Abstract
The erythrocyte membrane has been extensively studied, both as a model membrane system and to investigate its role in gas exchange and transport. Much is now known about the protein components of the membrane, how they are organised into large multi-protein complexes and how they interact with each other within these complexes. Many links between the membrane and the cytoskeleton have also been delineated and have been demonstrated to be crucial for maintaining the deformability and integrity of the erythrocyte. In this study we have refined previous, highly speculative molecular models of these complexes by including the available data pertaining to known protein-protein interactions. While the refined models remain highly speculative, they provide an evolving framework for visualisation of these important cellular structures at the atomic level.
Keywords: erythrocyte, band 3, ankyrin, protein 4.1R, macrocomplex, junctional complex, protein modelling
Introduction
The erythrocyte membrane has long been studied, both as a model membrane system and for its unique importance as a gas exchange interface. The underlying cytoskeleton is also vital for erythrocyte function, maintaining the high surface-area-to-volume ratio of the biconcave disc while allowing massive, reversible deformation during passage through the vasculature. Cytoskeletal proteins are physically linked to the membrane by vertical interactions with integral membrane proteins, which in turn form multiprotein ‘macrocomplexes’ via horizontal interactions in the membrane [1]. A variety of adaptor proteins are involved in forming and maintaining these links. Disruption of the connections between cytoskeletal and membrane protein components results in misshapen red cells (spherocytes, elliptocytes or ovalocytes) illustrating their importance to the function of erythrocytes.
The major cytoskeleton-membrane linkages are focused at two key sites: the ankyrin associated complex, and the junctional complex. The ankyrin associated complex brings together proteins of both the band 3 tetrameric complex (band 3, glycophorin A (GPA), protein 4.2, carbonic anhydrase II) and the Rh complex (RhAG, RhCE, RhD, CD47, ICAM-4, glycophorin B (GPB)) using the direct association of band 3 [2], protein 4.2 [3] and RhAG [4] with ankyrin [5]. Protein 4.2, which binds both band 3 and ankyrin has been suggested to serve as a bridge between the two sub-complexes, by binding both band 3 and CD47 [6]. The whole macrocomplex is anchored to the cytoskeleton by the interaction of ankyrin to β-spectrin [7].
The junctional complex is focused around a hub or ‘junction’ arising from lateral connections between protein 4.1, actin and β spectrin (the first of which stabilises the actin spectrin association via direct binding to both proteins [8]). These comprise the junction with which a host of additional cytoskeletal adaptor proteins are associated including the actin binding proteins dematin, α and β adducin. Actin is further enclosed by a tropomysin dimer and capped by tropomodulin [9]. These junctional complexes are known to provide membrane cytoskeletal linkage via interaction of the integral membrane proteins glycophorin C and D (GPC and GPD) with p55 and protein 4.1 [10]. In recent years, increasing evidence has emerged implicating both dimeric band 3 and glucose transporter-1 GLUT1 as additional sites of membrane linkage at the junctional complexes. The membrane proteins DARC (Duffy), Kell, Kx and Rh were found to be reduced in a protein 4.1 knockout mouse suggesting they may also be found at the site of junctional complexes [11], these results were confirmed in a human patient completely deficient in protein 4.1, although no reduction in Rh was observed suggesting that this protein is part of the junctional complex in mice but not humans (unpublished data – referred to in [12]).
The erythrocyte cytoskeleton is formed from a hexagonal lattice of spectrin tetramers, with short actin filaments at the junctions. It has been hypothesized that each hexagon is composed of six triangular repeat elements [12]. Every triangular repeat would contain a junctional complex at each apex (three in total), an ankyrin associated complex on each side (three in total) and contains four free band 3 dimers (not associated with the cytoskeleton). The skeleton has previously been visualised by electron microscopy of spread out skeletons, but recent work has used cryo-electron tomography to visualise the skeleton in its native state [13]. This work confirmed the presumption that the length of a spectrin tetramer seen in early EM images (~190nm) is much longer than it is in situ in an erythrocyte (~46nm).
A previous study [12] amalgamated available data on the copy number, stoichiometry and molecular structures of the components of the ankyrin-associated and junctional complex. The culmination of which was the construction of highly speculative representations of the complexes, using molecular structures as opposed to cartoon schematics. Here we refine these representations, using all currently available data pertaining to the structure of the components and the nature of the intermolecular contacts.
Representing natively disordered regions
It is important to note that a key difference between the models presented in this paper and the previous models [12] is our attempt to represent protein regions which are predicted to be natively disordered. These regions would not be expected to adopt a single 3D structure and therefore any static representation is necessarily flawed. In addition, disordered regions often undergo a disordered-ordered transition upon formation of protein-protein interactions [14]. Despite the difficulty in generating meaningful approximations of their true structure, we feel it is informative to include these regions in our models. Their inclusion enables appreciation of the volume of protein that is present, but of unknown structure. In order to generate representative structures the sequences of the disordered regions were submitted to the RanCh routine within the EOM software [15]. The routine generates random coil structures as alpha-carbon chains and calculates the radius of gyration (Rg) for each structure. One hundred structures were generated for each disordered sequence. It has been shown that the majority of disordered proteins are well-described by a power-law relationship between polymer length and ensemble average Rg [16]. Therefore, the RanCh-generated structure with the best fit to the theoretical average Rg was selected to represent the region in our models. All-atom models were not generated; disordered regions are represented as chains of spheres centred on the alpha-carbon atoms.
Ankyrin-associated complex
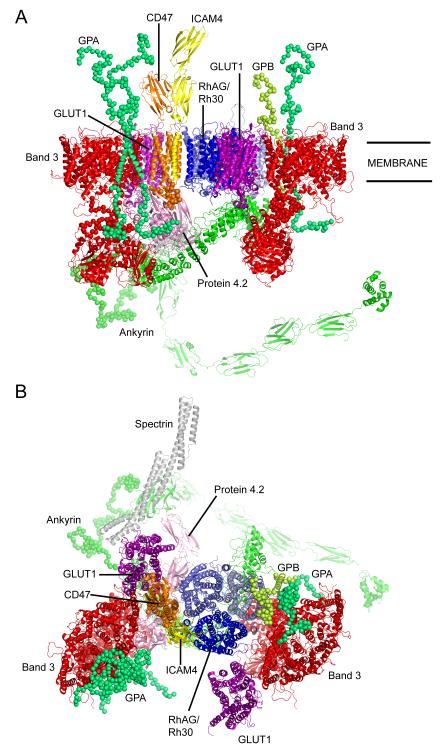
The ankyrin linked band 3 multiprotein complex, also referred to as the band 3 macrocomplex is comprised of two smaller subcomplexes, a band 3 complex, consisting of a tetramer of band 3 interacting with GPA, protein 4.2 and ankyrin and the Rh complex, including RhD, RhCE, RhAG, ICAM-4, GPB and CD47. These components and the known interactions between them are summarised in Table 1. The refined model of the entire ankyrin-associated complex is shown in Figure 1 and its assembly is discussed below.
Table 1. Ankyrin complex: components and known interactions.
| Protein | Oligomeric state | Number present in ankyrin complex | Interacting partners included in model |
|---|---|---|---|
| Band 3 | Dimer | 2 | GLUT1 Glycophorin A RhAG Ankyrin Protein 4.2 |
| GLUT1 | Monomer | 2 | Band 3 |
| Glycophorin (A/B) | Homodimer (A2) Heterodimer (A/B) |
1 1 |
Band 3 RhAG |
| RhAG/Rh30 | Heterotrimer (RhAG-Rh2) | 1 | Glycophorin B CD47 ICAM-4 Band 3 |
| CD47 | Monomer | 1 | Rh/RhAG Protein 4.2 |
| ICAM-4 | Monomer | 1 | Rh/RhAG |
| Ankyrin | Monomer | 1 | Band 3 RhAG Protein 4.2 |
| Protein 4.2 | Monomer | 1 | Band 3 CD47 Ankyrin |
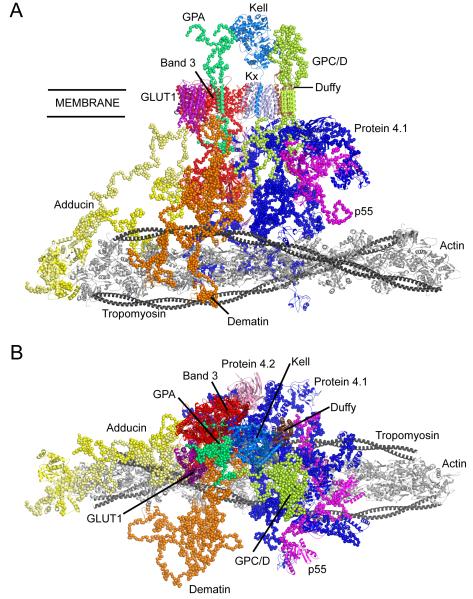
Figure 1. Overall model of ankyrin complex.
