Significance
Serotype V group B Streptococcus (GBS) infection rates in humans have steadily increased during the past several decades. We determined that 92% of bloodstream infections caused by serotype V GBS in Houston and Toronto are caused by genetically related strains called sequence type (ST) 1. Whole-genome analysis of 202 serotype V ST-1 strains revealed the molecular relationship among these strains and that they are closely related to a bovine strain. Moreover, we found that a subset of GBS genes is under selective evolutionary pressure, indicating that proteins produced by these genes likely contribute to GBS host–pathogen interaction. These data will assist in understanding how bacteria adapt to cause disease in humans, thereby potentially informing new preventive and therapeutic strategies.
Keywords: Streptococcus agalactiae, pathogenesis, evolution, single nucleotide polymorphisms, surface protein
Abstract
The molecular mechanisms underlying pathogen emergence in humans is a critical but poorly understood area of microbiologic investigation. Serotype V group B Streptococcus (GBS) was first isolated from humans in 1975, and rates of invasive serotype V GBS disease significantly increased starting in the early 1990s. We found that 210 of 229 serotype V GBS strains (92%) isolated from the bloodstream of nonpregnant adults in the United States and Canada between 1992 and 2013 were multilocus sequence type (ST) 1. Elucidation of the complete genome of a 1992 ST-1 strain revealed that this strain had the highest homology with a GBS strain causing cow mastitis and that the 1992 ST-1 strain differed from serotype V strains isolated in the late 1970s by acquisition of cell surface proteins and antimicrobial resistance determinants. Whole-genome comparison of 202 invasive ST-1 strains detected significant recombination in only eight strains. The remaining 194 strains differed by an average of 97 SNPs. Phylogenetic analysis revealed a temporally dependent mode of genetic diversification consistent with the emergence in the 1990s of ST-1 GBS as major agents of human disease. Thirty-one loci were identified as being under positive selective pressure, and mutations at loci encoding polysaccharide capsule production proteins, regulators of pilus expression, and two-component gene regulatory systems were shown to affect the bacterial phenotype. These data reveal that phenotypic diversity among ST-1 GBS is mainly driven by small genetic changes rather than extensive recombination, thereby extending knowledge into how pathogens adapt to humans.
The recent increase in large-scale DNA sequencing feasibility has allowed for significant advances in understanding the population genetics of bacteria that cause disease in humans (1, 2). A major outcome of the rapid expansion of available bacterial genomes has been the appreciation of the marked intraspecies genetic variability present in a wide variety of human bacterial pathogens (3, 4). This genetic variation can have profound impact on host–pathogen interaction by affecting transmissibility, infection severity, and antimicrobial resistance (2, 5, 6). The observed genetic intraspecies variability can arise via many distinct mechanisms including large-scale events such as recombination and bacteriophage-mediated horizontal gene transfer as well as small-scale genetic changes such as short insertions, deletions, and/or single nucleotide changes (2, 3, 5).
Group B Streptococcus (GBS) is a common colonizer of humans that emerged in the 1970s as the leading cause of invasive bacterial disease in neonates and infants less than 3 mo of age (7). GBS is divided into 10 serotypes based on the carbohydrate composition of its sialic acid containing capsule, but gene content at the genomic level does not necessarily correlate with capsular serotype (8, 9). A seven-gene multilocus sequence typing (MLST) allows for the classification of the majority of GBS strains isolated from humans into five major clonal complexes (CCs) with a recent study by Da Cunha et al. showing that the major GBS CCs are primarily derived from a limited number of tetracycline-resistant clones, suggesting a key role of tetracycline resistance in GBS strain emergence (10, 11). CC-17 GBS strains have been particularly well studied given their role as the major cause of severe, invasive infant disease (10, 12). In contrast, serotype V strains cause a larger percentage of invasive disease in nonpregnant adults compared with neonates (13–15). Importantly, rates of invasive GBS disease have been increasing during the past 25 y in nonpregnant adults, with a significant part of the rise resulting from serotype V GBS strains (15–17).
Despite the clear and increasing impact of serotype V strains, data are limited regarding molecular epidemiology of serotype V GBS causing invasive disease in nonpregnant adults (14, 15, 18). Only a few studies of serotype V strains have investigated the noncapsular genetic makeup of the strains, and those that have done so have included colonizing and invasive GBS strains isolated from infants or have not described the clinical origin of the tested strains (18, 19). Thus, we sought to analyze a large cohort of clinically well-defined, geographically distinct, and temporally disparate GBS isolates by using a whole-genome approach to elucidate the population structure of serotype V GBS causing invasive disease in nonpregnant adults. Data using non–genome-wide level approaches found that many serotype V strains were closely related, suggesting that a particular clone, rather than a genetically diverse array of strains arising from large-scale recombination, might be responsible for the majority of serotype V disease (18). Thus, we specifically sought to test the hypothesis that genetic diversity among invasive serotype V GBS strains is driven by small genetic changes at loci that are critical to GBS host–pathogen interaction.
Results
The Vast Majority of GBS Serotype V Bloodstream Isolates from Nonpregnant Adults Are Multi-Locus Sequence Type 1.
We determined the MLST of 229 serotype V GBSs isolated from the blood of unique nonpregnant adults in Houston, TX, and Toronto, ON, Canada (SI Appendix, Table S1). A total of 200 isolates were sequence type (ST) 1 and an additional 10 isolates were single-loci variants that we will consider ST-1 for the purposes of this manuscript (Materials and Methods provides further details on these 10 strains), for a total of 210 of the 229 strains (92%). The non–ST-1 serotype V strains were a mixture of other STs, with the most common being ST-19. Thus, the overwhelming majority of invasive serotype V GBS strains causing bacteremia in nonpregnant adults in Houston and Toronto belong to a single ST.
