Significance
Peroxisomal fission is crucial for cell viability because peroxisome fission defects cause severe disease. The initial step in peroxisomal fission, membrane elongation, requires the membrane remodeling protein Peroxin 11 (Pex11p). Here, we identify an additional function for Pex11p, demonstrating that Pex11p also plays a crucial role in the final step of peroxisomal fission: membrane separation. We show that Pex11p functions as a GTPase activating protein (GAP) for Dynamin-related 1 (Dnm1p) and that this GAP activity is conserved from yeast to mammalians. This work identifies a previously unknown requirement for a GAP in dynamin-like protein function.
Keywords: dynamin-like protein, organelle fission, GTPase activating protein, DLP, GAP
Abstract
The initial phase of peroxisomal fission requires the peroxisomal membrane protein Peroxin 11 (Pex11p), which remodels the membrane, resulting in organelle elongation. Here, we identify an additional function for Pex11p, demonstrating that Pex11p also plays a crucial role in the final step of peroxisomal fission: dynamin-like protein (DLP)-mediated membrane scission. First, we demonstrate that yeast Pex11p is necessary for the function of the GTPase Dynamin-related 1 (Dnm1p) in vivo. In addition, our data indicate that Pex11p physically interacts with Dnm1p and that inhibiting this interaction compromises peroxisomal fission. Finally, we demonstrate that Pex11p functions as a GTPase activating protein (GAP) for Dnm1p in vitro. Similar observations were made for mammalian Pex11β and the corresponding DLP Drp1, indicating that DLP activation by Pex11p is conserved. Our work identifies a previously unknown requirement for a GAP in DLP function.
Peroxisomes are ubiquitous, single-membrane–bounded cell organelles that harbor enzymes involved in a large number of metabolic processes. Common functions are the β-oxidation of fatty acids and hydrogen peroxide metabolism. Specialized functions include the metabolism of various carbon and organic nitrogen sources in fungi and the production of plasmalogens and bile acids in mammals, to name but a few (1). Their importance is underlined by the severe, often lethal human disorders caused by defects in peroxisome biogenesis or metabolism (2). Importantly, defects in peroxisome multiplication, caused by mutations in genes that control peroxisome fission, also result in severe human disorders (3, 4).
Based on data from yeast and mammals, the current model for peroxisomal fission describes a three-step process, consisting of (i) organelle elongation, (ii) constriction, and (iii) the actual scission step (5–7). So far, Peroxin 11 (Pex11p), a highly conserved and abundant peroxisomal membrane protein, is the only protein known to play a crucial role in the first step (8). Its vital role in peroxisome multiplication is illustrated by the observation that in all organisms studied so far, Pex11p overproduction results in enhanced peroxisome proliferation, whereas PEX11 deletion causes a decrease in number, together with an increase in peroxisome size (8). The function of Pex11p in organelle elongation is mediated by the extreme N-terminal region of Pex11p, which can adopt the structure of an amphipathic helix, which upon insertion into membranes induces their curvature, resulting in organelle tubulation (9).
The molecular mechanisms of peroxisome constriction are poorly understood. In contrast, several proteins required for the final stage of the fission process are known. The first protein shown to be involved in this process was Saccharomyces cerevisiae Vps1p, a dynamin-like protein (DLP) (10). Later studies revealed that in this organism the DLP Dynamin-related 1 (Dnm1p) is also involved in peroxisome fission, especially under peroxisome-inducing growth conditions (11). Dnm1p forms a fission machinery together with the tail-anchored fission protein Fis1p and (in S. cerevisiae) the accessory proteins Mdv1p and Caf4p (12). Interestingly these proteins are also responsible for mitochondrial fission in yeast (13).
Dnm1p (Drp1 in mammals) (11, 14) is a large GTPase that achieves membrane fission by forming oligomeric, ring-like structures around constricted sites on organelle membranes (15). Powered by GTP hydrolysis, these ring-like structures then tighten further until the membrane severs. Interestingly, Dnm1p is recruited to Pex11p-enriched elongated peroxisomal membranes, suggesting that Pex11p and Dnm1p are functionally linked (16, 17).
Here, we identify a previously unknown role for Pex11p in peroxisomal fission. We show that Pex11p directly interacts with Dnm1p and that this interaction stimulates the GTPase activity of Dnm1p, establishing Pex11p as a GTPase activating protein (GAP) that plays a crucial role in the last step of the peroxisome fission process.
Results
Pex11p Is Required for Dnm1p-Mediated Peroxisomal Fission.
