Abstract
Objective
HCV infection has a chronicity rate of about 70%, several studies have shown that interleukin-18 (IL-18) was associated with etiology and progression of hepatitis C virus (HCV) infections. However, the association between single-nucleotide polymorphisms − 607C/A (rs1946518) and − 137G/C (rs187238) located in the IL-18 gene promoter and chronic hepatitis C virus infections was still controversial and ambiguous. To derive a more precise effect on the association between these polymorphisms and chronic hepatitis C virus infections, we performed this first meta-analysis based on the currently available evidence of the literature.
Methods
A total of 4 studies with 1222 cases and 1115 controls for − 607C/A polymorphism and 3 studies with 959 cases and 987 controls for − 137G/C polymorphism were identified to perform a meta-analysis. Summary ORs and corresponding 95% CIs for IL-18 polymorphisms and chronic hepatitis C virus infections were estimated using fixed- and random-effects models when appropriate. Heterogeneity, sensitivity analysis, and publication bias were evaluated.
Results
We found a significant association between − 137G/C polymorphism and chronic hepatitis C virus infections (CG + CC versus GG: OR = 2.157, 95% CI [1.822, 2.553]; CC versus CG + GG: OR = 2.007, 95% CI [1.441, 2.797]). However, no significant association was observed between − 607C/A polymorphism and chronic hepatitis C virus infections under different contrast models.
Conclusions
The present meta-analysis suggested that IL-18 − 137G/C polymorphism in promoter region was associated with chronic hepatitis C virus infections, but no evidence indicate association between − 607C/A polymorphism and chronic hepatitis C virus infections. High-quality studies with larger sample size and larger number are warranted.
Keywords: Interleukin-18, IL-18, HCV, Hepatitis C virus, Polymorphism, Meta-analysis
Introduction
Hepatitis C virus (HCV), a member of the Flaviviridae family, is one of the leading causes of chronic liver disease worldwide with an estimated number of 170 million chronically infected individuals (WHO Consulatation, 1999). HCV is transmitted parenterally by contaminated blood or other body fluids through blood vessels, skin, or mucous membranes (Thursz and Fontanet, 2014). The natural history of HCV infection varies from spontaneous recovery post-infection, to chronic asymptomatic carrier, to decompensated cirrhosis, and hepatocellular carcinoma (HCC) (Lauer and Walker, 2001, Poynard et al., 1997). Chronic hepatitis C virus infection is a well-recognized risk factor for occurrence of hepatocellular carcinoma, and among patients with hepatitis C and cirrhosis (Thomas et al., 2000), the annual incidence rate of HCC ranges between 1 and 8%, being higher in Japan (4–8%) intermediate in Italy (2–4%) and lower in USA (1.4%) (Fassio, 2010). Liver diseases related to HCV infections have caused enormous economic and health burden (Averhoff et al., 2012, Davis et al., 2011, Jhaveri et al., 2006, McHutchison and Bacon, 2005), it's estimated that from the year 2010 through 2019 in the United States HCV may lead to 720,700 years of decompensated cirrhosis and hepatocellular carcinoma and to the loss of 1.83 Ma of life in those younger than 65 at a societal cost of $21.3 and $54.2 billion (Wong et al., 2000).
Numerous viral and host-related factors have been shown to accelerate the progression, immunological factors, especially cytokines and some host genetic variations, rather than direct HCV action, seem to play a critical role in the pathogenesis of HCV infection (Choi and Ou, 2006, Rauch et al., 2010, Zeremski et al., 2007). IL-18, first identified as an IFN-g inducing factor (IGIF) (Dao et al., 1996), plays a strategic role in inflammation and liver injury (Chikano et al., 2000, Dinarello, 2000, Tsutsui et al., 1997). Elevated levels of serum interleukin-18 were described previously for chronically (HCV)-infected patients (McGuinness et al., 2000, Sharma et al., 2009), resulted in large interest in the study of IL-18 genetic polymorphisms that might play a role in the pathogenesis and influence the outcomes of HCV infections.
