Abstract
The 15-kDa selenoprotein (Sep15) is a selenoprotein residing in the lumen of the endoplasmic reticulum (ER) and implicated in quality control of protein folding. Herein, we established an inducible RNAi cell line that targets Sep15 mRNA in Chang liver cells. RNAi-induced Sep15 deficiency led to inhibition of cell proliferation, whereas cell growth was resumed after removal of the knockdown inducer. Sep15-deficient cells were arrested at the G1 phase by upregulating p21 and p27, and these cells were also characterized by ER stress. In addition, Sep15 deficiency led to the relocation of focal adhesions to the periphery of the cell basement and to the decrease of the migratory and invasive ability. All these changes were reversible depending on Sep15 status. Rescuing the knockdown state by expressing a silent mutant Sep15 mRNA that is resistant to siRNA also reversed the phenotypic changes. Our results suggest that SEP15 plays important roles in the regulation of the G1 phase during the cell cycle as well as in cell motility in Chang liver cells, and that this selenoprotein offers a novel functional link between the cell cycle and cell motility.
Keywords: 15-kDa selenoprotein, cell cycle arrest, cell proliferation, cyclin E, selenium
INTRODUCTION
Selenium is an essential trace element that has been reported to provide many health benefits when it is obtained from the diet in adequate amounts in humans and many other life forms. For example, selenium is known to play roles in decreasing the incidence of cancer, delaying aging, augmenting the immune system, preventing heart disease, and supporting male reproduction and development (Boosalis, 2008; Flohé, 2007; Hatfield and Gladyshev, 2002; Papp et al., 2007; Roman et al., 2014 and references therein). Most of the benefits of selenium are mediated by selenoproteins, which contain selenocysteine (Sec) as a selenium-containing amino acid (Hatfield and Gladyshev, 2002; Roman et al., 2014). The 15-kDa selenoprotein (Sep15) was initially identified as a strongly [75Se]-labeled selenoprotein in human T cells, and it shares homology with selenoprotein M (SelM; Gladyshev et al., 1998; Korotkov et al., 2002). Similar to all other eukaryotic selenoprotein mRNAs, Sep15 mRNA uses UGA as a Sec codon, and a Sec insertion sequence (SECIS) element in the 3′-untranslated region (Kumaraswamy et al., 2000; Low and Berry, 1996). Although Sep15 is evolutionarily conserved in most animals and plants, only the vertebrate Sep15 homologs contain Sec. Similar to many other selenoproteins such as thioredoxin reductases and glutathione peroxidases, Sep15 has redox activity and belongs to the thiol oxidoreductase class of proteins (Ferguson et al., 2006; Kumaraswamy et al., 2000; Labunskyy et al., 2007). Sep15 contains a signal peptide at its N-terminus and is localized in the lumen of the ER (Korotkov et al., 2002; Labunskyy et al., 2007). It has been proposed that Sep15 in the ER participates in quality control of protein folding either through the rearrangement of disulfide bonds (isomerase function) or through the reduction of incorrectly formed disulfide bonds (reductase function) in misfolded glycoproteins bound to UDP-glucose:glycoprotein glucosyltransferase (UGGT) (Korotkov et al., 2002; Labunskyy et al., 2007).
Sep15 has been implicated in cancer development, although its mechanistic role is not yet elucidated. The human SEP15 gene is located at the 1p31 locus, a locus where mutations and deletions have been observed in various human cancer cells (Gladyshev et al., 1998; Nasr et al., 2003). The expression of Sep15 is decreased in liver, prostate, and lung cancers (Kumaraswamy et al., 2000), and in several human malignant mesothelioma cell lines (Apostolou et al., 2004). There are two single nucleotide polymorphisms (SNPs) at nucleotides 811 (C/T) and 1125 (G/A) in the SECIS element of Sep15 (Gladyshev et al., 1998), and these SNPs were found to be associated with various cancers, including colorectal cancer (Davis et al., 2012; Sutherland et al., 2010), malignant mesothelioma (Apostolou et al., 2004), and lung cancer (Jablonska et al., 2008).
Recently, it has been reported that inhibition of Sep15 expression in in vitro and in vivo models of colon carcinogenesis reversed the cancer phenotypes. The knockdown of Sep15 mRNA in a colon cancer cell line led to the inhibition of colony formation, tumor growth, and lung metastasis (Irons et al., 2010; Tsuji et al., 2011). SEP15 knockout in mice prevented chemically induced aberrant crypt formation presumably by regulating guanylate binding protein-1 (Tsuji et al., 2012).
To obtain insights into the molecular function of Sep15 in human cells, we constructed a Chang liver cell line that inducibly expressed short hairpin RNA (shRNA) targeting Sep15 mRNA, and analyzed the effect of Sep15-deficiency on cell proliferation and motility. Sep15 deficiency inhibited cell growth by arresting cells in the G1 phase and decreased migratory and invasive ability of these cells. This study provides a possible mechanism of how Sep15 regulates cell proliferation and motility.
MATERIALS AND METHODS
Materials
Chang liver cells were purchased from ATCC (#CCL-13). G418 sulfate was purchased from AG Scientific. Anti-paxillin antibody, doxycycline, and Matrigel-coated invasion chambers with 8.0 μm pore size were purchased from BD Biosciences. Transwell chambers containing polycarbonate membrane with 8.0 μm pore size was purchased from Corning. Alexa Fluor 488 goat anti-mouse IgG antibody, pcDNA6/TR vector, blasticidin and TRIZOL reagent were purchased from Invitrogen. Rhodamin phalloidin was purchased from Life Technologies. pSuperior.neo vector was purchased from OligoEngine. Mo-MuLV reverse transcriptase was purchased from Promega. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), aphidicolin, blebbistatin, bovine serum albumin (BSA), cycloheximide, 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI), eosin Y, hematoxylin solution, nocodazole, propidium iodide, protease inhibitor mixture, Y-27632, and RNase A were purchased from Sigma. DNAs were synthesized from Cosmogenetech (Korea). The His-tagged Tat-C3 transferase exoenzyme (pHis-Tat-C3) expression vector was provided by Jae Bong Park and the recombinant C3 transferase was prepared as previously described (Park et al., 2003). Anti-MAD2 antibody (Santa Cruz) and anti-p-27 antibody (Santa Cruz) were obtained from H.S. Lee, and anti-p21 (Santa-Cruz) antibody, and anti-cyclin E1 antibody (Santa-Cruz) from N.V. Kim. Control siRNA and siSep15 RNA that has the same sequences as the stem region of shSep15 RNA were purchased from Dharmacon.