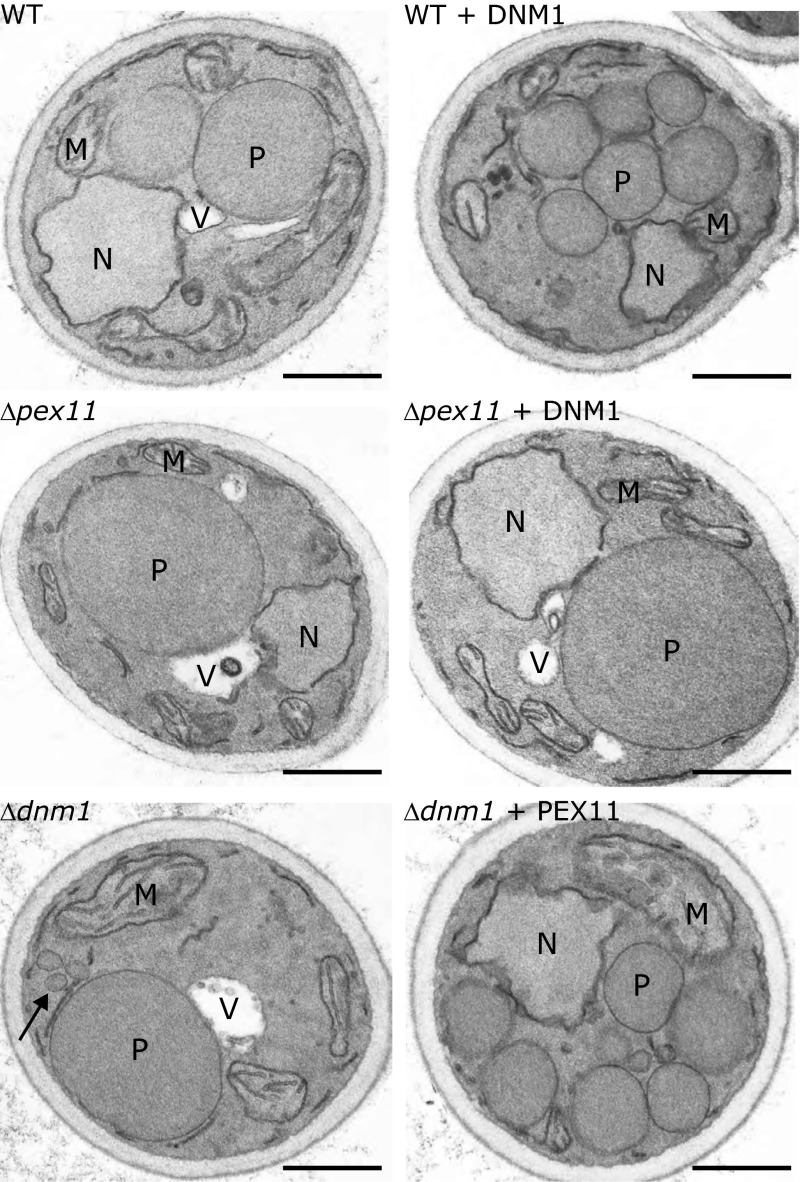
Deletion of PEX11 or DNM1 in the yeast Hansenula polymorpha affects peroxisome fission (18), resulting in strongly reduced peroxisome numbers relative to wild-type (WT) controls (17, 19). Furthermore, in budding H. polymorpha dnm1Δ cells, the single enlarged peroxisome in the mother cell forms a long tubular extension that protrudes into the bud (17), supporting the conclusion that fission is blocked at the final step. These extensions are not observed in H. polymorpha pex11Δ cells, in line with the model that Pex11p catalyzes the elongation step. To assess whether Pex11p is essential for Dnm1p-mediated peroxisome fission, we asked if the peroxisome fission defect of pex11Δ cells could be restored by overproducing Dnm1p. Electron microscopy (EM) analysis of thin sections revealed increased numbers of peroxisomal profiles upon overproduction of Dnm1p in WT cells (Fig. 1 and SI Appendix, Fig. S1A), confirming our previous observation that Dnm1p overproduction in WT cells stimulates peroxisome proliferation (20). However, no increase in peroxisomal profiles was observed in thin sections of pex11Δ cells overproducing Dnm1p relative to pex11Δ controls (Fig. 1 and SI Appendix, Fig. S1A), indicating that in these cells peroxisome fission was not enhanced. This conclusion was confirmed by the quantification of average numbers of peroxisomes per cell using serial sectioning in pex11Δ and pex11Δ cells overproducing Dnm1p (SI Appendix, Fig. S1B).
Fig. 1.
Pex11p is required for Dnm1p-mediated peroxisomal fission. H. polymorpha WT cells (WT), WT cells overproducing Dnm1-GFP (WT + DNM1), pex11Δ cells (Δpex11), pex11Δ cells overproducing Dnm1-GFP (Δpex11 + DNM1), dnm1Δ cells (Δdnm1), or dnm1Δ cells overproducing Pex11p (Δdnm1 + PEX11) were grown for 16 h on methanol-containing media and analyzed by EM. (Scale bars, 1 μm.) M, mitochondria; N, nucleus; P, peroxisome; V, vacuole. Arrow represents the peroxisomal vesicle associated with the mother organelle.
Previous 3D reconstruction of serial sections of H. polymorpha dnm1Δ cells revealed that at the base of the peroxisome extension of the single enlarged organelle, small vesicular extensions are formed, which still adhere to the mother organelle (21) (Fig. 1). This explains the presence of multiple peroxisomal profiles in thin sections of dnm1Δ cells. EM analysis of thin sections of dnm1Δ cells overproducing Pex11p revealed an increase in peroxisomal profiles per cell section (Fig. 1 and SI Appendix, Fig. S1A). These profiles were smaller than the mother peroxisomes in dnm1Δ cells but much larger than the vesicular structures that associate with dnm1Δ organelles. To study whether these structures also still adhered to the mother organelle, we performed electron tomography. This revealed the presence of numerous constricted tubules (average diameter, 30 ± 4 nm; n = 13) connecting individual vesicular structures (Fig. 2 and Movie S1), confirming that the peroxisomal profiles observed in the dnm1Δ strain overproducing Pex11p are still interconnected. Hence, the total number of peroxisomes per cell does not increase upon overproduction of Pex11p, although the number of peroxisomal profiles is enhanced in thin sections of these cells. Taken together, these data establish that Pex11p is essential for Dnm1p-mediated peroxisome fission.
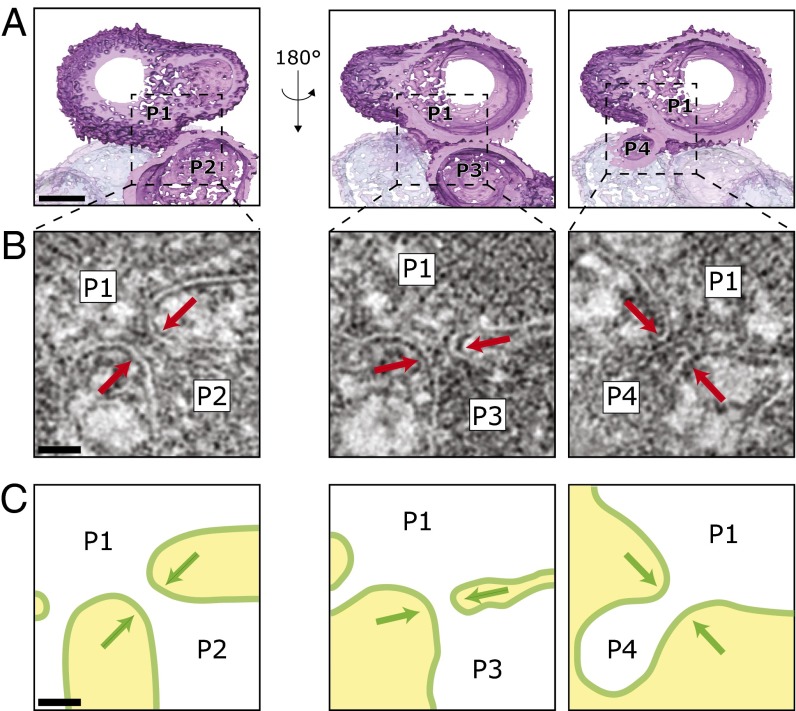
Fig. 2.