Many SNPs in the IL-18 gene region have been researched, such as − 607C/A and − 137G/C in the IL-18 promoter regions, 148G/C and 105A/C in regulatory gene sequences, etc. The two SNPs (rs187238 and rs1946518) in the promoter region of the IL-18 gene have been repeatedly found to be associated with the IL-18 gene promoter transcription activity and may be associated with chronic hepatitis C virus infections.
Given the lack of previous meta-analyses in this area, our main purpose was to assess the association between Interleukin-18 gene − 607C/A (rs1946518) and − 137G/C (rs187238) polymorphisms and chronic hepatitis C virus infections by summarizing the results of published cohort and case-control studies.
Materials and methods
Publication search
A comprehensive literature search for studies reporting on the association of Interleukin-18 gene − 607C/A (rs1946518) and − 137G/C (rs187238) polymorphisms with chronic hepatitis C virus infections was conducted in the MEDLINE, EMBASE, Cochrane Library, CBM (Chinese Biomedical Literature), and CNKI (China National Knowledge Infrastructure) databases. The following search terms and keywords were used: (“Interleukin-18” OR “IL-18”) AND (polymorphism OR variant OR variation OR mutation) AND (“hepatitis C virus” OR “HCV”). Only studies published in English or in Chinese were included.
Inclusion and exclusion criteria
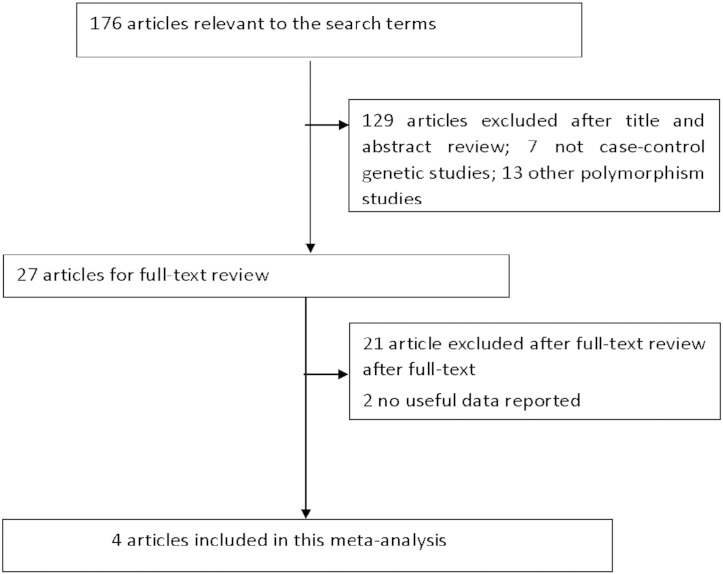
The following inclusion standards were used to select potential studies for this meta-analysis: (1) evaluated the interleukin-18 gene promoter polymorphisms with chronic hepatitis C virus infections risks; (2) all patients in the candidate studies met the diagnostic criteria for chronic HCV infections; (3) sufficient available genotype data for estimating ORs with their corresponding 95% CIs. The major reasons for the exclusion of studies are as follows: (1) not related to the interleukin-18 gene promoter polymorphisms with chronic hepatitis C virus infections risks; (2) studies only examining case populations; (3) repeated studies; (4) no usable data reported. The studies inclusion and exclusion procedures are shown in Fig. 1.
Fig. 1.
Flow diagram of study selection process.
Data extraction
Two investigators independently extracted the data from all qualified studies using the selection standard listed above. Discrepancies were solved through discussion until agreement was reached. The following information were extracted: the first author's name, year of publication, the country in which the study was conducted, the source of control groups evidence of Hardy–Weinberg equilibrium (HWE) in controls, the sample size, number of cases and controls with the CC, CA, and AA genotypes (− 607C/A (rs1946518)); GG, GC, and CC genotypes (− 137G/C (rs187238)).