Cell culture and establishment of cell lines
Cell culture and transfection of cells were carried out as described previously (Kim et al., 2010). An inducible Sep15 knockdown cell line was constructed as described previously (Bang et al., 2014). To construct a Sep15 rescue vector, two silent point mutations were introduced in the siRNA target sequence by performing two-step PCRs. In the first step, two DNA fragments (5′-half and 3′-half) were amplified from Chang liver cell cDNA prepared as described previously (Bang et al., 2014) using two sets of primers; the forward primer1 5′-AAAATGGTAGCGATGGCG-3′ and the reverse primer1 5′-GTCTGAACCACGCACGTAC-3′, and the forward primer2 5′-GTACGTGCGTGGTTCAGAC-3′ and the reverse primer2 5′-GCTAGAATTCGGACTTTTCTGTAAGAATGTA-3′ (altered bases are underlined). The PCR products were subjected to nested PCR to amplify the final Sep15 rescue construct containing two silent mutations. The final Sep15 rescue construct was cloned into the BamHI and EcoRI restriction sites of pRV.IRES.Puro. The vector (pRV.Puro.Sep15-rescue) was transfected into the shSep15 cell line and 1 μg/ml puromycin was added to the media for selection. The expression of Sep15 rescue mRNA was measured by employing quantitative RT-PCR technology using PCR primers specific for the altered SEP15.
Measurement of knockdown efficiency
After induction of shRNA expression by doxycycline (Dox) treatment, total RNA was isolated from pSuperior.neo.Sep15i containing cells and subjected to northern blot analysis. The probe for detecting endogenous shSep15 mRNA was prepared by amplifying a region of the Sep15 coding sequence using the following primers: forward 5′-AGGACTGCAAATCAAGTATGTC-3′ and reverse 5′-ACAAATGCAGGAGGATGGAC-3′. GAPDH mRNA was used as an internal control. To detect the Sep15 rescue mRNA, Sep15-Rescue specific forward primer, 5′-ACAAATGCAGGAGGATGGAC-3′, and the same reverse primer employed for probe preparation were used for PCR. The PCR conditions were as follows: denaturation at 95°C for 5 min followed by cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s and a final extension at 72°C for 10 min.
Synchronization of cells and flow cytometry
After inducing expression of the shRNA targeting Sep15 mRNA for 3 days, cells were incubated with 5 μg/ml of aphidicolin or 100 ng/ml of nocodazole for 24 h. Flow cytometry was performed as described previously (Kim et al., 2010). Briefly, cells were released from the aphidicolin/nocodazole block by washing with fresh medium and incubated for appropriate time periods before analysis. Cells were harvested and washed once with phosphate buffered saline (PBS), fixed in 70% ethanol at 4°C for 1 h, washed with PBS twice and, then resuspended with PBS containing 200 μg/ml of RNase A. After 10 min incubation at room temperature, cells were further incubated with 100 μg/ml of propidium iodide in PBS for 30 min at room temperature. Stained cells were analyzed using a FACS Calibur (BD Biosciences) flow cytometer and FlowJo software (World’s Premier Cytometric Analytical Software).
Immunostaining and confocal microscopy
shSep15 cells were induced to express shRNA by adding 10 μg/ml of Dox overnight and then transferred onto a chambered glass slide. Immunocytochemistry was carried out as described previously (Shim et al., 2009) with minor modifications. After the cells were fixed, they were permeabilized, blocked, and incubated with anti-paxillin antibody (1:500) and 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature. The cells were subsequently incubated with Alexa 488 anti-mouse IgG (1:1,000) for 1 h at room temperature. For nuclear counterstaining, cells were stained with DAPI diluted in PBS (10 μg/ml) for 15 min at room temperature. Cells were imaged with an LSM 700 confocal microscope (Carl Zeiss). The excitation wavelength was 488 nm for Alexa 488, 540 nm for rhodamine phalloidin, and 350 nm for DAPI, respectively.
Western analysis
Western blot analysis was carried out as described previously (Kim et al., 2013) using anti-p21 (1:1,000), anti-p27 (1:1,000), anti-cyclinE1 (1:2,000) and anti-MAD2 (1:1,000) antibodies. Anti-α-tubulin antibodies were used as controls, after stripping with stripping buffer (62.5 mM Tris/HCl, pH 6.8, 2% SDS and 100 mM β-mercaptoethanol) at 50°C for 10 min.
Endoplasmic reticulum stress assays
ER stress was analyzed by measuring the mRNA levels of glucose-regulated protein 78 (GRP78) and C/EBP homology protein (CHOP) and monitoring the generation of alternatively spliced X-box binding protein 1 (XBP1) mRNA.
Measuring GRP78 and CHOP mRNA levels by quantitative RTPCR
After inducing knockdown in shSep15 cells for an appropriate time period, total RNA was isolated using TRIZOL reagent. First-strand cDNA was synthesized from total RNA and used as template in the subsequent PCR reactions. The cDNA levels were measured by quantitative RT-PCR technology using the following primers: GRP78 forward, 5′-GCTCGACTCGAATTCCAAAG-3′, GRP78 reverse 5′-GTCAGGCAGTTCTGGTCATT-3′, CHOP forward, 5′-ATGAGGACCTGCAAGAGGT-3′, and CHOP reverse, 5′-CCAATTGTTCATGCTTGGTG-3′. The PCR conditions were as follows; denaturation at 95°C for 5 min followed by 40 cycles of 95°C for 15 s, 55°C for 1 min, 72°C for 1 min and a final extension at 72°C for 10 min.
