Significance
The enigmatic millipede Xystocheir bistipita has been rediscovered after a half-century. The rediscovery unexpectedly reveals that the species is bioluminescent. By reconstructing its evolutionary history, we show that X. bistipita is the evolutionary sister of Motyxia, the only bioluminescent millipede genus in the order Polydesmida. We demonstrate that bioluminescence originated in the group’s common ancestor and incrementally grew brighter through evolutionary time. Luminescence in Motyxia may have initially evolved to cope with metabolic stress triggered by a hot, dry environment and was repurposed as a warning signal by species colonizing high-elevation habitats with greater predation risk. The discovery of bioluminescence in X. bistipita and its pivotal evolutionary location provides insight into repeated evolution of bioluminescence across the tree of life.
Keywords: photoprotein, Luminodesmus, Xystodesmidae, Polydesmida, cyanide
Abstract
The rediscovery of the Californian millipede Xystocheir bistipita surprisingly reveals that the species is bioluminescent. Using molecular phylogenetics, we show that X. bistipita is the evolutionary sister group of Motyxia, the only genus of New World bioluminescent millipedes. We demonstrate that bioluminescence originated in the group’s most recent common ancestor and evolved by gradual, directional change through diversification. Because bioluminescence in Motyxia has been experimentally demonstrated to be aposematic, forewarning of the animal’s cyanide-based toxins, these results are contrary to aposematic theory and empirical evidence that a warning pattern cannot evolve gradually in unpalatable prey. However, gradual evolution of a warning pattern is plausible if faint light emission served another function and was co-opted as an aposematic signal later in the diversification of the genus. Luminescence in Motyxia stem-group taxa may have initially evolved to cope with reactive oxygen stress triggered by a hot, dry environment and was repurposed for aposematism by high-elevation crown-group taxa colonizing new habitats with varying levels of predation. The discovery of bioluminescence in X. bistipita and its pivotal phylogenetic location provides insight into the independent and repeated evolution of bioluminescence across the tree of life.
The gradual evolution of complex structures by beneficial intermediates is a distinctive feature of evolution by natural selection. The origin of the cephalopod camera-type eye (1) or the miniature rotary electromotor of ATP synthase (2) by gradual changes quickly captured the attention of evolutionary biologists, including Darwin, who remarked, “Let us look to the great principle of gradation, and see whether Nature does not reveal to us her method of work” (3). Through detailed natural history observations, Darwin explored the incremental effects of natural selection with regard to the electric organs of fish, parachutes of flying squirrels, and luminous organs of insects. These novel innovations and “strange gradations in nature” provide exemplars of natural selection and fundamental arguments for biological evolution. By necessity, aposematic patterns are some of the most garish appearances in the living world. Toxic animals are frequently bright and highly contrasted to appear as dissimilar as possible to edible prey to avoid recognition errors and costly mistakes for both predator and prey. In addition, a conspicuous appearance hastens predator learning and retains memory of the unpleasant experience—e.g., a mouthful of spines or cyanide-induced fever (4). However, the gradual evolution of aposematic signals is problematic because positive frequency-dependent selection maintaining warning coloration predicts that naïve predators (those uneducated about the toxicity of a novel prey) should immediately eliminate rare mutant intermediates (5, 6). Here we show that, despite the expectation that a warning pattern cannot evolve gradually in unpalatable prey, bioluminescent aposematism in millipedes evolved by gradual and directional change through diversification of the genus.
Understanding the origins of complex traits is intuitively challenging, and several such traits have been considered especially difficult to explain by the theory of natural selection. However, through many steps of intermediate and beneficial forms (what Darwin referred to as “serviceable gradations”), these bizarre anatomies posed an elegant solution. One example noted by Darwin was the luminous organs of insects (3). There are 13 distantly related orders of arthropods that exhibit bioluminescence and at least that many evolutionary origins of the ability to emit light (7). Of the many functions of biological light, aposematism is a frequently cited role of bioluminescence both on land and in the sea (7, 8). One example of bioluminescent aposematism is found in Californian luminous millipedes (genus Motyxia), endemic to the southern Sierra Nevada Mountains of California. Millipedes in the genus Motyxia produce a blue-green light (λmax = 495 nm, FWHM = 38 nm) that can be seen in darkness (9, 10). Light emission from Motyxia originates in the integument and requires four components: (i) a photoprotein that contains a chromophore with ostensibly porphyrin as its functional group, (ii) O2, (iii) Mg2+, and (iv) ATP (11). The sequence and structure of the photoprotein remains uncertain, and its homology to proteins of other animals is unknown. Bioluminescence in Motyxia has been extensively studied in the species Motyxia sequoiae and, based on field experiments, serves as an aposematic signal to deter nocturnal mammalian predators—e.g., small neotomine rodents like the grasshopper mouse (12). Relative to diurnal species, these nocturnal mammals possess a suite of adaptations that enhance light sensitivity and permit the detection of just a few photons (13). These features include a higher density of rod photoreceptors per area, a reflective tapetum lucidum, and a compact architecture of heterochromatin in the photoreceptor nuclei that decreases light scattering (14). From detailed natural history observations, each Motyxia species emits light of varying intensity, but glows with a similar cyan hue (λmax = 495 nm). Notably, mammalian scotopic vision is extraordinarily sensitive to this wavelength, and empirical measurements of the rhodopsin absorption spectra (responsible for night vision) among mammals is remarkably uniform, with an average λmax of 497 and a nearly invariant shape among taxa (15, 16). The light of Motyxia species (even the weak luminescence of Xystocheir bistipita) is therefore well within the visual detection limits of nocturnal mammalian predators. Although many animals can see their own light, Motyxia themselves are blind and lack eyes and known photosensitive structures. Consequently, the role of emitted light for intraspecific communication is unlikely. Because light in Motyxia has a restricted use and limited set of functional possibilities, the group provides an excellent model system for investigating bioluminescence and evolution of complex phenotypic traits.
