Significance
In gram-negative bacteria, OxyR is the master peroxide sensor that regulates the transcription of defense genes in response to a low level of cellular H2O2 via a rapid kinetic reaction. In this study, we present the first, to our knowledge, full-length structures of peroxide-sensing transcription regulator OxyR together with an oxidation intermediate-mimicking structure. The structures show all of the structural features describing the tetrameric assembly and a bound H2O2 molecule near the conserved cysteine. Combining the structural results, we reveal a step-by-step molecular mechanism for OxyR from H2O2 sensing to structural changes for transcriptional activation. Our study provides a structural basis for potentially answering key questions about the role of the cysteine residue in other Cys-based sensors, even mammalian ones, in response to various oxidants.
Keywords: OxyR, hydrogen peroxide, conformational change, reaction mechanism, transcription regulator
Abstract
OxyR, a bacterial peroxide sensor, is a LysR-type transcriptional regulator (LTTR) that regulates the transcription of defense genes in response to a low level of cellular H2O2. Consisting of an N-terminal DNA-binding domain (DBD) and a C-terminal regulatory domain (RD), OxyR senses H2O2 with conserved cysteine residues in the RD. However, the precise mechanism of OxyR is not yet known due to the absence of the full-length (FL) protein structure. Here we determined the crystal structures of the FL protein and RD of Pseudomonas aeruginosa OxyR and its C199D mutant proteins. The FL crystal structures revealed that OxyR has a tetrameric arrangement assembled via two distinct dimerization interfaces. The C199D mutant structures suggested that new interactions that are mediated by cysteine hydroxylation induce a large conformational change, facilitating intramolecular disulfide-bond formation. More importantly, a bound H2O2 molecule was found near the Cys199 site, suggesting the H2O2-driven oxidation mechanism of OxyR. Combined with the crystal structures, a modeling study suggested that a large movement of the DBD is triggered by structural changes in the regulatory domains upon oxidation. Taken together, these findings provide novel concepts for answering key questions regarding OxyR in the H2O2-sensing and oxidation-dependent regulation of antioxidant genes.
All aerobic organisms are prone to being exposed to hydrogen peroxide (H2O2), which is generated from endogenous aerobic metabolism. The elevated level of H2O2 can be converted to deadly toxic hydroxyl radicals under certain circumstances and can deplete the cellular thiol pool (1, 2). In gram-negative bacteria, the OxyR transcriptional regulator senses low amounts of intracellular H2O2 and maintains H2O2 levels within safe limits. OxyR functions primarily as a global regulator of the peroxide stress response by activating the expression of a range of antioxidant defense genes (2–4). OxyR belongs to the LysR-type transcriptional regulator (LTTR) family, consisting of an N-terminal DNA-binding domain (DBD) with a winged helix-turn-helix motif and a C-terminal regulatory domain (RD) (5). The crystal structure of the Escherichia coli OxyR (EcOxyR) RD revealed that the intramolecular disulfide bond is formed in the homodimeric RDs between the conserved cysteine residues that are separated by an α-helix by 17 Å (6). The H2O2-dependent activation of OxyR begins with the rapid S-hydroxylation of the conserved Cys199 (Cys199-SOH) in the presence of H2O2, followed by the rapid formation of an intramolecular disulfide bond with the second conserved Cys208 (7).
A structural comparison of the reduced and oxidized states of the EcOxyR RD revealed that H2O2 induces a large structural change within the RD dimers (6). The protomers of the OxyR RD dimer in the oxidized state have a relative rotation of ∼30° compared with that in the reduced state (6). OxyR also functions as a transcriptional repressor for some genes under normal growth conditions by binding to a more extended region of the target promoters than in the oxidized state, occluding RNA polymerase binding (6, 8). However, it remains elusive how OxyR binds to the target genes depending on its redox state, mainly due to the lack of the full-length structure. Moreover, the exact mechanism of H2O2 sensing by OxyR in various bacteria is still a subject of active investigation. The opportunistic human pathogen Pseudomonas aeruginosa is a gram-negative bacterium that also deploys OxyR (PaOxyR) as the master peroxide-sensing regulator for antioxidant genes, such as katA, katB, and ahpC (9, 10). To answer these questions, we performed a structural and biochemical study on PaOxyR (9, 10), which contains only three conserved cysteine residues (11).
Results
Structural Determination and Overall Structures of OxyR.