Detection of alternatively spliced XBP1 mRNA
Total RNA was isolated and cDNA was synthesized as described above. PCR was performed using the following primers: XBP1 forward, 5′-CTGGAACAGCAAGTGGTAGA-3′ and XBP1 reverse, 5′-CTGGGTCCTTCTGGGTAGAC-3′. The PCR conditions were as follows; denaturation at 95°C for 5 min followed by 25 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 30 s and a final extension at 72°C for 10 min. PCR products were separated by 4% agarose gel electrophoresis.
Wound healing assay
Cells were grown to confluence in 6-well culture plates with or without Dox for 2 days and scraped with a yellow pipette tip to create a linear cell-free gap. After cells were washed with PBS 3 times, fresh medium with or without Dox was added. After incubation at 37°C for 4 days, the movement of cells into the wound area was photographed.
Migration and invasion assay
The cell migration assay was performed using Transwell chambers containing polycarbonate membrane with 8.0 μm pore size. Sep15 knockdown was induced by adding Dox for 2 days and, if necessary, the cells were preincubated with blebbistatin (10 μM), Y-27632 (5 μM) or C3 transferase (2.5 μg/ml) for 2 h, 1 h and 16 h, respectively, prior to transferring the cells onto the chamber. Cells were harvested and resuspended to make a final concentration of 1 × 105 cells/ml with serum free DMEM media, and 0.5 ml of cell suspension was added into each chamber. The media obtained after harvesting the cultured shSep15 cells were added to the lower chambers of the plate. After incubation at 37°C for 12 h, unmigrated cells were removed with a cotton plug. The migrated cells were fixed with methanol and stained with hematoxylin and eosin Y. Stained cells were counted from the images taken at 200× magnification. Cell invasion assay was performed using Matrigel-coated invasion chambers with 8.0 μm pore size with similar methods used in the migration assay. The major differences were the final concentration of cells (4 × 104 cells/ml) and incubation time (24 h).
Statistical analysis
Each experiment was performed in biological triplicate for statistical analysis. Statistical significance was tested by the Student’s t-test followed by an f-test to ensure the equality of the 2 variances.
RESULTS
Sep15 knockdown inhibits cell proliferation
Since Sep15 deficiency may be related to the suppression of colon carcinogenesis (Tsuji et al., 2011), there is a possibility that Sep15 participates in the regulation of cell proliferation. To examine the effect of Sep15 deficiency on cell proliferation, we constructed a cell line that can be induced to express a Sep15-targeted shRNA. As described in Materials and Methods, we first used the pcDNA6/TR plasmid to generate a Chang liver cell line that can inducibly express the TetR gene in response to Dox. The successful insertion of this vector into the host genome was previously confirmed by Southern blotting (Bang et al., 2014). pSuperior.neo.Sep15i, a plasmid that encodes Sep15 shRNA was then transfected into these TetR-expressing cells. Cell colonies that contained this vector were isolated and vector integration into the host genome was also verified previously by Southern blotting (Bang et al., 2014). Of these colonies, one clone was chosen arbitrarily, renamed as shSep15 and used for all subsequent knockdown analyses. To reduce the possibility of an off-target effect of siRNA, we also constructed a rescue cell line (shSep15/Rescue) by transfecting a plasmid that harbors SEP15 containing two silent mutations within the siRNA target sequence into shSep15 cells. The temporal knockdown efficiency of the shSep15 cell line was measured by northern blotting. Sep15 expression was significantly reduced one day after the induction of shSep15 expression by Dox (∼70%) and the knockdown efficiency reached over 90% by day 2 (Fig. 1A). Subsequently, the reduction of Sep15 mRNA was maintained. The shSep15 cells containing pRV.IRES.Puro (shSep15/mock) vector showed a similar pattern of expression of both endogenous and rescue mRNAs as the initial shSep15 cells. In contrast, although the levels of the endogenous Sep15 mRNA in shSep15/Rescue+Dox cells were reduced greatly, the levels of the rescue-specific Sep15 mRNA were not affected by Dox treatment (Fig. 1B).
Fig. 1.
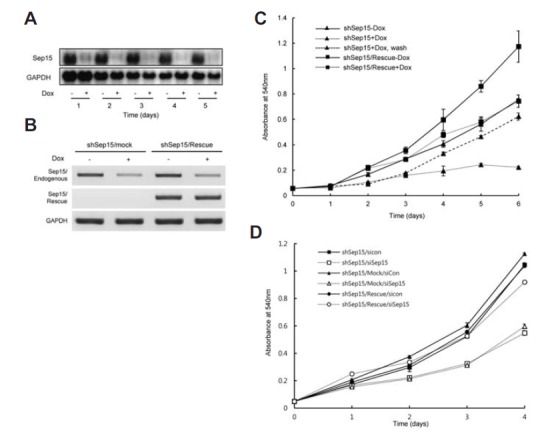
Sep15 deficiency leads to the inhibition of cell proliferation. (A) Efficiency of Sep15 knockdown after the induction of shRNA expression was measured by northern hybridization. GAPDH was used as an internal control. (B) Measuring the endogenous and rescue-specific Sep15 mRNAs by RT-PCR. cDNAs were amplified using specific primer sets as described in Materials and Methods. (C) Cell proliferation in shSep15-Dox cells (triangles connected with a solid line), shSep15+Dox (triangles connected with a dotted line), removal of Dox 3 days after induction of knockdown (triangles connected with a discontinuous line), shSep15/Res- cue-Dox cells (squares connected with a solid line), and shSep15/Res- cue+Dox cells (squares connected with a dotted line) were measured by a MTT assay. Data shown are representative of at least three independent experiments. (D) The effect of Sep15 knockdown using siRNA on cell proliferation. shSep15 cells were transfected with a solid line) or siSep15 (open squares with a dotted line), shSep15/Mock cells containing both shSep15 and rescue backbone vector (shSep15/Mock) with siCon (closed triangles with a solid line) or siSep15 (open triangles with a dotted line), and shSep15/Rescue cells with siCon (closed circles with a solid line) or siSep15 (open circles with a dotted line). Cell proliferation was measured by a MTT assay. Data shown are representative of at least three independent experiments and error bars stand for standard deviations.