In 1967, two small xystodesmid millipedes were discovered near San Luis Obispo, California. Nearly 30 y later, these specimens were described as the new species X. bistipita, a geographical outlier of its genus located 120 km south of its congeners (17). X. bistipita was described on the basis of two teneral males preserved for nearly three decades in an alcohol jar. Female representatives, its species distribution, color in life, and phylogenetic placement were unknown. For nearly 50 y, these type specimens remained the sole representatives of the species. However, in 2013, several specimens of X. bistipita were rediscovered in the foothills of San Luis Obispo, CA. Several live males and females were captured and examined in the laboratory. Their color is a light tan, the color of a dry oak leaf, with salmon pink spots on the paranota (Fig. 1C). However, one of the most intriguing features of this diminutive millipede—and only now observed in these rediscovered individuals—is its bioluminescence, which was previously undocumented in Xystocheir (Fig. 1D). Based on its ability to bioluminesce and close phylogenetic relationship with Motyxia species, we transfer X. bistipita to the genus Motyxia and establish the new combination Motyxia bistipita. When viewed in a darkroom with the unaided eye and after 30 min of light adaptation, the luminescence appears as a soft blue-green glow, which is much reduced compared with the brighter species M. sequoiae (Fig. 1B). In photographs exposed solely with its bioluminescence, light appears to originate from crystalline grains on the cuticle, which differs in appearance from the more uniform and homogenous photic tissue of all other Motyxia species (Fig. 1D, Inset).
Fig. 1.
Bioluminescent millipedes: M. sequoiae (A and B) and M. bistipita (C and D). (Upper) Millipedes photographed under incident white light. (Lower) Millipedes photographed with light emitted from the cuticle. (Inset) A 5× magnification of the granular photocytes of M. bistipita. (Scale bars: 5.0 mm.)
Here, we document the unexpected discovery of bioluminescence in M. bistipita and infer the evolutionary history of the species in the context of a comprehensive molecular phylogeny of the Xystocheirini. Based on the tree, and phylogenetic trait mapping, faint light originated in the most recent common ancestor of Motyxia and gradually intensified through the evolutionary diversification of the genus. The linear escalation in luminescent intensity as a function of tree depth reverses in some crown group species and may indicate a shift in the functional role of luminescence over time.
Results
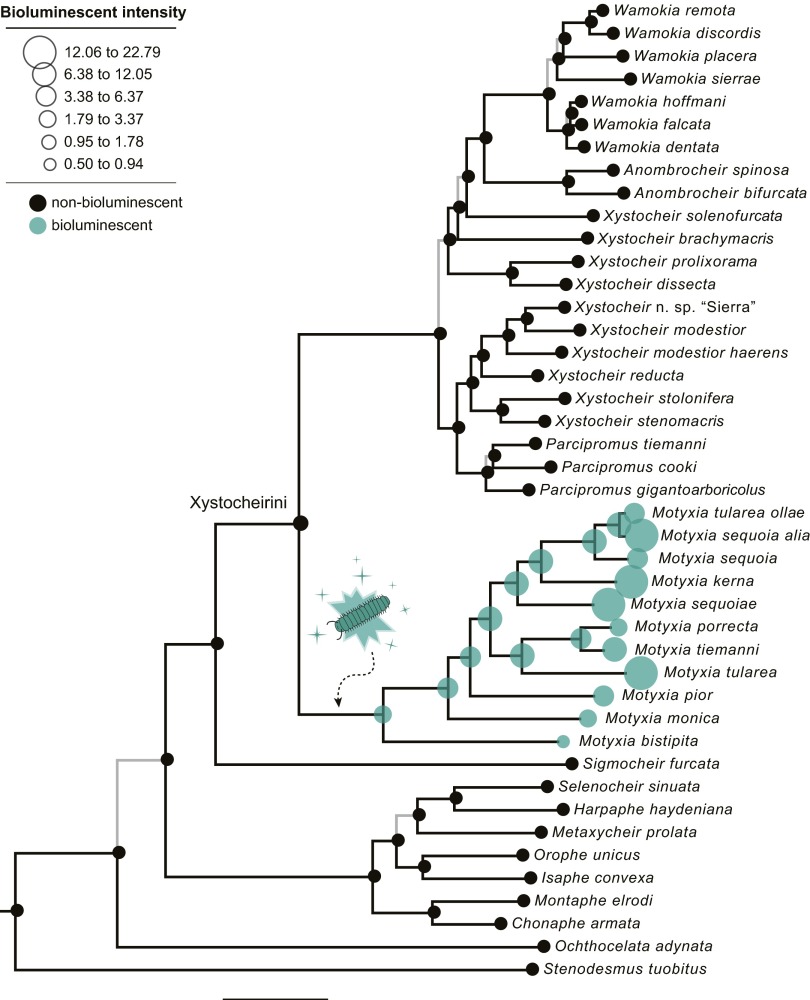
We included every species in the tribe Xystocheirini (29 species) to estimate the evolutionary history of bioluminescent millipedes and test a sister-group relationship between M. bistipita and Motyxia species. To infer the phylogeny, we used 4 kb of nuclear and mitochondrial DNA from 54 species, including the 10 nominative taxa of Motyxia and all nine species of the genus Xystocheir. We also included a broad sampling of other taxa in Xystodesmidae—which comprised representatives from California, Appalachia, East Asia, and the Mediterranean Basin—and rooted the tree with two closely related families, Paradoxosomatidae and Trichopolydesmidae. Our concatenated matrix contained sequence data from five genes (two nDNA and three mtDNA loci), which were divided into 12 partitions. We used a mixed-model partitioned Bayesian analysis to simultaneously estimate the phylogeny, nodal support values, and branch lengths across the tree (Fig. 2). To accommodate the variation of our five gene trees and incorporate within-species polymorphism, we estimated the Motyxia species tree using the multispecies coalescent model (18). Individual gene trees were also estimated by using maximum likelihood (ML) to show varying degrees of resolution among topologies (Fig. S1). Our phylogenetic analysis recovered most historical groupings as