Determination of a Complete Genome Sequence of an ST-1 GBS Strain Causing Invasive Human Infection.
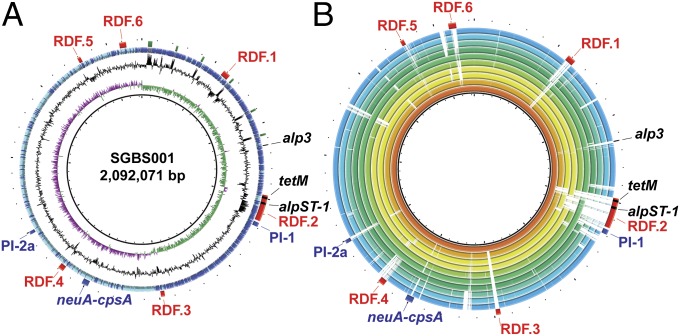
As a complete genome of an ST-1 GBS strain isolated from a human had not been determined previously, we next used a combination of long reads by using a PacBio instrument and paired-end Illumina short-read data to completely assemble the genome of the ST-1 strain SGBS001 (20). The genome was 2,092,071 bp with 2,061 predicted ORFs and contains genes predicted to encode alpha-like protein (Alp) 3 (Alp3) and pilus 1 and 2a (Fig. 1A) (21, 22). Compared with GBS strains whose complete genomes are available from the National Center for Biotechnology Information (NCBI), strain SGBS001 was most similar to the serotype V Swedish cow mastitis strain 09mas018883 (also ST-1; Fig. 1B) (23). Strain SGBS001 is >99% similar and has >99% coverage compared with strain 09mas018883, with the major difference being the presence of a ∼40 kb region uniquely in strain 09mas018883 (located between homologs of rdf_1233 and rdf_1234), which includes genes encoding proteins involved in lactose utilization (the lac.2 operon; Fig. 1B). The lac.2 operon has previously been shown to be preferentially found in GBS isolated from cattle (24). Given that the presence of the lac.2 operon in bovine GBS strains has been suggested to occur via lateral gene transfer (25), our findings indicate that a single genetic event could account for the majority of differences observed between these two strains.
Fig. 1.
Whole-genome analysis of an ST-1 serotype V GBS strain isolated from the bloodstream of a nonpregnant adult. (A) Genome atlas for the reference serotype V ST-1 strain SGBS001. Genome scale in megabases is given in the innermost circle (circle 1). Circle 2 shows GC skew, calculated as (G − C)/(G + C) and averaged over a moving window of 10,000 bp showing excess G (green) and C (purple). GC content is displayed in circle 3 with values above average (outward directed) or below average (inward directed) indicated. Annotated coding sequences are shown in dark blue (forward, clockwise) and light blue (reverse, counterclockwise) in circle 4. Reference genome landmarks are displayed in circle 5 and include ribosomal operons (green), MGEs (red, RDF.1–6), pilus islands [blue, pilus island 1 (PI-1) and 2a (PI-2a)], capsule biosynthesis operon (blue, neuAcpsA), and Alp-encoding genes. (B) BLASTN comparisons of the reference genome SGBS001 and publically available completed GBS genomes as labeled in the legend. The innermost ring shows the genome scale (in megabases) and subsequent rings (inner- to outermost) show BLASTN comparisons in order of decreasing homology (ring, serotype, NCBI accession no.) for 09mas018883 (1, V, NC_ 021485), A909 (2, 1a, NC_007432), GD201008-001 (3, 1a, NC_018646), 2603V/R (4, V, NC_004116), NEM316 (5, III, NC_004368), ILRI112 (6, VI, NC_021507), ILRI005 (7, V, NC_021486), 2–22 (8, Ia, NC_021195), SA20-06 (9, Ia, NC_019048), and 138P (10, Ib, CP007482). Reference genome landmarks for SGBS001 are shown as in A.
Description of a Novel Alp Present in SGBS001.
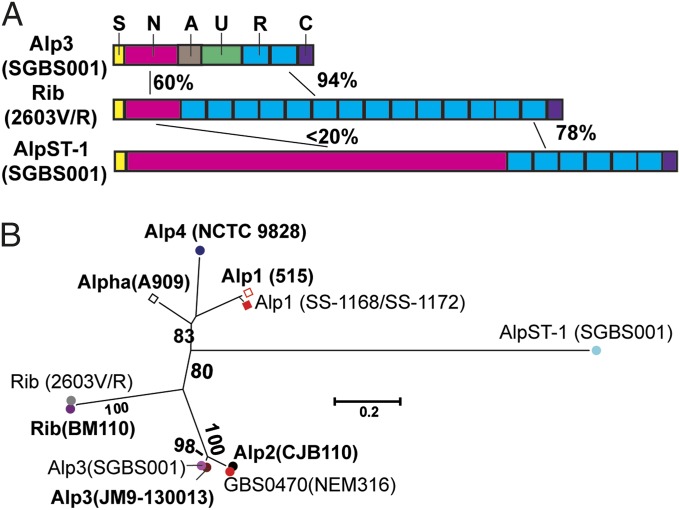
To begin to study why ST-1 GBS strains have a specific predilection for causing disease in nonpregnant adult humans, we searched the SGBS001 genome for cell-surface or actively secreted proteins that were unique to ST-1 GBS. We found a single gene, rdf_0594, present on the mobile genetic element (MGE) RDF.2 (Fig. 1A and SI Appendix, Fig. S1), which encodes a cell-surface protein that was absent in non–ST-1 GBS as determined by BLAST analysis. RDF_0594 is predicted to be 1,769 aa long and to contain an N-terminal secretion signal sequence and a C-terminal LPXTG cell-wall localization motif consistent with cell-surface localization (Fig. 2A). The C-terminal portion of RDF_0594 contains six repeats of 79 aa that are 77% similar to the repeats found in the alpha-like Rib protein (21). Alpha and Alps are major components of the GBS cell surface, although their precise role in GBS pathogenesis is unknown (26). Although the N-terminal signal sequence and C terminus of RDF_0594 place it in the Alp family, the N-terminal 1,187 aa of RDF_0594 have no readily identifiable homology to other GBS Alps. By homology modeling using I-TASSER and GenTHREADER (27, 28), these 1,187 aa were predicted to consist of a repeat β-sheet structure typically found in proteins involved in cell adhesion or binding to host proteins, such as fibrinogen or complement factor binding proteins (29). Interestingly, strains SS-1168 and SS-1172, which were among the first serotype V GBS strains isolated from humans (in 1977 and 1978, respectively) and were confirmed to be ST-2, which differs by only one SNP in the atr allele from ST-1, lacked the RDF.2 MGE containing rdf_0594 and encoded Alp1 rather than Alp3 (Fig. 2B). Given that RDF_0594 is quite distinct from other Alps (Fig. 2B) and has not previously been described outside of ST-1 GBS strains, we have named it AlpST-1 and suggest that further study of the role of this protein in GBS pathogenesis is warranted.