The effect of Sep15 deficiency on the cell proliferation rate was measured using a MTT assay. We found that the proliferation rate of Sep15-deficient cells (shSep15+Dox) was significantly reduced from day 2 compared to Dox-untreated control cells (shSep15-Dox, Fig. 1C). Six days after knockdown, Sep15-deficient cells (shSep15+Dox) showed greater than 3.5-fold reduction in cell proliferation compared to control cells (shSep15-Dox). In contrast, the growth slope was significantly increased in Dox-treated rescue cells (shSep15/Rescue+Dox) compared to shSep15+Dox cells, but was similar to that of shSep15-Dox cells. These results suggest that cell proliferation was recovered by rescuing SEP15. It should be noted that the growth rate was increased in shSep15/Rescue-Dox cells compared to that of shSep15-Dox cells. This result indicates that the Sep15 status plays an important role in the regulation of cell proliferation. Interestingly, cell proliferation was resumed after Dox was removed from the medium at day 3 (shSep15+Dox, wash). Although the growth of shSep15 cells was recovered by Sep15 rescue, the growth rate of the Sep15-rescued cells was decreased when Dox was added (compare shSep15/Rescue-Dox with shSep15/Rescue+Dox). It seems that this difference is due to the Dox effect on cell growth. It, however, should be noted that the growth rate of shSep15/Rescue+Dox cells was not decreased under the control of shSep15-Dox. We also examined the effect of Dox on proliferation in control cells that express TetR. Although the addition of Dox to these cells affected cell growth negatively, they still showed a high growth rate compared to that of shSep15+Dox cells (Fig. S1). The growth rate of TetR+Dox was similar with that of shSep15/Rescue+Dox cells suggesting that Dox has a limited effect on cell growth. To exclude the effect of Dox on cell proliferation, synthetic siRNA was transfected into shSep15, shSep15/Mock and shSep15/Rescue cells, and cell proliferation was measured using the MTT assay in the absence of Dox. The backbone vector was used in preparing the rescue vector construct which was then used as the mock vector. Control siRNA served as a knockdown control. Control cell proliferation was not affected by siRNA treatment in shSep15/Rescue cells, while control cells (shSep15 and shSep15/Mock) showed a significant decrease in cell proliferation by siRNA treatment (Fig. 1D). At Day 4, the growth rates of control cells treated by siSep15 RNA were slightly increased. This appears to be due to the decrease of the RNAi effect at Day 4. These results suggest that Sep15 participates in the regulation of cell proliferation in Chang liver cells.
Sep15-deficient cells are arrested at the G1 phase
Cell proliferation is usually inhibited by arresting cells at a specific stage of the cell cycle. To examine if Sep15 deficiency causes cell cycle arrest, we performed fluorescence-activated cell sorting (FACS) analysis. In the asynchronized steady state, the relative amount of G1 phase cells compared to G2/M phase cells in Sep15-deficient cells (shSep15+Dox) was greater than in control cells (shSep15-Dox, compare the top two panels designated Asy in Fig. 2A), suggesting that Sep15-deficient cells are arrested at the G1 phase. To examine this in more detail, both normal and Sep15 knockdown cells were subjected to synchronization by treating these cells with aphidicolin for 24 h, and cell cycle progression was examined after release. When the cells were arrested at the G1 phase with aphidicolin, most of the control cells (shSep15-Dox) moved to the S phase 3 h after release and returned to the steady state within 24 h. In contrast, most of the Sep15-deficient cells (shSep15+Dox) remained in the arrested state (G1 phase), while only a small population entered the S phase approximately 6 h after the release and progressed through the cell cycle (see the right panels of Fig. 2A). TetR expressing control cells with or without Dox treatment showed a similar cell cycle progression pattern to shSep15 cells not treated with Dox (Fig. S2). In addition to the G1 phase arrest with aphidicolin, shSep15 cells were treated with nocodazole, which is known to arrest cells at the G2/M phase, and cell cycle progression was examined. Most of the shSep15+Dox cells remained in the G1 phase even during nocodazole treatment and only a small population progressed through the cell cycle after release (see the right panels of Fig 2B). However, the control cells (shSep15-Dox) were arrested at the G2/M phase by nocodazole treatment and moved normally to the G1 phase after release. Since many cell cycle-specific proteins such as cyclins and CDKs have been known to be regulated in their expression during cell cycle progression, we measured the mRNA levels of these proteins to determine whether Sep15 deficiency affects expression of cell cycle-specific genes. The levels of cyclin E1, p21 and p27 were examined by Western blot analysis. As shown in Fig. 2C, both p21 and p27 expression levels were increased significantly only upon Sep15 deficiency. Notably, the levels of p21 and p27 in shSep15/Rescue+Dox cells were similar to those in control cells (shSep15-Dox and shSep15/Dox-Dox), suggesting that Sep15 participates in cell cycle regulation, presumably by controlling the expression of p21 and p27. We also examined the expression of MAD2 by Western blot analysis. Mad2 mRNA has been known to be highly susceptible to off-targeting by siRNAs, when the inhibition of cell cycle progression was observed (Sigoillot et al., 2012). We found that the levels of MAD2 were not changed in both knockdown and rescue cells indicating that cell cycle arrest was not caused by off-targeting effect (Fig. 2D). These results, together with the MTT assay, suggest that Sep15 participates in cell proliferation and cell cycle progression by regulating p21 and p27 expression.
