predicted, the ependymal cells lining the cerebral aqueduct of double transgenic mice were mostly negative for acetylated tubulin antibody staining (Figs. 5A–5J). This result strongly suggests that ependymal cells in the midbrain are defective for proper development of multi-cilia. Taken together, these findings support our hypothesis that Odin plays a role in the development of ependymal cells in the diencephalon and mesencephalon during early postnatal brain development.
Fig. 4.
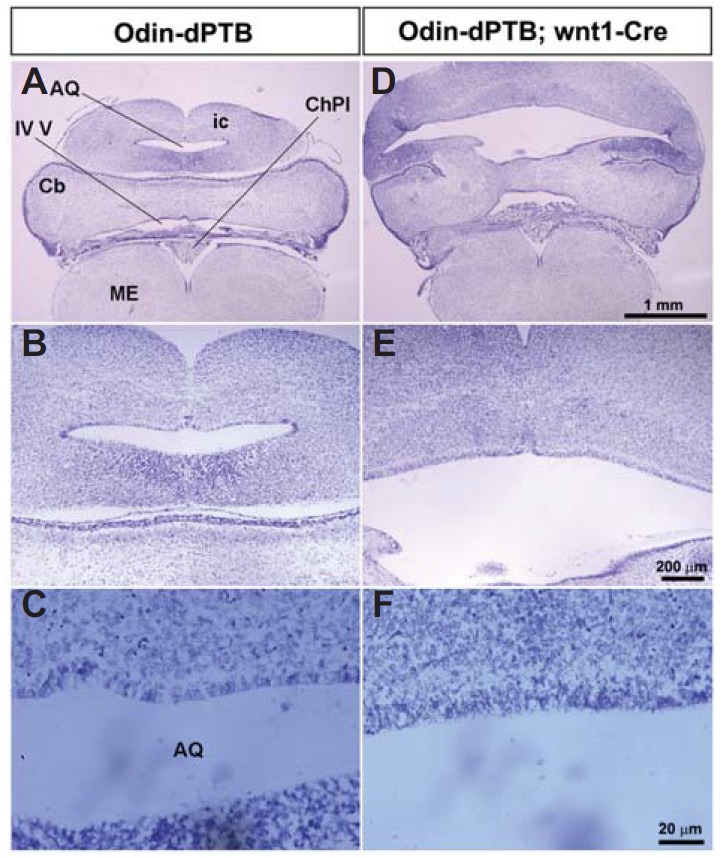
The midbrain of Odin-dPTB mice becomes severely expanded upon Cre expression. (A and D) The coronal sections of the indicated brains were analyzed by cresyl violet staining. Note that both the cerebral aqueduct and inferior colliculus (ic) are severely expanded. ME, medulla. (B and E) Enlarged view of the cerebral aqueduct regions in A and D, respectively. (C and F) High magnification of the ependymal cells lining the cerebral aqueduct in B and E. Note that the ependymal cells of control mice have a regular and well organized morphology, whereas those of the double transgenic mice do not.
Fig. 5.
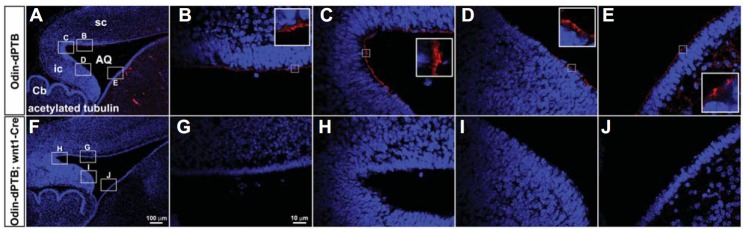
The ependymal cells lining the cerebral aqueduct are defective for development of multi-cilia. (A and F) The mid-sagittal section of the indicated mice was subjected to immunohistochemical staining using anti-acetylated antibody to reveal multi-ciliated ependymal cells. (B–E and G–J) High magnification of the brain regions corresponding to boxes in A and F, respectively.