Statistical analysis
Statistical analyses were performed using the STATA software for windows (version 13.0). All statistical assessments were two-sided and statistical significance was accepted at a probability value < 0.05. For the control groups, the observed genotype frequencies of the IL-18 polymorphism were evaluated for Hardy–Weinberg equilibrium (Cannings and Edwards, 1969). The strength of the association between interleukin-18 gene promoter polymorphisms and chronic hepatitis C virus infections was assessed by the odds ratios (ORs) with 95% confidence intervals (CIs). The pooled ORs (− 607C/A) were calculated for the homozygote model (AA versus CC), heterozygote model (CA versus CC), dominant model (CA + AA versus CC), recessive model (AA versus AC + CC), and an additive model (A versus C). Similarly, the pooled ORs (− 137G/C) were calculated for the homozygote model (CC versus GG), heterozygote model (CG versus GG), dominant model (CG + CC versus GG), recessive model (CC versus CG + GG), and an additive model (G versus C) (Fig. 2, Fig. 3, Fig. 4, Fig. 5).
Fig. 2.
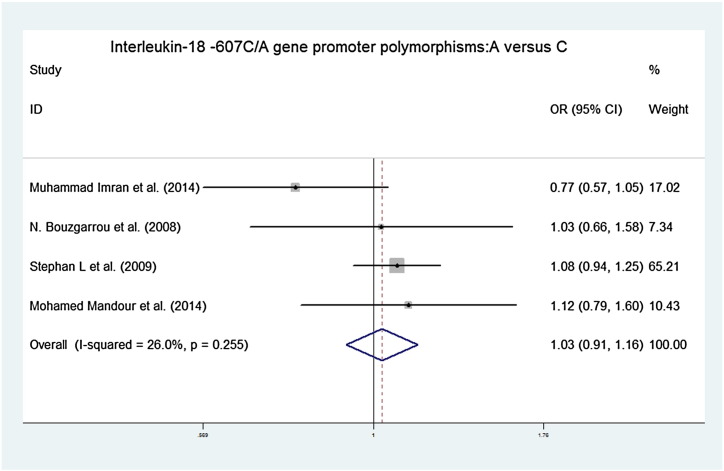
Forest plots of the association between IL-18 − 607C/A polymorphisms and chronic hepatitis C virus infections in an additive contrast model (A versus C). OR = 1.029, 95% CI [0.915, 1.158] chi-squared = 4.06 (d.f. = 3) p = 0.255, I-squared = 26.0%, z = 0.48, p = 0.634.
Fig. 3.
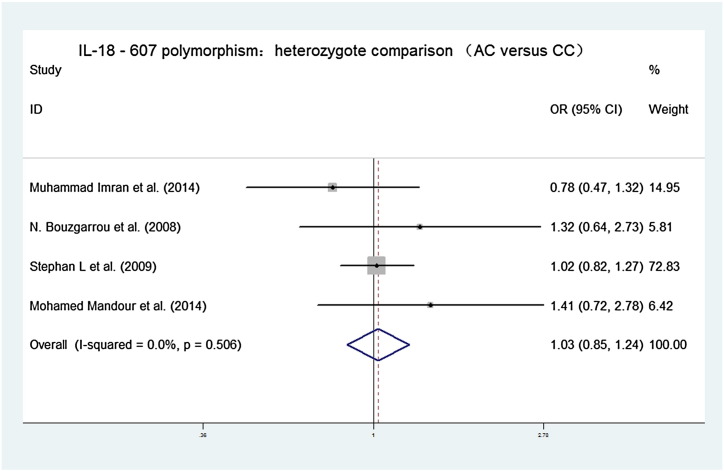
Forest plots of the association between IL-18 − 607C/A polymorphisms and chronic hepatitis C virus infections in heterozygote contrast model (AC versus CC). OR = 1.029, 95% CI [0.854, 1.241], chi-squared = 2.33 (d.f. = 3), p = 0.506, I-squared = 0.0%, z = 0.30, p = 0.764.