Fig. 2.
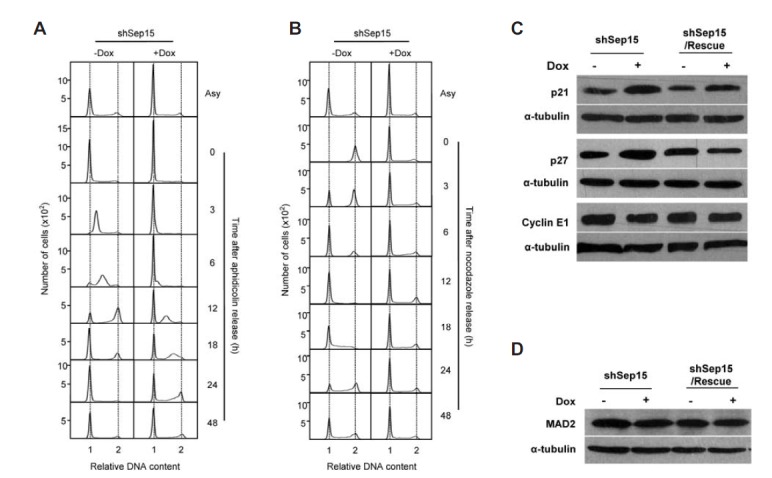
Sep15 deficiency arrests cells at the G1 phase by downregulating cyclin E1 and E2 expression. (A–B) Sep15 knockdown caused cell cycle arrest at the G0/G1 phase. After the cell cycle was arrested by the treatment with aphidicolin (A) or nocodazole (B), cell cycle analysis was performed as described in Materials and Methods. The numbers 1 and 2 on the bottom of each panel represent the relative DNA contents of the G0/G1 and G2/M phases, respectively. The number of cells and release time after arrest are marked on the left and right side of Y-axis, respectively. Asy designates asynchronized cells. Data shown are representative of at least three independent experiments. (C) Western blot analyses to measure the levels of p21, p27 and cyclin E1 in shSep15+Dox versus -Dox cells and in shSep15/Rescue+Dox versus -Dox cells. (D) Levels of MAD2 in shSep15+Dox versus -Dox cells and in shSep15/Rescue+Dox versus -Dox cells were measured by Western blot analysis.
Sep15 knockdown induces a mild ER stress response
Because Sep15 is localized in the ER lumen and it has been proposed to participate in quality control of protein folding (Korotkov et al., 2002; Labunskyy et al., 2007), we tested if Sep15 deficiency leads to ER stress. Specifically, we examined the relationship between ER stress and Sep15 deficiency by analyzing several established biomarkers of ER stress in our cell line. As shown in Figs. 3A and 3B, the mRNA levels of ER stress marker genes, such as GRP78 and CHOP, were both significantly increased by Sep15 knockdown; however, the level of induction differed for these two genes. Four days after knockdown of Sep15, the expression of GRP78 and CHOP was increased 2.5-fold and 9.5-fold, respectively. Notably, the mRNA level of CHOP in Sep15-deficient cells was similar to that induced by the ER stress activator, tunicamycin (Fig. 3B). However, the expression level of GRP78 was greater in cells treated with tunicamycin or dithiothreitol (DTT) than those with Sep15 deficiency (Fig. 3A). CHOP expression was also increased to a greater extent in cells treated with DTT. Dox addition to the TetR control cell line did not result in the induction of expression of these genes, indicating that Dox has no effect on their expression. The levels of CHOP mRNA were differentially increased in response to tunicamycin and DTT treatment. Tunicamycin increased CHOP mRNA levels 7-fold, a lesser extent than that induced by Sep15 deficiency, whereas DTT increased the expression of CHOP by approximately 43-fold (Fig. 3B). Next, we examined the alternative splicing of the XBP1 transcript in Sep15-deficient cells (Fig. 3C). The active form of XBP1, the smaller splice variant (lowest band, 398 bases), appeared 4 days after shSep15 cells were induced with Dox. However, the levels of the XBP1 active form (398 bases) were less than that generated in positive control treated cells. Interestingly, Sep15 mRNA levels were increased by tunicamycin and DTT treatment 2-fold and 1.5-fold, respectively (Fig. S3). This result is consistent with a previous report (Labunskyy et al., 2009) suggesting ER stress occurs normally and the pathway governing ER stress is similar in both systems. Together, these data indicate that the knockdown of Sep15 in Chang liver cells leads to the activation of the unfolded protein response (UPR), a measure of ER stress. However, the UPR state in shSep15 cells is mild compared to that induced by treatment with tunicamycin or DTT.
Fig. 3.
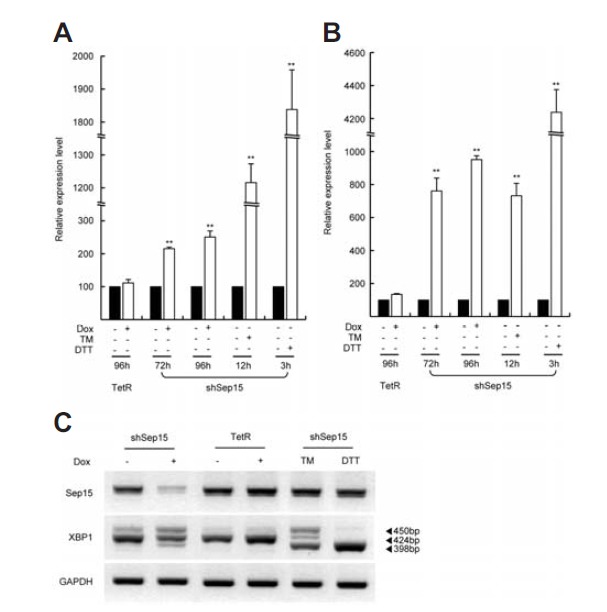
Sep15 knockdown induces the expression of ER stress response markers. (A) Relative levels of GRP78 mRNA in shSep15+Dox versus -Dox cells were measured. Treatment times and cell lines are indicated on the bottom of the graph. (B) Relative levels of CHOP mRNA. (C) Alternative splicing of XBP1 mRNA was induced by Sep15-deficiency. The 424 bp, 450 bp, and 398 bp XBP1 bands represent the unspliced, hybrid, and spliced forms, respectively. As a control, cells were treated with tunicamycin (TM) or dithiothreitol (DTT). ** indicates significance at p < 0.001.