DISCUSSION
Here, we report that Odin, a PTB-domain containing adaptor protein, may play a role in the development of ependymal cells in the SCO and midbrain during early postnatal brain development. Odin-dPTB was ectopically expressed in ependymal cells by wnt1-Cre, and we subsequently showed that Odin-dPTB plays a causative role in the severe midbrain hydrocephalic phenotype. This hydrocephalic phenotype was likely due to agenesis of the SCO and/or immature development of ependymal cells lining the cerebral aqueduct (Picketts, 2006). As the hydrocephalic phenotype was rarely found in the midbrain of Odin null-mutant mice, we postulate that Odin-related PTB adaptors may supplement the deficient Odin function to develop ependymal cells in these mice. For example, AIDA-1b is highly homologous to Odin, and these two members constitute the Anks family proteins containing six ankyrin repeats at their NH2-terminus, two SAM domains, and a PTB domain at their COOH-terminus (Ghersi et al., 2004). Further experiments will be needed to determine the role of AIDA-1b in the development of ependymal cells during early postnatal bran development. In particular, it would be interesting to generate double knockout mice lacking both Odin and AIDA-1b to examine whether these mice display the hydrocephalic phenotype. However, it would be far more complex and time-consuming to generate AIDA-1b null-mutant mice because they have various splicing variants and an enormous genome size of > 1000 kb.
The SCO is one of the secretory organs in the circumventricular system (Cottrell and Ferguson, 2004). Although the main function of the SCO is not clearly defined, some evidence suggests that it plays a role as a neuroendocrine organ circulating CSF. In particular, the ependymal cells of the SCO are specialized to secrete glycoproteins, such as spondin, a giant glycoprotein, and they play a role in homeostasis (Huh et al., 2009; Picketts, 2006; Rodriguez et al., 1998). It has been proposed that malfunction of the SCO in various genetically mutant mice is related with the development of congenital hydrocephalus (Picketts, 2006). Our transgenic mice ectopically expressing Odin-dPTB also had an immaturely developed SCO with a severe midbrain hydrocephalic phenotype. Thus, Odin may have an essential role in SCO cell secretory function during early postnatal brain development. For example, the lack of secretory function in SCO cells may result from ectopic expression of the Odin-dPTB protein, and this defect may play a causative role in immature development or malformation of the SCO in Odin-dPTB mice. The secretory function of SCO cells will need to be examined to determine if they are partially abnormal in Odin null-mutant mice.
Our results strongly suggest that the PTB-deleted Odin protein may be effective as a dominant-negative mutant against the endogenous Odin protein. However, it is unclear how Odin-dPTB disturbs the wild-type Odin-mediated signaling pathway. Odin-dPTB contains six Ank repeats and two SAM domains, so it may interact with other proteins critically involved in the Odin-mediated signaling cascade. These large and stable protein complexes involving Odin-dPTB may prevent the endogenous Odin protein from binding to those signaling proteins in the pathway. However, we cannot rule out the possibility that Odin-dPTB may not be restricted to Odin but may also disturb other PTB-domain containing adaptors in various ways. A recent ependymal cell culture technique (Guirao et al., 2010; Paez-Gonzalez et al., 2011) will be an essential tool to analyze the mechanism by which Odin-dPTB disrupts endogenous Odin in cultured cells. Although the mechanism underlying Odin function remains to be analyzed in the future, our findings highlight the potential role of Odin in the proper development of the SCO in the third ventricle and in ependymal cells in the cerebral aqueduct.
