Significance
The molybdenum cofactor (Moco) is an enzyme cofactor critical for the survival of almost all organisms from all kingdoms of life, and its biosynthesis is associated with various medical conditions such as inheritable human diseases and bacterial pathogenesis. The characteristic pyranopterin backbone of Moco is formed by the action of two enzymes, MoaA and MoaC (molybdenum cofactor biosynthesis protein A and C, respectively). Conventionally, MoaA was considered responsible for the majority of the transformation. In contrast to this view, the combined studies reported here revealed that it is MoaC that is responsible for the majority of the rearrangement reaction required for Moco pyranopterin backbone formation. Based on the conservation of MoaC and its catalytic residues, the mechanism of pyranopterin ring formation is likely conserved among all organisms.
Keywords: molybdenum cofactor, pterin biosynthesis, radical SAM enzyme, enzymatic rearrangement, cPMP synthase
Abstract
The molybdenum cofactor (Moco) is essential for all kingdoms of life, plays central roles in various biological processes, and must be biosynthesized de novo. During Moco biosynthesis, the characteristic pyranopterin ring is constructed by a complex rearrangement of guanosine 5′-triphosphate (GTP) into cyclic pyranopterin (cPMP) through the action of two enzymes, MoaA and MoaC (molybdenum cofactor biosynthesis protein A and C, respectively). Conventionally, MoaA was considered to catalyze the majority of this transformation, with MoaC playing little or no role in the pyranopterin formation. Recently, this view was challenged by the isolation of 3′,8-cyclo-7,8-dihydro-guanosine 5′-triphosphate (3′,8-cH2GTP) as the product of in vitro MoaA reactions. To elucidate the mechanism of formation of Moco pyranopterin backbone, we performed biochemical characterization of 3′,8-cH2GTP and functional and X-ray crystallographic characterizations of MoaC. These studies revealed that 3′,8-cH2GTP is the only product of MoaA that can be converted to cPMP by MoaC. Our structural studies captured the specific binding of 3′,8-cH2GTP in the active site of MoaC. These observations provided strong evidence that the physiological function of MoaA is the conversion of GTP to 3′,8-cH2GTP (GTP 3′,8-cyclase), and that of MoaC is to catalyze the rearrangement of 3′,8-cH2GTP into cPMP (cPMP synthase). Furthermore, our structure-guided studies suggest that MoaC catalysis involves the dynamic motions of enzyme active-site loops as a way to control the timing of interaction between the reaction intermediates and catalytically essential amino acid residues. Thus, these results reveal the previously unidentified mechanism behind Moco biosynthesis and provide mechanistic and structural insights into how enzymes catalyze complex rearrangement reactions.
Moco is an essential enzyme cofactor that mediates redox reactions in the active sites of enzymes. Moco-dependent enzymes play central roles in purine and sulfur catabolism in mammals, anaerobic respiration in bacteria, and nitrate assimilation in plants (1, 2). Importantly, Moco must be synthesized de novo in cells because it is chemically unstable, particularly under aerobic conditions, and cannot be taken up as a nutrient (1, 2).
During Moco biosynthesis, the characteristic pyranopterin ring is constructed by a complex rearrangement of GTP into cPMP (3). This unusual transformation involves the insertion of the guanine C-8 between C-2′ and C-3′ of ribose (Fig. 1A) (4). Although this conversion has been shown to require two proteins, MoaA and MoaC (molybdenum cofactor biosynthesis protein A and C, respectively) (4–6), their individual contributions have remained elusive and are the subject of the current study. MoaA belongs to the radical S-adenosyl-l-methionine (SAM) superfamily, of which members catalyze unique free-radical reactions (7). By contrast, MoaC shows no significant sequence or structural similarities to any functionally characterized enzyme. Therefore, the predominant view of the field has been that MoaA catalyzes the majority or all of the rearrangement of GTP to form the pterin structure, with MoaC playing little to no catalytic role in this process (2). In line with this view, studies identifying a putative MoaA product after chemical derivatization suggested that MoaA catalyzed the transformation of GTP to pyranopterin triphosphate (Fig. 1B) (8, 9). This conventional view was challenged by a recent in vitro characterization of MoaA, where formation of 3′,8-cyclic-7,8-dihydro-GTP (3′,8-cH2GTP; Fig. 1B) rather than pyranopterin triphosphate was proposed (10). This finding suggested the possibility of a novel mechanism in which MoaC, and not MoaA, may ultimately be responsible for pyranopterin ring formation. However, it was also proposed that 3′,8-cH2GTP could simply be a transient intermediate of MoaA (8, 9), leaving ambiguity about the functions of MoaA and MoaC.