Deficiency of Sep15 inhibits cell migration and invasion
Since colony formation and lung metastasis were found to be inhibited in Sep15-deficient colon cancer cell lines (Irons et al., 2010; Tsuji et al., 2011), and the cytoskeleton reorganization was induced by Sep15 deficiency (Bang et al., 2014), we assumed that cell migration and invasion would be inhibited by Sep15 deficiency. The effect of Sep15 knockdown on cell migration was examined with both wound healing and Boyden chamber assays. A wound was introduced into a field of cells by scraping cells, and cell migration was assayed by measuring the width of the gap created over time. In control shSep15-Dox cells and Dox-washed cells, the gap gradually narrowed and was almost completely filled within 3 days of scraping (Fig. 4A). On the other hand, in shSep15+Dox cells, the gap width was only slightly reduced over the course of the same time period. In TetR-expressing cells, the gap filled rapidly regardless of Dox treatment. In this experiment, the initial density of Dox-treated cells was two-fold higher than that of untreated cells. This adjustment was necessary to ensure that the final cell densities 3 days after scraping were comparable in the 2 groups. Otherwise, the decreased migration could be considered to be due to the difference in cell proliferation and not the result of impaired migration. To address this question, we also performed a Boyden chamber assay. Sep15-deficient cells showed an approximately 25% decrease in migration compared to the shSep15-Dox control cells (Fig. 4B). Another striking feature of Sep15 deficiency was membrane bleb formation (Bang et al., 2014 and Fig. S4). In many cases, membrane blebbing occurs through the RhoA/ROCK pathway. Interestingly, Y-27632, a ROCK inhibitor, and C3 transferase, a RhoA inhibitor, both significantly increased migration of Sep15-deficient cells to the level of control Sep15-Dox cells (Fig. 4C). Notably, blebbistatin, a MLCK inhibitor, increased the migration of Sep15-deficient cells greater than Y-27632 and C3 transferase. Although it is not clear how the migration of Sep15-deficient cells was inhibited, these data suggest that membrane blebbing inhibits cell migration.
Fig. 4.
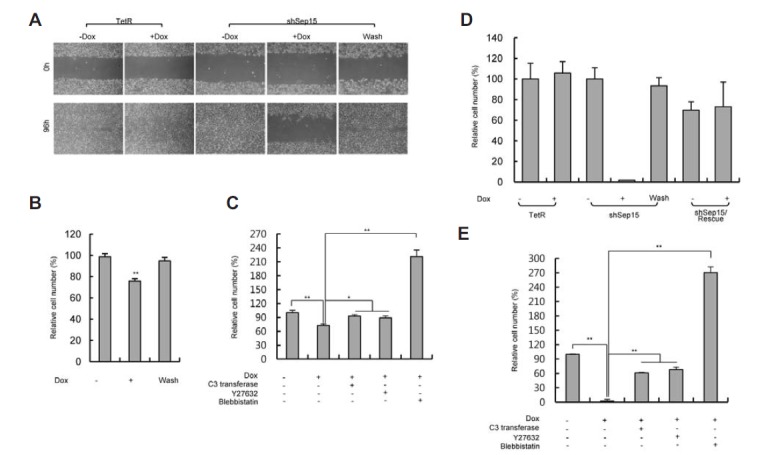
Effects of Sep15 knockdown on cell migration and invasion. (A) Measuring cell migration by the wound healing assay. After incubating the scratched cells for 4 days, cells were photographed at 40× magnification. (B–C) Cell migration assay using Boyden chamber. After staining, the migrated cells were photographed (200×) and counted. (D–E) Invasion assay using Matrigel-coated invasion chamber. After staining, the invaded cells were photographed (200×) and counted. After counting, relative cell numbers of each testing group against shSep15-Dox were calculated. * and ** indicate significance at P < 0.05 and 0.001, respectively. Error bars represent standard deviation obtained from 3 independent experiments.
Because cell migration and invasion are closely related, we investigated the effect of Sep15 knockdown on the invasion of cells using a chamber coated with Matrigel (an extracellular matrix, ECM). The invasive behavior of Sep15-deficient cells was decreased greater than 30-fold relative to the control (Fig. 4D). On the other hand, the invasiveness of TetR or shSep15/Rescue cells was not affected by Dox addition indicating that Sep15 deficiency caused the loss of invasiveness. In addition, the invasion was recovered in Dox-treated shSep15 cells, when Dox was removed. Like migration, the addition of C3 transferase and Y27632 recovered the invasive ability to levels near that of normal cells, while blebbistatin increased the invasive ability much more than normal cells (Fig. 4E). Because the cells were incubated for a short time during the assay, the invasion rate would not be significantly affected by cell proliferation and, therefore, the number of cells that invaded the ECM is a measure of invasive ability. Remarkably, the knockdown of Sep15 inhibited cell invasion to a greater extent than cell migration.