Acknowledgments
This study was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare grant (A111706).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Baas D, Meiniel A, Benadiba C, Bonnafe E, Meiniel O, Reith W, Durand B. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur. J. Neurosci. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- Blatt EN, Yan XH, Wuerffel MK, Hamilos DL, Brody SL. Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am. J. Respir. Cell Mol. Biol. 1999;21:168–176. doi: 10.1165/ajrcmb.21.2.3691. [DOI] [PubMed] [Google Scholar]
- Breunig JJ, Arellano JI, Rakic P. Cilia in the brain: going with the flow. Nat. Neurosci. 2010;13:654–655. doi: 10.1038/nn0610-654. [DOI] [PubMed] [Google Scholar]
- Carlen M, Meletis K, Goritz C, Darsalia V, Evergren E, Tanigaki K, Amendola M, Barnabe-Heider F, Yeung MS, Naldini L, et al. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat. Neurosci. 2009;12:259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- Cottrell GT, Ferguson AV. Sensory circumventricular organs: central roles in integrated autonomic regulation. Regul. Pept. 2004;117:11–23. doi: 10.1016/j.regpep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Dietrich P, Shanmugasundaram R, Shuyu E, Dragatsis I. Congenital hydrocephalus associated with abnormal subcommissural organ in mice lacking huntingtin in Wnt1 cell lineages. Hum. Mol. Genet. 2009;18:142–150. doi: 10.1093/hmg/ddn324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi E, Noviello C, D’Adamio L. Amyloid-beta protein precursor (AbetaPP) intracellular domain-associated protein-1 proteins bind to AbetaPP and modulate its processing in an isoform-specific manner. J. Biol. Chem. 2004;279:49105–49112. doi: 10.1074/jbc.M405329200. [DOI] [PubMed] [Google Scholar]
- Guirao B, Meunier A, Mortaud S, Aguilar A, Corsi JM, Strehl L, Hirota Y, Desoeuvre A, Boutin C, Han YG, et al. Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat. Cell Biol. 2010;12:341–350. doi: 10.1038/ncb2040. [DOI] [PubMed] [Google Scholar]
- Huh MS, Todd MA, Picketts DJ. SCO-ping out the mechanisms underlying the etiology of hydrocephalus. Physiology (Bethesda) 2009;24:117–126. doi: 10.1152/physiol.00039.2008. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–686. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development. 2009;136:4021–4031. doi: 10.1242/dev.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiho H, Fukushima S, Kontani K, Katada T. RINL, guanine nucleotide exchange factor Rab5-subfamily, is involved in the EphA8-degradation pathway with odin. PLoS One. 2012;7:e30575. doi: 10.1371/journal.pone.0030575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Song E, Choi S, Park S. Engineering lacZ Reporter gene into an ephA8 bacterial artificial chromosome using a highly efficient bacterial recombination system. J. Biochem. Mol. Biol. 2007;40:656–661. doi: 10.5483/bmbrep.2007.40.5.656. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee H, Kim Y, Yoo S, Park E, Park S. The SAM domains of Anks family proteins are critically involved in modulating the degradation of EphA receptors. Mol. Cell. Biol. 2010;30:1582–1592. doi: 10.1128/MCB.01605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen TZ, Nielsen MM, Blagoev B, Pandey A, Mann M. Mouse embryonic fibroblasts derived from Odin deficient mice display a hyperproliiferative phenotype. DNA Res. 2004;11:285–292. [PubMed] [Google Scholar]