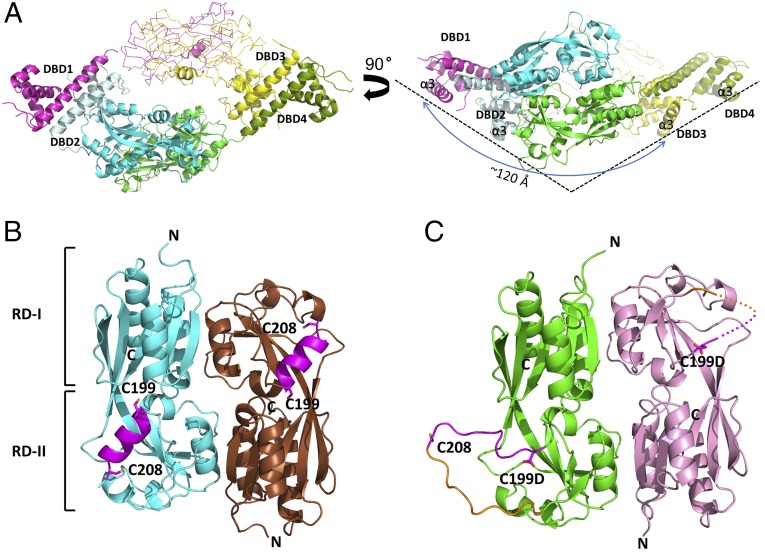
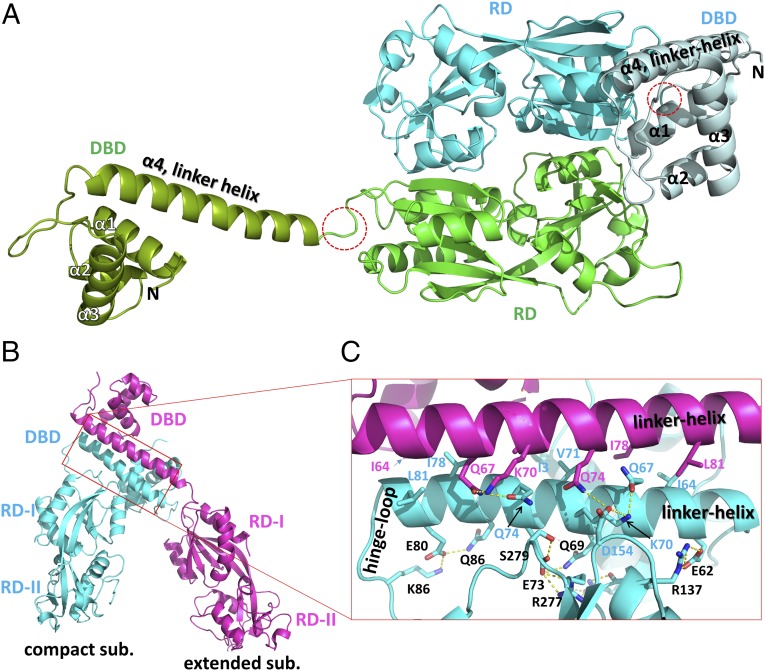
To investigate the structural features of PaOxyR, the crystal structures of both the full-length (FL) protein and RD (residues 88–310) of PaOxyR and its C199D mutant [PaOxyR (C199D)] that might mimic the intermediate state with Cys199-SOH during the oxidation cycle were determined in the presence of a reducing agent (Fig. 1, Table 1, Fig. S1, and Table S1). The overall structures of the FL proteins of PaOxyR and PaOxyR (C199D) show structural features that are typical of other tetrameric LysR-type proteins (Fig. 1A and Fig. S1) (6, 8, 12, 13). The tetrameric assembly is consistent with gel-filtration data during the purification of PaOxyR FL proteins (Fig. S2) and with other OxyR proteins (6, 8, 12, 13). The PaOxyR FL tetramer consists of two compact subunits and two extended subunits, with two different dimeric interfaces (RD and DBD). Each subunit is composed of a DBD and an RD that are connected by a short hinge region (Fig. 2). Four DBDs are arranged in the bottom of the tetramer in a side view of the OxyR tetramer, and the distance between DBD1 and DBD3 is ∼120 Å (Fig. 1A). Polar interactions between the DBD and RD were observed in the compact subunits, whereas none were observed between those in the extended subunits. The dimeric interface at the DBDs is formed by hydrophobic interactions together with some polar interactions, suggesting that the DBD dimers are relatively stable even upon structural changes in the RDs (Fig. 2B). The hinge motions of the DBD dimers with respect to the RD dimers are thus expected with different propensity between the compact and extended subunits.
Fig. 1.
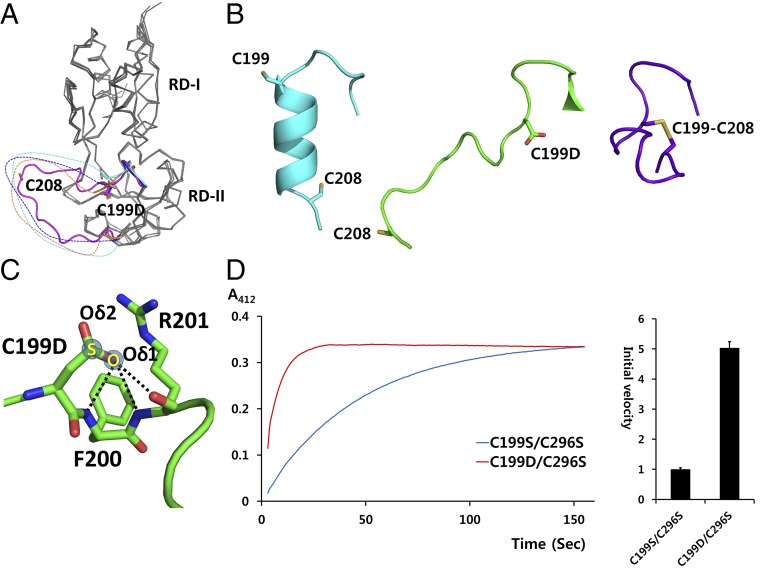
Overall structures of PaOxyR. (A) Two orthogonal views of the PaOxyR (C199D) FL protein (Left, bottom view; Right, side view). Each protomer is in magenta, cyan, yellow, or green, except for DBD2 (pale cyan) and DBD4 (dark green). The RDs of protomer 3 and protomer 1 are drawn in Cα tracing representation, but the intercysteine regions are in the ribbon diagram. Cys199 and Cys208 are displayed in stick representation. DNA-recognition helices α3 are labeled in the side view. (B) PaOxyR (WT) RD structure. Each protomer is in cyan or brown, except the intercysteine α-helical regions (magenta). The conserved cysteine residues are in stick representation. (C) PaOxyR (C199D) RD structure. Each protomer is colored in green or pink, except the intercysteine regions (residues 199–208 in magenta and residues 208–218 in orange). Asp199 and Cys208 are in stick representations. The disordered region is described in dotted line.