Fig. 4.
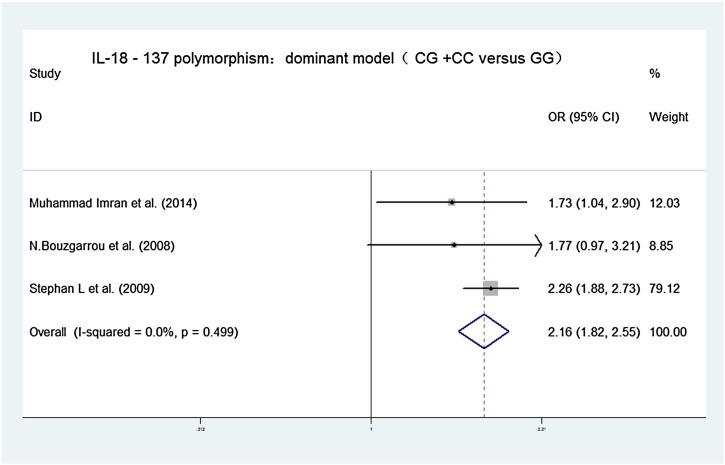
Forest plots of the association between IL-18 − 137G/C polymorphisms and chronic hepatitis C virus infections in dominant contrast model (CG + CC versus GG). OR = 2.157, 95% CI [1.822, 2.553], chi-squared = 1.39 (d.f. = 2) p = 0.499, I-squared = 0.0%, z = 8.93, p = 0.000.
Fig. 5.
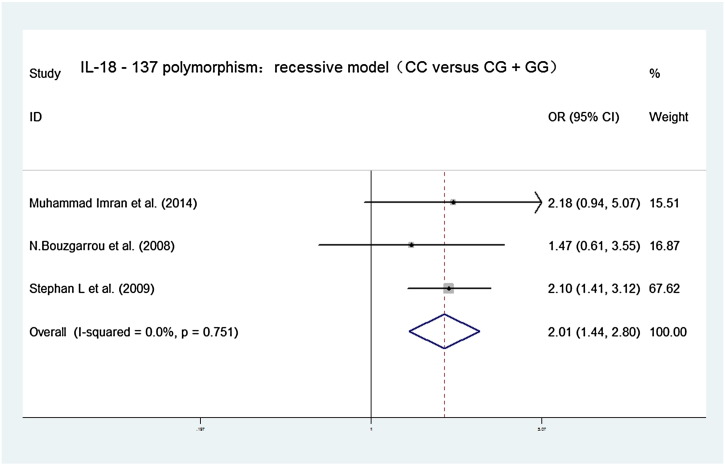
Forest plots of the association between IL-18 − 137G/C polymorphism and chronic hepatitis C virus infections in recessive contrast model (CC versus CG + GG). OR = 2.007, 95% CI [1.441, 2.797], chi-squared = 0.57 (d.f. = 2) p = 0.751, I-squared = 0.0% z = 4.12, p = 0.000.
A test for heterogeneity, a sensitivity analysis, and an assessment of publication bias were performed in our meta-analysis. Considering possible heterogeneity between studies, Cochran's Q statistic and the I2 metric were conducted (Higgins and Thompson, 2002, Jackson et al., 2012), P < 0.10 and I2 > 50% were considered to indicate the existence of significant heterogeneity. The pooled ORs were analyzed using the random-effects model (DerSimonian and Laird, 1986), if the heterogeneity test result was P > 0.1. Or else, if the heterogeneity test result was P < 0.1, the fixed effects model was used (Mantel and Haenszel, 1959). Sensitivity analyses were also performed and the Begg's funnel plot and Egger's test were performed to examine the publication bias (Peters et al., 2006).