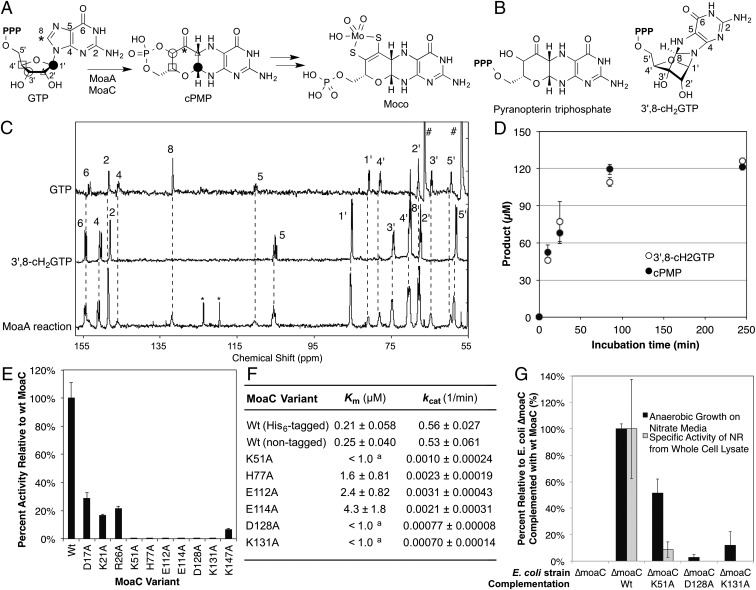
Fig. 1.
In vivo and in vitro functional characterization of MoaC. (A) Moco biosynthesis. Symbols indicate the fate of each atom (4). cPMP may also be in a hydrate form in solution (14). (B) Previously proposed structures for the MoaA product. (C) In situ 13C NMR characterization of the MoaA product. Shown are the 13C NMR spectra of [U-13C]GTP (Top), purified [U-13C]3′,8-cH2GTP (Middle), and the MoaA (0.4 mM) reaction using [U-13C]GTP (1 mM) as the substrate (Bottom). Numbers are the signal assignments for atoms labeled in A and B. Signals highlighted by * and # are derived from toluensulfonate and glycerol, respectively. (D) Timecourse of the formation of the biosynthetically relevant MoaA product based on the quantitation of 3′,8-cH2GTP before (open circles) or after (filled circles) its conversion to cPMP. The assay solution contained 65 μM WT-MoaA, 0.2 mM GTP, and 1 mM SAM. (E) In vitro activity of WT and variants of MoaC determined by a coupled assay with MoaA. (F) Steady-state kinetic parameters for WT and variants of MoaC. The assays were performed in the absence of MoaA by using purified 3′,8-cH2GTP as a substrate. a, only the upper limit (1.0 μM) was determined because the reaction rate became impractically low below this substrate concentration. (G) Moco production in E. coli ΔmoaC expressing WT or variants of MoaC based on anaerobic growth rates (black bars) or NR activity (gray). All data in D–G are average of at least three replicates, and the errors are based on SDs.
Most of these previous studies have focused on the characterization of MoaA; the contribution of MoaC has been largely unexplored. Here, we report a comprehensive functional and structural study that clarifies these issues and provides strong evidence that MoaC is the enzyme responsible for pyranopterin backbone formation and that MoaA provides 3′,8-cH2GTP as the MoaC substrate. Further structural and functional studies revealed that MoaC catalyzes the complex rearrangement of 3′,8-cH2GTP into cPMP via a unique mechanism involving dynamic conformational changes of substrate and enzyme.
Results
Characterization of MoaA Product.