monophyletic, except Xystocheir, which is polyphyletic and distributed in three separate locations in the tree (Fig. 2 and Fig. S1). The species M. bistipita is sister to Motyxia and supported by a Bayesian posterior probability equal to 1 (0.971 in the coalescent tree). Trees are congruent between the concatenated Bayesian phylogeny and the multispecies coalescent. Like in previous phylogenetic studies of US millipedes and many wingless arthropods, geographical proximity is a more accurate predictor of evolutionary history than prior classification (19, 20). For example, Xystocheir species endemic to the Sierra Nevada are monophyletic, and a paraphyletic grade of species occurs in the northern Coast Ranges. The high-elevation genus Parcipromus (endemic to giant Sequoia forests) is monophyletic, as are Anombrocheir and Wamokia. Across the phylogeny, low-elevation taxa are generally paraphyletic with respect to monophyletic high-elevation clades (e.g., Xystocheir is rendered paraphyletic with respect to Parcipromus, Anombrocheir, and Wamokia). The species phylogeny of Motyxia is geographically structured with clades endemic to the foothills of the Great Western Divide, Greenhorn, and Piute Mountains, and a paraphyletic group in the eastern Tehachapi and Santa Lucia Mountains. Overall, the shape of the Motyxia clade is asymmetrical and pectinate, and the pattern resembles a nonadaptive radiation where species diversified from a single common ancestor into an assemblage of ecologically similar parapatric replacements of one another (21). The biogeographical pattern of Motyxia possesses a resemblance with other codistributed fauna in the mountains of southern California, characterized by complexes of species with predominately exclusive (parapatric) distributions and slight microhabitat differentiation (22, 23).
Fig. 2.
Species phylogeny of xystocheirine millipedes showing a single origin of bioluminescence in the most recent common ancestor of Motyxia species. Tree was estimated by using a partitioned Bayesian analysis of the concatenated dataset of five genes. Gray branches indicate < 0.95 posterior probability. (Scale bar: 0.1 expected substitutions per site.) Bioluminescent intensity was back-transformed from log-scale.
To infer the evolution of emitted light, we mapped bioluminescent intensity onto the phylogeny of Xystocheirini. We measured light emissions from 284 live millipedes representing the 43 species subsampled in the tree. Between 5 and 15 specimens per species were examined to gauge intraspecific variation. Luminescence was measured photographically in a darkroom by placing individuals in a collimating light tube at a constant distance from the camera lens (12). Bioluminescent intensity was quantified as the green triplet value from each pixel and was natural-log-transformed and averaged within species. Next, we visually examined each raw image and, while ignoring red pixels caused by electrical noise, adjusted pixel brightness until a blue-green image formed to confirm the presence or absence of emitted light. We observed luminescence restricted to Motyxia species (mean = 12.789, SD = 12.796, n = 88). Although some non-Motyxia xystocheirines possessed green triplet values greater than M. bistipita, luminescence was not detected in these and all of the other species based on visual examination of the brightness-adjusted images (mean = 0.757, SD = 0.235, n = 196). The species M. sequoiae exhibited the brightest luminescence (mean = 29.722). In contrast, M. bistipita showed the faintest (mean = 0.789). Using the phylogeny, we tracked the evolution of luminescent intensity using a squared change parsimony method of reconstructing ancestral states (Fig. 2). The reconstructed luminescent intensity at the ancestral node of the entire tree of Xystocheirini is 1.126 and at the ancestral node of Motyxia is 1.830. The luminescent intensity of Motyxia species—ranked from faintest to brightest—strongly tracks the nested branching pattern of the phylogeny.