Fig. 2.
Characteristics of novel alpha-like protein from ST-1 GBS. (A) Comparison of structure of Alps from strain 2603V/R and SGBS001. Schematic of “S” (signal), “N” (N terminus), “A” (A repeat), “U” (unknown), “R” (repeat), and “C” (C terminus) regions are from Lachenauer et al. (21). Numbers refer to percent amino acid identify. (B) Phylogenetic tree of alpha and Alps was created by using MEGA following alignment via ClustalW. In bold type are alpha and alpha-like type sequences derived from Lachenauer et al. (21) with strain source indicated in parentheses. In regular type are alpha and Alps from the fully sequenced strains 2603V/R (serotype V), NEM316 (serotype III), and SGBS001 (serotype V) and the early serotype V isolates SS-1168 and SS-1172. Numbers refer to confidence of branching as determined by bootstrap analysis (1,000 replicates). Bracket refers to genetic distance.
Large-Scale Recombination Is Not the Major Factor Driving Genetic Diversity Among ST-1 GBS.
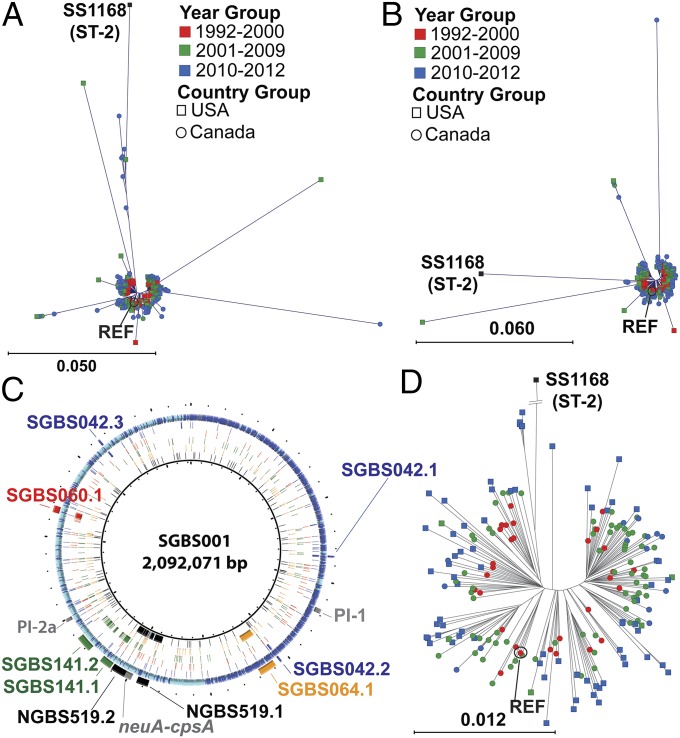
We next sought to determine the genetic relationship of the invasive ST-1 strains at the whole-genome level. Polymorphisms in the genome sequences of 201 ST-1 strains were identified relative to strain SGBS001 by using two independent pipelines [variant ascertainment algorithm (VAAL) (30) and CLC Genomics Workbench version 7; Materials and Methods], and 9,876 unique SNP loci were used to determine phylogenetic relationships (Fig. 3A). After accounting for differences in MGE content, eight strains showed greater phylogenetic divergence compared with the main phylogenetic cluster of ST-1 strains (Fig. 3B). By using Bayesian analysis of recombination (BRATNextGen) (31), we identified discrete regions of recombination with non–ST-1, non-serotype V GBS in these outlying strains (SI Appendix, Fig. S2 and Table S2). For example, strain SGBS064 contains a 41-kb region that most closely resembles DNA from the ST-23, serotype III strain NEM316 (labeled in orange as SGBS064.1 in Fig. 3C). Importantly, however, evidence of such recombination was quite rare among the ST-1 strains in this cohort, indicating that recombination is not a major force driving genetic diversity among serotype V ST-1 GBS. Given that we analyzed only serotype V strains, we would not have detected recombination involving the capsular polysaccharide synthesis (cps) locus, but a recent study of 229 GBS isolates identified only one ST-1 strain that was not capsular type V, suggesting that cps recombination is not common among ST-1 strains (10). The eight ST-1 strains identified as having significant recombination were excluded from the remainder of our analysis, leaving a total of 194 sequenced ST-1 strains.
Fig. 3.