To clarify the roles of MoaA and MoaC in pyranopterin ring formation, we first determined the product of MoaA. The difficulty in identifying the MoaA product is associated with its limited chemical stability under aerobic conditions (t1/2 ∼ 80 min) (10). As a result, previous studies relied on derivatization of possible MoaA products to more chemically stable forms (8). To detect the authentic and nonderivatized MoaA product, we performed in situ 13C NMR studies that followed the MoaA reaction by using uniformly 13C-labeled GTP as a substrate in an anaerobically sealed NMR tube. This method allowed the unbiased detection of all possible MoaA product(s) derived from GTP without any treatment that could potentially alter the chemical structure of any unstable MoaA product(s). The observed NMR spectrum for the MoaA reaction (Fig. 1C, Bottom) contained signals from [U-13C]GTP and [U-13C]3′,8-cH2GTP (Fig. 1C, Top and Middle, respectively). No signals from pyranopterin molecules were observed, as judged by a comparison with the reported 13C NMR spectra of related pyranopterin molecules, which have characteristic signals at 90–91 ppm (11) (lower detection limit is 5% of 3′,8-cH2GTP). To investigate the relevance of 3′,8-cH2GTP in Moco biosynthesis, the MoaA reaction was quenched at multiple time points and 3′,8-cH2GTP amounts were measured or the resulting products were incubated with MoaC, and the amount of cPMP generated was quantified. At all time points, the quantity of 3′,8-cH2GTP, as determined by its conversion to the fluorescent derivative, coincided with the quantity of cPMP detected upon incubation of the MoaA product with MoaC (Fig. 1D). These results indicate that 3′,8-cH2GTP is likely the physiologically relevant product of MoaA and that it is used as a substrate by MoaC, which then generates cPMP.
A previous study suggested a different compound, pyranopterin triphosphate, as a possible product of MoaA. This proposal was based on the generation of a derivatized molecule with fluorescent properties analogous to oxidized pterins (8). In our assay, we also observed the formation of a molecule with such properties (Fig. S1). However, we found that its estimated amount was 0.4–3% that of 3′,8-cH2GTP, consistent with the absence of corresponding signals in our in situ 13C NMR studies. Importantly, the amount of this fluorescent molecule was not affected by its incubation with MoaC (Fig. S1B), suggesting that it is likely an off-pathway compound formed under in vitro assay conditions. Thus, these combined data indicate that 3′,8-cH2GTP is the only product of MoaA relevant to Moco biosynthesis.
Identification of MoaC Active-Site Amino Acid Residues.
Because MoaC shows no significant sequence similarity to any previously characterized enzyme, the mechanism by which it catalyzes the conversion of 3′,8-cH2GTP to cPMP is unclear. Thus, to obtain functional and mechanistic insight into this process, we next sought to identify residues that when substituted, affected MoaC catalytic activity. Because we hypothesized that MoaC likely uses general acid/base catalysis, we performed Ala scanning on 19 acidic and basic residues that are strictly conserved among MoaC homologs from all kingdoms of life (Fig. S2). The resulting variants of Escherichia coli MoaC were expressed, purified to >90% homogeneity, and assayed for activity by three methods. In the first method, the activities of the MoaC variants were tested by using a previously established coupled assay with MoaA, where the conversion of GTP into cPMP was monitored (6, 9, 10). Using this assay, six MoaC variants were found to exhibit less than 1% activity of the WT protein (Fig. 1E).
In the second assay, steady-state kinetic parameters were determined for WT and variants of MoaC in the absence of MoaA by using purified 3′,8-cH2GTP as a substrate (>95% purity based on 1H NMR). These analyses revealed that WT-MoaC catalyzes the conversion of 3′,8-cH2GTP to cPMP with high specificity (Fig. 1F and Fig. S3), consistent with our hypothesis that 3′,8-cH2GTP is the physiological substrate of MoaC. The six MoaC variants that showed low-level activities in the coupled assay were found to exhibit diminished activity in this study as well (<0.1% kcat/Km of WT MoaC). Three of these variants (H77A-, E112A-, and E114A-MoaC) exhibited 8- to 20-fold increases in Km.
In the third assay, the in vivo activity of MoaC variants were examined by the functional complementation of a Moco-deficient E. coli strain lacking the WT moaC gene (E. coli ΔmoaC). The E. coli ΔmoaC strain does not express functional nitrate reductase (NR) because NR requires Moco as a cofactor. Hence, the E. coli ΔmoaC strain exhibits significant growth defects under conditions of anaerobic nitrate respiration (12, 13). Genetic complementation of this strain with WT-MoaC rescued Moco biosynthesis, as assessed by NR activity and bacterial growth under conditions of anaerobic nitrate respiration (Fig. 1G and Fig. S4). By contrast, complementation with variants of moaC genes resulted in poor growth under the same conditions and very low or no detectable NR activity. These observations indicate that MoaC residues K51, D128, and K131 are important for the in vivo synthesis of Moco. The excellent agreement in the results from our three distinct assays indicates that the MoaC residues selected for substitution are critical for the catalytic function of MoaC in vitro and in vivo. The results also clearly demonstrate the physiological relevance of 3′,8-cH2GTP as the substrate of MoaC.