Table 1.
X-ray diffraction and refinement statistics for the PaOxyR RD (C199D) variant and FL PaOxyR (C199D), whose crystal was exposed to 20 mM H2O2 vapor
| Statistics | PaOxyR RD (C199D) | PaOxyR FL (C199D) exposed to H2O2 vapor |
| Data collection | ||
| Space group | P61 | P1211 |
| Cell dimensions | ||
| a, b, c, Å | 129.9, 129.9, 135.7 | 81.6, 151.1, 141.6 |
| α, β, γ, ° | 90, 90, 120 | 90, 97.28, 90 |
| Resolution, Å | 50.0–3.0 (3.05–3.00) | 50.0–2.0 (2.03–2.00) |
| Rmerge | 0.099 (0.359) | 0.068 (0.386) |
| I/σI | 25.5 (3.5) | 13.2 (1.9) |
| Completeness, % | 98.1 (99.8) | 95.3 (89.4) |
| Redundancy | 12.1 (7.2) | 4.2 (2.5) |
| Refinement | ||
| Resolution, Å | 19.99–3.0 | 20.0–2.0 |
| No. of reflections | 25,434 | 172,391 |
| Rwork/Rfree | 0.2293/0.2604 | 0.1856/0.2366 |
| No. of total atoms | 6,176 | 19,805 |
| No. of ligands | 0 | 8 |
| No. of water molecules | 0 | 1,142 |
| Average B factor, Å | 60.40 | 36.10 |
| Rms deviations | ||
| Bond lengths, Å | 0.003 | 0.013 |
| Bond angles, ° | 0.84 | 1.5 |
| Ramachandran plot | ||
| Favored, % | 94.58 | 96.93 |
| Allowed, % | 5.42 | 2.94 |
| Outliers, % | 0.00 | 0.13 |
| PDB ID code | 4XWS | 4X6G |
Values in parentheses are for the highest resolution shell.
Fig. 2.
Two dimeric interfaces of the PaOxyR FL protein. (A) Two protomers are assembled by the dimeric interface at the RDs. The extended subunit is in green (RD) and dark green (DBD), whereas the compact subunit is in cyan (RD) and pale cyan (DBD). The hinge regions of an extended subunit are indicated by dotted red circles. The α-helices in the DBDs are labeled. The angles between the α4 linker helix of the DBD and the long axis to a line connecting two Cα carbons of residues 88 and 175 of the RD are 25° in the compact subunit and 155° in the extended subunit. (B) The dimeric interface at the DBDs between the compact subunit (cyan) and the extended subunit (magenta). The red rectangle indicates the interfaces for DBD–DBD and DBD–RD. (C) A close-up view of the red rectangular region of B. The residues that are involved in the DBD–DBD interaction are labeled in magenta or cyan, and the residues for the RD–DBD interaction in the compact subunits are in black.
The RD region of the PaOxyR (C199D) FL protein exhibited a similar structure to that of the wild-type PaOxyR RD with the typical reduced conformation (rmsd 0.329 Å for 176 Cα atoms) (Fig. S3). However, this structure was significantly different from the PaOxyR (C199D) RD, especially in the Cys208-containing region (see below for details). When the four DBDs are designated DBD1, DBD2, DBD3, and DBD4 from the left in the side view, DBD1 and DBD2 form one dimeric pair, and DBD3 and DBD4 form the other pair. The DNA-recognition helices (α3) from the four DBDs are perpendicular to the axis across the four DBDs, and the DNA-recognition helices from DBD1 and DBD3 are nearly parallel (Fig. 1A).
H2O2-Binding Site.