Whole genome-level phylogenetic analysis of invasive ST-1 GBS strains. (A) A rooted neighbor joining tree representing 10,365 unique SNP loci from 201 serotype V ST-1 GBSs relative to the reference genome SGBS001. The outgroup ST-2 strain SS1168 is labeled for reference. (B) Rooted neighbor joining tree after exclusion of MGEs (RDF.1–6) representing 8,459 unique SNP loci from 201 GBS isolates as in A. (C) Genome atlas showing SNP distribution of strains NGBS519 (black, innermost, ring 1), SGBS064 (orange, ring 2), SGBS141 (green, ring 3), SGBS060 (red, ring 4), and SGBS042 (blue, ring 5). Ring 6 shows selected reference genome landmarks including pilus islands [pilus island 1 (PI-1) and 2a (PI-2a)] and capsule biosynthesis genes (neuA-cpsA). Also displayed are regions of putative recombination as identified by BratNextGen for each strain. (D) Rooted neighbor joining tree representing 6,375 unique SNP loci from 194 serotype V ST-1 GBSs after excluding strains with potential recombination. For A, B, and D, the year and location of strain isolation is as shown in the legend.
Genetic Relationship Among ST-1 GBS Strains Lacking Evidence of Recombination.
By using the method of Harris et al. (1), we determined that the core genome of the 194 ST-1 strains without evidence of recombination was 1,931 genes, and included the gene encoding the AlpST-1 protein. Moreover, the majority of gene variance compared with SGBS001 was observed in only one or two strains (SI Appendix, Table S3). A total of 2,001 genes, or 97% of the SGBS001 genome, were present in at least 192 of the 194 strains, demonstrating that there were minimal differences in terms of gene presence or absence among the ST-1 strains.
The elucidation of the ST-1 core genome and the finding that only a few strains had significant recombination provided the opportunity to determine the genetic relationship of ST-1 strains. Phylogenetic analysis for the 194 strains was performed by using 5,880 unique SNP loci (Fig. 3D). The average number of genetic polymorphisms separating any two ST-1 strains was 97, which is consistent with the relatively small number of SNPs separating strains of serotype M1 or M3 group A Streptococcus (4, 5). When temporal and geographic factors were considered, we observed a radial pattern of divergence that appeared temporally but not geographically dependent (Fig. 3D shows a radial phylogenetic tree whereas SI Appendix, Fig. S3A, shows a phylogram of the same dataset). For example, when strains were divided into early (1992–2000; Fig. 3D, red), middle (2001–2009; Fig. 3B, green), and late time groupings (2010–2012; Fig. 3D, blue), strains from the latter time group tended to be located on the periphery of the phylogenetic tree whereas strains form the early period were primarily located closest to center. Although one possible explanation for this appearance is that the Canadian strains were exclusively isolated in the later time period, this relationship was replicated when only Houston strains were analyzed (SI Appendix, Fig. S3B). Moreover, strains that were isolated from Toronto were interspersed among strains from Houston (Fig. 3D) despite the fact that two cities are separated by ∼2,100 km. To statistically test our visual observation of temporal divergence, we calculated the number of SNPs that separated strains within each group and found a statistically significant, progressive increase (i.e., late > middle > early) in the average number of intragroup SNPs (P < 0.05, Mann–Whitney U test; SI Appendix, Fig. S3C). Given the finding of temporal divergence, we used Path-O-Gen to estimate the origin of serotype V ST-1 strains and arrived at a date of 1963, which is relatively close to the first reported isolation of serotype V from humans in 1975 (18) (SI Appendix, Fig. S3D). We additionally analyzed 24 serotype V ST-1 strains from three different continents and for which whole-genome data were recently reported (10) and found that the strains were interspersed among our North American isolates, suggesting the global dissemination of the ST-1 clone (SI Appendix, Fig. S4).
High Rates of Antimicrobial Resistance Genes Present in ST-1 Strains.
Consistent with a recent report regarding the key role of tetracycline resistance in GBS evolution (10), tetracycline resistance elements were found ubiquitously throughout our collection (91%), with the most common determinant being tetM (SI Appendix, Fig. S5A). When present, tetM was invariably located in the transposable element Tn916 inserted in RDF.2 (SI Appendix, Fig. S1) at the location reported for the Tn916–1 lineage by Da Cunha et al. (10). Macrolide resistance factors were present at a relatively higher rate in the present study (59%; SI Appendix, Fig. S5A) than what has been reported in other GBS studies (10, 17). The majority of macrolide resistance elements were ermB or ermTR (SI Appendix, Fig. S5A), with ermB being colocalized with tetM in Tn3872 as recently described (10). The vast majority of strains carrying ermB alleles clustered on the same phylogenetic branch, indicating that single insertion events resulted in most of the macrolide resistant strains (SI Appendix, Fig. S5B). The two serotype V strains from the 1970s lacked tetM or erm genes, in accord with the absence of RDF.2.
Identification of GBS Genes Undergoing Positive Selection.