Focal adhesions were moved to the periphery of the cell basement
The polarity of focal adhesions is frequently used as an indicator of cell migration. Interestingly, we found that the focal adhesions in Sep15-deficient cells became localized to the basolateral plasma membrane (+Dox of Fig. 5A), suggesting that the cells were only attached at the peripheral part of their membrane. In contrast, control shSep15-Dox cells had focal adhesions throughout their basal surface. The focal adhesions were enriched at the tip of each lamellipodium of control cells. However, Sep15-deficient cells, which are characterized by the lack of lamellipodia, lost this focal adhesion polarity. These data support the observed inhibitory effect of Sep15 knockdown on cell migration and invasion, because both lamellipodia formation and the polarity of focal adhesions are disrupted in Sep15-deficient cells. Moreover, the inhibition of RhoA and ROCK activity restored the polarity and distribution of focal adhesions, and filopodia/lamellipodia formation (Figs. 5B and 5C). These results together with the migration and invasion assay suggest that Sep15 regulates cell motility through controlling localization of focal adhesions and formation of filopodia/lamellipodia.
Fig. 5.

Rearrangement of focal adhesion sites by Sep15 knockdown. (A) Cells were stained for paxillin (green) and F-actin (red). (B) Effect of inhibiting RhoA and ROCK signaling on focal adhesion distribution. Cells were treated with Y-27632 or C3 transferase were stained for paxillin. Scale bars represent 10 μm. (C) Lamellipodia formation was inhibited by Sep15 knockdown at the leading edge of migrating cells and was restored by the inhibition of RhoA and ROCK activity. The morphology of migrating cells was examined under a light microscope and imaged. Arrows indicate lamellipodia. Scale bars represent 20 μm. Where indicated, cells were preincubated with Y-27632 or C3 transferase for 1 h and 16 h, respectively, before seeding.
DISCUSSION
Sep15 has been implicated in cancer development (Gladyshev et al., 1998; Nasr et al., 2003; Kumaraswamy et al., 2000; Apostolou et al., 2004; Tsuji et al., 2011; 2012). However, the mechanistic role of Sep15 during cancer development is not fully understood. In this study, we constructed an inducible Sep15 knockdown cell line from Chang liver cells and examined the phenotypic changes that occurred after the induction of Sep15 deficiency. Chang liver cells were originally reported as an immortalized human liver cell line, but later it was reported that these cells manifest many characteristics of HeLa cells which are cancer cells. We found that the inhibition of cell proliferation by Sep15 deficiency occurs through G1 phase arrest and that the expression of p21 and p27 was increased in Sep15-deficient cells. Both p21 and p27 proteins have been shown to inhibit cell cycle progression by interacting with cyclins and cyclin-dependent kinases (CDKs), and by affecting Rb phosphorylation (Sherr and Roberts, 1999). When p21 was upregulated in the cells containing functional pRb, cells showed a tendency to be arrested at the G1 phase. However, G2 arrest was more prominent in Rb-negative cells (Niculescu III et al., 1998). Since cell cycle was arrested at the G1 phase, Chang liver cells seemed to contain active pRb. In addition to cell cycle regulation, p21 has been known to regulate cell motility. Synthetic p21 peptides inhibited cell spreading that was mediated by integrin signaling which dissociated the integrin receptor from focal-adhesion contacts. Moreover, p21 can interact with Rho kinase and inhibit stress fiber formation (Coqueret, 2003). When Sep15 deficiency was induced in Chang liver cells, cell motility (cell spreading, cell migration and invasion) was inhibited, focal adhesions were formed only on the periphery of the cell basement and lamellipodial formation was also inhibited. Therefore, these effects of Sep15 deficiency on cell motility seem to be correlated with intracellular p21 levels. It was also reported that N-terminal phosphorylation of p27 and cytoplasmic translocation induced cell motility (Reed, 2002). However, whether both p21 and p27 are required for cell motility and how they participate in cell motility remain to be determined.
The activation of the ER stress response by Sep15 deficiency supports its previously reported function as a participant in protein folding in the ER. An open question remains, which is how does Sep15, which is localized in the lumen of ER, regulate cell proliferation and motility? Because Sep15 participates in the folding of proteins in the ER, the deficiency of Sep15 may increase the population of misfolded proteins in the ER and/or cause the impairment of ER function. The accumulation of mis-folded proteins or malfunction of ER may inhibit cyclin expression and cell motility. It would be interesting to identify the protein(s) in the ER that regulate(s) cyclin E expression and cell motility.
The fact that focal adhesions were mislocalized to the lateral part of the basal plasma membrane, and that focal adhesions lost their polarity in Sep15-deficient cells supports the observation that cell migration/invasion was inhibited by Sep15 deficiency. The decrease in lamellipodia formation also provides evidence that Sep15 deficiency inhibits the migration/invasion of these cells. Blebbing cells treated with the inhibitors of RhoA, ROCK, or MLCK reverted to their normal spindle shape, restored lamellipodia and filopodia formation, and these cells had restored migration/invasion ability (see Figs. 4 and 5). The data provide insights into how Sep15 knockdown-induced membrane blebbing inhibits migration as well as invasion. Recently, it was reported that Sep15 deficiency causes the reorganization of cytoskeletal proteins and membrane blebbing through a RhoA/ROCK pathway (Bang et al., 2014). Inhibitors of the RhoA/ROCK pathway recovered the impairment of cell motility suggesting that Sep15 is involved in cell motility by controlling the organization of cytoskeletal proteins. The results obtained in this study provide an interesting perspective on how Sep15 deficiency may contribute to the reversal of the cancer phenotype.
