Groundwater contamination near hydraulic fracturing wells
Foaming drinking water from a contaminated well.
High-volume hydraulic fracturing (HVHF) employs volumes of water, sand, and chemicals under extreme pressure to create cracks in deep rock formations, releasing natural gas deposits. Garth Llewellyn et al. (pp. 6325–6330) investigated potential groundwater contamination in the vicinity of five HVHF Marcellus Shale gas wells in Bradford County, Pennsylvania. The authors analyzed water from drinking water wells on several properties in which gas contamination was documented by the Pennsylvania Department of Environmental Protection and attributed to the nearby gas wells. An analytical technique that is sensitive to low chemical concentrations, called comprehensive 2D gas chromatography coupled with time-of-flight mass spectrometry, identified complex mixtures of organic contaminants and 2-n-Butoxyethanol, a chemical associated with HVHF and drilling fluids, in the drinking water wells. Samples from the five Marcellus Shale gas wells were not available for analysis, but the compounds found in the residential well water were similar to compounds contained in so-called flowback water—HVHF fluid that returns to the surface of a drilling site after it has been pumped below the surface—collected from other Marcellus Shale HVHF drilling sites. According to the authors, one or more Marcellus Shale gas wells may have likely introduced natural gas, foam, and organic contaminants into the aquifer used for drinking water by nearby residents. — J.P.J.
Water diffusion and neurodegenerative disease
Water mobility increases when tau protein forms amyloid fibers.
The human protein tau helps stabilize microtubules, which form a part of the cellular backbone. However, in pathological conditions marked by neurodegeneration, such as Alzheimer’s disease (AD), the tau protein can aggregate into fibers that are broadly organized into a rigid core and a flexible coat. Because water is thought to influence protein aggregation, Yann Fichou et al. (pp. 6365–6370) used neutron scattering and molecular dynamics simulations to compare the mobility of water molecules on the surface of tau monomers and experimentally induced tau fibers. The authors report that the fraction of water molecules undergoing diffusion around the fibers was 25% greater than that around the monomers, suggesting increased water entropy, which favors fiber formation. Further, the authors report that the increased movement of water around the fibers likely occurs around the coat rather than around the core of the fibers. Increases in water diffusion in the hippocampus in the brain of AD patients have been previously proposed as a potential early biomarker of AD that is detectable through diffusion MRI. According to the authors, the reported increases in water diffusion may be partly attributable to enhanced water mobility around tau fibers, thus providing a potential basis for the clinical utility of the biomarker. — P.N.
Natural selection on the X chromosome
The inheritance pattern for the X chromosome, which occurs in a single copy in males, differs from that of the 22 pairs of autosomes. Researchers have proposed that this unique inheritance pattern may expose the X chromosome to natural selection in a way that differs from that of the autosomes, potentially resulting in accelerated evolution. Kiwoong Nam et al. (pp. 6413–6418) explored how natural selection differs on the X chromosome versus the autosomes. The authors studied polymorphism patterns in 10 species of great apes, including humans, using 87 high-coverage whole genomes. The findings uncover megabase-sized regions on the X chromosomes of most species that display strikingly little variation, whereas no such regions were found on the autosomes. The regions of low variation were found to partially overlap among species and share certain genetic aberrations that set them apart from the rest of the X chromosome. The authors report that the reduction in diversity cannot be explained by direct selection, background selection, or soft selective sweeps. Rather, the findings suggest that hard, strong selective sweeps have independently targeted specific genomic regions. According to the authors’ hypothesis, such sweeps might target multicopy testis-expressed genes while engaging in a genetic conflict with the Y chromosome for transmission to the next generation. — A.G.