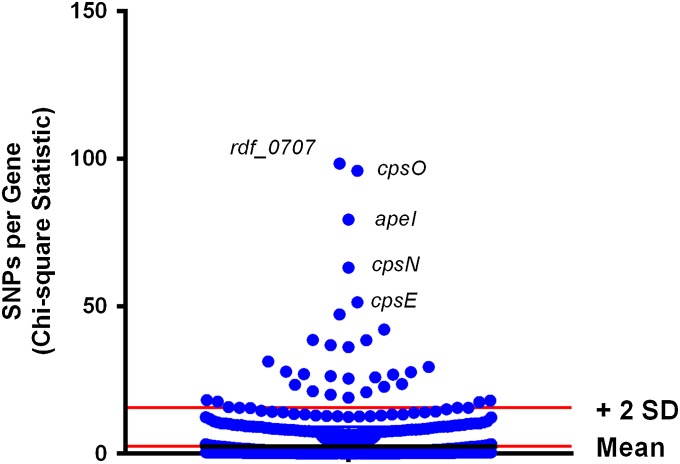
We next determined the degree of genetic variation at each of the 1,931 loci present in the core genome for the 194 ST-1 strains to ask the question whether particular GBS genes have high levels of genetic variation, as such loci would be expected to participate in host–pathogen interaction (32). After accounting for multiple comparisons, 31 genes had significantly higher genetic variation levels compared with the remainder of the genome (Fig. 4, Table 1, and SI Appendix, Table S4). These highly polymorphic loci included genes encoding proteins involved in polysaccharide capsule synthesis, regulators of pilus production, and members of two-component gene regulatory systems (TCS) (Table 1). There was a strong bias toward nonsynonymous SNPs in these genes with a nonsynonymous:synonymous ratio of 5, suggesting that these genes are undergoing adaptive evolution as a result of selective pressure (32) (Table 1).
Fig. 4.
Identification of GBS genes with high levels of genetic variation. Shown is the distribution of χ2 statistics for observed vs. expected numbers of SNPs for the 1,931 genes in the serotype V ST-1 core genome. The genes with the five lowest corrected P values are labeled.
Table 1.
Examples of GBS genes with significantly increased genetic variation
| Gene location | Gene name | Mutations* | Putative function of encoded protein |
| rdf_1161 | cpsO | 17/0 | Capsule synthesis protein |
| rdf_0654 | ape1 | 15/2 | Pilus 1 regulator |
| rdf_1162 | cpsN | 13/2 | Capsule synthesis protein |
| rdf_1167 | cpsE | 17/0 | Capsule synthesis protein |
| rdf_0315 | 7/1 | Response regulator | |
| rdf_1166 | cpsF | 7/0 | Capsule synthesis protein |
| rdf_1410 | rga | 9/0 | Pilus 2a regulator |
| rdf_0973 | 12/3 | Oligopeptide transporter | |
| rdf_1359 | rogB | 10/2 | Pilus 2a regulator |
| rdf_1119 | skzL | 10/3 | Streptokinase B |
| rdf_1169 | cpsC | 4/1 | Capsule synthesis protein |
Nonsynonymous/synonymous mutations.
Small Genetic Changes Are Associated with Significant Changes in Capsule and Pilus Production.
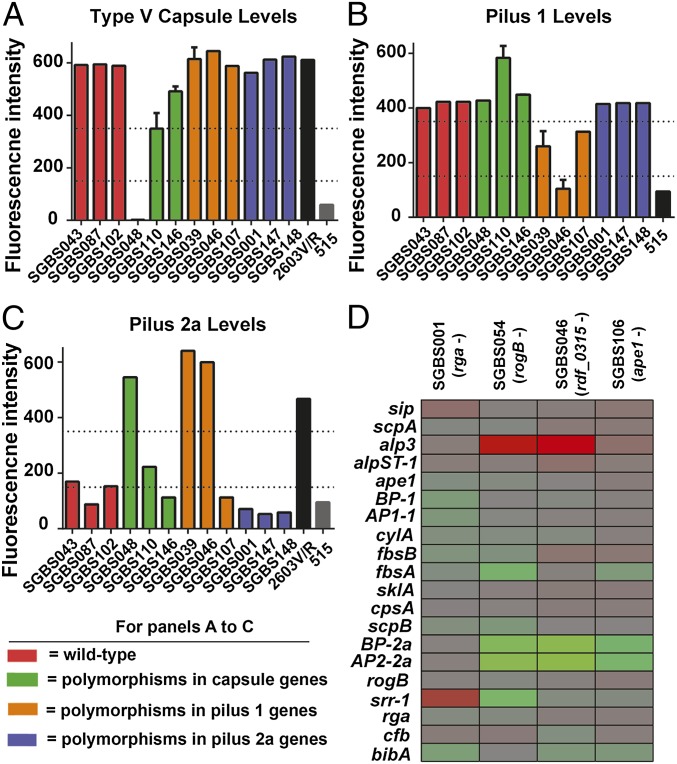
The genetic variation data suggested that alterations in polysaccharide capsule and pilus production figure prominently in ST-1 interstrain variation. Capsule and pilus proteins are the major targets of proposed GBS vaccines (33). To confirm that the observed genetic alterations were associated with phenotypic diversity in capsule and pilus levels, we tested the ability of strains with defined genetic alterations (SI Appendix, Table S1) to produce type V capsule and type 1 and 2a pilus proteins by using monoclonal antibodies and FACS analysis (22). Consistent with the genetic data, the only strains to produce low or undetectable levels of type V capsule were those with insertions in the capsule biosynthesis encoding genes cpsG and cpsO (Fig. 5A). Similar to the capsule data, pilus 1 levels were low only in the three strains that contained genetic alterations encoding for pilus 1 accessory or backbone proteins (Fig. 5B). Compared with pilus 1, pilus 2a levels were more variable across the strains but were undetectable in the three strains that contained predicted deleterious polymorphisms in genes encoding pilus 2a encoding proteins (Fig. 5C).
Fig. 5.