We assessed the relationship of luminescence with toxicity by measuring the size of individual cyanide glands. These exocrine glands, lining the sides of the millipede, comprise two compartments: a lateral reaction chamber and a medial precursor reservoir (Fig. S2). Mandelonitrile, a biologically stable chemical housed in the precursor reservoir, is enzymatically disassociated through a cyanohydrin reaction to produce a mixture of benzaldehyde liquid and hydrogen cyanide gas (24). The cyanogenic reaction takes place in the heavily armored reaction chamber and is discharged from pores on the side of the body. The area of the reaction chamber and diameter of the pore are invariant across equally sized xystodesmid taxa. In contrast, the area of the precursor chamber varies considerably (range = 0.023–0.529, SD = 0.372, n = 292), and, in an analysis of the defense chemistry of three Motyxia species, Duffey et al. (25) found that the secretions of M. sequoiae and Motyxia tularea tularea possessed a 2- to 10-fold greater concentration of the reaction product benzaldehyde than Motyxia tiemanni—a less luminescent species with smaller cyanide glands—thereby indicating a functional link between gland volume and toxicity. Using our tree as a guide, and controlling for nonindependence due to phylogenetic relationship, we found that millipedes with a brighter bioluminescence possessed larger cyanide glands (r = 0.842), indicating that the conspicuous warning displays of Motyxia are quantitatively honest signals of their toxicity.
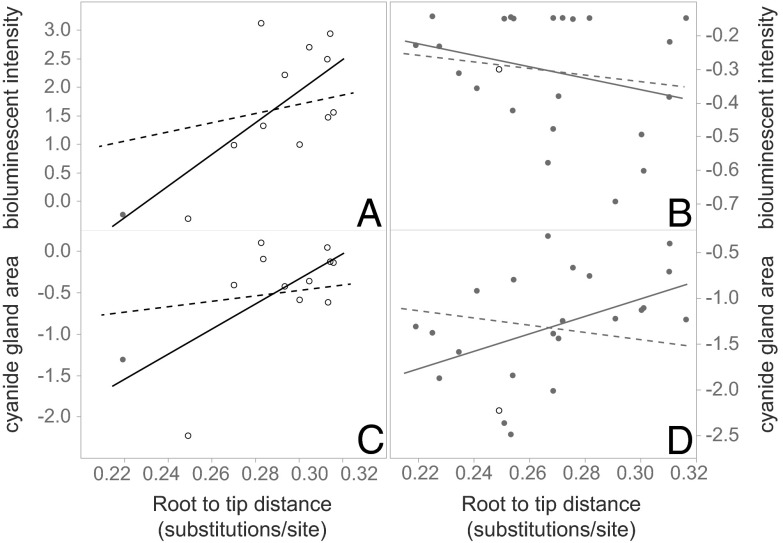
We then tested for a directional trend of luminescence through evolutionary diversification by estimating the relationship between light intensity and phylogenetic distance. If the hypothesis of a phylogenetic escalation of brightness is true, we predicted a strong positive correlation between a node’s luminescent intensity and its distance from the root, as opposed to the null hypothesis of no correlation (one-tailed test, α = 0.05). Although trends can occur in both directions, in cases where novel predator/herbivore defense traits (e.g., plant secondary metabolites) promote speciation or a shift into a new adaptive zone, than the quantity or intensity of these features will escalate and evolve in a directional and positive manner (26–28). Because the ecological phenomenon of defense-trait escalation underlying our hypothesis predicts that the sign of a relationship should be positive, we formulated a one-tailed hypothesis (the two-tailed probability is also shown in parentheses). Using the comparative framework afforded by the phylogeny, we also tested the null hypothesis with nonluminescent Xystocheirini, predicting conformity with the null expectation. First, we estimated phylogenetic conservatism of luminescent trait values under a Brownian motion model of character evolution with Pagel’s λ (29), where λ = 0 indicates a lack of fit between the data and the tree. We found that phylogeny strongly influences our observed luminescent data with a Pagel’s λ = 0.718 (not significantly different from 1, with a Bayes factor = 0.656; compare with λ = 0, Bayes factor = 3.53). We then used the branch length scaling factor κ to differentiate between a gradual vs. punctuational mode of continuous trait evolution on the tree, where κ = 0 indicates a punctuational model with change associated with speciation events (i.e., clumped at nodes), as opposed to a gradual change model (κ = 1) where character evolution is proportional to estimated branch lengths of the phylogram (27). When the models are compared by using a Bayes factor test, the empirical κ = 0.544 is not significantly different from κ = 1 (Bayes factor = 1.59; compare with κ = 0, Bayes factor = 3.49). Therefore, a gradual model of evolution better fits our observed luminescent trait values, given the phylogeny. We then assessed whether bioluminescence underwent a directional change on the phylogeny by testing a correlation between luminescent intensity and a node’s path length from the root (calculated from branch lengths). Using phylogenetically independent contrasts, we estimated the phenotype of the internal nodes of the tree and compared these values with their root-to-node distance using the Pearson product moment rho (r). We conducted the same analyses and tested for a correlation between toxicity (estimated from cyanide gland area) and phylogenetic distance. The tests were then repeated with the nonluminescent xystocheirine taxa. In Motyxia, we found a strong positive linear relationship between luminescent values and their root-to-node heights, with r = 0.712. We also observed this relationship between a taxon’s raw luminescent intensity and its root-to-tip height, r = 0.725 (Fig. 3A). Millipede toxicity appears linked to the intensification of luminescence, and we detected a strong positive linear correlation between cyanide gland area and phylogenetic distance (root-to-node r = 0.742 and root-to-tip r = 0.685; Fig. 3C). In contrast, we did not observe these patterns in the nonluminescent millipedes and instead found a weak negative correlation or no evidence of a correlation between luminescence or toxicity with phylogenetic distance (luminescence/root-to-node r = −0.556; luminescence/root-to-tip r = −0.282; toxicity/root-to-node r = 0.177; toxicity/root-to-tip r = 0.453; Fig. 3 B and D). Lastly, to test whether the observed correlation could have been due to the random association of traits with species, we generated a probability distribution by repeating the processes of node state reconstruction and correlation tests of light intensity (or gland area) and path length from the root for 10,000 datasets after shuffling the luminescent intensity data among the tips. We then evaluated whether our empirical Pearson r was within the range of permuted values representative of the null distribution of