Structural Determination of MoaC in Complex with 3′,8-cH2GTP.
To obtain structural insight into the MoaC catalytic mechanism, the K51A-MoaC variant, which exhibited significantly reduced kcat without apparent perturbation in Km, was used to solve the structure of the MoaC–3′,8-cH2GTP complex. K51A-MoaC was crystallized without bound substrate, and the crystals were incubated with purified 3′,8-cH2GTP. The resulting K51A-MoaC structure revealed a trimer of homodimers (Fig. 2A; 1.78 Å resolution, Rwork/Rfree = 17.4%/19.4%, PDB ID code 4PYA; Table S1). Although the overall oligomeric state was similar to the previously reported WT apo MoaC structure (12), data from the soaked crystals revealed clear electron density surrounded by five conserved amino acid residues (H77, E112, E114, D128, and K131; Fig. 2B). Importantly, all these residues were shown in the above-described assays to be essential for the in vitro and in vivo catalytic activity of MoaC (Fig. 1 E–G), suggesting that the 3′,8-cH2GTP binding pocket is likely the active site of MoaC. Indeed, the observed electron density in the pocket is well fit by 3′,8-cH2GTP (Fig. 2D): No significant discrepancy between the model and the electron density, nor clashes of the model with surrounding residues, were observed (Fig. S5). Moreover, the modeled 3′,8-cH2GTP makes H-bonding interactions with surrounding conserved and catalytically essential amino acids (Fig. 2B). The presence of 3′,8-cH2GTP in the crystals was also confirmed by HPLC analysis of the crystals following data collection (Fig. S6). All these observations suggest the ligand to be 3′,8-cH2GTP.
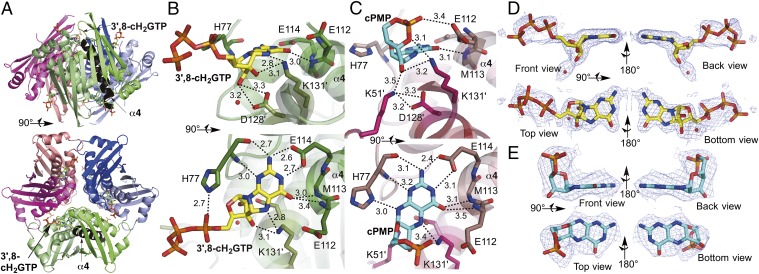
Fig. 2.
Crystal structure of MoaC in complex with 3′,8-cH2GTP or cPMP. (A) The hexameric structure (a trimer of homodimers) of K51A-MoaC with 3′,8-cH2GTP (shown in sticks) bound at the N terminus of helix α4 (highlighted in black). (B and C) The active sites of K51A-MoaC in complex with 3′,8-cH2GTP (B) and WT-MoaC with cPMP (C). Ligands (3′,8-cH2GTP in yellow; cPMP in blue) and the catalytically essential residues are shown as sticks. H-bond interactions are indicated by dashed lines with distances in angstroms. (D and E) Simulated annealing Fo−Fc electron density maps (mesh, contoured at 3 σ). Shown is the electron density in the active-sites of K51A-MoaC•3′,8-cH2GTP (D) and WT-MoaC•cPMP (E).
When other structures previously proposed as the MoaA product were modeled into the electron density, none provided a reasonable fit. In particular, attempts to model pyranopterin triphosphate resulted in significant deviations, and much of the electron density could not be explained by this model (Fig. S5). Specifically, whereas the aminopyrimidinone moiety of pyranopterin triphosphate can be fit to the density and anchored by the H bonds from the E114 side chain, the remainder of the electron density does not accommodate the third ring whatsoever. These crystallographic observations, in combination with the results of the in situ 13C NMR experiments and in vivo and in vitro assays (Fig. 1), support 3′,8-cH2GTP as the likely physiologically relevant substrate of MoaC.
Structural Determination of MoaC in Complex with cPMP.