Small genetic changes lead to significant phenotypic variation. (A–C) Indicated strains are color coded as follows: red strains are WT at capsule and pilus loci; green strains have polymorphisms in capsule encoding genes; orange strains have polymorphisms in pilus 1 encoding genes; purple strains have polymorphisms in pilus 2a encoding genes; and black/gray strains are controls. Strains that have mutations in one loci are WT at the other two loci (e.g., strains with capsule locus mutations are WT at pilus 1 and 2a loci), and the same 12 ST-1 strains are tested in A–C. Data graphed are fluorescence above baseline for indicated monoclonal antibody. Lower dashed line is cutoff between positive and negative, whereas upper dashed line is cutoff between low and high expression (33). (A) Type V capsule: strain 2603V/R (serotype V) is positive control whereas strain 515 (serotype 1a) is negative control. (B) Type 1 pilus: strain 515 is negative control. (C) Type 2a pilus: strain 515 is positive control and strain 2603V/R is negative control. (D) Heat map of RNA-Seq data for selected genes. Log2 fold transcript level is shown relative to strain SGBS102. Strain SGB102 is WT at the loci encoding regulatory proteins which are mutated in strains as indicated in the figure.
Polymorphisms in Regulatory Protein Encoding Genes Are Associated with Distinct Transcriptomes.
Given that several regulator encoding genes contained high levels of genetic polymorphisms (Table 1), we next tested the hypothesis that strains with polymorphisms in known or putative regulatory genes have distinct transcriptomes. To this end, we used RNA sequencing (RNA-Seq) to compare the transcriptome of four strains with defined mutations in regulatory genes that were identified as being highly polymorphic in our genetic variation analysis. The genotypes of the strains with defined mutations in four distinct regulatory proteins are shown in Fig. 5D (genotype details are provided in SI Appendix, Table S1), whereas strain SGBS102 is WT at these four regulatory loci and thus was chosen as the control strain for our analysis. In concert with the idea that the observed genetic polymorphisms resulted in significant phenotypic variation, principal component analysis of transcriptome data showed that the global gene expression profiles of the tested strains were quite distinct (Fig. 5D and SI Appendix, Fig. S6).
The minimum number of genes with differential transcript levels (defined as mean transcript level at least twofold and final, corrected P ≤ 0.05) between the test and control strains (e.g., SGBS001 vs. SGBS102 or SGBS106 vs. SGBS102) was 25 for strain SGBS106, whereas the highest number was 92 for strain SGBS046, which encodes a truncated form of the TCS regulator RDF_0315 (SI Appendix, Table S5). Multiple genes known to contribute to GBS host–pathogen interaction were included among the differentially transcribed genes including srr-1, pilus encoding genes, Alp encoding genes, and bibA, which encodes an cell-surface adhesion critical to GBS survival in human blood (Fig. 5D) (34). An example of the diversity observed among the studied strains comes from the alp3 gene, whose transcript level was ∼25-fold lower in strains SGBS054 and SGBS046 compared with strain SGBS102 (Fig. 5D). In concert with the capsule and pilus expression data, the transcriptome analyses show that small genetic variations can be associated with significant phenotypic variation among ST-1 GBS strains.
Discussion
The increasing feasibility of whole-genome sequencing of large cohorts of clinical bacterial isolates is beginning to elucidate mechanisms of pathogen evolution, which, in turn, is providing key insights into a broad range of medically important issues such as occurrence of epidemics and transmission of drug-resistant pathogens (2, 4, 6). Most studies that involve high density genomic sequencing of bacterial pathogens investigated bacteria that have been prevalent causes of human disease for centuries (1, 4, 6). In contrast, GBS has only recently emerged as a significant cause of human infection, perhaps as a result of the proliferation of tetracycline-resistant clones during an era of widespread tetracycline use, as suggested by Da Cunha et al. (10, 15, 16, 18).
A key finding of the present work was that ST-1 strains that are highly similar at the whole-genome level accounted for >90% of the invasive infections in Houston and Toronto caused by serotype V GBS in nonpregnant adults. Moreover, diversity in the ST-1 strains was primarily driven by small genetic events rather than recombination. This finding was somewhat unexpected, as recombination has been found to be a major driver of GBS genetic diversity when diverse serotypes are analyzed (3, 10). Together with previous studies, these data suggest that GBS evolution may be viewed as similar to the antigenic shift/antigenic drift model of influenza in which recombination drives the emergence of new GBS subtypes (as may have occurred for serotype V in the mid-1970s), which then slowly accumulate new genetic polymorphisms over time. A recent report by Da Cunha et al. analyzing a broad array of GBS serotypes also found that the majority of GBS CCs comprised strains that were highly clonal in nature, as exemplified by a 3% recombination rate among ST-17 strains, suggesting that a similar population structure may hold for other major GBS STs as we describe herein for ST-1 (10).
Intriguingly, analysis of two of the first serotype V strains causing disease in humans (strains ST-1168 and ST-1172) determined that these strains were a single loci variant of ST-1, but lacked the Alp3 and AlpST-1 proteins ubiquitous in our 1992–2013 cohort. It has been shown that serotype V strains were present in humans for approximately 15 y before expanding in the early 1990s and that the pulsed-field gel electrophoresis (PFGE) pattern of serotype V strains responsible for the majority of the expansion observed in the 1990s is the same as serotype V strains isolated as early as 1975 (18). Our MLST findings are in concert with the previously published PFGE data, but the increased sensitivity of our whole-genome sequencing approach reveals that acquisition of Alp3 and AlpST-1 by ST-1 strains occurred sometime between the first known isolation of serotype V GBS from humans in 1975 and the expansion of serotype V strains observed in the early 1990s. It was recently suggested that acquisition of tetracycline resistance has been a major force in the emergence of GBS, and the finding that ST-1168 and ST-1172 lack tetM is consistent with this proposal (10). Importantly, however, tetM is present in the RDF.2 MGE, which also contains the gene encoding AlpST-1 along with numerous other proteins (SI Appendix, Fig. S1). Thus, at least for ST-1 strains, it remains possible that the widespread nature of tetM is a result of its colocalization with other key genes rather than tetracycline resistance per se being the driving force for clone emergence. Similarly to Da Cunha et al., we found that a significant proportion of our ST-1 strains contained the ermB macrolide resistance element present along with tetM in Tn3872 (10). Taken together, these data appear most consistent with the idea that expansion of ST-1 serotype V GBS since 1990 was facilitated by acquisition of genetic determinants that allowed for an increased capacity to cause disease in nonpregnant adults.