Pex11p overproduction in dnm1Δ cells remodels peroxisomes into interconnected compartments. H. polymorpha dnm1Δ cells overproducing Pex11p were grown for 16 h on methanol-containing media and analyzed by electron tomography. (A) Surface rendering of four adjacent peroxisomes (P1–P4). (B) The 5.5-nm-thick digital slices illustrating the neck-like tubules (arrows) that connect the indicated peroxisomes. Slices have been slightly tilted through the tomographic volume at the indicated position in A to maximize clarity. (C) Schematic drawing of the peroxisomal membranes shown in B. [Scale bars, (A) 100 nm and (B and C) 25 nm.]
Pex11p Interacts with Dnm1p Both in Vivo and in Vitro.
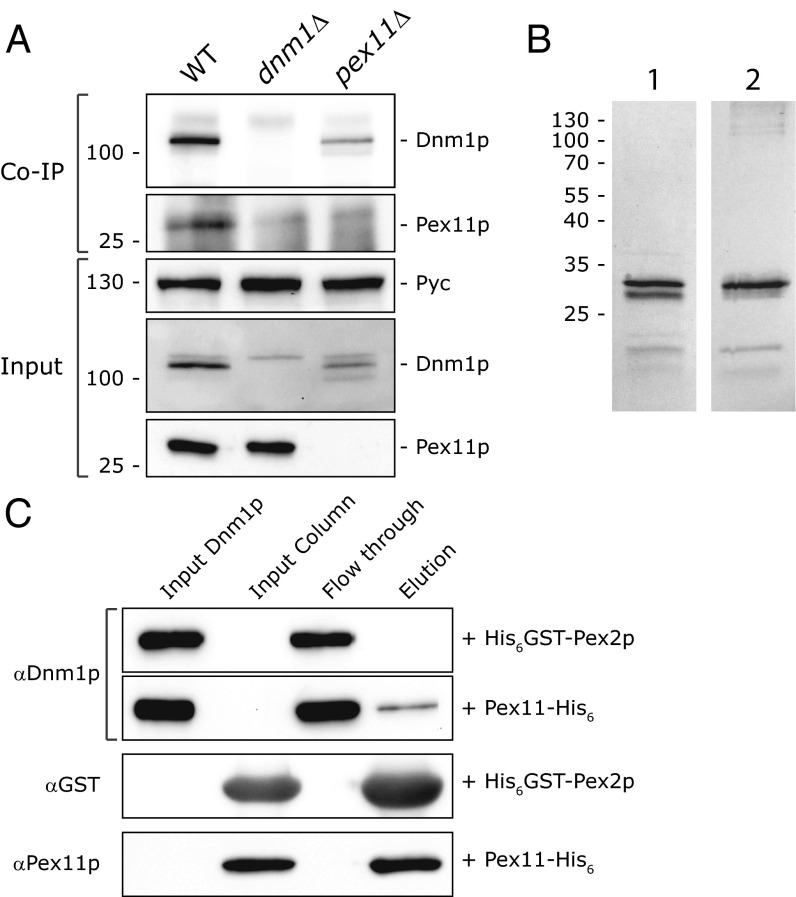
Previous data from mammalian cells suggested that Pex11β binds indirectly to the Dnm1p homolog Drp1 (14). To gain further insight into the relationship between Pex11p and Dnm1p in H. polymorpha, we subjected lysates of WT, pex11Δ, and dnm1Δ cells to coimmunoprecipitation analysis using anti-Dnm1p antibodies. As shown in Fig. 3A, Pex11p can be coprecipitated with Dnm1p antibodies from WT cell lysates, but not from lysates from both mutant strains, indicating that both proteins interact in vivo. Taking our analysis further, we used purified Dnm1p from E. coli, together with Pex11-His6, isolated from yeast (Fig. 3B), to demonstrate that the Dnm1p–Pex11p interaction is direct (Fig. 3C).
Fig. 3.
Pex11p interacts with Dnm1p both in vivo and in vitro. (A) Coimmunoprecipitation analysis (Upper panels) of cell lysates (Lower panels) derived from WT, pex11Δ, and dnm1Δ cells, using antibodies raised against H. polymorpha Dnm1p. Samples were analyzed by SDS/PAGE and Western blotting using antibodies raised against Pex11p, Dnm1p, and Pyruvate carboxylase (Pyc, loading control). (B) SDS/PAGE and coomassie staining (1) and Western blot probed with Pex11p antibodies (2) of Pex11-His6 purified from yeast. (C) Pull-down assays using Dnm1p purified from E. coli with purified Pex11-His6 or His6GST-Pex2 really interesting new gene (RING) domain bound to Ni-NTA resin. Samples were analyzed by SDS/PAGE and Western blotting using antibodies raised against S. cerevisiae Dnm1p, GST, and Pex11p. Equal portions were loaded per lane.
Large, Hydrophobic Residues in the N Terminus of Pex11p Control Dnm1p Binding.