(A) view from within the plane of the membrane; (B) view from outside of the cell. Components shown in the figure are: two band 3 homodimers (red), one Rh/RhAG heterotrimer (RhAG light blue, RhCE mid-blue, RhD dark blue), one GPA homodimer (mint-green), one GPA/GPB heterodimer (mint-green/lime-green), one ICAM-4 monomer (yellow), one CD47 monomer (orange), two GLUT1 monomers (purple), one protein 4.2 monomer (pink) and one ankyrin monomer (green). All components are shown with secondary structure elements highlighted as cartoons and residues predicted to be disordered displayed as spheres centred on the Cα atoms. For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.
Anchoring to spectrin
Erythroid ankyrin-1 mediates the association of the spectrin cytoskeleton to band 3 in the erythrocyte plasma membrane. Ankyrin-1 is composed of three domains, an 89kDa N-terminal membrane binding domain, a central 62kDa spectrin binding domain and a 55kDa C-terminal regulatory domain. Ankyrin binding to spectrin involves the 14th and 15th repeat element of β spectrin and a fragment of the spectrin binding domain of ankyrin comprising residues 911-1068 (ZU5-ANK) [17]. A crystal structure has been solved for the complex of spectrin repeats 13-15 and ZU5-ANK; the interface displays significant charge complementarity between positive-charged residues on the ZU5 domain and negatively-charged residues on spectrin repeat 14 [17]. Our model includes this crystal structure but we have not extended the spectrin molecule beyond repeats 13-15. It should be noted that the ZU5 domain is connected to the membrane binding domain of ankyrin by a 100-residue disordered region. This would allow considerable potential flexibility in the relative orientation of these domains and therefore the location of spectrin relative to the membrane linkage. Spectrin is known to bind protein 4.2 [18; 19; 20] and hence we have located the ankyrin-bound spectrin repeats proximal to protein 4.2. However, it is not known which spectrin repeats interact with protein 4.2, precluding further refinement of the model by consideration of tripartite ankyrin-spectrin-protein 4.2 interactions.
Further ankyrin interactions: band 3 and protein 4.2
The N-terminal membrane binding domain of ankyrin is composed of 24 consecutive 33 amino acid tandem ‘ANK repeats’ and in addition to containing two binding sites for band 3, located in Domain 2 (D2) of this region and across the boundary of Domains 3 and 4 (D3-4) [2; 21], also contains binding sites for a number of other proteins including CD44, Na+K+ ATPase, protein 4.2 [22] and RhAG [4]. Crystal structures are available for ANK D3-4 [23] and the cytoplasmic domain of band 3 (cdb3) [24]. These structures have been combined with spectroscopic data to delineate reliable models for the cdb3-ANK D3-4 complex [21]. These co-ordinates have been included in our model in combination with the theoretically-modelled extension of the N-terminal region of ankyrin which includes all 24 ANK repeats. As discussed by the authors, the model of Kim and co-workers [21] precludes the binding of a single tetrameric band 3 simultaneously to ANK D2 and ANK D3-4 binding sites. Given this, we concur with the view that band 3 dimers are spatially separated and ‘tetramerisation’ is only mediated via ankyrin.
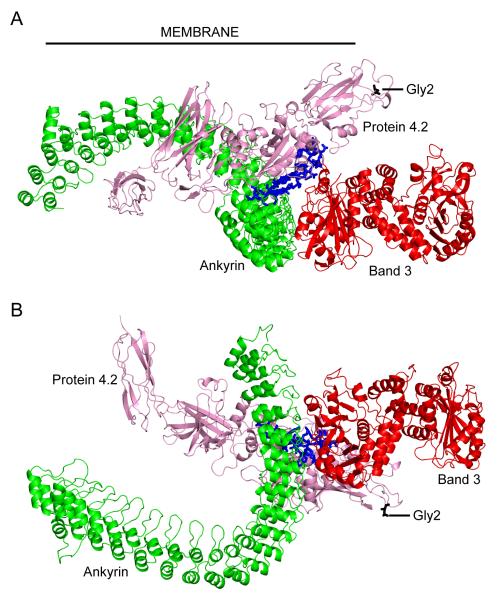
In addition to its interactions with band 3 and spectrin, there is evidence that ankyrin also binds protein 4.2 [3], the absence of protein 4.2 causes hereditary spherocytosis (HS) [6; 25; 26] and a reduction in the association of band 3 with ankyrin [27]. Protein 4.2 binds to cdb3 and ANK D3-4 [28; 29], strengthening their interaction. A stable complex containing band 3, ankyrin and protein 4.2 can also be co-purified from erythrocytes [30] and a recombinant fragment of protein 4.2 has been shown to associate with residues 402-827 of ankyrin [22] confirming the capacity for ankyrin binding to protein 4.2. Studies using recombinant fragments of protein 4.2 have suggested that residues 187-200 are involved in the interaction with ANK D3-4 [22]. Protein 4.2 residues 187-211 have also been shown to contain a binding site for cdb3 [22; 31]. The proximity of the ankyrin and band 3 binding sites on protein 4.2 suggests a close association between the three proteins (Figure 2). The proposed orientation of protein 4.2 relative to the experimentally-determined structure of the ankyrin-cdb3 complex is highly speculative. However, assembly of this model demonstrates the structural plausibility of binding sites suggested by the previous studies. The orientation shown in Figure 2 is also consistent with anchoring of protein 4.2 to the membrane via myristoylation at residue 2 [32].
Figure 2. Ankyrin-band 3-protein 4.2 complex.
(A) view from within the plane of the membrane, approximate location of the inner face of the cell membrane is indicated with a black line; (B) view from the cytoplasm. Proteins are displayed as Cα cartoon, ANK repeat region of ankyrin in green, cytoplasmic domain of band 3 (disordered regions omitted) in red and protein 4.2 in pink. Protein 4.2 residues 187-211 (associated with ankyrin and band 3 binding) are coloured blue, and displayed as sticks; residue 2 (myristolyation site) is coloured black and displayed as sticks. For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.
Further band 3 interactions: GLUT1, GPA, Rh
The N-terminal cytoplasmic domain of band 3 (cdb3, amino acids 1-400) provides the binding site for a host of peripheral membrane proteins and forms the site of its interaction with ankyrin [33; 34; 35; 36; 37], protein 4.2 [38], and the junctional complex proteins, protein 4.1R [39] and adducin [40] (see below). The cytoplasmic domain of band 3 has also been shown to interact with the C-terminal cytoplasmic region of GLUT1 [41], two copies of which are suggested to be present within the ankyrin-associated complex [12]. Hence close proximity of these regions has been maintained in our model. The transmembrane region and C-terminus of band 3 is involved in the interaction with GPA, with residues E658 and S667 of the band 3 membrane domain and GPA residues 59-70 implicated in the interaction [42; 43; 44]. Close proximity of these residues is also included in our model. For one copy of band 3 this involves a GPA homodimer, for the other it involves a GPA/GPB heterodimer. The GPA homodimer is known to dimerise through its transmembrane α-helix [45; 46; 47].GPB is believed to have some form of association with RhAG due to the presence of hyperglycosylated RhAG in GPB null individuals [48] and a reduction in GPB expression in Rh null individuals [49]. Inclusion of the GPA/GPB heterodimer at an interface between band 3 and the RhAG/Rh30 trimer is easily accommodated within our model.