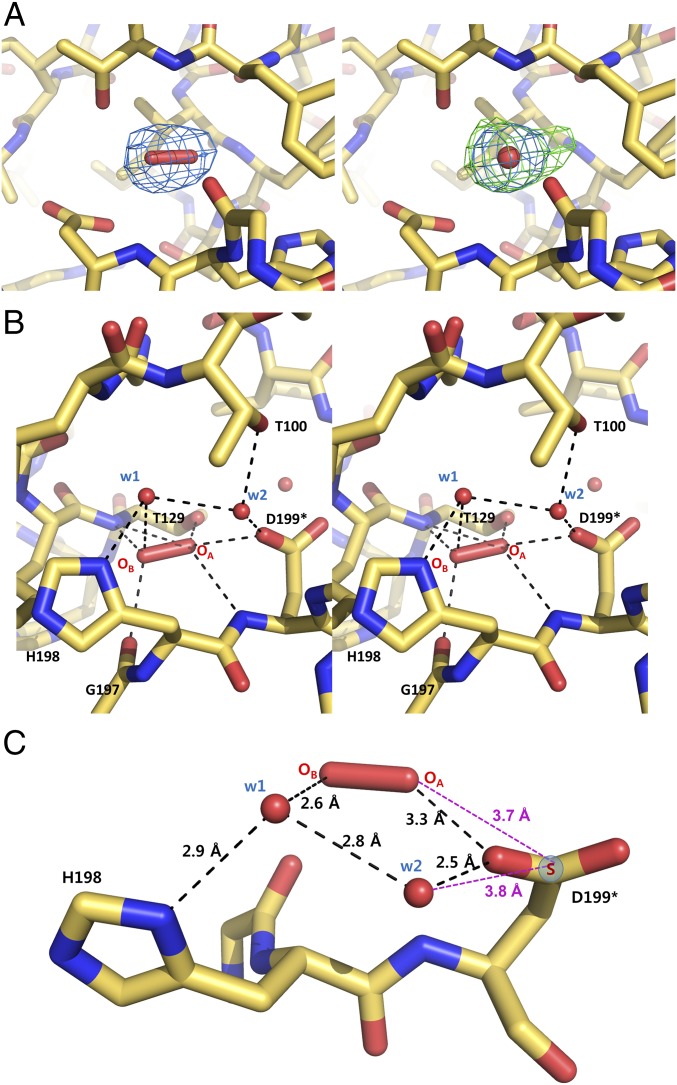
We initially solved the structure of the FL PaOxyR (C199D) at 2.3-Å resolution, where eight protomers were present in the asymmetric unit. Remarkably, ovoid-shaped electron density maps were found near Asp199 in the RD region of three protomers in the asymmetric unit of the FL OxyR (C199D) structure, which coincided well with H2O2 (Fig. S4A). To better define the ovoid-shaped electron density map, the crystals were incubated in the vapor from 20 mM H2O2 in the crystallization solution and the structure was solved at 2.0-Å resolution (Fig. 3A). The electron density maps were well-defined in all eight protomers of the crystal structure (Fig. 3A). We could exclude the possibility of one or two water molecules or a Cl− based on swap experiments in the different Fourier maps in the high-resolution crystal structure (Fig. 3A and Fig. S4B). We found a good parallel in the H2O2-binding environment of peroxiredoxin having an oxidized peroxidatic cysteine residue as well (14). Furthermore, the local environment surrounding the electron density map is also quite likely to hold a hydrogen peroxide molecule. The backbone NH and carbonyl groups and the side-chain hydroxyl group of Thr129 and the carboxylic group of Asp199 are involved in its binding (Fig. 3B). Thus, we concluded that the PaOxyR (C199D) FL protein captures an H2O2 molecule near the mutated aspartic acid residue. Interestingly, the surface representation of OxyR revealed that the H2O2 molecule is in a small pocket that is accessible to the solvent (Fig. S5).
Fig. 3.
H2O2-binding site. (A) The 2.0-Å-resolution structures are shown around the putative H2O2, superposed onto 2Fo − Fc (blue) and Fo − Fc (green) electron density maps contoured at 1.0 σ and 3.0 σ, respectively, when assuming an H2O2 molecule (Left) and when assuming a water molecule instead of H2O2 (Right). (B) Stereoview of the structure around the putative H2O2 and two water molecules. The broken lines indicate the polar interactions whose distances are within 3.3 Å. (C) A circular hydrogen-bonding network [Cys199-H2O2-w1-w2 (-Cys199)] and a hydrogen bond between His198 and w1. Sγ (blue circle) of Cys199 replaces the interaction of Asp199. The asterisk indicates a mutated residue.
In addition to H2O2, two water molecules were also trapped in the conserved residues Thr100, Thr129, His198, and Asp199 (Fig. 3B and Figs. S6 and S7). Moreover, three water molecules (the two water molecules, and the other water molecule bound in place of the OA atom of H2O2) are also present at this site of the PaOxyR RD structure and the other OxyR structures at high resolution (15, 16) (Fig. S7). We exchanged Asp199 with a cysteine residue in silico to determine whether the H2O2 and water molecules are still retained in the wild-type OxyR. As shown in Fig. 3C, the Sγ of Cys199 is within distance to form potential hydrogen bonds with H2O2 and a water molecule (w2), forming a circular hydrogen-bonding network.
H2O2-Driven Oxidation of Cys199.