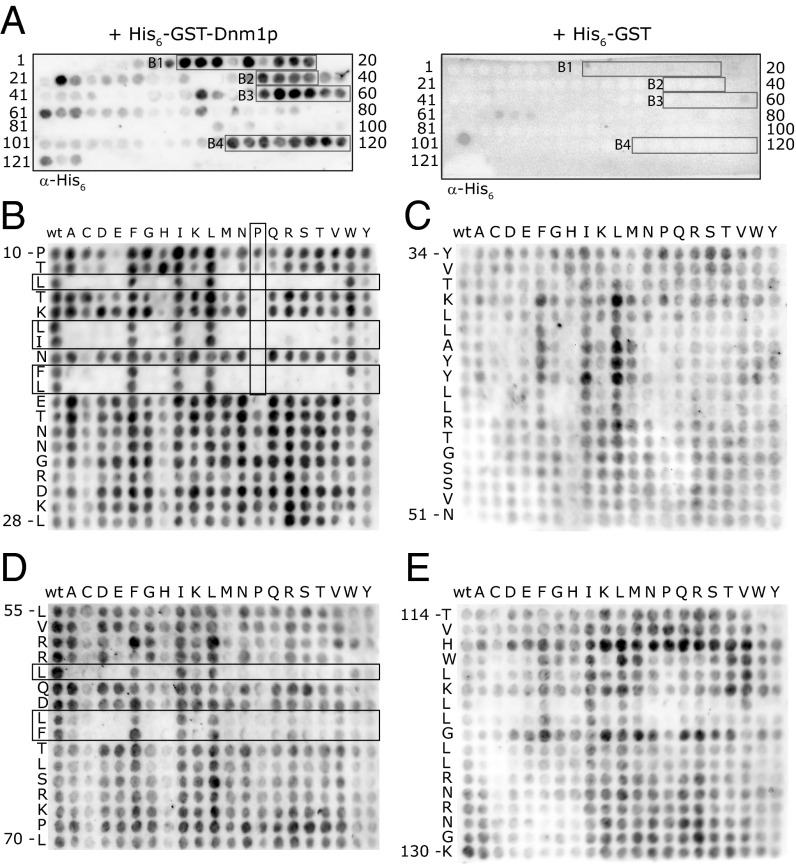
To identify regions in Pex11p that are involved in Dnm1p binding, we performed a peptide blot analysis using overlapping 12-mer peptides from the N-terminal 134 amino acids of Pex11p and purified His6GST-Dnm1p (Fig. 4A). This region of Pex11p was deemed most likely to bind Dnm1p because it is exposed to the cytosol (SI Appendix, Fig. S2). Our peptide blot analysis identified four potential binding sites (termed B1–B4) for His6GST-Dnm1p in Pex11p (Fig. 4A, boxed). These regions did not display binding to His6GST alone (Fig. 4A).
Fig. 4.
Detailed analysis of the Pex11p–Dnm1p interaction. (A) Peptide blots of overlapping 12-mer peptides covering the N-terminal 134 amino acids of Pex11p were incubated with His6GST-Dnm1p (Left panel) or His6GST (Right panel). Bound protein was detected using antibodies raised against the His6 tag. The four potential interacting regions, corresponding to amino acids 10–29 (B1), 35–51 (B2), 55–70 (B3), and 113–130 (B4), are boxed. (B–E) Substitution analysis with an array of Pex11p peptide variants derived from residues 10–28 (B), 34–51 (C), 55–70 (D), and 114–130 (E). Each spot corresponds to a version of the WT peptide (sequence shown in the left margin; the first and last amino acid are numbered) where one amino acid was changed to one of the 20 amino acids (shown across the top of the blot). Spots in the first column represent replicas of the WT sequence. Peptides were spot-synthesized on cellulose membranes, incubated with His6GST-Dnm1p, and probed as above. Substitutions that disturb binding (B and D) are boxed.
To determine which residues control the Dnm1p–Pex11p interaction, a peptide array was produced, where each residue of the interacting regions B1–B4 was substituted with every other amino acid. These peptides were spotted onto cellulose membranes, and the ability of these peptides to bind His6GST-Dnm1p was tested (Fig. 4 B–E). Introduction of alternative amino acids into the peptides covering regions B2 (Fig. 4C) and B4 (Fig. 4E) did not significantly disturb the ability of His6GST-Dnm1p to bind, which suggests that either these regions bind nonspecifically or no single amino acid substitution can disturb binding. In contrast, the peptides covering regions B1 and B3 were sensitive to single amino acid substitutions (Fig. 4 B and D). Specifically, changing Leu12, Leu15, Ile16, Phe18, or Leu19 in B1 or Leu59, Leu62, or Phe63 in B3 into Alanines inhibited the interaction. These residues are largely conserved within the Pex11p family (SI Appendix, Fig. S3B). Furthermore, introduction of a Proline into B1 also inhibited the interaction, suggesting that Dnm1p binding requires an intact alpha helix. Secondary structure predictions propose that residues 14–19 of Pex11p are indeed alpha helical, whereas the B3 region is part of a larger amphipathic helix that extends from residue 56–86 (SI Appendix, Fig. S3A).
Mutations in the DLP Binding Site of Pex11p Inhibit Peroxisome Fission in Yeast and Mammals.
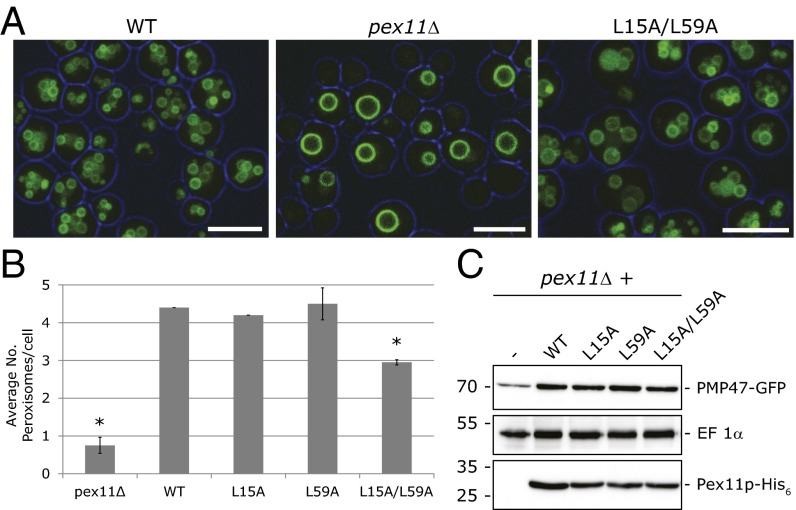
Disturbances to peroxisomal fission result in lower numbers of peroxisomes per cell (17, 19). Therefore, we determined the effect of inhibiting the Pex11p–Dnm1p interaction on peroxisome numbers in H. polymorpha cells using PMP47-GFP as a peroxisomal membrane marker and fluorescence microscopy (Fig. 5). The Pex11p–Dnm1p interaction was inhibited by mutating the Leucine residues at positions 15 and 59 in Pex11p to Alanines (L15A and L59A), mutations which disturbed the Dnm1p–Pex11p interaction (Fig. 4). Expression of the L15A or L59A forms of Pex11p in pex11Δ cells did not compromise peroxisome fission (Fig. 5B). However, the two putative Dnm1p binding sites in Pex11p display similar amino acid sequences, as well as similar secondary structural properties (SI Appendix, Fig. S3), which could suggest a level of redundancy. To investigate this, we produced a version of Pex11p where both Leu15 and Leu59 were mutated to Alanines (L15A/L59A) (Fig. 5). pex11Δ cells producing this mutant contained significantly reduced average numbers of peroxisomes per cell (Fig. 5A and SI Appendix, Table S1), as well as an increased percentage of cells with no peroxisomes or one peroxisome, typical for pex11Δ cells (SI Appendix, Fig. S4A). As expected, Pex11 L15A/L59A displayed a reduced ability to bind Dnm1p in vitro (SI Appendix, Fig. S4B). Introduction of the L15A/L59A mutations into Pex11p did not alter the levels of Pex11p (Fig. 5C) nor compromise the ability of the strain expressing this mutant to grow on methanol (SI Appendix, Fig. S4C), indicating that the observed fission defect results from disturbing the Pex11p–Dnm1p interaction.