Inclusion of the Rh subcomplex
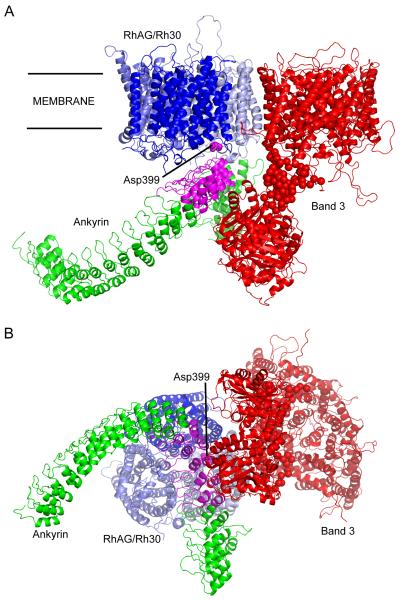
The Rh30 proteins, RhCE and RhD, are two highly similar (92% sequence identity) 30-32kDa unglycosylated proteins. Rh30 associates with Rh-associated glycoprotein (RhAG) [5], upon which its expression in erythrocytes is completely dependent [50; 51; 52; 53]. A crystal structure has been determined for RhCG, a kidney isoform of RhAG, and clearly shows a trimeric architecture [54]. The high degree of sequence similarity between RhCG and the erythroid isoforms strongly suggests that the oligomerisation state would be conserved, as predicted by previous models of the erythroid isoforms [55]. Consideration of the copy number of each protein suggests a predominance of RhAG1Rh302 trimers [12], although modelling studies suggest RhAG2Rh301 trimers would be more stable. In addition to RhAG/Rh30, CD47, ICAM4 and GPB form the Rh complex of proteins that are absent or markedly reduced in Rh null individuals [56]. While no direct interaction between band 3 and Rh proteins has been proven, an association is suggested by co-immunoprecipitation assays and the sub-complexes have been shown to be linked [5]. It has also been demonstrated that there is a RhAG binding site in the ANK D2 region [4]. RhAG residue D399 is believed to be involved in the interaction. The structure of the RhAG/Rh30 trimer may be confidently modelled based on the RhCG crystal structure [54], and no regions of disorder are predicted in the vicinity of D399. This constrains the placement of RhAG/Rh30 with respect to both ankyrin and band 3, and suggests that the proteins are likely to be in close proximity even in the absence of a direct interaction (Figure 3).
Figure 3. Ankyrin-band 3-RhAG/Rh30 complex.
(A) view from within the plane of the membrane; (B) view from the cytoplasm. Proteins are displayed as Cα cartoon, ANK repeat region of ankyrin in green (D2 region in magenta), band 3 in red, RhAG in light blue, RhCE in mid-blue and RhD in dark blue. RhAG residue D399 (associated with ankyrin binding) is coloured magenta, and displayed as spheres. For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.
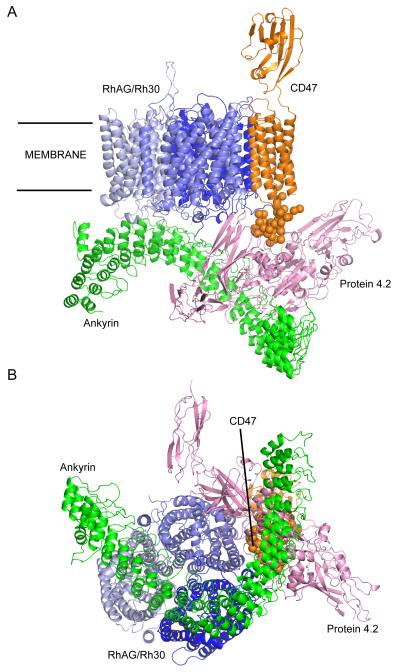
Protein 4.2 has been proposed as a crucial linkage between the band 3- and Rh-based subcomplexes, due to the dependence of CD47 on protein 4.2 for expression [6; 27; 57]. Interestingly in patients with the -D- phenotype, lacking RhCe specifically, CD47 expression is severely reduced [57], no reduction is observed in erythrocytes specifically lacking RhD indicating that RhCe is the component of the core Rh complex that is likely to associate with CD47.. As neither protein has a large cytoplasmic region the membrane domains must be in close proximity. Our model demonstrates that this is structurally plausible, while maintaining an interaction between CD47 and protein 4.2. No details of the CD47-protein 4.2 interaction are available, so our model is limited to placing the two proteins adjacent to one another (Figure 4). Reduction of CD47 expression in patients lacking either RhCe or protein 4.2 raises the possibility of a CD47 binding interface formed by both proteins; this would be consistent with our model. ICAM-4 is also placed next to the RhAG/Rh30 trimer, in an arbitrary orientation, as no details are known beyond the inclusion of ICAM-4 in the Rh subcomplex [58; 59].
Figure 4. Ankyrin-Rh/RhAG-CD47-protein 4.2 complex.
(A) view from within the plane of the membrane; (B) view from the cytoplasm. Proteins are displayed as Cα cartoon, ANK repeat region of ankyrin in green, RhAG in light blue, RhCE in mid-blue RhD in dark blue, CD47 in organe and protein 4.2 in pink. For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.
Discussion
As discussed above, assembly of our speculative model for the overall structure of the ankyrin-associated complex (Figure 1), leads us to favour the view that band 3 ‘tetramers’ are actually two spatially separated dimers, which appear tetrameric due to confinement within the complex by association with other proteins. As previously noted [21], it is physically impossible to bind a single band 3 tetramer to one ankyrin molecule, occupying both the D2 and D3-4 binding sites. However, perhaps surprisingly, this was the only insurmountable steric conflict encountered during assembly of the complex. Admittedly, there are still many degrees of freedom, with many interactions poorly understood – notably those within the Rh subcomplex. It will be fascinating to keep the model updated as new data becomes available – allowing delineation of structures and associations more precisely.
Junctional complex
The junctional complex is centred on the junction of spectrin tetramers which bind to a short actin filament. The adaptor proteins: adducin, dematin, protein 4.1 and p55 connect the cytoskeleton to the integral membrane components: band 3, GLUT1, GPC/D, Duffy, GPA, Kell and Kx. These components and the known interactions between them are summarised in Table 2. The refined model of the entire junctional complex is shown in Figure 5 and its assembly is discussed below.
Table 2. Junctional complex, components and known interactions.
| Protein | Oligomeric state | Number present in junctional complex | Interacting partners included in model |
|---|---|---|---|
| Band 3 | Dimer | 1 | GLUT1 GPA Adducin Protein 4.1R |
| GLUT1 | Monomer | 1 | Band 3 Adducin Dematin |
| GPA | Dimer | 1 | Band 3 |
| GPC/D | Cluster (5 monomers) | 1 | Protein 4.1R p55 |
| Kell | Monomer | 1 | Kx Protein 4.1R |
| Kx | Monomer | 1 | Kell Protein 4.1R |
| DARC | Monomer | 1 | Protein 4.1R |
| Protein 4.1R | Monomer | 5 | Actin Band 3 Kell/Kx DARC GPC/D p55 |
| Protein 4.2 | Monomer | 1 | Band 3 Protein 4.1 |
| p55 | Monomer | 2 | Protein 4.1R GPC/D |
| Adducin (α/β) | Heterodimer (α/β) | 1 | Band 3 GLUT1 Actin |
| Dematin | Trimer | 1 | GLUT1 Actin |
| Actin | Filament (15 monomers) | Tropomyosin Dematin Adducin Protein 4.1R |
|
| Tropomyosin | Homodimer | 2 | Actin |
Figure 5. Overall model of junctional complex.
(A) view from within the plane of the membrane; (B) view from outside of the cell. The components shown in the figure are: one band 3 homodimer (red), five protein 4.1R monomers (dark blue), one GPA homodimer (mint-green), a cluster of five GPC/D monomers (lime-green), one DARC monomer (brown), one α/β-adducin heterodimer (bright/pale yellow, respectively), one dematin trimer (orange), one GLUT1 monomer (purple), one protein 4.2 monomer (pink), one Kell monomer (mid-blue), one Kx monomer (light blue), two p55 monomers (magenta), one actin filament (light grey) and two tropomyosin homodimers (dark grey). All components are shown with secondary structure elements highlighted as cartoons and residues predicted to be disordered displayed as spheres centred on the Cα atoms. For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.