random assignments between traits and species. If <5% of our permuted values were greater than our empirical value (r = 0.712), then we rejected the null hypothesis that luminescent intensity or gland area is negatively related or unrelated to taxon assignment. Because the proportion of instances in the randomized distribution where r is greater than our observed value is <5%, we rejected the null hypothesis, P = 0.045 (significant as a one-tailed test α = 0.05, but not as a two-tailed test, P = 0.101). Although we also reject the null hypothesis with the toxicity dataset in the luminescent millipedes (one-tailed, P = 0.049; two-tailed, P = 0.069), we fail to reject the null hypothesis with the nonluminescent millipedes, which is consistent with our prediction of no escalation of either luminescence or toxicity with phylogenetic distance (luminescence, one-tailed P = 0.965, two-tailed P = 0.065; toxicity, one-tailed P = 0.307, two-tailed P = 0.614). Based on these results—and in contrast with related nonluminescent taxa—we conclude that luminescent intensity in Motyxia is remarkably conserved phylogenetically and exhibits evolutionary escalation through species diversification.
Fig. 3.
Tests of phylogenetic escalation of bioluminescent intensity and cyanide gland area. (A) In luminescent millipedes, a positive linear relationship exists between a taxon’s luminescent intensity and its phylogenetic distance from the root. (C) A similar relationship is observed between its toxicity and phylogenetic distance. (B and D) A weak negative or no evidence of a correlation is evident between luminescence and toxicity with phylogenetic distance in nonluminescent taxa. Raw luminescent data points are shown for clarity of illustration. Open circles denote luminescent taxa, and dots indicate nonluminescent taxa. Regression lines: phylogenetically corrected values (dotted), uncorrected raw values (solid).
Discussion
Luminous millipedes experienced a gradual escalation of bioluminescent intensity through evolutionary time. However, a few species exhibited a reduction in luminescent intensity compared with their closest relatives (e.g., M. bistipita, Motyxia porrecta, and Motyxia tularea ollae). These changes in direction of trait evolution may indicate a shift in the function of luminescence associated with a modification of habitat. From detailed natural history observations (30), an abiotic feature that is consistently related with bioluminescent intensity in Motyxia is elevation, which is a significant indicator of habitat variation in California due to important ecological correlates such as rainfall and temperature (31). With the biotic variables of phylogeny and toxicity, elevation is a reliable predictor of bioluminescent intensity (multiple regression, R2 = 0.75). Species at higher elevations exhibit the brightest emissions of light and inferred reversals of luminescent intensities are coincident with lower elevations. These lower-elevation taxa are even distinctive in body size and color and are similar to the plesiomorphic appearance of xystocherine millipedes, which include small cryptic forms and individuals with small yellow to red spots (Fig. S3). These results signify a link between bioluminescent aposematism with elevation and correspond with prior studies that have demonstrated a substantial increase in small-mammal alpha diversity with elevation (32)—small neotomine rodents are the most ubiquitous predators of M. sequoiae (12). Although our observations indicate a gradual escalation of bioluminescent intensity with phylogenetic descent and elevation, the early evolutionary jump required to overcome steep selection against rare mutants remains uncertain. However, if the faint luminescence of millipedes like M. bistipita initially originated to serve a different role, especially in lower-elevation habitats with potentially a relaxed burden of predation (e.g., San Luis Obispo localities, mean 215 m elevation), then novel, rare intermediates would be allowed to flourish and overcome these initial obstacles. Based on natural history observations of P.E.M., lower-elevation Motyxia species occur in some of the driest and warmest habitats of any species in the family Xystodesmidae. These species, like all animals, are susceptible to heat and oxidative stress due to the accumulation of reactive oxygen species (ROS) such as peroxides. This burden is particularly severe in Motyxia because the group is blind and wingless and has extremely low dispersal capability. However, many bioluminescent animals depend exclusively on superoxide anions and ROS as a critical element for the production of light (e.g., the clam Pholas, scaleworm Harmothoe, and tubeworm Chaetoperus) (10). These animals are often in extreme environments (deep in anoxic sediments or near hot hydrothermal vents). Their light-generating reactions, which are catalyzed by either a luciferase or photoprotein, alternatively undergo oxidative luminescence in the presence of peroxides, superoxide anions, and other ROS. Oxidative luminescence upon in vitro addition of hydrogen peroxide was recently demonstrated in a close nonluminescent relative of Motyxia, Parafontaria laminata (33) (Fig. S4). This finding implies a similar oxidative luminescence system in Motyxia. Although superoxide anions and ROS are toxic and typically removed to prevent oxidative stress in most life forms, in many bioluminescent taxa, they are harnessed for the production of light for a diversity of roles, including many nondefense functions that may be evolutionarily co-opted for different tasks—including aposematism (10). Based on this scenario, the evolution of oxidative luminescence in Motyxia stem-group taxa to cope with ROS triggered by a hot, dry environment may have been repurposed for aposematism by high-elevation crown-group taxa colonizing new habitats with higher levels of predation.