Fig. 5.
Mutations in the Dnm1p binding sites of Pex11p disturb peroxisome fission. (A) Fluorescence microscopy analysis of methanol-grown pex11Δ cells (pex11Δ) and pex11Δ cells producing WT Pex11p (WT) or Pex11p L15A/L59A (L15A/L59A). Cells coproduced the peroxisomal membrane marker PMP47-GFP. (Scale bar, 5 μm.) (B) Quantification of peroxisome numbers in pex11Δ cells producing WT or mutant forms of Pex11-His6. All strains coexpressed PMP47-GFP, the readout of which was used to quantify peroxisome numbers. Values represent the mean ± SD of two independent experiments (*P < 0.05). (C) Western blots of cell lysates from pex11Δ cells producing WT or mutant forms of Pex11p, coproducing PMP47-GFP. Blots were probed with antibodies raised against elongation factor (EF) 1α (loading control), GFP, and the His6 tag.
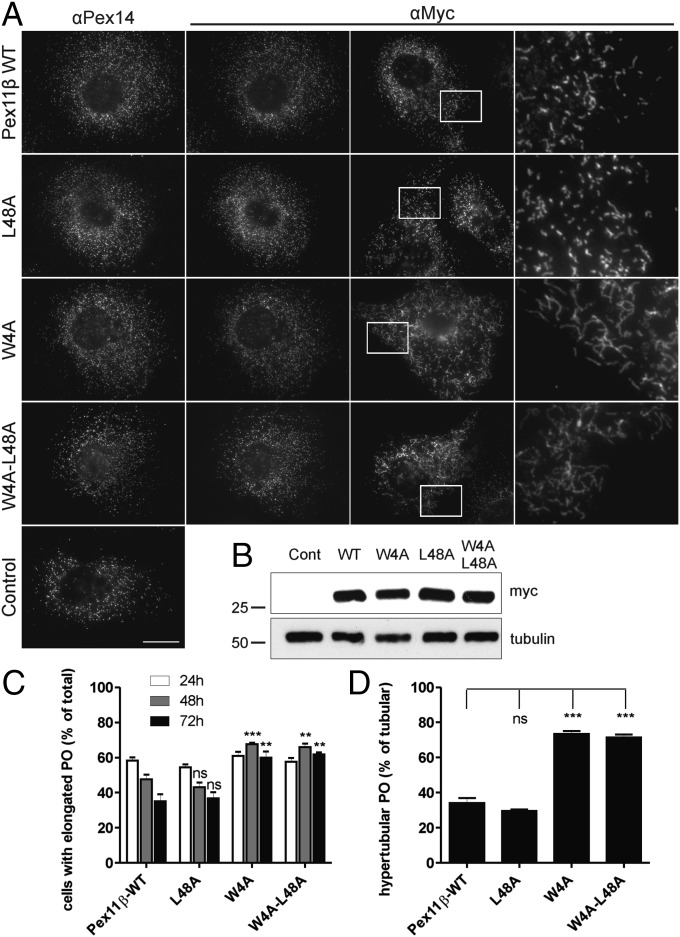
As in H. polymorpha, peroxisome fission in mammals requires the action of a DLP, the Dnm1p homolog Drp1 (22). Sequence alignment analysis indicates that many of the residues that regulate the Dnm1p–Pex11p interaction in yeast are conserved in mammalian Pex11β, whereas the domain structure of Pex11β is also highly similar (SI Appendix, Figs. S3 and S7). To investigate the link between Drp1 and Pex11β further, we mutated the residues corresponding to Leu15 and Leu59 in mammalian Pex11β (Trp4 and Leu48) and followed the effect on peroxisome fission in COS-7 cells (Fig. 6). Expression of Pex11β L48A induced peroxisome proliferation in a similar manner to WT Pex11β (23). Conversely, altering Trp4 of Pex11β to an Alanine (W4A) inhibited peroxisome fission and resulted in the formation of hypertubular peroxisomes (Fig. 6), a sign that Drp1 activity is inhibited (22). Immunofluorescence studies demonstrated that Drp1 localization is not altered by these mutations (SI Appendix, Fig. S8A). Significantly, introduction of the W4A mutation into a peptide derived from Pex11β disturbed the ability of Drp1 to bind to this peptide in vitro (SI Appendix, Fig. S8B). These observations support the conclusion that, similar to yeast, Drp1-mediated peroxisomal fission can be compromised by disturbing the Drp1–Pex11β interaction.
Fig. 6.
Pex11β W4A results in the accumulation of tubular peroxisomes. (A) COS-7 cells were transfected with Pex11β-Myc, Pex11β-L48A-Myc, Pex11β-W4A-Myc, or Pex11β-W4A/L48A-Myc. Fixed cells were labeled with anti-Pex14p and anti-Myc antibodies. Higher magnification view of boxed regions is shown in the Right panel. (B) Immunoblotting of cell lysates with anti-Myc and anti-Tubulin (loading control) antibodies. (C) Quantitative analysis of peroxisome morphology in cells expressing Pex11β mutants at several time points after transfection. (D) The percentage of cells with hypertubular peroxisomes (48 h). For quantitative evaluation of peroxisome morphology, peroxisomes were categorized as tubular (<5 μm) or unusually long, hypertubular (>5 μm). Values represent the mean ± SD of at least three independent experiments (**P < 0.01, ***P < 0.001). (Scale bar, 10 μm.)
The Amphipathic Helix of Pex11p Functions as a GAP for Dnm1p in Vitro.