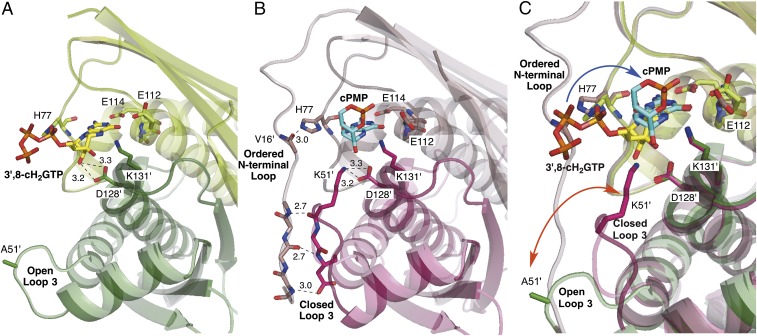
The MoaC structure was also solved in complex with the product, cPMP (Fig. 2C; PDB ID code 4PYD). In this structure, electron density corresponding to cPMP (Fig. 2E) was located in the same pocket as that which binds 3′,8-cH2GTP (Fig. 2C). Moreover, the orientation of the pyrimidinone ring was identical to that of 3′,8-cH2GTP (compare Fig. 2 B and C). These observations further support that this pocket is the MoaC active site. Although the overall oligomeric structures of the WT-MoaC•cPMP and K51A-MoaC•3′,8-cH2GTP complexes are essentially identical, significant differences were observed around their active sites (Fig. 3). In the K51A-MoaC•3′,8-cH2GTP structure (Fig. 3A), loop 3 points away from the active site and the N-terminal loop is disordered. By contrast, in the WT-MoaC•cPMP complex (Fig. 3B), loop 3 points toward the active site, and the N-terminal loop folds and closes off the edge of the active site. These observations suggest that these loops may undergo significant conformational changes (Fig. 3C) during catalysis to account for the structural alterations in the reaction intermediate and to control the accessibility of the catalytically essential K51 to the substrate or reaction intermediate.
Fig. 3.
Coupled conformational dynamics of MoaC and its ligands. (A and B) The crystal structures of K51A-MoaC•3′,8-cH2GTP (A) and WT-MoaC•cPMP (B). Two subunits within a dimer are shown with different shades. Key H-bonding interactions in the N-terminal loop, loop 3, K51, and H77 are indicated by dashed lines with distances in angstroms. (C) Overlay of the structures of K51A-MoaC•3′,8-cH2GTP (green), and WT-MoaC•cPMP (magenta). The conformational change of loop 3 and phosphate moiety of the ligands are indicated by orange and blue arrows, respectively. The phosphate movement is likely irreversible because it is coupled to cyclic phosphate formation, hence the single-headed nature of the blue arrow.
Entrapment of a Reaction Intermediate by Using Active-Site Variants.
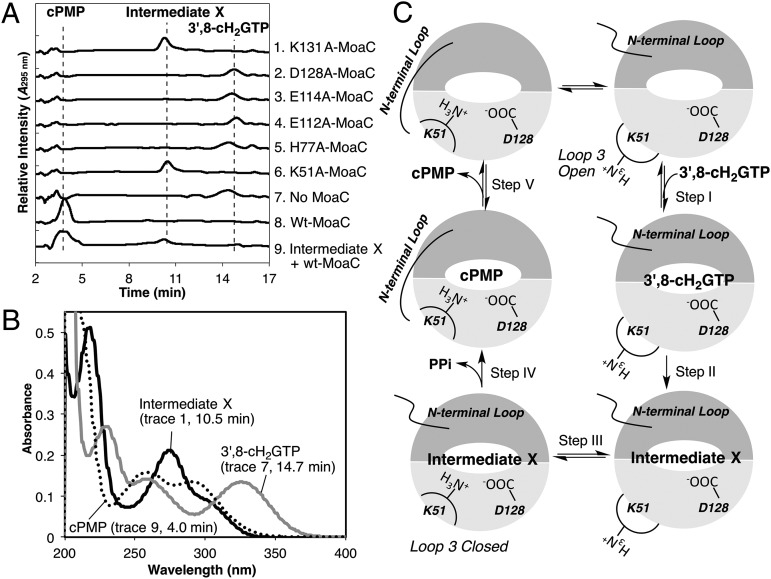
To obtain further mechanistic insight in MoaC catalysis, we analyzed the active-site variants of MoaC for their potential accumulation of reaction intermediates. In this analysis, MoaC variants were incubated with 3′,8-cH2GTP and the resulting product(s) were analyzed by HPLC under strictly anaerobic conditions. Consistent with the aforementioned assay results, these variants did not generate detectable amounts of cPMP (<1% of that observed in the WT-MoaC reaction; Fig. 4A, traces 1–6 and 8). Instead, when K131A- and K51A-MoaC were used, a unique peak (Intermediate X; Fig. 4A, traces 1 and 6) was formed with concomitant loss of 3′,8-cH2GTP. Intermediate X exhibited a molecular mass 18 Da larger than 3′,8-cH2GTP, and a UV-vis absorption spectrum with a λmax at 272 nm and a shoulder at 290 nm, distinct from either 3′,8-cH2GTP (10) or cPMP (3) (Fig. S7 A and B). The comparison of the UV-vis absorption spectra with known compounds (Fig. S7A) in addition to analysis by chemical derivatization (Fig. S7C) suggested that Intermediate X has an acid-labile triaminopyrimidinone base and not a pterin structure. Notably, when Intermediate X was separated from the proteins, and incubated with WT-MoaC, the formation of cPMP with concomitant loss of Intermediate X was observed (Fig. 4A, trace 9). These observations suggest that Intermediate X is the on-pathway reaction intermediate of the MoaC-catalyzed conversion of 3′,8-cH2GTP to cPMP (see Discussion for the possible structures of Intermediate X). The accumulation of the same Intermediate X in the two MoaC variants may also suggest that K131 and K51 act together as a general acid/base catalyst pair and, hence, both are essential for the transformation of Intermediate X to cPMP.