Results
Study characteristics
A total of 4 studies with 1222 cases and 1115 controls for − 607C/A polymorphism and 3 studies with 959 cases and 987 controls for − 137G/C polymorphism were included in this meta-analysis (Bouzgarrou et al., 2008, Haas et al., 2009, Imran et al., 2014, Mandour et al., 2014). The genotype distributions in the controls for all of the included studies were consistent with Hardy–Weinberg equilibrium. The characteristics of all reports on the association between interleukin-18 gene promoter polymorphisms and chronic hepatitis C virus infections risks are shown in Table 1.
Table 1.
Characteristics of studies included in the meta-analysis.
| Author | Year | Region | Total | Case GG |
Case GC |
Case CC |
Total | Con GG |
Con GC |
Con CC |
H-W (P) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-18 − 607C/A polymorphism | |||||||||||
| Muhammad Imran et al. | 2014 | Pakistan | 260 | 87 | 113 | 60 | 120 | 32 | 53 | 35 | 0.203 |
| N. Bouzgarrou et al. | 2008 | Tunisian | 82 | 21 | 44 | 17 | 81 | 24 | 38 | 19 | 0.601 |
| Stephan L et al. | 2009 | Germany | 757 | 276 | 347 | 134 | 791 | 300 | 369 | 122 | 0.628 |
| Mohamed Mandour et al. | 2014 | Egypt | 123 | 20 | 63 | 40 | 123 | 26 | 58 | 39 | 0.608 |
| IL-18 − 137G/C polymorphism | |||||||||||
| Muhammad Imran et al. | 2014 | Pakistan | 120 | 43 | 61 | 16 | 115 | 48 | 57 | 10 | 0.226 |
| N. Bouzgarrou et al. | 2008 | Tunisian | 82 | 35 | 35 | 12 | 81 | 38 | 31 | 12 | 0.187 |
| Stephan L et al. | 2009 | Germany | 757 | 386 | 315 | 56 | 791 | 439 | 299 | 53 | 0.829 |
Results of the overall meta-analysis
Our main results on the association between interleukin-18 gene promoter polymorphisms and chronic hepatitis C virus infections are listed in Table 2. The association between interleukin-18 − 607C/A gene promoter polymorphisms and chronic hepatitis C virus infections showed pooled ORs and 95% CI for the homozygote comparison (AA versus CC: 1.073, [0.846, 1.360]), heterozygote comparison (CA versus CC: 1.029, [0.854, 1.241]), dominant model (CA + AA versus CC: 1.039, [0.872, 1.237]), recessive model (AA versus AC + CC: 1.036, [0.843, 1.274]), and an additive model (A versus C: 1.029, [0.915, 1.158]). The association between interleukin-18 − 137G/C gene promoter polymorphisms and chronic hepatitis C virus infections showed pooled ORs and 95% CI for the homozygote comparison (CC versus GG: 1.257, [0.896, 1.763]), heterozygote comparison (CG versus GG: 1.200, [0.995, 1.447]), dominant model (CG + CC versus GG: 2.157, [1.822, 2.553]), recessive model (CC versus CG + GG 2.007, [1.441, 2.797]), and an additive model (G versus C: 1.152, [1.002, 1.324]).
Table 2.
Main results on association between interleukin-18 gene promoter (− 607C/A and − 137G/C) polymorphisms and chronic hepatitis C virus infections.