GTP hydrolysis by Dnm1p is needed to achieve organelle fission (11, 16, 24). Because Pex11p is required for Dnm1p function, we evaluated whether Pex11p binding alters the kinetic properties of Dnm1p. Purified Dnm1p (Fig. 7A) was able to hydrolyze GTP in a time-dependent manner (Fig. 7B) and displayed a catalytic activity (Kcat) of 2.16 ± 0.42 min−1 and a substrate affinity (Km) of 381 ± 6.5 µM (Fig. 7D). These values are similar to those of mammalian Drp1, isolated from S. cerevisiae (25). The addition of purified Pex11-His6 resulted in a small increase in the GTPase activity of Dnm1p (SI Appendix, Fig. S5). However, addition of detergents to Dnm1p, which were essential to solubilize full-length Pex11-His6, strongly reduced its activity (SI Appendix, Fig. S5), which hampered the analysis of the effect of full-length Pex11-His6 on Dnm1p. Therefore, we continued our analysis using peptides of Pex11p. Peptides corresponding to the binding regions B1 or B3 in Pex11p did not alter the activity of Dnm1p (SI Appendix, Fig. S6A). However, the B3 binding site resides in a larger amphipathic helix, extending from residues 56 to 86 (SI Appendix, Fig. S2). Addition of the complete amphipathic helix to Dnm1p significantly enhanced the catalytic activity (Kcat = 4.14 ± 0.32 min−1) as well as substrate affinity (Km = 237 ± 13 µM). This stimulation was also observed when reactions were performed in the presence of high salt (SI Appendix, Fig. S6B), conditions known to block Dnm1p oligomerization (25). No increase in activity was observed when a similar peptide was added where Leu59, Leu62, and Phe63 were altered to Alanine residues (SI Appendix, Fig. S6C). This peptide is unable to bind to Dnm1p in vitro (SI Appendix, Fig. S6D), indicating that the interaction between the amphipathic helix and Dnm1p is crucial for the observed stimulation. Finally, we established that the peptide from Pex11β that is able to bind to Drp1 (SI Appendix, Fig. S8) can also stimulate its GTPase activity in vitro (SI Appendix, Fig. S7), clearly demonstrating that Pex11p, in addition to its role in membrane remodeling (9), can function as a GAP for DLPs during peroxisome fission.
Fig. 7.
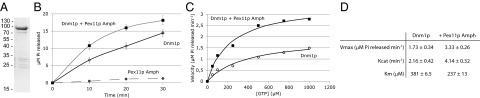
The amphipathic helix of Pex11p functions as a GAP for Dnm1p in vitro. (A) SDS/PAGE and coomassie staining analysis of Dnm1p purified from E. coli. (B) Time course of GTP hydrolysis by Dnm1p (open circles), Dnm1p together with a peptide of the amphipathic helix of Pex11p (closed squares), or the Pex11p peptide alone (open diamonds). (C) Steady-state kinetics of GTP hydrolysis by Dnm1p in the presence (closed squares) and absence (open circles) of the Pex11p amphipathic helix peptide. (D) Kinetic parameters of Dnm1p in the presence and absence of the Pex11p amphipathic helix peptide. Values represent the mean ± SD of three separate measurements.
Discussion
Pex11p initiates peroxisomal fission by catalyzing organelle elongation (9, 26). In the present study, we identify a new role for yeast Pex11p in the final stage of peroxisomal fission, namely as a GAP for Dnm1p. Similar observations were made for the mammalian homologs Pex11β and Drp1.
Our data clearly demonstrate that Pex11p and Dnm1p are required for different aspects of peroxisomal fission, as the lack of one protein cannot be suppressed by overproduction of the other. Pex11p overproduction in H. polymorpha dnm1Δ cells results in tubulation, but not scission, of the peroxisomal membrane, whereas pex11Δ cells contain a single peroxisome irrespective of whether Dnm1p is overproduced or not. These data fit a model where Pex11p and Dnm1p act together in the final step of peroxisomal fission. Pex11p also functions as an initiator of the fission process through insertion of an amphipathic helix into the peroxisomal membrane, with Dnm1p-mediated scission of the organelle following. However, the observation that Pex11p interacts directly with Dnm1p, plus that this interaction is crucial for peroxisomal fission, establishes that Pex11p performs an additional yet vital role in the last stage of peroxisomal fission. How can these data be integrated into the prevailing model describing peroxisomal fission? The binding sites for Dnm1p reside in the N-terminal region of Pex11p, as does the membrane remodeling amphipathic helix of Pex11p (9). Furthermore, one Dnm1p binding site (B3, residues 55–70) is present in this amphipathic helix, suggesting the existence of a synergistic relationship between the membrane remodeling and GTPase activating roles of Pex11p. Our previous data establish that this amphipathic helix adopts an unstructured conformation in aqueous solution (9), suggesting that this region of Pex11p can undergo transition from an unfolded to helical conformation. This transition may facilitate insertion of the amphipathic helix into the peroxisomal membrane, to initiate fission. As the introduction of helix-breaking mutations can disturb the Dnm1p interaction with Pex11p peptides (Fig. 4), we conclude that helix formation is required for Pex11p to bind Dnm1p. Hence, Dnm1p binding, and consequently stimulation of Dnm1p GTPase activity, is likely to occur after Pex11p has remodeled the peroxisomal membrane, suggesting a spatiotemporal relationship. On the other hand, our data on mammalian Pex11β, which suggest that a different region of Pex11β binds to and stimulates the activity of Drp1 (Fig. 6 and SI Appendix, Figs. S7 and S8), indicate that although the GAP activity of Pex11p is conserved in several species, the underlying mechanisms may differ.
In S. cerevisiae several Dnm1p binding partners are known to regulate peroxisomal fission, including the membrane protein Fis1p and the cytosolic proteins Mdv1p and Caf4p (12, 27). Originally these proteins were proposed to facilitate Dnm1p recruitment to membranes, yet recent data on mitochondrial fission identified additional, postrecruitment roles. Importantly, these binding partners do not stimulate Dnm1p GTPase activity, instead appearing to modulate Dnm1p oligomer assembly (25). In mammals Drp1 binds not only to Fis1p (28) but additionally to the membrane protein Mff (29), and although the role of the Drp1–Fis1p interaction in organelle fission remains unclear, binding to Mff allows Drp1 to associate with organellar membranes (30). Significantly, Mff binding does not enhance the GTPase activity of Drp1 (25). Consequently, the available data on DLP interacting proteins suggest that DLP-mediated organelle fission is coordinated by a range of binding partners that provide different functions at different stages of the fission process, including recruitment factors that govern localization, adaptor proteins that control oligomerization, and based on our data, modulators that stimulate activity during membrane separation.