Actin bundling and capping
An actin filament consisting of 15-20 monomers is bundled by tropomyosin and capped at the barbed and pointed ends by adducin and tropomodulin, respectively. Tropomodulin interacts with both tropomyosin and actin and is believed to stabilise the junctional complex [60]. The N-terminal half of tropomodulin interacts with tropomyosin [61] and contains a tropomyosin dependant actin capping site [62]. Indeed the affinity of tropomodulin for actin doubles in the presence of tropomyosin [63]. The N-terminal half of tropomodulin contains a short length of α-helical structure but is mainly unstructured until it is bound to tropomyosin [64]. The C-terminal half is a leucine-rich repeat domain consisting of alternate α-helices and β-strands arranged in a horseshoe shape [65] and contains the principle, tropomyosin independent, pointed end actin capping site [62]. A structural model has been proposed as to how tropomodulin and actin directly interact [65; 66]. Adducin, which will be discussed in more detail below, has been shown to preferentially bind actin at the fast-growing (barbed) end [67; 68] but also bind lower-affinity lateral sites along the actin filament. Barbed end capping by adducin is believed to be crucial for regulation of the length of the actin filament. Capping activity has been shown to be regulated by calcium[67], and require the adducin C-terminal myristoylated alanine-rich protein kinase C substrate-related (MARCKS) domain [68] Tropomyosin is a 40nm long coiled coil that contains seven tandem repeating units designed to bind seven successive actin monomers [69; 70]. The N-terminus of tropomyosin contains the site of interaction with tropomodulin [71]. Recent EM data have enabled the construction of a model of an actin-tropomyosin complex revealing an overall strong interaction which comprises of weak individual local associations [70]. It may be seen from the overall model of the junctional complex (Figure 5) that the actin-tropomyosin filament dominates the complex. The availability of a well-defined structure for the actin-tropomysin complex forms a strong base on which to construct a model of the whole junctional complex.
Protein 4.1R linkages
The best characterised interactions of the junctional complex are those that link the cytoskeletal proteins β-actin and β-spectrin to the membrane proteins, traditionally band 3, GPC and GPD, but now also thought to be DARC, Kell and Kx by protein 4.1R [11; 72]. Protein 4.1R interactions are mediated by calmodulin, PIP2 and phosphorylation [72; 73; 74; 75]. The structure of the 30kDa FERM domain of protein 4.1R (residues 210-488) has been determined by X-ray crystallography [76], revealing a cloverleaf structure which comprises three lobes (the three leaves) joined by a central domain containing two calmodulin binding sites [76]. Each of the lobes is involved with inter-protein interactions: the N-terminal or N-lobe binds to cytoplasmic domain of band 3 (cdb3), the α-lobe binds GPC/D and the C-terminal or C-lobe binds to p55 [76]. Each lobe is structurally distinct: the N-lobe consists of two β sheets connected by an α-helix, the α-lobe consists of four α-helices and the C-lobe contains two β sheets and a C-terminal α-helix [76]. The 10kDa spectrin-actin binding (SAB) domain of protein 4.1R (residues 615-713) includes two β-spectrin binding sites [77] which flank an actin binding site [78]. The SAB domain lies within a large portion of protein 4.1R which is predicted to be natively disordered (residues 489-758).
The protein 4.1R FERM domain binds the cytoplasmic tail of GPC/D via residues 82-98 (GPC) and residues 61-77 (GPD) [10]. It has also been proposed that p55 interacts with both proteins to form a trimeric complex [10]. NMR spectroscopy has been used to delineate the structure of the p55 PDZ domain in complex with a peptide corresponding to residues 112-128 of GPC [79]. The p55-protein 4.1R interaction is mediated by residues 221-265, which are immediately C-terminal of the p55 SH3 domain [80]. Based on the available data we have constructed a model of the heterotrimeric assembly of the 4.1R FERM domain, GPC/D cytoplasmic domain and p55 (Figure 6). The details of the complex are highly speculative but the model does demonstrate that the proposed trimeric assembly is possible. In order to incorporate these assemblies into our overall model of the junctional complex (Figure 5) they are connected to the membrane by joining of the GPC cytoplasmic and transmembrane regions and to the cytoskeleton by the large disordered C-terminal region of protein 4.1R. We have placed the SAB region of the protein 4.1R molecules in close proximity to the actin filament, in an arbitrary arrangement as no further information regarding the details of this interaction are available. Two more protein 4.1R molecules are assumed to interact with the two copies of band 3 found within the junctional complex (as one band 3 dimer). Therefore we have placed the FERM domains of these protein 4.1R molecules in close proximity to the two band 3 cytoplasmic domains. A further protein 4.1R molecule has been included connecting actin to the Kell-Kx complex and Duffy, as discussed below.
Figure 6. Protein 4.1R-GPC-p55 complex.
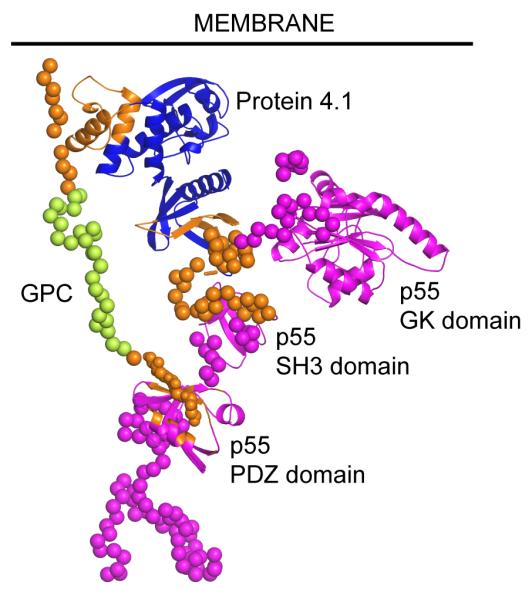
Trimeric complex of protein 4.1R FERM domain (blue) with GPC cytoplasmic domain (lime-green) and p55 (magenta). Approximate location of the inner face of the cell membrane is indicated with a black line. All components are shown with secondary structure elements highlighted as cartoons and residues predicted to be disordered displayed as spheres centred on the Cα atoms. Residues shown to be involved in inter-protein interactions are coloured orange. For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.
Adducin interactions
Another link between actin and band 3 is formed by adducin. The major form of adducin in erythrocytes is comprised of an α and β subunit in either heterodimers or heterotetramers [81]. Adducin interacts with actin, spectrin and band 3 forming a link between the three proteins, it stabilises the actin-β-spectrin interaction and is critical for erythrocyte membrane stability [40]. Actin increases the affinity of adducin for spectrin [82] promoting the recruitment of spectrin to actin at the site of the junctional complex [81; 83]. Like protein 4.1, adducin also binds calmodulin [83]. Adducin consists of three domains: an N-terminal globular head domain, a neck domain and a C-terminal natively disordered tail domain [84]. The head domain of adducin has been proposed to bind to the fast growing (barbed) end of the actin filament [67]. Accordingly, our model places the dimeric head region of α/β adducin at the barbed end of the actin filament, although none of the molecular details of the interaction are known. The interaction of adducin with spectrin, actin, calmodulin and band 3 all involve the tail domain of adducin [84]. The affinity of adducin for the cdb3 varies between α and β adducin and a band 3 dimer only contains one adducin binding site [40]. As α adducin was found to have a higher affinity for band 3 we have placed the α adducin tail in close proximity with cdb3, in an arbitrary conformation. The C-terminal MARCKS domain of β-adducin has been shown to be required for actin-capping activity, suggesting an association with the barbed end of actin. While this has not been included in our model, it could easily be accommodated due to the potential flexibility of the large, disordered β-adducin tail domain.