The relatively low rate of recombination among ST-1 strains allowed for elucidation of discrete GBS loci under positive selection, only some of which have been previously identified as important for GBS host–pathogen interaction (22, 26, 35). Thus, in conjunction with our Alp findings, these data provide a new platform for investigating why ST-1 GBS strains have emerged as a major cause of invasive disease in nonpregnant adults, such as analysis of the previously unstudied TCS encoded by rdf_0314-0315. Interestingly, although the exact genes under positive selection are distinct between GBS and GAS, the GBS findings are thematically similar to those previously reported for GAS in terms of the high prevalence of nonsynonymous polymorphisms predicted to alter the bacterial cell surface through variation in regulatory proteins controlling cell-surface composition or through changes in cell-surface proteins themselves (4, 5, 36).
In summary, we have discovered that the emergence of serotype V GBS causing invasive disease in nonpregnant adults is primarily driven by ST-1 strains that are highly similar at the whole-genome level but possess significant phenotypic diversity as a result of small genetic changes. These data provide a new platform for investigating factors that contribute to invasive GBS disease in adult humans and shed new light into the molecular mechanisms by which pathogenic bacteria adapt to the human host.
Materials and Methods
Bacterial Strains and Serotyping.
The strains used in the present work are shown in SI Appendix, Table S1. The United States strains comprised consecutive serotype V GBSs isolated from the bloodstream of nonpregnant adults as part of an ongoing surveillance program at the Texas Medical Center in Houston, TX, between 1992 and 2013. Canadian strains with the same characteristics were isolated as part of a surveillance program for invasive streptococcal infection in Toronto from 2011 to 2012 as described previously (13). Identification of GBS serotype was performed by using latex agglutination. SS-1168 and SS-1172 are serotype V strains isolated in 1977 and 1978, respectively, and were obtained from the Centers for Disease Control and Prevention from the collection of H. W. Wilkinson (37).
MLST and Whole-Genome Sequencing.
MLST was performed as described previously (11) and verified directly from short-read whole-genome sequence by using SRST2 (https://github.com/katholt/srst2). Ten strains differed from ST-1 by a single polymorphism. Whole-genome sequencing showed that these strains clustered tightly with ST-1 strains, and they were considered as ST-1 strains for the purposes of this manuscript. Details on whole-genome sequencing of SGBS001 using a combination of PacBio and Illumia HiSeq data are presented in SI Appendix, SI Materials and Methods. The GenBank accession number for the SGBS001 genome is CP010867. Of the remaining 208 ST-1 strains, whole-genome sequencing of 201 GBS strains (seven random strains were not sequenced for multiplexing convenience reasons) was performed by using Illumina HiSeq or MiSeq instruments with an average sequencing depth of 120× (38). Sequence data for the 208 genomes have been deposited in the NCBI Short Read Archive database (BioProject PRJNA274384). Noncore aspects of the ST-1 genome were defined as all sequences > 1 kb that were not present in all sequenced isolates, as previously described (1). Polymorphisms in the core genome were called against the reference SGBS001 genome by using VAAL (30) and the CLC Genomics Workbench, version 7.0.3. Phylogenetic analyses and trees were generated by using the CLC Genomics Workbench, version 7.0.3. Regions of recombination were identified by using BratNextGen (31). By using the neighbor-joining tree of 194 ST-1 GBS strains, we generated a chronogram to estimate the time to most recent common ancestor by using Path-O-Gen (tree.bio.ed.ac.uk/software/pathogen). Genes with an excess of polymorphisms were identified as detailed in SI Appendix, SI Materials and Methods, using the Bonferroni method to correct for multiple comparisons (5). Genomic visualizations were generated by using BRIG (39).
Phylogenetic Analysis of the AlpST-1 Protein.
Following alignment using CLUSTALW, sequences were analyzed in MEGA, version 5.2, to create radial trees by using the neighbor-joining statistical method and the maximum-likelihood composite model. The robustness of the nodes was evaluated via bootstrapping (1,000 replicates).
Capsule and Pilus Expression Level Analysis.
Genotypes of tested strains are listed in SI Appendix, Table S1. Capsule and pilus expression level were determined by using monoclonal antibodies and FACS analysis as described previously (22).
RNA Isolation and Transcriptome Analysis.
For RNA-Seq analysis, strains were grown in quadruplicate to midexponential phase in Todd–Hewitt broth (Difco) and RNA was isolated using a Qiagen RNeasy kit. RNA-Seq analysis was performed in quadruplicate per strain as described previously (40) and as detailed in SI Appendix, SI Materials and Methods. Transcript levels were considered significantly different if the mean transcript level difference was ≥ 2.0-fold and the final adjusted P value was less than 0.05.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Allergy and Infectious Diseases/National Institutes of Health Grant 5R01AI089891 (to S.A.S.), Chapman Foundation, and MD Anderson Cancer Center Support Grant P30 CA016672 (Bioinformatics Shared Resource).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology BioProject database, www.ncbi.nlm.nih.gov/bioproject (BioProject PRJNA274384). For a list of accession numbers, see Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504725112/-/DCSupplemental.