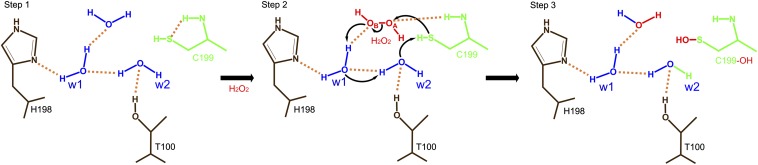
Based on the bound H2O2 and two water molecules (w1 and w2), we propose a novel mechanism by which Cys199 is specifically oxidized by the bound H2O2, resulting in Cys199-SOH, where the deprotonation of Cys199 and the donation of a proton to H2O2 are coupled, lowering the activation energy. We noted the circular hydrogen-bonding network of Cys199-H2O2-w1-w2 (-Cys199). Additionally, Thr100 and His198 hold w2 and w1 via hydrogen bonds, respectively (Fig. 4, step 1). According to the mechanism, the reactivity of Cys199-SH is largely increased only when H2O2 is bound to the site near Cys199. The reaction would begin with a nucleophilic attack of Cys199-SH on the close OA atom of the incoming H2O2, leading to the breakdown of H2O2 and the S-hydroxylation of Cys199 and OBH− (Fig. 4). Because OH− is an excellent base, the resulting OBH− could abstract a proton from Cys199-SH via w2 and w1. This proton transfer from Cys199-SH to OBH− can accelerate the reaction rate of these reactions. Because w1 and w2 play an important role in proton transfer, the reaction rate would be slower without w1 and w2. The nucleophilic attack of Cys199, the breakdown of H2O2, and the proton transfer occur simultaneously in this mechanism because the first step, the deprotonation of Cys199-SH, is facilitated by the last step.
Fig. 4.
Proposed mechanism for the S-hydroxylation of Cys199 as driven by bound H2O2. In the absence of H2O2, three water molecules are bound at the site near Cys199 by the residues His198 and Thr100 (step 1). The incoming H2O2 replaces a water molecule at the site, and H2O2 triggers the S-hydroxylation of Cys199 by the nucleophilic attack of the thiol group of Cys199 on the close OA atom of H2O2 (step 2). The resulting OBH− acquires a proton transferred from the thiol of Cys199 via the two water molecules (w1 and w2) (step 2), leading to S-hydroxylation of Cys199 (step 3).
To verify the importance of the residues that are presumably involved in binding to H2O2 and water molecules, we constructed PaOxyR variants and tested their susceptibility to H2O2 in vivo. The T100V mutation compromised PaOxyR function, similar to the C199S mutation. In contrast, the homologous mutation T100S slightly enhanced PaOxyR function (Fig. S8A). However, the H198A mutation displayed only partially decreased activity. Because w1 interacts with w2 and H2O2, together with His198 (Asn in Neisseria meningitidis OxyR), the H198A mutation appears not to abolish the function of OxyR. However, the lower occupancy of w1 by the H198A mutation would also diminish the affinity of H2O2 and w1 to the sites. In agreement, the H198A mutant OxyR showed a higher minimal concentration of H2O2 for oxidation in P. aeruginosa (Fig. S9). These results suggest the important role of Thr100 and His198 during the oxidation of PaOxyR. Thus, our proposed mechanism provides insight into how OxyR Cys199 is efficiently oxidized to Cys-SOH by the lowest level of H2O2.
Structure of the Reaction-Intermediate State.
The PaOxyR (C199D) RD structure exhibited mixed and unique structural features in the region containing the two conserved cysteine residues (Figs. 1C and 5A). The relative orientation of the subunits is closer to the conformation of the reduced state. However, the conformation of the Asp199-containing region (residues 196–200) resembles the structure of EcOxyR in the oxidized state (Fig. 5 A and B). Asp199 is located at the site of the disulfide bond between Cys199 and Cys208 of the EcOxyR RD in the oxidized state (Fig. 5B). Remarkably, the Cys208-containing loop was extended out or structurally disordered and was distinguishable from both the reduced and oxidized structures (Figs. 1D and 5B). We found new hydrogen bonds engaged in Oδ1 of Asp199 in the PaOxyR (C199D) RD, without an interaction involving Oδ2, indicating that Asp199 displaced Cys-SOH in this local environment. The Oδ1 atom of Asp199 (or the -OH group of Cys199-SOH in the wild-type protein) forms polar interactions with the adjacent backbone NH and the carbonyl groups (Fig. 5C). These observations indicate that the structure of the PaOxyR (C199D) RD may represent a reaction-intermediate state with Cys199-SOH. Given that the RD region in the PaOxyR (C199D) FL protein displayed a typical reduced structure, the PaOxyR (C199D) RD has at least dual conformations, which may be equilibrated between the two conformations in solution. We speculate that the new hydrogen bonding drives the equilibrium to the reaction-intermediate state with the exposed Cys199-SOH and disordered Cys208-containing loop, facilitating the disulfide bridge between the two cysteine residues.
Fig. 5.