Fig. 4.
Entrapment of the MoaC reaction intermediate (Intermediate X). (A) Anaerobic ion-pairing HPLC analysis of the reaction products of WT or variants of MoaC by using 3′,8-cH2GTP (traces 1–8) or Intermediate X (trace 9) as substrates. The peaks were identified based on the retention time, coinjection with authentic standards, and the UV-vis absorption spectra determined by an inline diode-array detector. Minor fluctuations in the retention time (<1 min) were typical for the ion pairing method. (B) UV-vis absorption spectra of Intermediate X (black solid line), 3′,8-cH2GTP (gray line), and cPMP (dotted line) determined based on diode-array detection of HPLC traces 1, 7, and 9 in A, respectively. (C) Model for loop 3 and N-terminal loop dynamics during the transformation of 3′,8-cH2GTP into cPMP. Circles represent the MoaC dimer units with the active site (white oval) at the dimer interface. Loop 3 and the N-terminal loop are shown in either their open or closed conformations.
Discussion
Our results provide comprehensive enzymological and structural insight into the molecular mechanism of pyranopterin ring formation in Moco biosynthesis. First, our in situ 13C NMR, chemical, and enzymological characterization of the MoaA product provide strong support that 3′,8-cH2GTP is the only product relevant to Moco biosynthesis. Subsequent structural and functional characterization of MoaC reveals that 3′,8-cH2GTP binds to a conserved pocket and forms extensive H-bonding interactions with catalytically essential amino acid residues. Furthermore, cPMP binds in the same pocket with a comparable orientation. These observations all support 3′,8-cH2GTP as the physiologically relevant substrate of MoaC. Based on these observations, we propose that MoaA and MoaC now be designated as GTP 3′,8-cyclase and cPMP synthase, respectively.
The crystal structures of MoaC in complex with 3′,8-cH2GTP and cPMP revealed significant conformational differences of loops adjacent to the active-site: Loop 3 containing the catalytically essential K51 and the N-terminal loop interacting with the catalytically essential H77 both change their conformation depending on the bound ligand. Furthermore, the K51A- and K131A-MoaC variants were found to accumulate the on-pathway reaction intermediate X. The combined structural and enzymological evidence suggest that MoaC catalyzes the complex rearrangement of 3′,8-cH2GTP to cPMP by using conformationally flexible loops as a mechanism to account for the structural change in the reaction intermediates (Fig. 4C). In this model, 3′,8-cH2GTP binds to MoaC with loop 3 and the N-terminal loop in the open conformation (step I). This conformation is capable of converting 3′,8-cH2GTP to Intermediate X (step II). Intermediate X is likely less bulky than 3′,8-cH2GTP around D128, allowing loop 3 closure and incorporation of K51 into the active site (step III). Therefore, we propose that loop 3 serves as a sensor to detect the formation of Intermediate X and triggers the subsequent transformation. Loop 3 closure will also induce folding of the N-terminal loop because these loops make extensive interactions (Fig. 3B). Because the N-terminal loop competes with the triphosphate moiety for the binding site (H77), the folding of the N-terminal loop should accompany the relocation of the triphosphate moiety and facilitate cyclic phosphate formation (step IV). Therefore, the conformational dynamics of the loops are likely the key determinants in directing the complicated and multistep rearrangement of 3′,8-cH2GTP into cPMP.