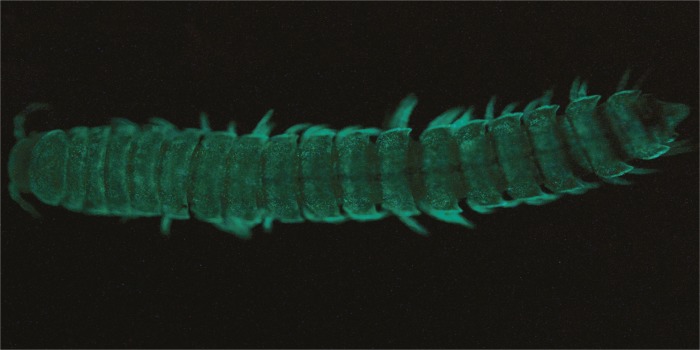
Gradual evolution of bioluminescence in millipedes
The millipede Xystocheir bistipita, now referred to as Motyxia bistipita, is bioluminescent.
The rediscovery of the California millipede Xystocheir bistipita after 50 years unexpectedly revealed that the species is bioluminescent. Paul Marek and Wendy Moore (pp. 6419–6424) demonstrate that X. bistipita is the evolutionary sister of Motyxia, the only genus of New World bioluminescent millipedes, and that bioluminescence originated in the group’s common ancestor and incrementally grew in brightness through evolutionary time. Millipedes with bioluminescence of increased brightness possessed larger cyanide glands, indicating that millipede toxicity appears linked to the intensification of luminescence. In addition, the authors report that species at higher elevations exhibit the brightest luminescence. The authors suggest that luminescence in Motyxia may have initially evolved to allow the animals to cope with metabolic stress triggered by a hot, dry environment at low elevations and may have been repurposed as a warning signal by species colonizing high-elevation habitats with increased predation risk. The authors conclude that luminescent intensity in Motyxia exhibits evolutionary escalation through species diversification, and suggest that the discovery of bioluminescence in X. bistipita provides insights into the evolution of bioluminescence. — S.R.
Bird-feeding pastime might ruffle species balance
Native silvereye (Zosterops lateralis) feeding on bread.
In 2002 alone, more than 450 million kg of seeds were fed to wild birds as a pastime in the United States, by some estimates. Because urban bird feeding can potentially cause malnutrition, alter bird species richness, and help transmit bird-borne diseases, Josie Galbraith et al. (pp. E2648–E2657) performed an 18-month-long bird-feeding experiment at 23 residential gardens in Auckland, New Zealand, nearly half of which provided a few slices of bread and a metric cup of seed—determined through a nationwide survey to be the most common food items offered by bird feeders—each morning at feeding stations, while the other half did not feed birds. Covering more than 18,000 birds of 33 species through 597 surveys, the study found that feeding reversibly affected the community structure of common garden bird species, occasionally favoring introduced species, which tend to be omnivorous, over native ones, which are often nectarivorous, insectivorous, or frugivorous. For example, during experimental feeding, the feeding gardens had 2.4 times more house sparrows and 3.6 times more spotted doves—both introduced species—than nonfeeding gardens. By contrast, feeding was negatively associated with the abundance of the native grey warbler. Despite its limited size, the study uncovers effects of urban bird feeding on the balance between native and introduced species, according to the authors. — P.N.
Improving brain disease modeling
Induced pluripotent stem cell (iPSC) technology enables a patient’s disease to be modeled in a dish, facilitating the discovery of personalized drug treatments for brain disorders. However, the clinical relevance of in vitro models has remained unclear because the basic culture conditions for neurons have not been tested for the ability to support fundamental neuronal functions. Using electrophysiological techniques, Cedric Bardy et al. (pp. E2725–E2734) found that widely used tissue culture media such as DMEM, Neurobasal, and serum, impair the neurophysiological functions of iPSC-derived human neurons. The authors designed a neuronal medium called BrainPhys, which closely mimics the chemical environment of the healthy living brain. In contrast to widely used culture media, BrainPhys contains natural levels of glucose, amino acids, and inorganic salts crucial for the generation and propagation of electrical signals called action potentials. As a result, BrainPhys increased the number of iPSC-derived human neurons that fired a high frequency of action potentials, improved the long-term stability of electrical activity, and supported neuronal maturation and growth of functional neural networks. According to the authors, BrainPhys is likely to recapitulate the dysfunctions occurring in patients’ brains and might thus enable the discovery of effective treatments for neurological and psychiatric disorders. — J.W.