The recent observation that a 30% increase in the activity of a DLP, as measured with in vitro approaches, can cause a significant increase in membrane remodeling capabilities of the protein in vivo (31) supports our observation that a twofold increase in the activity of Dnm1p in vitro translates into efficient peroxisome fission in vivo. Binding of Pex11p to a DLP occurs at its site of action, the peroxisomal membrane, guaranteeing that the resulting increase in GTPase activity occurs at the right time and right place for efficient membrane fission. Leading on from this, recent data indicate that the activity of Drp1 is enhanced by cardiolipin (32). This lipid, which plays a role in mitochondrial fission (33), is also a potent stimulator of membrane tubulation (32). Cardiolipin is present in the peroxisomal membrane (34), whereas the amphipathic helix of Pex11p can only tubulate liposomes that contain cardiolipin (9). Taken together, these observations support the suggestion that DLP activity and, consequently, the final act of organelle fission can be fine-tuned by factors on the organelle membrane. Identifying other factors that could control DLP activity in similar ways will provide invaluable insight into how membrane scission is achieved.
Our understanding of peroxisomal fission was advanced considerably by the observation that Pex11p directly controls membrane elongation (9, 26). In addition, previous work from our group (21) suggests that Pex11p also directs the redistribution of peroxisomal membrane proteins during fission, supporting the idea that the function of Pex11p goes much further than membrane remodeling. We propose that Pex11p is in fact a master regulator of peroxisomal fission, providing vital contributions to a range of events that occur during fission.
Materials and Methods
For details of plasmids, oligonucleotides, and H. polymorpha strains used in this study, see SI Appendix.
Culture Conditions.
H. polymorpha cells were grown in batch cultures at 37 °C on mineral media supplemented with 0.25% glucose or 0.5% methanol, with or without 0.05% glycerol as the carbon source and 0.25% ammonium sulfate or methylamine as the nitrogen source. Leucine, when required, was added to a final concentration of 30 μg/mL COS-7 cells (ECACC 87021302) and was maintained in DMEM supplemented with 10% (wt/vol) FCS (Life Technologies) at 37 °C in a 5% CO2-humidified incubator. Cells were transfected by electroporation (BTX ECM 630; BTX Instrument Division, Harvard Apparatus; settings, 230 V, 1500 µF, 125 Ω) as described (22).
Biochemical Techniques.
Details on cell extracts, protein expression/purification, and binding assays can be found in SI Appendix.
Peptide Blot Assays.
The peptide arrays corresponding to the first 134 amino acids of Pex11p were synthesized on amino-modified cellulose membranes (β-alanine membrane) according to SPOT synthesis protocols (35). The design and the peptide spot arrangement for the substitutional analyses of variant peptides derived from the sequence were carried out using the in-house software LISA. For further details, see SI Appendix.
GTPase Activity Measurements.
The GTP hydrolyzing activity of purified Dnm1p (0.8 µM), in the absence or presence of Pex11p peptides (8 µM), was followed by measuring the release of inorganic phosphate using the Pi ColorLock Gold kit (Innova Biosciences) according to the manufacturer’s instructions. For further details, see SI Appendix. The Vmax, Kcat, and Km were calculated in Microsoft Excel using nonlinear regression curve fitting. The data presented represent the average ± SD of three separate measurements.
Fluorescence Microscopy.
All yeast fluorescence microscopy images were acquired using a Zeiss Plan-Neofluar 100×/1.3 oil objective. Confocal images of yeast cells were captured using a Zeiss LSM510, using photomultiplier tubes (Hamamatsu Photonics); images were acquired using Zen 2009 software. GFP fluorescence was analyzed by excitation of the cells with a 488-nm argon ion laser (Lasos), and emission was detected using either a 500–530-nm or a 500–550-nm bandpass emission filter. For quantification of peroxisomes, Z stack images were made using an interval of 0.6 μm. Wide-field fluorescence microscopy was performed using a Zeiss Axioscope50 fluorescence microscope. Images were acquired with a Princeton Instruments 1300Y CCD camera (Roper Scientific). GFP fluorescence was visualized with a 470/40-nm bandpass excitation filter, a 495-nm dichromatic mirror, and a 525/50-nm bandpass emission filter. Image analysis was carried out using ImageJ (rsb.info.nih.gov/nih-image/) and Adobe Photoshop (Adobe Systems). For immunofluorescence microscopy on COS-7 cells, cells grown on glass coverslips were fixed with 4% (vol/vol) paraformaldehyde in PBS pH 7.4, permeabilized with 2.5 μg/mL digitonin, blocked with 1% BSA, and incubated with primary and secondary antibodies as described (36). Information on quantification and statistical analysis can be found in SI Appendix.
EM and Electron Tomography.
For EM, cells were fixed in 1.5% (wt/vol) KMnO4 and prepared for EM as described (37). For electron tomography, H. polymorpha dnm1Δ overproducing Pex11p were cryo-fixed in liquid ethane using the sandwich plunge freezing method (38). Cells were freeze-substituted in 1% osmium tetroxide, 0.5% uranyl acetate, and 5% (vol/vol) distilled water in acetone using the fast low-temperature dehydration and fixation method (39). Cells were infiltrated overnight with Epon and polymerized for 48 h at 60 °C. We cut 200-nm-thick sections and overlayed them with 10 nm of fiducial gold particles. Two single-axis tilt series, each containing 131 images with 1° tilt increments, were acquired on a FEI Tecnai20 running at 200 kV using the FEI automated tomography acquisition software and a cooled slow-scan charge-coupled device camera (Ultrascan 4000; Gatan) in 2 × 2 binned mode with a final pixel size of 1.1 nm. The tilt series were aligned and reconstructed by the simultaneous iterative reconstruction technique (SIRT) algorithm using the IMOD software package (40) and analyzed using the AMIRA visualization package (TGS Europe). To generate three-dimensional surface-rendered models in AMIRA, masks of peroxisomes were drawn manually and afterward improved by thresh-holding. The movie was shot in AMIRA as sequential TIF images, which subsequently were merged into a movie using FFmpeg (www.ffmpeg.org).