Inclusion of GLUT1 via band 3, adducin and dematin
As in the ankyrin-associated complex, a horizontal interaction has been established between the C-terminal cytoplasmic loop of GLUT1 and cdb3 [41], although no further details have been defined. This interaction is accounted for in the model by ensuring that these regions are adjacent. GLUT1 is also involved in vertical links between the membrane and cytoskeleton by binding adducin and dematin [85]. Dematin is a trimeric villin-type headpiece protein which consists of a small, conserved actin-binding headpiece domain at the end of a larger, disordered, more varied “core” domain, which mediates trimerisation and is proposed to act as a scaffold around which other components of the junctional complex are organised [86; 87]. One dematin trimer is believed to be present per junctional complex. This trimeric assembly is represented in our model by ensuring that the three core domains are in close proximity, but the specific arrangement is arbitrary, as no further details are available. Dematin headpiece knockout mice have spheroidal osmotically fragile erythrocytes indicating a role for dematin in the stability of erythrocyte membranes [88]. The dematin headpiece is expressed as two spliceforms, one with and one without a 22 amino-acid insert, resulting in trimers containing two 48kDa and one 52kDa subunits [89]. Dematin is the only one of the villin-type headpiece proteins that is phosphorylated with the phosphorylation believed to cause a conformational change within the dematin headpiece [90] that brings the core domain closer to the headpiece domain [41] and results in reduced affinity for actin [87]. Structural analysis of villin-type headpiece proteins has revealed a ‘variable length’ or V-loop that is present only in those that bind actin [91]. This loop does not contain amino-acids that comprise the actin binding site; however, it is the site of phosphorylation on dematin [41; 90]. The actin binding residues within the headpiece domain of dematin consist of a hydrophobic cap (W64) surrounded by the alternative charges of K65, K70, and the C-terminal carboxylate along with an adjacent positive patch formed by R35 [91]. The structure of the complex formed between actin and the dematin headpiece has been delineated by electron microscopy [92], and this structure has been incorporated into our model. There is no data identifying which actin monomer(s) mediates the interaction with dematin, or how many of the dematin subunits bind actin. Given that the large disordered core domain of dematin would permit large conformational flexibility we assume that all three dematin headpieces are in complex with actin, and this is reflected in the model. The dematin core domain is the point of interaction with the large cytoplasmic loop of GLUT1 [85], but beyond this no details of the interaction are known. One of the dematin core domains is situated in close proximity to the large cytoplasmic loop of GLUT1, to reflect that linkage.
Protein 4.2, Kell-Kx and DARC
A molecule of protein 4.2 has also been included, accounting for known interactions with both cdb3 and protein 4.1 [3; 29]. Inclusion of protein 4.2 at the junctional complex is also consistent with its known association with the carboxy-terminal EF hands of α spectrin [93]. The cytoplasmic regions of the Kell-Kx complex and DARC are known to be associated with the junctional complex via interactions with lobe B of the protein 4.1R FERM domain [94]. Therefore Kell-Kx and DARC have been included in the model and the molecule of protein 4.1R not associated with GPC/p55 or band3 has been oriented such that these interactions could be maintained.
Discussion
Inspection of our speculative assembly of the junctional complex (Figure 5) reveals some striking features. One is the visual display of the natively disordered regions, represented in our model by Cα chains. Not only do these regions make up a significant fraction of the mass, they also mediate many of the inter-protein interactions upon which the complex is based. The large degree of flexibility inherent within disordered regions made the assembly of the model technically challenging, due to handling such large areas of uncertainty, but conceptually easy as no insurmountable structural conflicts were encountered due to the conformational flexibility. A traditional view of protein function – adhering to the concept of folded protein domains being the active species – would largely ignore the disordered regions. However, they clearly demand to be, and increasingly are, the focus of research [95]. In the context of the erythrocyte, it may be that large disordered regions mediating protein-protein interactions represent an important attribute which contributes to the extraordinary deformability of the membrane. Another feature of this ‘membrane protein macrocomplex’ is the dearth of membrane proteins relative to the mass of intracellular proteins. The junctional complex appears to be more a cytoskeletal complex which is anchored to the membrane, rather than a membrane complex which is anchored to the cytoskeleton.
Conclusion
With their easy availability and relative simplicity erythrocytes have traditionally been the cell of choice for studying the plasma membrane. The development of a method for removing the cytoplasmic content and acquiring purified erythrocyte plasma membrane [96] lead to the purification and preliminary characterization of spectrin [97]. From these roots the erythrocyte membrane has become the most characterized of any cell type, however, at the atomic level there is still a great deal to learn.
The assignment of proteins into protein complexes in the erythrocyte membrane has expanded our knowledge of the composition, structure and functions of the erythrocyte and is providing exciting new avenues of research. However, identification of these new interactions must be placed in the context of existing data. As additional proteins and interactions are identified within both the ankyrin dependent and junctional complexes, thought must be given to where they would integrate given the steric limitations between the protein constituents already known and within the plasma membrane itself. Complications arise by the likelihood that multiple similar complexes may exist with similar core components that may be decorated to different extents with low copy number proteins. In the case of band 3, the oligomeric configuration has been thought to differ between populations of the same protein – with tetramers incorporated into the ankyrin-associated complex and dimers into the junctional complex. However, evidence increasingly suggests that the tetramers exist as a spatially separated loosely interacting dimer of dimers [2; 21; 98]. An arrangement of band 3 dimers with proteins of the Rh complex enclosed between two dimers forming a tetramer mediated by ankyrin binding has been suggested [2; 4] and is supported by the absence of tetrameric band 3 in full length ankyrin deficient nb/nb mice [98]. Importantly, this is the arrangement of band 3 subunits that arose, without being imposed, while constructing our model and it is consistent with maintaining the known interactions of band 3 (Figure 1).
Many gaps remain in the structural information available for components of the ankyrin dependent and junctional complexes. For instance, while high-resolution, experimental structural data is available for the protein 4.1R FERM domain [76], the N-terminal domain is characterised only by a very speculative homology model [12]. In addition many proteins have significant regions predicted to be natively disordered. Indeed, excluding the actin-tropomyosin filament, over two thirds of the residues which make up the junctional complex are predicted to be disordered. Even regions whose structures are considered to be known may contain appreciable regions of disorder. For instance, the cytoplasmic region of band 3 consists of approximately 400 residues, of which only 293 are resolved within the crystal structure [24]. The study of these regions is challenging by traditional structural biology methods, e.g. X-ray crystallography, hence novel approaches must be adopted. These regions may also undergo disordered-ordered transitions upon complex formation, and therefore their structures may alter more significantly than would be expected for a globular protein.
Our current knowledge of the protein-protein interactions within the ankyrin dependent and junctional complexes is wide but not exhaustive and there are numerous interactions for which data is scarce. For example, in the ankyrin dependent complex little is known about the associations between protein 4.2, CD47 and RhCe/D. What are the different interactions that bring RhD, RhCe RhAG, ICAM4, CD47 and GPB together in the Rh complex and indeed are all involved within this complex? Within the junctional complex the interactions between GLUT1 and band 3 and adducin and GLUT1 are uncharacterised and it is unknown exactly how and where DARC, Kell and Kx fit into the complex. Studying multi-protein assemblies is inherently more challenging than individual proteins, but is essential if we are to fully understand the nature of these complexes. This may be especially true for the natively disordered regions.
Due to the presence of regions of undefined structure and poorly understood protein-protein interactions our current models are necessarily speculative. However, we have come a long way from the early diagrams of the erythrocyte membrane [99]. Looking to the future, data from structural biology and other biophysical techniques, in combination with molecular modelling, has an important role to play in validating our current view of these complexes and incorporating newly discovered components and interactions. In this article we have further refined the atomic visualisations of Burton and Bruce [11] to compose as detailed a representation of the two most abundant membrane protein complexes found within the erythrocyte as current data allows.
Acknowledgments
The work was supported by the Department of Health, England (NMB, TJM) and a Wellcome Trust Project Grant (TJS). We would like to thank Dr Lesley Bruce for providing inspiration for the project and critical reading of the manuscript and Dr Ashley Toye for critical reading of the manuscript. We also thank the following people for kindly providing atomic co-ordinates: Professor William Lehmann and Dr Xiaochuan (Edward) Li for the actin-tropomyosin complex, Professor Nikolaus Grigorieff and Dr James Chen for the actin-dematin complex and Professor Albert Beth and Dr Sunghoon Kim for the band 3-ankyrin complex.