Acknowledgments
This work was supported by the Priority Research Centers Program (Grant No. 2010-0029694) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant No. 2011-0012947), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (2012M3A9D1054622) to BJL, and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research to DLH and NIH CA080946 to VNG.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Apostolou S, Klein JO, Mitsuuchi Y, Shetler JN, Poulikakos PI, Jhanwar SC, Kruger WD, Testa JR. Growth inhibition and induction of apoptosis in mesothelioma cells by selenium and dependence on selenoprotein SEP15 genotype. Oncogene. 2004;23:5032–5040. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- Bang J, Jang M, Huh JH, Na J, Shim M, Carlson BA, Tobe R, Tsuji PA, Gladyshev VN, Hatfield DL, et al. Deficiency of the 15-kDa selenoprotein led to cytoskeleton remodeling and non-apoptotic membrane blebbing through a RhoA/ROCK pathway. Biochem. Biophys. Res. Commun. 2014. in press. [DOI] [PMC free article] [PubMed]
- Boosalis MG. The role of selenium in chronic disease. Nutr. Clin. Pract. 2008;23:152–160. doi: 10.1177/0884533608314532. [DOI] [PubMed] [Google Scholar]
- Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Davis CD, Tsuji PA, Milner JA. Selenoproteins and cancer prevention. Annu. Rev. Nutr. 2012;32:73–95. doi: 10.1146/annurev-nutr-071811-150740. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Labunskyy VM, Fomenko DE, Arac D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J. Biol. Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- Flohe L. Selenium in mammalian spermiogenesis. Biol. Chem. 2007;388:987–995. doi: 10.1515/BC.2007.112. [DOI] [PubMed] [Google Scholar]
- Gladyshev VN, Jeang KT, Wootton JC, Hatfield DL. A new human selenium-containing protein. Purification, characterization, and cDNA sequence. J. Biol. Chem. 1998;273:8910–8915. doi: 10.1074/jbc.273.15.8910. [DOI] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons R, Tsuji PA, Carlson BA, Ouyang P, Yoo MH, Xu XM, Hatfield DL, Gladyshev VN, Davis CD. Deficiency in the 15-kDa selenoprotein inhibits tumorigenicity and metastasis of colon cancer cells. Cancer Prev. Res. (Phila) 2010;3:630–639. doi: 10.1158/1940-6207.CAPR-10-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska E, Gromadzinska J, Sobala W, Reszka E, Wasowicz W. Lung cancer risk associated with selenium status is modified in smoking individuals by Sep15 polymorphism. Eur. J. Nutr. 2008;47:47–54. doi: 10.1007/s00394-008-0696-9. [DOI] [PubMed] [Google Scholar]
- Kim JY, Lee KH, Shim MS, Shin H, Xu XM, Carlson BA, Hatfield DL, Lee BJ. Human selenophosphate synthetase 1 has five splice variants with unique interactions, subcellular localizations and expression patterns. Biochem. Biophys. Res. Commun. 2010;397:53–58. doi: 10.1016/j.bbrc.2010.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Chen Z, Shim MS, Lee MS, Kim JE, Kwon YE, Yoo TJ, Kim JY, Bang JY, Carlson BA, et al. SUMO modification of NZFP mediates transcriptional repression through TBP binding. Mol. Cells. 2013;35:70–78. doi: 10.1007/s10059-013-2281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Novoselov SV, Hatfield DL, Gladyshev VN. Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol. Cell. Biol. 2002;22:1402–1411. doi: 10.1128/mcb.22.5.1402-1411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaraswamy E, Malykh A, Korotkov KV, Kozyavkin S, Hu Y, Kwon SY, Moustafa ME, Carlson BA, Berry MJ, Lee BJ, et al. Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology. J. Biol. Chem. 2000;275:35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- Labunskyy VM, Hatfield DL, Gladyshev VN. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59:1–5. doi: 10.1080/15216540601126694. [DOI] [PubMed] [Google Scholar]
- Labunskyy VM, Yoo MH, Hatfield DL, Gladyshev VN. Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute ER stresses. Biochemistry. 2009;48:8458–8465. doi: 10.1021/bi900717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- Nasr MA, Hu YJ, Diamond AM. Allelic loss at the Sep15 locus in breast cancer. Cancer Therapy. 2003;1:293–298. [Google Scholar]
- Niculescu AB, III, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21Cip1/Waf1 at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol. Cell. Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid. Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- Park J, Kim JS, Jung KC, Lee HJ, Kim JI, Kim J, Lee JY, Park JB, Choi SY. Exoenzyme Tat-C3 inhibits association of zymosan particles, phagocytosis, adhesion, and complement binding in macrophage cells. Mol. Cells. 2003;16:216–223. [PubMed] [Google Scholar]
- Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI. Cell cycling? Check your brakes. Nat. Cell Biol. 2002;4:E199–E201. doi: 10.1038/ncb0802-e199. [DOI] [PubMed] [Google Scholar]
- Roman M, Jitaru P, Barbante C. Selenium biochemistry and its role for human health. Metallomics. 2014;6:25–54. doi: 10.1039/c3mt00185g. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Shim MS, Kim JY, Jung HK, Lee KH, Xu XM, Carlson BA, Kim KW, Kim IY, Hatfield DL, Lee BJ. Elevation of glutamine level by selenophosphate synthetase 1 knockdown induces megamitochondrial formation in Drosophila cells. J. Biol. Chem. 2009;284:32881–32894. doi: 10.1074/jbc.M109.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. A bioinformatics method identifies prominent off-targeted transcript in RNAi screens. Nat. Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland A, Kim DH, Relton C, Ahn YO, Hesketh J. Polymorphisms in the selenoprotein S and 15-kDa selenoprotein genes are associated with altered susceptibility to colorectal cancer. Genes Nutr. 2010;5:215–223. doi: 10.1007/s12263-010-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji PA, Carlson BA, Naranjo-Suarez S, Yoo MH, Xu XM, Fomenko DE, Gladyshev VN, Hatfield DL, Davis CD. Knockout of the 15 kDa selenoprotein protects against chemically-induced aberrant crypt formation in mice. PLoS One. 2012;7:e50574. doi: 10.1371/journal.pone.0050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji PA, Naranjo-Suarez S, Carlson BA, Tobe R, Yoo MH, Davis CD. Deficiency in the 15 kDa selenoprotein inhibits human colon cancer cell growth. Nutrients. 2011;3:805–817. doi: 10.3390/nu3090805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