Materials and Methods
Collections and Phylogenetics.
Specimens sampled for this study are listed in Table S1 and are deposited as museum specimens in the Virginia Tech Insect Collection (collection.ento.vt.edu). We extracted and purified genomic DNA from leg tissue archived in RNAlater using a Qiagen DNeasy kit. Amplification of gene regions using PCR followed standard techniques previously implemented for millipedes (34). Approximately 4 kb of DNA from the following genes were sequenced for our phylogenetic analysis: 12S (including tRNA-Val), 16S, EF1α, 28S, and COI. Raw chromatograms were cleaned and trimmed by Phred and Phrap in the Mesquite module Chromaseq (Version 1.01) (35, 36). Sequences were sequentially aligned by using a parsimony guide tree in Prank (Version 140110) (37). Sequences were partitioned by gene, codon position, and intron boundaries in Mesquite. Alternative partitioning schemes and models of nucleotide substitution were evaluated in PartitionFinder (Version 1.1.1) (38). We used the best-fit partitioning scheme determined under a Bayesian Information Criterion as a dataset for phylogenetic analysis in MrBayes (Version 3.2.2) (39). Trees were evaluated by using a Markov chain Monte Carlo method with a simultaneous estimation of branch lengths, topology, and other parameters. Stabilization among four concurrent chains was reached at 10 million generations, and one quarter of the trees were discarded as a burnin phase. Branch lengths and other parameters were averaged, and a consensus topology was calculated from the posterior distribution of trees (Fig. S4). Taxa missing bioluminescence data were then pruned from the trees for subsequent analyses of trait evolution. We estimated the tree with the multispecies coalescent model in *BEAST (18, 40) and genealogies separately for each locus using ML in RAxML (Version 7.5.3) (Fig. S1). We used one to three individuals per species and applied a Yule process species tree prior and unlinked substitution, clock, and tree models among loci (except mitochondrial genes, which are effectively linked). The BEAST MCMC analysis was run for 10M generations, sampling every 2,000 states, and half of the trees were discarded as burnin to generate a maximum clade credibility tree. A general time-reversible model with gamma-distributed rate heterogeneity of nucleotide sites was used in the ML analysis, and support values were generated for each clade by 500 rapid bootstrap replicates.
Trait Measurements.
Bioluminescence was measured from images captured from live millipedes in a darkroom with a Nikon D40 camera and a 60-mm macro lens set at a working distance of 100 mm and at a 3.5 f-stop and 120-s shutter speed. We first tested the presence of bioluminescence by increasing overall image brightness in ImageJ (Version 1.48; imagej.nih.gov/ij/) until a discernable body outline appeared. If an outline did not appear, we then scored bioluminescence as absent, denoting it with a black circle; in contrast, if present, then bioluminescence was denoted with a green circle, sized proportional to its luminescent intensity (Fig. 2). Next, we quantified the magnitude of light emitted from the body by measuring red, green, and blue pixel intensity values extracted from the images. The bioluminescence of Motyxia is entirely green and blue, and these colors are strongly correlated, so green values were retained and averaged within individuals and species. We found that light emission is restricted to Motyxia species. However, the average green pixel value measured in M. bistipita is within the range of some nonluminescent taxa. This overlap in measurements, occurring at the boundary between faint and nonluminescent taxa, is due to the large SD of values in Motyxia and the smaller values in nonluminescent taxa. The overlap may also be due to the presence of a continuum of luminescence between Motyxia and other xystocheirine taxa. Although most organisms spontaneously emit ultraweak photon emissions from metabolic reactions—e.g., humans (41)—only those with light that is visible by others are functionally bioluminescent. We measured the size of individual cyanide glands to assess millipede toxicity. We removed the 10th body ring and digested it in warm trypsin to clear tissue and expand the precursor reservoir (Fig. S2). Both gland compartments were excised from the segment and photographed with a scale bar by using a Canon 6D camera and MPE 65-mm macrolens. The precursor reservoirs were outlined, and their areas were calculated in ImageJ. Bioluminescent intensity and gland area were natural-log-transformed based on examination of the frequency distribution of individual measurements (42). Because gland area and luminescent intensity are coupled with millipede body size, data transformation provided a means to exclude body-size-related variation and standardize comparison of traits from the range of individual sizes that occur among millipedes.