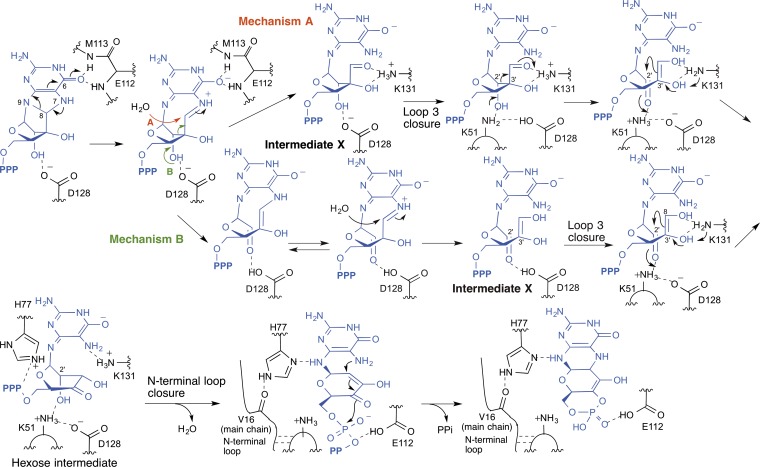
This conformational gating model aligns with the possible chemical mechanism of MoaC catalysis (Fig. 5). In this model, MoaC initiates the catalysis by cleaving the C8-N9 bond by stabilizing the anionic charge on O6 via H-bonding interactions with the amide NH of E112 and M113. The resulting imine intermediate will be hydrolyzed (mechanism A) or cleaved at the C2′-C3′ bond (mechanism B). Mechanism A is based on the general susceptibility of an aminal functional group to acid hydrolysis (10), and the solvent exposure of C8 in the K51A-MoaC•3′,8-cH2GTP complex crystal structure. In this mechanism, Intermediate X is a nucleotide with an aldehyde functionality at the 3′ position. This intermediate would be less bulky around 2′-OH compared with 3′,8-cH2GTP, allowing the incorporation of K51 into the active site and the closure of loop 3. The presence of K51 in the active site is essential for the subsequent ring expansion reaction by retroaldol-aldol rearrangement to yield the hexose intermediate. Mechanism B is based on the presence of D128 in H-bonding distance from 2′-OH and the absence of Intermediate X accumulation in the D128A-MoaC reaction. In this mechanism, Intermediate X is a nucleotide without the C2′-C3′ bond, which leaves significant flexibility around D128, and allows loop 3 closure and incorporation of K51. Subsequent aldol condensation would yield the common hexose intermediate. The conversion of the hexose intermediate to cPMP is common to both mechanisms and proceeds via the concerted formation of the pyranopterin and the cyclic phosphate rings. This reaction is likely facilitated by the folding of the N-terminal loop, induced by the closure of loop 3. The N-terminal loop competes with the triphosphate moiety for interaction with H77. Thus, its folding would induce the relocation of the triphosphate moiety and subsequent cyclization reactions. Therefore, the chemical reaction catalyzed by MoaC is likely coupled to the dynamic motions of the active-site loops.
Fig. 5.
Proposed chemical mechanism of MoaC catalysis. MoaC initiates the reaction by stabilizing the anionic charge on O6, which results in either aminal hydrolysis (mechanism A), or C2′-C3′ bond cleavage (mechanism B). Shown are the structures of possible intermediates, including Intermediate X and the hexose intermediate, and the amino acid residues relevant to the chemistry of each step. See main text for details about the mechanism.
In conclusion, our enzymological and structural characterizations provided strong evidence for the physiological functions of MoaA (GTP 3′,8-cyclase) and MoaC (cPMP synthase). The structure guided mechanistic studies of MoaC revealed the importance of conformational changes within the MoaC active-site during catalysis. Considering the strict conservation of the MoaC catalytic residues identified in this study, the function and mechanism used by MoaC homologs are likely conserved among all kingdoms of life, from bacteria to humans.