| Contrast | OR, 95% CI | Heterogeneity | Z and P |
|---|---|---|---|
| IL-18 − 607C/A polymorphism | |||
| AA versus CC | 1.073, [0.846, 1.360] | Chi-squared = 4.07 (d.f. = 3), p = 0.254, I-squared = 26.2% | z = 0.58, p = 0.563 |
| AC versus CC | 1.029, [0.854, 1.241] | Chi-squared = 2.33 (d.f. = 3), p = 0.506, I-squared = 0.0% | z = 0.30, p = 0.764 |
| AA + AC versus CC | 1.039, [0.872, 1.237] | Chi-squared = 3.21 (d.f. = 3), p = 0.361, I-squared = 6.5% | z = 0.42, p = 0.672 |
| AA versus AC + CC | 1.036, [0.843, 1.274] | Chi-squared = 3.16 (d.f. = 3), p = 0.368, I-squared = 5.0% | z = 0.34, p = 0.736 |
| A versus C | 1.029, [0.915, 1.158] | Chi-squared = 4.06 (d.f. = 3), p = 0.255, I-squared = 26.0% | z = 0.48, p = 0.634 |
| IL-18 − 137G/C polymorphism | |||
| CC versus GG | 1.257, [0.896,1.763] | Chi-squared = 0.74 (d.f. = 2), p = 0.690, I-squared = 0.0% | z = 1.32, p = 0.186 |
| CG versus GG | 1.200, [0.995,1.447] | Chi-squared = 0.00 (d.f. = 2), p = 0.998, I-squared = 0.0% | z = 1.91, p = 0.057 |
| CG + CC versus GG | 2.157, [1.822,2.553] | Chi-squared = 1.39 (d.f. = 2), p = 0.499, I-squared = 0.0% | z = 8.93, p = 0.000 |
| CC versus CG + GG | 2.007, [1.441,2.797] | Chi-squared = 0.57 (d.f. = 2) p = 0.751, I-squared = 0.0% | z = 4.12, p = 0.000 |
| C versus G | 1.152, [1.002,1.324] | Chi-squared = 0.27 (d.f. = 2) p = 0.874, I-squared = 0.0% | z = 1.99, p = 0.047 |
Test for heterogeneity and sensitivity analysis
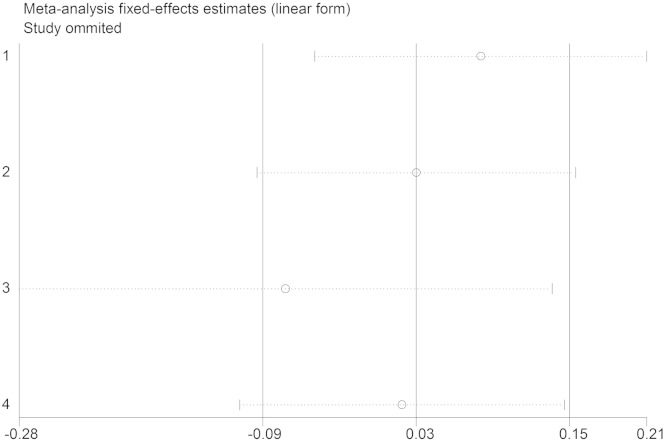
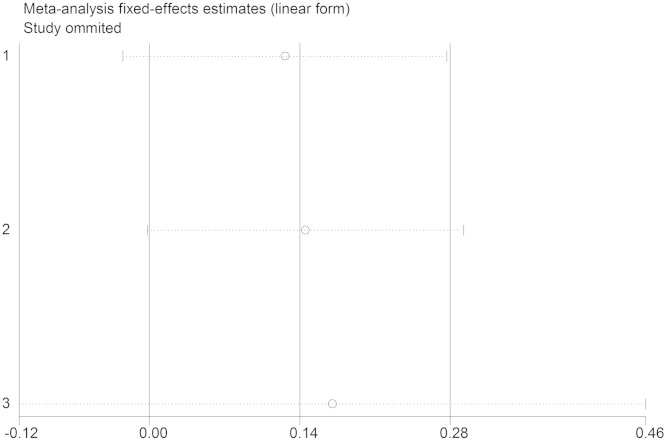
We assessed the source of heterogeneity by region, publication year, control source, and sample size. However, we did not observe any sources that contribute to the substantial heterogeneity (Table 2). Sensitivity analyses were conducted to ascertain the primary origin of the heterogeneity. NO independent study affected the heterogeneity in 2 groups (Fig. 6, Fig. 7).
Fig. 6.
Sensitivity analysis (IL-18 − 607C/A).
Fig. 7.
Sensitivity analysis (IL-18 − 137G/C).