Trait Evolution.
We inferred the evolution of continuous traits on the tree using a least-squares parsimony method of estimating ancestral states where the cost of a state change is equivalent to the squared difference in values of the states, (x – y)2. To correct for the phylogenetic nonindependence of species, we used the PDAP module in Mesquite to produce a set of independent contrasts for statistical analysis (43). If light and toxicity evolved in a directional manner, we expected a positive correlation between the trait value at a node and the distance of the node from the root of the tree. We calculated the nodal values of contrasts and the node’s path length from the root and tested for a correlation using the Pearson product-moment correlation coefficient r.
The results of a stepwise multiple regression with bioluminescent intensity as the dependent Y-variable and root-to-tip distance (RT), cyanide gland area (CN), and elevation (EL) as independent predictor variables—and with a minimum Akaike Information Criterion as a stopping rule to select the best model—indicate that cyanide gland area contributes most to the multiple regression equation of Y = 18.000xRT + 0.722xCN + 0.0004xEL - 3.690, with an R2 = 0.75. To evaluate millipede body width as a possible confounding factor, because we expect it to covary with cyanide gland area as a result of isometric growth, we conducted a second multiple regression including width as a fourth predictor variable and found that cyanide gland remains the most significant contributor to the regression model.
We evaluated alternative models of trait evolution using BayesTraits (44) and the posterior set of burnin trees. To determine whether a random-walk Brownian model of trait evolution best explains the distribution of observed continuous characters on the phylogeny, we estimated λ (29) and evaluated whether our empirical value is significantly different from λ = 0 (indicating no relationship between the tree and the observed trait data and a default star phylogeny) vs. λ = 1 (indicating a strong relationship between tip data and the tree and phylogenetic conservatism). We compared these competing hypotheses using a Bayes factor comparison of harmonic mean likelihoods (44). We estimated κ using the same framework and compared our empirical value against κ = 0 (indicating trait changes accumulate at speciation events instead of proportional to branch lengths) and κ = 1 (indicating gradualism and continuous phenotypic change proportional to branch lengths).
Supplementary Material
Acknowledgments
Two anonymous reviewers, Charity Hall, and Jackson Means provided improvements to earlier drafts. We appreciate the careful work of Tim McCoy and Elizabeth Francis with dissections. Tsutomu Tanabe, Deren Ross, Pavel Stoev, Michael Jorgensen, Derek Hennen, Avery Lane, Brent Hendrixson and Rob Marek helped collect specimens for the project. This work was supported by National Science Foundation Grant DEB1410911 and National Geographic Society Grant EC0564-12.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KR135885–KR136081).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500014112/-/DCSupplemental.
References
- 1.Land MF, Nilsson D-E. Animal Eyes. 2nd Ed. Oxford Univ Press; Oxford: 2012. p. xiii, 271, 274. [Google Scholar]
- 2.Yoshida M, Muneyuki E, Hisabori T. ATP synthase—a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2(9):669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 3.Darwin C. On the Origin of Species by Means of Natural Selection; or, The Preservation of Favored Races in the Struggle for Life. 5th Ed. D. Appleton and Co.; New York: 1872. p. 447. [Google Scholar]
- 4.Mallet JLB, Joron M. Evolution of diversity in warning color and mimicry: Polymorphisms, shifting balance, and speciation. Annu Rev Ecol Syst. 1999;30:201–233. [Google Scholar]
- 5.Lindström L, Alatalo RV, Lyytinen A, Mappes J. Strong antiapostatic selection against novel rare aposematic prey. Proc Natl Acad Sci USA. 2001;98(16):9181–9184. doi: 10.1073/pnas.161071598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joron M. Aposematic coloration. In: Cardé RT, Resh VH, editors. Encyclopedia of Insects. 2nd Ed. Academic; New York: 2009. pp. 33–38. [Google Scholar]
- 7.Haddock SHD, Moline MA, Case JF. Bioluminescence in the sea. Annu Rev Mar Sci. 2010;2:443–493. doi: 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- 8.Sivinski J. The nature and possible functions of luminescence in Coleoptera larvae. Coleopt Bull. 1981;35(2):167–179. [Google Scholar]
- 9.Hastings JW, Davenport D. The luminescence of the millipede, Luminodesmus sequoiae. Biol Bull. 1957;113(1):120–128. [Google Scholar]
- 10.Shimomura O. Bioluminescence: Chemical Principles and Methods. World Scientific; Hackensack, NJ: 2006. p. xxvii. [Google Scholar]
- 11.Shimomura O. Porphyrin chromophore in Luminodesmus photoprotein. Comp Biochem Physiol B. 1984;79(4):565–567. [Google Scholar]
- 12.Marek P, Papaj D, Yeager J, Molina S, Moore W. Bioluminescent aposematism in millipedes. Curr Biol. 2011;21(18):R680–R681. doi: 10.1016/j.cub.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sterling P. How retinal circuits optimize the transfer of visual information. In: Chalupa L, Werner J, editors. Visual Neurosciences. MIT Press; Cambridge, MA: 2003. pp. 234–259. [Google Scholar]