Putative intermediate structure given by the PaOxyR (C199D) RD. (A) Structural superposition of four protomers in the asymmetric unit of the PaOxyR (C199D) RD. The two-cysteine-containing regions are in magenta, orange, cyan, or blue. Asp199 and Cys208 are in stick representations. The disordered regions are drawn arbitrarily in broken lines. (B) Structural comparison of OxyR in different states. The PaOxyR RD in the reduced state (Left; cyan), the PaOxyR (C199D) RD (Middle; green), and the EcOxyR RD in the oxidized state (Right; purple) are displayed. (C) The interaction of Asp199 Oδ1. Asp199 Oδ1 forms hydrogen bonds with the backbone NH and/or carbonyl groups of Phe200 and Arg201, indicated by broken lines. In contrast, no interaction was observed with Asp199 Oδ2. S and O are in semitransparent blue circles in the Cys199-SOH structure. (D) Kinetic measurement of the thiol reactivity of PaOxyR RD variants [PaOxyR (C199D/C296S) and PaOxyR (C199S/C296S)] with DTNB. Both of the PaOxyR variants were at 35 µM concentration in 0.1 M Tris buffer (pH 7.5) containing 1% DMSO and 100 µM DTNB. The absorbance of the liberated TNB2− was measured at 412 nm. Five independent experiments were performed and the averaged lines are displayed (Left). The relative initial velocities were calculated from the slopes of the lines between 3 and 4 s (Right). The initial value of PaOxyR (C199S/C296S) is set to 1. The error bars indicate SE (n = 5).
The structural comparison between the reduced (wild type) and the intermediate state (C199D) rationalized the destabilization of the intercysteine α-helix. In the reduced state, Phe200 in the intercysteine helix is anchored into a hydrophobic pocket of the main body of the RD, stabilizing the α-helix (Fig. S10A). In the intermediate state, Phe200 is in another hydrophobic pocket, due to the newly made interactions that are mediated by the S-hydroxylation of Cys199. Taken together, we hypothesize that Phe200 is moved by the movement of an adjacent residue Cys199-SOH, leading to destabilization of the intercysteine α-helix (Fig. S10B).
We verified the increase in the flexibility of the Cys208-containing loop by the S-hydroxylation of Cys199 based on the kinetic measurement of the reactivity of Cys208 toward the free thiol-reactive agent 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) (17) (Fig. 5D). The chemical reaction rates of Cys208 with the disulfide-containing oxidizing agent DTNB were measured using the RDs of the C199S/C296S and C199D/C296S variants (15). PaOxyR (C199D/C296S) showed a faster reaction with DTNB than PaOxyR (C199S/C296S) in the kinetic experiments (Fig. 5D) (17), confirming the observations from the crystal structures. Moreover, we observed that PaOxyR (C199D) was nonfunctional: P. aeruginosa harboring PaOxyR (C199D) displayed a serial-dilution defect even in the absence of H2O2 stress (Fig. S8B). This result agrees well with the dual conformation of the C199D variants that we observed in this study, because neither conformation obeyed what is presumed in the oxidized (i.e., activated) state.
Discussion
The structural characterization of OxyR and OxyR (C199D) proteins in the present study along with the modeling studies shown in Fig. 6 have provided insight into the differential DNA-binding and gene-regulation behaviors depending on the redox states of OxyR, which is an old conundrum regarding the oxidation and function of OxyR. The asymmetric hinge motions at the joints of the DBD and RD between compact and extended subunits would allow structural change in the regulatory domains upon oxidation to convert the inward motion of the DBD dimers.
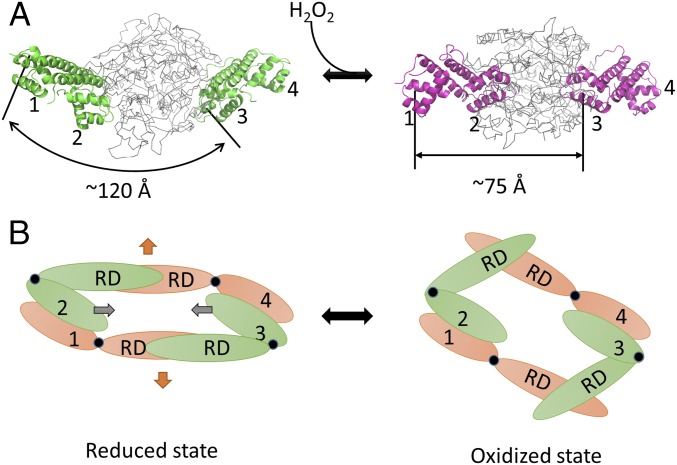
Fig. 6.
Modeling studies of PaOxyR in the reduced and oxidized states. (A) Modeling of the DNA binding-competent structures of OxyR in the reduced (Left) and oxidized (Right) states. The crystal structure of PaOxyR is shown (Left). DBDs are colored in green and magenta, whereas RDs are colored in gray. Each α3 of the DBDs is indicated by number. (B) Schematic drawings of the OxyR tetramer in bottom views. The reduced form of OxyR (Left) and the oxidized form of OxyR (Right) are shown. Compact subunits are colored in green, and extended subunits are in orange. Hinge regions are indicated by black circles. Each DBD is numbered, and the regulatory domain is labeled. The distances between DBD1 and DBD3 are indicated by a double-headed arrow. The kink motion within the RD dimers (orange arrows) is converted to the inward motion of the DBD dimers (gray arrows).