References
- [1].van den Akker E, Satchwell TJ, Williamson RC, Toye AM. Band 3 multiprotein complexes in the red cell membrane; of mice and men. Blood Cells Mol Dis. 2010;45:1–8. doi: 10.1016/j.bcmd.2010.02.019. [DOI] [PubMed] [Google Scholar]
- [2].Michaely P, Bennett V. The ANK repeats of erythrocyte ankyrin form two distinct but cooperative binding sites for the erythrocyte anion exchanger. J Biol Chem. 1995;270:22050–7. doi: 10.1074/jbc.270.37.22050. [DOI] [PubMed] [Google Scholar]
- [3].Rybicki AC, Musto S, Schwartz RS. Decreased content of protein 4.2 in ankyrin-deficient normoblastosis (nb/nb) mouse red blood cells: evidence for ankyrin enhancement of protein 4.2 membrane binding. Blood. 1995;86:3583–9. [PubMed] [Google Scholar]
- [4].Nicolas V, Le Van Kim C, Gane P, et al. Rh-RhAG/ankyrin-R, a new interaction site between the membrane bilayer and the red cell skeleton, is impaired by Rh(null)-associated mutation. J Biol Chem. 2003;278:25526–33. doi: 10.1074/jbc.M302816200. [DOI] [PubMed] [Google Scholar]
- [5].Bruce LJ, Beckmann R, Ribeiro ML, et al. A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood. 2003;101:4180–8. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
- [6].Bruce LJ, Ghosh S, King MJ, et al. Absence of CD47 in protein 4.2-deficient hereditary spherocytosis in man: an interaction between the Rh complex and the band 3 complex. Blood. 2002;100:1878–1885. doi: 10.1182/blood-2002-03-0706. [DOI] [PubMed] [Google Scholar]
- [7].La-Borde PJ, Stabach PR, Simonovic I, Morrow JS, Simonovic M. Ankyrin recognizes both surface character and shape of the 14-15 di-repeat of beta-spectrin. Biochem Biophys Res Commun. 2010;392:490–4. doi: 10.1016/j.bbrc.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Becker PS, Schwartz MA, Morrow JS, Lux SE. Radiolabel-transfer cross-linking demonstrates that protein 4.1 binds to the N-terminal region of beta spectrin and to actin in binary interactions. Eur J Biochem. 1990;193:827–36. doi: 10.1111/j.1432-1033.1990.tb19406.x. [DOI] [PubMed] [Google Scholar]
- [9].Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin Caps the Pointed Ends of Actin-Filaments. Journal of Cell Biology. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hemming NJ, Anstee DJ, Staricoff MA, Tanner MJ, Mohandas N. Identification of the membrane attachment sites for protein 4.1 in the human erythrocyte. J Biol Chem. 1995;270:5360–6. doi: 10.1074/jbc.270.10.5360. [DOI] [PubMed] [Google Scholar]
- [11].An X, Mohandas N. Disorders of red cell membrane. Br J Haematol. 2008;141:367–75. doi: 10.1111/j.1365-2141.2008.07091.x. [DOI] [PubMed] [Google Scholar]
- [12].Burton NM, Bruce LJ. Modelling the structure of the red cell membrane. Biochem Cell Biol. 2011;89:200–15. doi: 10.1139/o10-154. [DOI] [PubMed] [Google Scholar]
- [13].Nans A, Mohandas N, Stokes DL. Native ultrastructure of the red cell cytoskeleton by cryo-electron tomography. Biophys J. 2011;101:2341–50. doi: 10.1016/j.bpj.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Uversky VN, Dunker AK. Understanding protein non-folding. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. Journal of the American Chemical Society. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- [16].Kohn JE, Millett IS, Jacob J, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ipsaro JJ, Mondragon A. Structural basis for spectrin recognition by ankyrin. Blood. 2010;115:4093–4101. doi: 10.1182/blood-2009-11-255604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Golan DE, Corbett JD, Korsgren C, et al. Control of band 3 lateral and rotational mobility by band 4.2 in intact erythrocytes: release of band 3 oligomers from low-affinity binding sites. Biophys J. 1996;70:1534–42. doi: 10.1016/S0006-3495(96)79717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mandal D, Moitra PK, Basu J. Mapping of a spectrin-binding domain of human erythrocyte membrane protein 4.2. Biochem J. 2002;364:841–7. doi: 10.1042/BJ20020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Korsgren C, Peters LL, Lux SE. Protein 4.2 binds to the carboxyl-terminal EF-hands of erythroid alpha-spectrin in a calcium- and calmodulin-dependent manner. J Biol Chem. 2010;285:4757–70. doi: 10.1074/jbc.M109.056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim S, Brandon S, Zhou Z, et al. Determination of structural models of the complex between the cytoplasmic domain of erythrocyte band 3 and ankyrin-R repeats 13-24. J Biol Chem. 2011;286:20746–57. doi: 10.1074/jbc.M111.230326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Su Y, Ding Y, Jiang M, et al. Associations of protein 4.2 with band 3 and ankyrin. Mol Cell Biochem. 2006;289:159–66. doi: 10.1007/s11010-006-9159-x. [DOI] [PubMed] [Google Scholar]
- [23].Michaely P, Tomchick DR, Machius M, Anderson RG. Crystal structure of a 12ANK repeat stack from human ankyrinR. EMBO J. 2002;21:6387–96. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang DC, Kiyatkin A, Bolin JT, Low PS. Crystallographic structure and functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. Blood. 2000;96:2925–2933. [PubMed] [Google Scholar]
- [25].Yawata Y. Band 4.2 abnormalities in human red cells. Am J Med Sci. 1994;307:190–203. doi: 10.1097/00000441-199403000-00006. [DOI] [PubMed] [Google Scholar]
- [26].Dahl KN, Parthasarathy R, Westhoff CM, Layton DM, Discher DE. Protein 4.2 is critical to CD47-membrane skeleton attachment in human red cells. Blood. 2004;103:1131–6. doi: 10.1182/blood-2003-04-1331. [DOI] [PubMed] [Google Scholar]
- [27].van den Akker E, Satchwell TJ, Pellegrin S, et al. Investigating the key membrane protein changes during in vitro erythropoiesis of protein 4.2 (-) cells (mutations Chartres 1 and 2) Haematologica. 2010;95:1278–86. doi: 10.3324/haematol.2009.021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Korsgren C, Cohen CM. Purification and properties of human erythrocyte band 4.2. Association with the cytoplasmic domain of band 3. J Biol Chem. 1986;261:5536–43. [PubMed] [Google Scholar]
- [29].Korsgren C, Cohen CM. Associations of human erythrocyte band 4.2. Binding to ankyrin and to the cytoplasmic domain of band 3. J Biol Chem. 1988;263:10212–8. [PubMed] [Google Scholar]
- [30].Kumpornsin K, Jiemsup S, Yongkiettrakul S, Chookajorn T. Characterization of band 3-ankyrin-Protein 4.2 complex by biochemical and mass spectrometry approaches. Biochemical and Biophysical Research Communications. 2011;406:332–335. doi: 10.1016/j.bbrc.2011.02.026. [DOI] [PubMed] [Google Scholar]
- [31].Bhattacharyya R, Das AK, Moitra PK, et al. Mapping of a palmitoylatable band 3-binding domain of human erythrocyte membrane protein 4.2. Biochemical Journal. 1999;340:505–512. [PMC free article] [PubMed] [Google Scholar]
- [32].Risinger MA, Dotimas EM, Cohen CM. Human Erythrocyte Protein 4.2, a High Copy Number Membrane-Protein, Is N-Myristylated. Journal of Biological Chemistry. 1992;267:5680–5685. [PubMed] [Google Scholar]
- [33].Bennett V. Isolation of an ankyrin-band 3 oligomer from human erythrocyte membranes. Biochimica Et Biophysica Acta. 1982;689:475–84. doi: 10.1016/0005-2736(82)90305-4. [DOI] [PubMed] [Google Scholar]
- [34].Davis L, Lux SE, Bennett V. Mapping the ankyrin-binding site of the human erythrocyte anion exchanger. J Biol Chem. 1989;264:9665–72. [PubMed] [Google Scholar]
- [35].Willardson BM, Thevenin BJ, Harrison ML, et al. Localization of the ankyrin-binding site on erythrocyte membrane protein, band 3. J Biol Chem. 1989;264:15893–9. [PubMed] [Google Scholar]