Publication bias
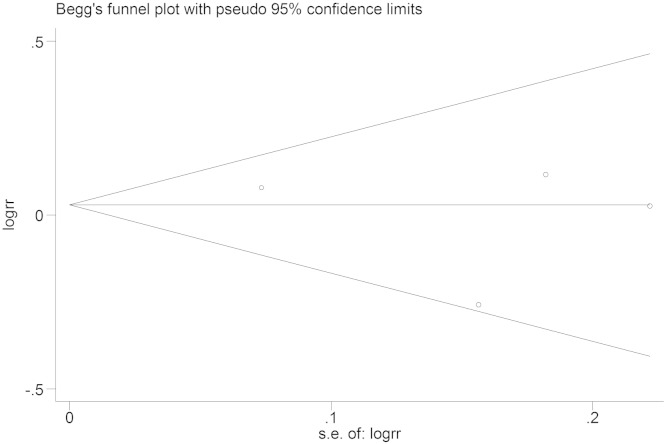
Funnel plots were used to evaluate publication bias, and Begg's Test was performed to statistically assess funnel plot symmetry. The results showed no significant publication bias (Fig. 8, Fig. 9), Begg's Test (− 607C/A): Pr > |z| = 0.734 (continuity corrected); Begg's Test (− 137G/C): Pr > |z| = 0.602 (continuity corrected).
Fig. 8.
Tests for publication bias (IL-18 − 607C/A), Begg's Test: Pr > |z| = 0.734.
Fig. 9.
Tests for publication bias (IL-18 − 137G/C), Begg's Test: Pr > |z| = 1.000.
Discussion
Hepatitis C virus (HCV) is a major cause of hepatic disease worldwide and a potential cause of substantial morbidity and mortality in the future (Shepard et al., 2005). There is no vaccine and no post-exposure prophylaxis for HCV, HCV infection has a chronicity rate of about 70% (Stadhouders and Cooreman, 1997). Prognosis may be quite different among individuals infected with hepatitis C virus (HCV): a chronic liver disease is believed to occur in half the patients while in the other half there are no signs of histologic progression of liver damage (Peano et al., 1994). Patients who acquire the infection early in life have a markedly increased mortality even when cirrhosis is absent at diagnosis and disease duration is short (Niederau et al, 1998). The immune response is critical in determining the outcome of hepatitis C virus (HCV) infection. Interleukin (IL)-18 is a pivotal mediator of the Th1/Th2-driven immune response. Two IL-18 promoter polymorphisms (− 607C/A and − 137G/C) and their haplotypes were known to affect IL-18 expression (Rehermann, 2009, An et al., 2008).
Despite the potential implication of IL-18 promoter polymorphisms (− 607C/A and − 137G/C) in the pathogenesis of chronic hepatitis C virus infections, the association between IL-18 promoter polymorphisms (− 607C/A and − 137G/C) and chronic hepatitis C virus infections remains unclear. Thus, in this study, we conducted a comprehensive meta-analysis integrating data from previous publications to derive a more precise assessment of the relationship between the IL-18 promoter polymorphisms (− 607C/A and − 137G/C) and chronic hepatitis C virus infections. To the best of our knowledge, this study is the first meta-analysis to comprehensively assess the association of IL-18 promoter polymorphisms (− 607C/A and − 137G/C) and chronic hepatitis C virus infections.
A total of 4 studies with 1222 cases and 1115 controls for − 607C/A polymorphism and 3 studies with 959 cases and 987 controls for − 137G/C polymorphism were included in this meta-analysis. No other polymorphisms were described in those studies except for one study (Imran et al., 2014 also described Osteopontin — 442 Polymorphism in their study). We found a significant association between − 137G/C polymorphism and chronic hepatitis C virus infections (CG + CC versus GG: OR = 2.157, 95% CI [1.822, 2.553]; CC versus CG + GG: OR = 2.007, 95% CI [1.441, 2.797]). However, no significant association was observed between − 607C/A polymorphism and chronic hepatitis C virus infections under different contrast models (AA versus CC OR = 1.073, [0.846,1.360]; AC versus CC OR = 1.029, 95% CI, [0.854,1.241]; AA + AC versus CCOR = 1.039, 95% CI [0.872,1.237]; AA versus AC + CC OR = 1.036, 95% CI [0.843,1.274]; A versus C OR = 1.029, 95% CI [0.915,1.158]).