The modeling study of the structural change of OxyR upon H2O2 exposure has revealed a significant structural change that is required to reposition the DNA contact for H2O2-oxidized OxyR (8). The oxidized OxyR protein has greater affinity to the sequence ATAGntnnnanCTAT-N7-ATAGntnnnanCTAT (8). According to previous results, the distance between DBD1 and DBD3 should be 75 Å (22 nt), to bind to the OxyR-binding sites at the major grooves (O1, O2, O3, and O4 in Fig. S11) in the oxidized state (8). Given that the distance between DBD1 and DBD3 is 120 Å in the FL PaOxyR structure, the DBD1–DBD2 dimer and the DBD3–DBD4 dimer should get closer by 45 Å during the transition to the oxidized state (Fig. 6A). We have modeled the RD dimers of PaOxyR in the active state by superposition onto the EcOxyR RD dimer in the oxidized state and applied the model of the RD dimers to the FL PaOxyR structure. The RD dimer became kinked in a side view due to the 30° rotation, and the distance between the RD dimers in the central chamber of the tetramer increased due to the transition from a “facing each other” arrangement to a “bending and pushing each other back” arrangement along the twofold axis between the two RD dimers. Given the stable dimeric interface of the DBD dimers, this structural change in the RD dimers would result in the mutual backward movements of the two RD dimers with a hinge motion at the joints between the DBD and RD (Fig. 6B). The proposed mechanism in this study provides comprehensive molecular details for the previous model for the activation of OxyR as proposed by Toledano et al. (8). However, this mechanism still uses the FL OxyR structure in the oxidized form to elucidate the regulatory mechanism depending on the redox state.
In this study, we found mechanistic parallels with peroxiredoxins. OxyR and peroxiredoxins have an H2O2-binding pocket and react with H2O2, leading to the breakdown of H2O2 and cysteine S-hydroxylation (14). However, we believe that there must be a difference between OxyR and peroxiredoxins. OxyR is an H2O2-sensor protein that senses a low amount of H2O2, and peroxiredoxins are enzymes that rapidly remove H2O2. In the proposed mechanism for peroxiredoxins, the pKa value of peroxidatic cysteine residues is lowered by the adjacent basic residues (18, 19). The pKa lowering is a key step to increase the reactivity toward H2O2 and alkylperoxides, which are another substrate of peroxiredoxins. However, OxyR should not be promiscuous, and specifically senses H2O2. According to the mechanism that we propose in this study, the reactivity of the peroxidatic cysteine residue increases only when H2O2 is bound. This mechanism would explain the specific nature of OxyR. Although both proteins have evolved to react H2O2 with cysteine residues, the evolutionary directions might be different: OxyR as a sensor and peroxiredoxins as scavenging enzymes.
In summary, OxyR-mediated regulation is an elaborate maneuver for bacteria to finely tune the expression of relevant antioxidant genes by sensing the lowest level of cellular H2O2, which is also important in understanding the defense mechanism of all organisms living in aerobic environments. The structures in this study reveal three significant aspects of OxyR functions upon oxidation: the H2O2- and water-binding sites that are connected by the hydrogen-bonding network to enhance the reactivity of H2O2, such as the peroxide sensor; the local structural disruption by Cys-SOH–mediated interactions to facilitate disulfide-bond formation; and the subsequent conformational rearrangement to alter the DNA-binding affinity. Furthermore, we suggest that the substitution of a cysteine residue with aspartic acid would be a technical tactic to help investigate the role of the cysteine residue in other Cys-based sensors, even mammalian ones, in response to various oxidants.
Experimental Procedures
Construction of Plasmids.
To express the FL PaOxyR (residues 1–310), we used the previously described vector pET15H-oxyR (20). To express the PaOxyR RD (residues 88–310), DNA encoding the PaOxyR RD was PCR-amplified from pET15H-oxyR. The resulting PCR product was inserted between the NcoI and XhoI sites of the pProEx-HTa vector (Invitrogen), resulting in pProEX-HT-oxyRRD. The resulting plasmid encodes the hexahistidine tag and tobacco etch virus (TEV) protease cleavage site at the N terminus of the PaOxyR RD. Site-directed mutagenesis was performed by two subsequent PCR reactions (21). Protein expression and purification steps are described in SI Experimental Procedures.
Crystallization, Data Collection, and Structural Determination of the FL OxyR Protein.
Crystallization, data collection, and structural determination of OxyR RDs (wild type and C199D) are described in SI Experimental Procedures. The FL PaOxyR (C199D) protein was crystallized in a precipitation solution containing 0.2 M sodium citrate (pH 8.3), 18% (wt/vol) PEG 3350, and 2 mM Tris(2-carboxyethyl)phosphine (TCEP) by hanging-drop vapor diffusion at 14 °C. The FL PaOxyR (C199D) crystals were flash-frozen using crystallization solution with 30% (vol/vol) glycerol as a cryoprotectant in a nitrogen stream at −173 °C. The crystal belonged to space group P1211 with unit-cell dimensions of a = 81.3, b = 151.0, c = 141.5 Å, and β = 97.7°. The coordinates for the wild-type PaOxyR and the DNA-binding domain of BenM [Protein Data Bank (PDB) ID code 3M1E] were used as search models for the molecular replacement method. The structure was refined at a 2.3-Å resolution, resulting in an R factor of 20.7% and an Rfree of 25.2% [PaOxyR FL (C199D) in Table S1]. To obtain FL PaOxyR (C199D)–H2O2 complex crystals, when FL PaOxyR (C199D) crystals were grown well, 20 mM H2O2 was added to the reservoir solution and not to the hanging-drop solution. After 2 wk, the H2O2-soaked FL PaOxyR (C199D) crystals were flash-frozen using the same cryoprotectant as FL PaOxyR (C199D) crystals in a nitrogen stream at −173 °C. The space group and unit-cell dimensions corresponded to those of FL PaOxyR (C199D) crystals. The FL PaOxyR (C199D)–H2O2 complex structure was determined by the molecular replacement method using the coordinates of FL PaOxyR (C199D) as an initial model. The structure was refined at a 2.0-Å resolution, resulting in an R factor of 18.6% and Rfree of 23.7% (Table 1).