- Lacar B, Young SZ, Platel JC, Bordey A. Imaging and recording subventricular zone progenitor cells in live tissue of postnatal mice. Front. Neurosci. 2010;4 doi: 10.3389/fnins.2010.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B, Song B, Davidson W, MacKenzie A, Smith N, McCaig CD, Harmar AJ, Shen S. Expression of the human PAC1 receptor leads to dose-dependent hydrocephalus-related abnormalities in mice. J. Clin. Invest. 2006;116:1924–1934. doi: 10.1172/JCI27597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortloff AR, Vio K, Guerra M, Jaramillo K, Kaehne T, Jones H, McAllister JP, 2nd, Rodriguez E. Role of the subcommissural organ in the pathogenesis of congenital hydrocephalus in the HTx rat. Cell Tissue Res. 2013;352:707–725. doi: 10.1007/s00441-013-1615-9. [DOI] [PubMed] [Google Scholar]
- Paez-Gonzalez P, Abdi K, Luciano D, Liu Y, Soriano-Navarro M, Rawlins E, Bennett V, Garcia-Verdugo JM, Kuo CT. Ank3-dependent SVZ niche assembly is required for the continued production of new neurons. Neuron. 2011;71:61–75. doi: 10.1016/j.neuron.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Blagoev B, Kratchmarova I, Fernandez M, Nielsen M, Kristiansen TZ, Ohara O, Podtelejnikov AV, Roche S, Lodish HF, et al. Cloning of a novel phosphotyrosine binding domain containing molecule, Odin, involved in signaling by receptor tyrosine kinases. Oncogene. 2002;21:8029–8036. doi: 10.1038/sj.onc.1205988. [DOI] [PubMed] [Google Scholar]
- Park E, Kim Y, Noh H, Lee H, Yoo S, Park S. EphA/ephrin-A signaling is critically involved in region-specific apoptosis during early brain development. Cell Death Differ. 2013;20:169–180. doi: 10.1038/cdd.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Figares JM, Jimenez AJ, Rodriguez EM. Subcommissural organ, cerebrospinal fluid circulation, and hydrocephalus. Microsc. Res. Tech. 2001;52:591–607. doi: 10.1002/1097-0029(20010301)52:5<591::AID-JEMT1043>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Picketts DJ. Neuropeptide signaling and hydrocephalus: SCO with the flow. J. Clin. Invest. 2006;116:1828–1832. doi: 10.1172/JCI29148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Liu M, Niu W, Zhang CL. Dysregulation of Kruppel-like factor 4 during brain development leads to hydrocephalus in mice. Proc. Natl. Acad. Sci. USA. 2011;108:21117–21121. doi: 10.1073/pnas.1112351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C, Fernandez-Llebrez P, Bach A, Robert B, Soriano E. Msx1 disruption leads to diencephalon defects and hydrocephalus. Dev. Dyn. 2004;230:446–460. doi: 10.1002/dvdy.20070. [DOI] [PubMed] [Google Scholar]
- Rodriguez EM, Rodriguez S, Hein S. The subcommissural organ. Microsc. Res. Tech. 1998;41:98–123. doi: 10.1002/(SICI)1097-0029(19980415)41:2<98::AID-JEMT2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Shin J, Gu C, Park E, Park S. Identification of phosphotyrosine binding domain-containing proteins as novel downstream targets of the EphA8 signaling function. Mol. Cell. Biol. 2007;27:8113–8126. doi: 10.1128/MCB.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Sydorskyy Y, St-Germain JR, Taylor P, Tsao MS, Moran MF. Odin (ANKS1A) modulates EGF receptor recycling and stability. PLoS One. 2013;8:e64817. doi: 10.1371/journal.pone.0064817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 2005;345:1–20. doi: 10.1016/j.jmb.2004.10.038. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Tichelaar JW. Forkhead transcription factor HFH-4 and respiratory epithelial cell differentiation. Am. J. Respir. Cell Mol. Biol. 1999;21:153–154. doi: 10.1165/ajrcmb.21.2.f159. [DOI] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- Zhang D, Stumpo DJ, Graves JP, DeGraff LM, Grissom SF, Collins JB, Li L, Zeldin DC, Blackshear PJ. Identification of potential target genes for RFX4_v3, a transcription factor critical for brain development. J. Neurochem. 2006;98:860–875. doi: 10.1111/j.1471-4159.2006.03930.x. [DOI] [PubMed] [Google Scholar]
- Zhao C, Suh H, Gage FH. Notch keeps ependymal cells in line. Nat. Neurosci. 2009;12:243–245. doi: 10.1038/nn0309-243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