- 14.Solovei I, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137(2):356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 15.Endler JA, Mielke PW. Comparing entire colour patterns as birds see them. Biol J Linn Soc Lond. 2005;86(4):405–431. [Google Scholar]
- 16.Zhao H, et al. Rhodopsin molecular evolution in mammals inhabiting low light environments. PLoS ONE. 2009;4(12):e8326. doi: 10.1371/journal.pone.0008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shelley RM. A re-evaluation of the milliped genus Motyxia Chamberlin, with a re-diagnosis of the tribe Xystocheirini and remarks on the bioluminescence (Polydesmida: Xystodesmidae) Insecta Mundi. 1997;11:331–351. [Google Scholar]
- 18.Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27(3):570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedin M, Tsurusaki N, Macías-Ordóñez R, Shultz JW. Molecular systematics of sclerosomatid harvestmen (Opiliones, Phalangioidea, Sclerosomatidae): Geography is better than taxonomy in predicting phylogeny. Mol Phylogenet Evol. 2012;62(1):224–236. doi: 10.1016/j.ympev.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Marek PE. A revision of the Appalachian millipede genus Brachoria Chamberlin, 1939 (Polydesmida: Xystodesmidae: Apheloriini) Zool J Linn Soc. 2010;159(4):817–889. [Google Scholar]
- 21.Rundell RJ, Price TD. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol Evol. 2009;24(7):394–399. doi: 10.1016/j.tree.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Jockusch EL, Wake DB. Falling apart and merging: Diversification of slender salamanders (Plethodontidae: Batrachoseps) in the American West. Biol J Linn Soc Lond. 2002;76(3):361–391. [Google Scholar]
- 23.Satler JD, Starrett J, Hayashi CY, Hedin M. Inferring species trees from gene trees in a radiation of California trapdoor spiders (Araneae, Antrodiaetidae, Aliatypus) PLoS ONE. 2011;6(9):e25355. doi: 10.1371/journal.pone.0025355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisner T, Eisner HE, Hurst JJ, Kafatos FC, Meinwald J. Cyanogenic glandular apparatus of a millipede. Science. 1963;139(3560):1218–1220. doi: 10.1126/science.139.3560.1218. [DOI] [PubMed] [Google Scholar]
- 25.Duffey SS, et al. Benzoyl cyanide and mandelonitrile benzoate in defensive secretions of millipedes. J Chem Ecol. 1977;3(1):101–113. [Google Scholar]
- 26.Vermeij GJ. The evolutionary interaction among species—selection, escalation, and coevolution. Annu Rev Ecol Syst. 1994;25:219–236. [Google Scholar]
- 27.Agrawal AA, Fishbein M. Phylogenetic escalation and decline of plant defense strategies. Proc Natl Acad Sci USA. 2008;105(29):10057–10060. doi: 10.1073/pnas.0802368105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrlich P, Raven PH. Butterflies and plants: A study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 29.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401(6756):877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 30.Causey NB, Tiemann DL. A revision of the bioluminescent millipedes in the genus Motyxia (Xystodesmidae: Polydesmida) Proc Am Philos Soc. 1969;113:14–33. [Google Scholar]
- 31.Raven PH, Axelrod DI. Origin and Relationships of the California Flora. Univ of California Press; Berkeley: 1978. p. viii. [Google Scholar]
- 32.McCain CM. Elevational gradients in diversity of small mammals. Ecology. 2005;86(2):366–372. [Google Scholar]
- 33.Kuse M, Yanagi M, Tanaka E, Tani N, Nishikawa T. Identification of a fluorescent compound in the cuticle of the train millipede Parafontaria laminata armigera. Biosci Biotechnol Biochem. 2010;74(11):2307–2309. doi: 10.1271/bbb.100171. [DOI] [PubMed] [Google Scholar]
- 34.Marek PE, Bond JE. Phylogenetic systematics of the colorful, cyanide-producing millipedes of Appalachia (Polydesmida, Xystodesmidae, Apheloriini) using a total evidence Bayesian approach. Mol Phylogenet Evol. 2006;41(3):704–729. doi: 10.1016/j.ympev.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 35.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 36.Maddison WP, Maddison DR. 2010 Mesquite: A Modular System for Evolutionary Analysis, Version 2.73. Available at mesquiteproject.org.
- 37.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102(30):10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 39.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi M, Kikuchi D, Okamura H. Imaging of ultraweak spontaneous photon emission from human body displaying diurnal rhythm. PLoS ONE. 2009;4(7):e6256. doi: 10.1371/journal.pone.0006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald JH. Handbook of Biological Statistics. Sparky House; Baltimore: 2010. Available at www.biostathandbook.com. [Google Scholar]
- 43.Midford PE, Garland T, Maddison WP. 2010 PDAP Package of Mesquite, Version 1.15. Available at mesquiteproject.org/pdap_mesquite/. Accessed January 24, 2014.
- 44.Pagel M, Meade A. 2013 BayesTraits, Version 2.0 (Beta). Available at evolution.rdg.ac.uk/BayesTraits.html. Accessed March 1, 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.