The FL wild-type PaOxyR protein was crystallized in a precipitation solution containing 0.2 M ammonium citrate (pH 7.0), 16% (wt/vol) PEG 3350, and 2 mM TCEP by hanging-drop vapor diffusion at 14 °C. The FL PaOxyR crystals were flash-frozen using crystallization solution with 30% (vol/vol) glycerol as a cryoprotectant in a nitrogen stream at −173 °C. The crystal belonged to space group P1211 with unit-cell dimensions of a = 70.8, b = 308.7, c = 96.2 Å, and β = 99.7°. The initial model was determined by molecular replacement using the tetramer structure of FL PaOxyR (C199D) and refined at a 5.0-Å resolution, resulting in an R factor of 25.7% and Rfree of 28.2% (Table S1). The program MOLREP in the CCP4 package (22) was used for molecular replacement, and Coot (23) and Phenix (24) were used to rebuild and refine the models.
Kinetic Study Using DTNB.
The two protein samples, PaOxyR RD (C199D/C296S) and (C199S/C296S) mutant, were concentrated to 350 µM in 20 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl. Each protein (35 µM) was reacted with 100 µM DTNB containing 0.1 M Tris⋅HCl (pH 7.5) and 1% DMSO at 4 °C. The absorbance of the liberated TNB2− was measured at 412 nm (25).
Supplementary Material
Acknowledgments
This research was supported by the R&D Convergence Center Support Program funded by the Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea (N.-C.H.) and by a National Research Foundation of Korea grant funded by the Korean government (to Y.-H.C.). We made use of beamline 5C at the Pohang Accelerator Laboratory.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4Y0M, 4XWS, and 4X6G).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424495112/-/DCSupplemental.
References
- 1.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14(6):1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storz G, Tartaglia LA, Farr SB, Ames BN. Bacterial defenses against oxidative stress. Trends Genet. 1990;6(11):363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- 4.Aslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA. 1999;96(11):6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 6.Choi H, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 7.Lee C, et al. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11(12):1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 8.Toledano MB, et al. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: A mechanism for differential promoter selection. Cell. 1994;78(5):897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 9.Panmanee W, Hassett DJ. Differential roles of OxyR-controlled antioxidant enzymes alkyl hydroperoxide reductase (AhpCF) and catalase (KatB) in the protection of Pseudomonas aeruginosa against hydrogen peroxide in biofilm vs. planktonic culture. FEMS Microbiol Lett. 2009;295(2):238–244. doi: 10.1111/j.1574-6968.2009.01605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hishinuma S, Yuki M, Fujimura M, Fukumori F. OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ Microbiol. 2006;8(12):2115–2124. doi: 10.1111/j.1462-2920.2006.01088.x. [DOI] [PubMed] [Google Scholar]
- 11.Bae HW, Cho YH. Mutational analysis of Pseudomonas aeruginosa OxyR to define the regions required for peroxide resistance and acute virulence. Res Microbiol. 2012;163(1):55–63. doi: 10.1016/j.resmic.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Kullik I, Stevens J, Toledano MB, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: Regions important for DNA binding and multimerization. J Bacteriol. 1995;177(5):1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubbs JM, Mongkolsuk S. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol. 2012;194(20):5495–5503. doi: 10.1128/JB.00304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura T, et al. Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide. J Biochem. 2010;147(1):109–115. doi: 10.1093/jb/mvp154. [DOI] [PubMed] [Google Scholar]
- 15.Sainsbury S, et al. The structure of a reduced form of OxyR from Neisseria meningitidis. BMC Struct Biol. 2010;10:10. doi: 10.1186/1472-6807-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svintradze DV, Peterson DL, Collazo-Santiago EA, Lewis JP, Wright HT. Structures of the Porphyromonas gingivalis OxyR regulatory domain explain differences in expression of the OxyR regulon in Escherichia coli and P. gingivalis. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 10):2091–2103. doi: 10.1107/S0907444913019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha S, Das KP. Structure and interactions in alpha-crystallin probed through thiol group reactivity. Adv Biol Chem. 2013;3(5):427–439. [Google Scholar]
- 18.Hall A, Parsonage D, Poole LB, Karplus PA. Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization. J Mol Biol. 2010;402(1):194–209. doi: 10.1016/j.jmb.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrer-Sueta G, et al. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24(4):434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 20.Heo YJ, et al. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J Bacteriol. 2010;192(2):381–390. doi: 10.1128/JB.00980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landt O, Grunert HP, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96(1):125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 22.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.DiMaio F, et al. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods. 2013;10(11):1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellman GL, Courtney KD, Andres V, Jr, Feather-Stone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.