- [36].Ding Y, Kobayashi S, Kopito R. Mapping of ankyrin binding determinants on the erythroid anion exchanger, AE1. J Biol Chem. 1996;271:22494–8. doi: 10.1074/jbc.271.37.22494. [DOI] [PubMed] [Google Scholar]
- [37].Bennett V, Stenbuck PJ. Association between ankyrin and the cytoplasmic domain of band 3 isolated from the human erythrocyte membrane. J Biol Chem. 1980;255:6424–32. [PubMed] [Google Scholar]
- [38].Rybicki AC, Musto S, Schwartz RS. Identification of a band-3 binding site near the N-terminus of erythrocyte membrane protein 4.2. Biochem J. 1995;309(Pt 2):677–81. doi: 10.1042/bj3090677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lombardo CR, Willardson BM, Low PS. Localization of the protein 4.1-binding site on the cytoplasmic domain of erythrocyte membrane band 3. J Biol Chem. 1992;267:9540–6. [PubMed] [Google Scholar]
- [40].Anong WA, Franco T, Chu H, et al. Adducin forms a bridge between the erythrocyte membrane and its cytoskeleton and regulates membrane cohesion. Blood. 2009;114:1904–12. doi: 10.1182/blood-2009-02-203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jiang WH, Ding Y, Su Y, et al. Interaction of glucose transporter 1 with anion exchanger 1 in vitro. Biochemical and Biophysical Research Communications. 2006;339:1255–1261. doi: 10.1016/j.bbrc.2005.11.138. [DOI] [PubMed] [Google Scholar]
- [42].Bruce LJ, Ring SM, Anstee DJ, et al. Changes in the Blood-Group Wright Antigens Are Associated with a Mutation at Amino-Acid-658 in Human Erythrocyte Band-3 - a Site of Interaction between Band-3 and Glycophorin-a under Certain Conditions. Blood. 1995;85:541–547. [PubMed] [Google Scholar]
- [43].Toye AM, Williamson RC, Khanfar M, et al. Band 3 Courcouronnes (Ser667Phe): a trafficking mutant differentially rescued by wild-type band 3 and glycophorin A. Blood. 2008;111:5380–5389. doi: 10.1182/blood-2007-07-099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fu G, Wang T, Yang B, et al. Purification and characterization of the human erythrocyte band 3 protein C-terminal domain. Biochemistry. 2004;43:1633–8. doi: 10.1021/bi035281c. [DOI] [PubMed] [Google Scholar]
- [45].Furthmayr H, Marchesi VT. Subunit Structure of Human Erythrocyte Glycophorin-A. Biochemistry. 1976;15:1137–1144. doi: 10.1021/bi00650a028. [DOI] [PubMed] [Google Scholar]
- [46].Bormann BJ, Knowles WJ, Marchesi VT. Synthetic Peptides Mimic the Assembly of Transmembrane Glycoproteins. Journal of Biological Chemistry. 1989;264:4033–4037. [PubMed] [Google Scholar]
- [47].Lemmon MA, Flanagan JM, Hunt JF, et al. Glycophorin-a Dimerization Is Driven by Specific Interactions between Transmembrane Alpha-Helices. Journal of Biological Chemistry. 1992;267:7683–7689. [PubMed] [Google Scholar]
- [48].Ridgwell K, Eyers SA, Mawby WJ, Anstee DJ, Tanner MJ. Studies on the glycoprotein associated with Rh (rhesus) blood group antigen expression in the human red blood cell membrane. J Biol Chem. 1994;269:6410–6. [PubMed] [Google Scholar]
- [49].Colin Y. Rh proteins: a family of structural membrane proteins with putative transport activity. Vox Sang. 2002;83(Suppl 1):179–83. doi: 10.1111/j.1423-0410.2002.tb05296.x. [DOI] [PubMed] [Google Scholar]
- [50].Cherif-Zahar B, Matassi G, Raynal V, et al. Rh-deficiency of the regulator type caused by splicing mutations in the human RH50 gene. Blood. 1998;92:2535–40. [PubMed] [Google Scholar]
- [51].Cherif-Zahar B, Raynal V, Gane P, et al. Candidate gene acting as a suppressor of the RH locus in most cases of Rh-deficiency. Nat Genet. 1996;12:168–73. doi: 10.1038/ng0296-168. [DOI] [PubMed] [Google Scholar]
- [52].Hyland CA, Cherif-Zahar B, Cowley N, et al. A novel single missense mutation identified along the RH50 gene in a composite heterozygous Rhnull blood donor of the regulator type. Blood. 1998;91:1458–63. [PubMed] [Google Scholar]
- [53].Huang CH. The human Rh50 glycoprotein gene. Structural organization and associated splicing defect resulting in Rh(null) disease. J Biol Chem. 1998;273:2207–13. doi: 10.1074/jbc.273.4.2207. [DOI] [PubMed] [Google Scholar]
- [54].Gruswitz F, Chaudhary S, Ho JD, et al. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci U S A. 2010;107:9638–43. doi: 10.1073/pnas.1003587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Burton NM, Anstee DJ. Structure, function and significance of Rh proteins in red cells. Current Opinion in Hematology. 2008;15:625–630. doi: 10.1097/MOH.0b013e328311f422. [DOI] [PubMed] [Google Scholar]
- [56].Cartron JP. RH blood group system and molecular basis of Rh-deficiency. Baillieres Best Pract Res Clin Haematol. 1999;12:655–89. doi: 10.1053/beha.1999.0047. [DOI] [PubMed] [Google Scholar]
- [57].Mouro-Chanteloup I, Delaunay J, Gane P, et al. Evidence that the red cell skeleton protein 4.2 interacts with the Rh membrane complex member CD47. Blood. 2003;101:338–44. doi: 10.1182/blood-2002-04-1285. [DOI] [PubMed] [Google Scholar]
- [58].Sistonen P. A Phenotypic Association between the Blood-Group Antigen Ne-a and the Rh Antigen-D. Medical Biology. 1981;59:230–233. [PubMed] [Google Scholar]
- [59].Booth PB, Serjeantson S, Woodfield DG, Amato D. Selective Depression of Blood-Group Antigens Associated with Hereditary Ovalocytosis among Melanesians. Vox Sanguinis. 1977;32:99–110. doi: 10.1111/j.1423-0410.1977.tb00612.x. [DOI] [PubMed] [Google Scholar]
- [60].An XL, Salomao M, Guo XH, Gratzer W, Mohandas N. Tropomyosin modulates erythrocyte membrane stability. Blood. 2007;109:1284–1288. doi: 10.1182/blood-2006-07-036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Babcock GG, Fowler VM. Isoform-specific interaction of tropomodulin with skeletal muscle and erythrocyte tropomyosins. J Biol Chem. 1994;269:27510–8. [PubMed] [Google Scholar]
- [62].Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. Journal of Biological Chemistry. 2003;278:40000–40009. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- [63].Weber A, Pennise CR, Fowler VM. Tropomodulin increases the critical concentration of barbed end-capped actin filaments by converting ADP.P(i)-actin to ADP-actin at all pointed filament ends. J Biol Chem. 1999;274:34637–45. doi: 10.1074/jbc.274.49.34637. [DOI] [PubMed] [Google Scholar]
- [64].Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophys J. 2005;88:372–83. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Krieger I, Kostyukova A, Yamashita A, Nitanai Y, Maeda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophys J. 2002;83:2716–25. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tsukada T, Kotlyanskaya L, Huynh R, et al. Identification of residues within tropomodulin-1 responsible for its localization at the pointed ends of the actin filaments in cardiac myocytes. J Biol Chem. 2011;286:2194–204. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kuhlman PA, Hughes CA, Bennett V, Fowler VM. A new function for adducin. Calcium/calmodulin-regulated capping of the barbed ends of actin filaments. J Biol Chem. 1996;271:7986–91. doi: 10.1074/jbc.271.14.7986. [DOI] [PubMed] [Google Scholar]
- [68].Li X, Matsuoka Y, Bennett V. Adducin preferentially recruits spectrin to fast-growing ends of actin filaments in a complex requiring the MARCKS related domain and an oligomerization domain. Molecular Biology of the Cell. 1997;8:1591–1591. doi: 10.1074/jbc.273.30.19329. [DOI] [PubMed] [Google Scholar]