Materials and Methods
S. aureus MoaA and E. coli MoaC (WT and variants) were expressed and purified as described in SI Materials and Methods. MoaC variants were prepared by site-directed mutagenesis using the primers in Table S2. MoaA and MoaC activity assays and the product analyses were carried out as described (10). The in situ 13C NMR characterization of MoaA products was performed by using [U-13C]GTP (Sigma-Aldrich) as a substrate under strict anaerobic conditions. The in vivo characterization of MoaC was performed by using E. coli ΔmoaC with a pBAD plasmid harboring WT or variants of MoaC. WT- and K51A-MoaC were crystallized by using the vapor-diffusion technique, and the ligands were introduced by the soaking method. The crystal structures were solved by using molecular replacement using the apo E. coli MoaC structures as a search model (PDB ID code: 1EKR). The details for all of the experimental protocols can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
X-ray intensity data were collected at beamline 22-ID at the Advanced Photon Source, Argonne National Laboratory through the Duke University X-ray Crystallography Shared Resource. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, and the Office of Basic Energy Sciences under Contract W-31-109-Eng-38. This work was supported by NIH Grant GM074815 (to M.A.S.) and Duke University Medical Center Department of Biochemistry start-up funds (to K.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.J.B. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4PYA and 4PYD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500697112/-/DCSupplemental.
References
- 1.Mendel RR, Schwarz G. Molybdenum cofactor biosynthesis in plants and humans. Coord Chem Rev. 2011;255:1145–1158. [Google Scholar]
- 2.Leimkühler S, Wuebbens MM, Rajagopalan KV. The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coord Chem Rev. 2011;255(9-10):1129–1144. doi: 10.1016/j.ccr.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wuebbens MM, Rajagopalan KV. Structural characterization of a molybdopterin precursor. J Biol Chem. 1993;268(18):13493–13498. [PubMed] [Google Scholar]
- 4.Wuebbens MM, Rajagopalan KV. Investigation of the early steps of molybdopterin biosynthesis in Escherichia coli through the use of in vivo labeling studies. J Biol Chem. 1995;270(3):1082–1087. doi: 10.1074/jbc.270.3.1082. [DOI] [PubMed] [Google Scholar]
- 5.Reiss J, et al. Mutations in a polycistronic nuclear gene associated with molybdenum cofactor deficiency. Nat Genet. 1998;20(1):51–53. doi: 10.1038/1706. [DOI] [PubMed] [Google Scholar]
- 6.Hänzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci USA. 2004;101(35):12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit Rev Biochem Mol Biol. 2008;43(1):63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 8.Mehta AP, Abdelwahed SH, Xu H, Begley TP. Molybdopterin biosynthesis: Trapping of intermediates for the MoaA-catalyzed reaction using 2′-deoxyGTP and 2′-chloroGTP as substrate analogues. J Am Chem Soc. 2014;136(30):10609–10614. doi: 10.1021/ja502663k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta AP, et al. Catalysis of a new ribose carbon-insertion reaction by the molybdenum cofactor biosynthetic enzyme MoaA. Biochemistry. 2013;52(7):1134–1136. doi: 10.1021/bi3016026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hover BM, Loksztejn A, Ribeiro AA, Yokoyama K. Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J Am Chem Soc. 2013;135(18):7019–7032. doi: 10.1021/ja401781t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinch K, et al. Synthesis of cyclic pyranopterin monophosphate, a biosynthetic intermediate in the molybdenum cofactor pathway. J Med Chem. 2013;56(4):1730–1738. doi: 10.1021/jm301855r. [DOI] [PubMed] [Google Scholar]
- 12.Wuebbens MM, Liu MT, Rajagopalan K, Schindelin H. Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC. Structure. 2000;8(7):709–718. doi: 10.1016/s0969-2126(00)00157-x. [DOI] [PubMed] [Google Scholar]
- 13.MacGregor CH, Schnaitman CA. Alterations in the cytoplasmic membrane proteins of various chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1971;108(1):564–570. doi: 10.1128/jb.108.1.564-570.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santamaria-Araujo JA, et al. The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J Biol Chem. 2004;279(16):15994–15999. doi: 10.1074/jbc.M311815200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.