References
- 1.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chewapreecha C, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46(3):305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brochet M, et al. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc Natl Acad Sci USA. 2008;105(41):15961–15966. doi: 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasser W, et al. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci USA. 2014;111(17):E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beres SB, et al. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA. 2010;107(9):4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casali N, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46(3):279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eickhoff TC, Klein JO, Daly AK, Ingall D, Finland M. Neonatal sepsis and other infections due to group B beta-hemolytic streptococci. N Engl J Med. 1964;271:1221–1228. doi: 10.1056/NEJM196412102712401. [DOI] [PubMed] [Google Scholar]
- 8.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci USA. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellais S, et al. Capsular switching in group B Streptococcus CC17 hypervirulent clone: A future challenge for polysaccharide vaccine development. J Infect Dis. 2012;206(11):1745–1752. doi: 10.1093/infdis/jis605. [DOI] [PubMed] [Google Scholar]
- 10.Da Cunha V, et al. DEVANI Consortium Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun. 2014;5:4544. doi: 10.1038/ncomms5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones N, et al. Multilocus sequence typing system for group B streptococcus. J Clin Microbiol. 2003;41(6):2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tazi A, et al. The surface protein HvgA mediates group B streptococcus hypervirulence and meningeal tropism in neonates. J Exp Med. 2010;207(11):2313–2322. doi: 10.1084/jem.20092594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teatero S, et al. Characterization of invasive group B streptococcus strains from the greater Toronto area, Canada. J Clin Microbiol. 2014;52(5):1441–1447. doi: 10.1128/JCM.03554-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phares CR, et al. Active Bacterial Core surveillance/Emerging Infections Program Network Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA. 2008;299(17):2056–2065. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 15.Lamagni TL, et al. Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991-2010. Clin Infect Dis. 2013;57(5):682–688. doi: 10.1093/cid/cit337. [DOI] [PubMed] [Google Scholar]
- 16.Skoff TH, et al. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin Infect Dis. 2009;49(1):85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- 17.Tazi A, et al. Invasive group B streptococcal infections in adults, France (2007-2010) Clin Microbiol Infect. 2011;17(10):1587–1589. doi: 10.1111/j.1469-0691.2011.03628.x. [DOI] [PubMed] [Google Scholar]
- 18.Elliott JA, Farmer KD, Facklam RR. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J Clin Microbiol. 1998;36(7):2115–2116. doi: 10.1128/jcm.36.7.2115-2116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohnsack JF, et al. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. Academic Centers from 1995 to 1999. J Clin Microbiol. 2008;46(4):1285–1291. doi: 10.1128/JCM.02105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren S, et al. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14(9):R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachenauer CS, Creti R, Michel JL, Madoff LC. Mosaicism in the alpha-like protein genes of group B streptococci. Proc Natl Acad Sci USA. 2000;97(17):9630–9635. doi: 10.1073/pnas.97.17.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margarit I, et al. Preventing bacterial infections with pilus-based vaccines: The group B streptococcus paradigm. J Infect Dis. 2009;199(1):108–115. doi: 10.1086/595564. [DOI] [PubMed] [Google Scholar]
- 23.Zubair S, et al. Genome sequence of Streptococcus agalactiae strain 09mas018883, isolated from a Swedish cow. Genome Announcements. 2013;1(4):e00456-13. doi: 10.1128/genomeA.00456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards VP, Choi SC, Pavinski Bitar PD, Gurjar AA, Stanhope MJ. Transcriptomic and genomic evidence for Streptococcus agalactiae adaptation to the bovine environment. BMC Genomics. 2013;14:920. doi: 10.1186/1471-2164-14-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards VP, et al. Comparative genomics and the role of lateral gene transfer in the evolution of bovine adapted Streptococcus agalactiae. Infect Genet Evol. 2011;11(6):1263–1275. doi: 10.1016/j.meegid.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindahl G, Stålhammar-Carlemalm M, Areschoug T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev. 2005;18(1):102–127. doi: 10.1128/CMR.18.1.102-127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones DT. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. J Mol Biol. 1999;287(4):797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- 29.Xiang H, et al. Crystal structures reveal the multi-ligand binding mechanism of Staphylococcus aureus ClfB. PLoS Pathog. 2012;8(6):e1002751. doi: 10.1371/journal.ppat.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusbaum C, et al. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat Methods. 2009;6(1):67–69. doi: 10.1038/nmeth.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marttinen P, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40(1):e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43(12):1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen VL, Avci FY, Kasper DL. A maternal vaccine against group B Streptococcus: Past, present, and future. Vaccine. 2013;31(suppl 4):D13–D19. doi: 10.1016/j.vaccine.2012.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santi I, et al. BibA: A novel immunogenic bacterial adhesin contributing to group B Streptococcus survival in human blood. Mol Microbiol. 2007;63(3):754–767. doi: 10.1111/j.1365-2958.2006.05555.x. [DOI] [PubMed] [Google Scholar]
- 35.Cieslewicz MJ, et al. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect Immun. 2005;73(5):3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shea PR, et al. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc Natl Acad Sci USA. 2011;108(12):5039–5044. doi: 10.1073/pnas.1016282108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson HW. Nontypable group B streptococci isolated from human sources. J Clin Microbiol. 1977;6(2):183–184. doi: 10.1128/jcm.6.2.183-184.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shelburne SA, et al. Streptococcus mitis strains causing severe clinical disease in cancer patients. Emerg Infect Dis. 2014;20(5):762–771. doi: 10.3201/eid2005.130953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horstmann N, et al. Dual-site phosphorylation of the control of virulence regulator impacts group a streptococcal global gene expression and pathogenesis. PLoS Pathog. 2014;10(5):e1004088. doi: 10.1371/journal.ppat.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.