- [69].Hitchcock-DeGregori SE. Tropomyosin: function follows structure. Adv Exp Med Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]
- [70].Li XE, Tobacman LS, Mun JY, et al. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100:1005–13. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Vera C, Sood A, Gao KM, et al. Tropomodulin-binding site mapped to residues 7-14 at the N-terminal heptad repeats of tropomyosin isoform 5. Arch Biochem Biophys. 2000;378:16–24. doi: 10.1006/abbi.2000.1802. [DOI] [PubMed] [Google Scholar]
- [72].Gauthier E, Guo X, Mohandas N, An X. Phosphorylation-dependent perturbations of the 4.1R-associated multiprotein complex of the erythrocyte membrane. Biochemistry. 2011;50:4561–7. doi: 10.1021/bi200154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].An X, Zhang X, Debnath G, Baines AJ, Mohandas N. Phosphatidylinositol-4,5-biphosphate (PIP2) differentially regulates the interaction of human erythrocyte protein 4.1 (4.1R) with membrane proteins. Biochemistry. 2006;45:5725–32. doi: 10.1021/bi060015v. [DOI] [PubMed] [Google Scholar]
- [74].Nunomura W, Takakuwa Y. Regulation of protein 4.1R interactions with membrane proteins by Ca2+ and calmodulin. Front Biosci. 2006;11:1522–39. doi: 10.2741/1901. [DOI] [PubMed] [Google Scholar]
- [75].Nunomura W, Takakuwa Y, Parra M, Conboy J, Mohandas N. Regulation of protein 4.1R, p55, and glycophorin C ternary complex in human erythrocyte membrane. J Biol Chem. 2000;275:24540–6. doi: 10.1074/jbc.M002492200. [DOI] [PubMed] [Google Scholar]
- [76].Han BG, Nunomura W, Takakuwa Y, Mohandas N, Jap BK. Protein 4.1R core domain structure and insights into regulation of cytoskeletal organization. Nat Struct Biol. 2000;7:871–5. doi: 10.1038/82819. [DOI] [PubMed] [Google Scholar]
- [77].Schischmanoff PO, Winardi R, Discher DE, et al. Defining of the minimal domain of protein 4.1 involved in spectrin-actin binding. J Biol Chem. 1995;270:21243–50. doi: 10.1074/jbc.270.36.21243. [DOI] [PubMed] [Google Scholar]
- [78].Gimm JA, An X, Nunomura W, Mohandas N. Functional characterization of spectrin-actin-binding domains in 4.1 family of proteins. Biochemistry. 2002;41:7275–82. doi: 10.1021/bi0256330. [DOI] [PubMed] [Google Scholar]
- [79].Kusunoki H, Kohno T. Structural insight into the interaction between the p55 PDZ domain and glycophorin C. Biochemical and Biophysical Research Communications. 2007;359:972–978. doi: 10.1016/j.bbrc.2007.05.215. [DOI] [PubMed] [Google Scholar]
- [80].Marfatia SM, Lue RA, Branton D, Chishti AH. Identification of the Protein 4.1 Binding Interface on Glycophorin-C and Glycophorin-P55, a Homolog of the Drosophila Disks-Large Tumor-Suppressor Protein. Journal of Biological Chemistry. 1995;270:715–719. doi: 10.1074/jbc.270.2.715. [DOI] [PubMed] [Google Scholar]
- [81].Hughes CA, Bennett V. Adducin: a physical model with implications for function in assembly of spectrin-actin complexes. J Biol Chem. 1995;270:18990–6. doi: 10.1074/jbc.270.32.18990. [DOI] [PubMed] [Google Scholar]
- [82].Gardner K, Bennett V. Modulation of spectrin-actin assembly by erythrocyte adducin. Nature. 1987;328:359–62. doi: 10.1038/328359a0. [DOI] [PubMed] [Google Scholar]
- [83].Mische SM, Mooseker MS, Morrow JS. Erythrocyte adducin: a calmodulin-regulated actin-bundling protein that stimulates spectrin-actin binding. J Cell Biol. 1987;105:2837–45. doi: 10.1083/jcb.105.6.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Franco T, Low PS. Erythrocyte adducin: a structural regulator of the red blood cell membrane. Transfus Clin Biol. 2010;17:87–94. doi: 10.1016/j.tracli.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Khan AA, Hanada T, Mohseni M, et al. Dematin and adducin provide a novel link between the spectrin cytoskeleton and human erythrocyte membrane by directly interacting with glucose transporter-1. J Biol Chem. 2008;283:14600–9. doi: 10.1074/jbc.M707818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chen L, Jiang ZG, Khan AA, Chishti AH, McKnight CJ. Dematin exhibits a natively unfolded core domain and an independently folded headpiece domain. Protein Sci. 2009;18:629–36. doi: 10.1002/pro.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Vardar D, Chishti AH, Frank BS, et al. Villin-type headpiece domains show a wide range of F-actin-binding affinities. Cell Motil Cytoskeleton. 2002;52:9–21. doi: 10.1002/cm.10027. [DOI] [PubMed] [Google Scholar]
- [88].Khanna R, Chang SH, Andrabi S, et al. Headpiece domain of dematin is required for the stability of the erythrocyte membrane. Proc Natl Acad Sci U S A. 2002;99:6637–42. doi: 10.1073/pnas.052155999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Azim AC, Knoll JH, Beggs AH, Chishti AH. Isoform cloning, actin binding, and chromosomal localization of human erythroid dematin, a member of the villin superfamily. J Biol Chem. 1995;270:17407–13. doi: 10.1074/jbc.270.29.17407. [DOI] [PubMed] [Google Scholar]
- [90].Frank BS, Vardar D, Chishti AH, McKnight CJ. The NMR structure of dematin headpiece reveals a dynamic loop that is conformationally altered upon phosphorylation at a distal site. J Biol Chem. 2004;279:7909–16. doi: 10.1074/jbc.M310524200. [DOI] [PubMed] [Google Scholar]
- [91].Brown JW, Vardar-Ulu D, McKnight CJ. How to Arm a Supervillin: Designing F-Actin Binding Activity into Supervillin Headpiece. Journal of Molecular Biology. 2009;393:608–618. doi: 10.1016/j.jmb.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Chen JZ, Furst J, Chapman MS, Grigorieff N. Low-resolution structure refinement in electron microscopy. Journal of Structural Biology. 2003;144:144–151. doi: 10.1016/j.jsb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- [93].Korsgren C, Peters LL, Lux SE. Protein 4.2 Binds to the Carboxyl-terminal EF-hands of Erythroid alpha-Spectrin in a Calcium- and Calmodulin-dependent Manner. Journal of Biological Chemistry. 2010;285:4757–4770. doi: 10.1074/jbc.M109.056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Salomao M, Zhang X, Yang Y, et al. Protein 4.1R-dependent multiprotein complex: new insights into the structural organization of the red blood cell membrane. Proc Natl Acad Sci U S A. 2008;105:8026–31. doi: 10.1073/pnas.0803225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Dunker AK, Gough J. Sequences and topology: intrinsic disorder in the evolving universe of protein structure. Current Opinion in Structural Biology. 2011;21:379–381. doi: 10.1016/j.sbi.2011.04.002. [DOI] [PubMed] [Google Scholar]
- [96].Dodge JT, Hanahan DJ, Mitchell C. Preparation and Chemical Characteristics of Hemoglobin-Free Ghosts of Human Erythrocytes. Archives of Biochemistry and Biophysics. 1963;100:119–&. doi: 10.1016/0003-9861(63)90042-0. [DOI] [PubMed] [Google Scholar]
- [97].Marchesi VT, Steers E. Isolation and Characterization of a Structural Protein from Red Cell Ghost Membranes. Journal of Cell Biology. 1967;35:A87–&. [Google Scholar]
- [98].Yi SJ, Liu SC, Derick LH, et al. Red cell membranes of ankyrin-deficient nb/nb mice lack band 3 tetramers but contain normal membrane skeletons. Biochemistry. 1997;36:9596–604. doi: 10.1021/bi9704966. [DOI] [PubMed] [Google Scholar]
- [99].Byers TJ, Branton D. Visualization of the Protein Associations in the Erythrocyte-Membrane Skeleton. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:6153–6157. doi: 10.1073/pnas.82.18.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]