Supplementary Material
Acknowledgments
The authors thank E. Boekema for access to advanced electron microscope facilities; R. de Boer, M. Mulder, and T. Schrader for technical assistance; L. Bosgraaf for help with the ImageJ plugin; D. Crane for Pex14p antibodies; J. Nunnari for S. cerevisiae Dnm1p antibodies; O. Daumke for the Drp1 plasmid and advice; and M. Veenhuis for discussions. C.W. is supported by Open Programme Grant 821.02.022 and Vidi Grant 723.013.004 from the Netherlands Organisation for Scientific Research (NWO). K.K. is supported by European Union Marie Curie Intra-European Fellowship FP7-330150. A. M. Kram is supported by the research program of the Kluyver Centre for Genomics of Industrial Fermentation, as part of the Netherlands Genomics Initiative/NWO. M.S. is supported by Biotechnology and Biological Sciences Research Council Grant BB/K006231/1 and Wellcome Trust Institutional Strategic Support Award WT097835MF. I.J.v.d.K. and M.S. are supported by Marie Curie Initial Training Networks Action PerFuMe Grant 316723.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418736112/-/DCSupplemental.
References
- 1.Gabaldón T. Peroxisome diversity and evolution. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):765–773. doi: 10.1098/rstb.2009.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wanders RJ, Waterham HR. Peroxisomal disorders: The single peroxisomal enzyme deficiencies. Biochim Biophys Acta. 2006;1763(12):1707–1720. doi: 10.1016/j.bbamcr.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Ebberink MS, et al. A novel defect of peroxisome division due to a homozygous non-sense mutation in the PEX11β gene. J Med Genet. 2012;49(5):307–313. doi: 10.1136/jmedgenet-2012-100778. [DOI] [PubMed] [Google Scholar]
- 4.Waterham HR, et al. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356(17):1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 5.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178(3):399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 7.Koch J, Brocard C. Membrane elongation factors in organelle maintenance: The case of peroxisome proliferation. Biomol Concepts. 2011;2(5):353–364. doi: 10.1515/BMC.2011.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdmann R, Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128(4):509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opaliński Ł, Kiel JA, Williams C, Veenhuis M, van der Klei IJ. Membrane curvature during peroxisome fission requires Pex11. EMBO J. 2011;30(1):5–16. doi: 10.1038/emboj.2010.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoepfner D, van den Berg M, Philippsen P, Tabak HF, Hettema EH. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol. 2001;155(6):979–990. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuravi K, et al. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119(Pt 19):3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 12.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121(Pt 10):1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim Biophys Acta. 2006;1763(5-6):531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Itoyama A, et al. Mff functions with Pex11pβ and DLP1 in peroxisomal fission. Biol Open. 2013;2(10):998–1006. doi: 10.1242/bio.20135298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18(1):20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem. 2003;278(19):17012–17020. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 17.Nagotu S, Saraya R, Otzen M, Veenhuis M, van der Klei IJ. Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim Biophys Acta. 2008;1783(5):760–769. doi: 10.1016/j.bbamcr.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Thoms S, Erdmann R. Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 2005;272(20):5169–5181. doi: 10.1111/j.1742-4658.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- 19.Krikken AM, Veenhuis M, van der Klei IJ. Hansenula polymorpha pex11 cells are affected in peroxisome retention. FEBS J. 2009;276(5):1429–1439. doi: 10.1111/j.1742-4658.2009.06883.x. [DOI] [PubMed] [Google Scholar]
- 20.Nagotu S, et al. Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission. Traffic. 2008;9(9):1471–1484. doi: 10.1111/j.1600-0854.2008.00772.x. [DOI] [PubMed] [Google Scholar]
- 21.Cepińska MN, Veenhuis M, van der Klei IJ, Nagotu S. Peroxisome fission is associated with reorganization of specific membrane proteins. Traffic. 2011;12(7):925–937. doi: 10.1111/j.1600-0854.2011.01198.x. [DOI] [PubMed] [Google Scholar]
- 22.Koch A, Schneider G, Lüers GH, Schrader M. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci. 2004;117(Pt 17):3995–4006. doi: 10.1242/jcs.01268. [DOI] [PubMed] [Google Scholar]
- 23.Schrader M, et al. Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem. 1998;273(45):29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- 24.Ingerman E, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170(7):1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koirala S, et al. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci USA. 2013;110(15):E1342–E1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch J, et al. PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J Cell Sci. 2010;123(Pt 19):3389–3400. doi: 10.1242/jcs.064907. [DOI] [PubMed] [Google Scholar]
- 27.Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J Biol Chem. 2007;282(46):33769–33775. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23(15):5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19(6):2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boissan M, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344(6191):1510–1515. doi: 10.1126/science.1253768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macdonald PJ, et al. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell. 2014;25(12):1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan R, Jones AD, Hu J. Cardiolipin-mediated mitochondrial dynamics and stress response in Arabidopsis. Plant Cell. 2014;26(1):391–409. doi: 10.1105/tpc.113.121095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wriessnegger T, et al. Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Biochim Biophys Acta. 2007;1771(4):455–461. doi: 10.1016/j.bbalip.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Volkmer R. Synthesis and application of peptide arrays: Quo vadis SPOT technology. ChemBioChem. 2009;10(9):1431–1442. doi: 10.1002/cbic.200900078. [DOI] [PubMed] [Google Scholar]
- 36.Bonekamp NA, et al. Self-interaction of human Pex11pβ during peroxisomal growth and division. PLoS ONE. 2013;8(1):e53424. doi: 10.1371/journal.pone.0053424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterham HR, Titorenko VI, Swaving GJ, Harder W, Veenhuis M. Peroxisomes in the methylotrophic yeast Hansenula polymorpha do not necessarily derive from pre-existing organelles. EMBO J. 1993;12(12):4785–4794. doi: 10.1002/j.1460-2075.1993.tb06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baba M. Electron microscopy in yeast. Methods Enzymol. 2008;451:133–149. doi: 10.1016/S0076-6879(08)03210-2. [DOI] [PubMed] [Google Scholar]
- 39.McDonald KL, Webb RI. Freeze substitution in 3 hours or less. J Microsc. 2011;243(3):227–233. doi: 10.1111/j.1365-2818.2011.03526.x. [DOI] [PubMed] [Google Scholar]
- 40.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.