Of course, we should be aware of that the hypothesis considering no association between IL-18 − 607C/A polymorphism and chronic hepatitis C virus infections merely on the basis of the negative results in this study, because the number of studies and sample size are relatively small. If a putative genetic association is of small magnitude with point estimates less than 1.5, the small and underpowered studies may be unable to identify true genetic associations (Hindorff et al., 2009, Ioannidis et al., 2006, Ioannidis, 2003). Although meta-analysis can increase the statistical power by combining all the eligible studies, it is limited in its effect estimation owing to small number of studies included. Thus, more evidence is needed to support or deny such an association between the IL-18 − 607C/A polymorphism and chronic hepatitis C virus infections.
By means of meta-analysis, a statistical technique for combining the results from independent studies, we drew a more reliable conclusion on the association between interleukin-18 gene promoter polymorphisms and chronic hepatitis C virus infections. However, chronic hepatitis C virus infections might be a result of the comprehensive interaction among numerous viral and host-related factors, immunological factors, cytokines and some host genetic variations.
Limitations
Meta-analysis is regarded as a useful method in synthesizing data from all the eligible studies to obtain greater statistical power. However, a few potential limitations of our meta-analysis we should pay attention: (1) Selection bias may have occurred because only studies in English or Chinese were included in this meta-analysis. (2) We did not perform subgroup analysis stratified by ethnicity. (3) The number of studies and sample size are relatively small. Despite the limitations listed above, our meta-analysis has some clear advantages: (1) this is the first meta-analysis that analyzed the association between IL-18 promoter polymorphisms (− 607C/A and − 137G/C) and chronic hepatitis C virus infections (2) sensitivity analysis did not show any single study strongly affecting the overall results, suggesting sufficient reliability and stability of the pooled results; (3) The strict search method, normative inclusion and exclusion Criteria significantly increased the statistical power of our meta-analysis;(4) No significant publication bias was detected, demonstrating that our overall results are reliable.
Conclusion
The present meta-analysis suggested that IL-18 − 137 G/C polymorphism in promoter region was associated with chronic hepatitis C virus infections, but no evidence indicates an association between − 607C/A polymorphism and chronic hepatitis C virus infections. High-quality studies with larger sample size and larger number are warranted.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mgene.2015.04.004.
Appendix A. Supplementary data
The following are the Supplementary related to this article.
PRISMA checklist.
Original Data.
References
- An P., Thio C.L., Kirk G.D., Donfield S., Goedert J.J., Winkler C.A. Regulatory polymorphisms in the interleukin-18 promoter are associated with hepatitis C virus clearance. J. Infect. Dis. 2008;198(8):1159–1165. doi: 10.1086/592047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averhoff F.M., Glass N., Holtzman D. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin. Infect. Dis. 2012;55:S10–S15. doi: 10.1093/cid/cis361. [DOI] [PubMed] [Google Scholar]
- Bouzgarrou N., Hassen E., Schvoerer E. Association of interleukin-18 polymorphisms and plasma level with the outcome of chronic HCV infection. J. Med. Virol. 2008;80(4):607–614. doi: 10.1002/jmv.21079. [DOI] [PubMed] [Google Scholar]
- Cannings C., Edwards A.W. Expected genotypic frequencies in a small sample: deviation from Hardy–Weinberg equilibrium. Am. J. Hum. Genet. 1969;21(3):245–247. [PMC free article] [PubMed] [Google Scholar]
- Chikano S., Sawada K., Shimoyama T. IL-18 and IL-12 induce intestinal inflammation and fatty liver in mice in an IFN-gamma dependent manner. Gut. 2000;47(6):779–786. doi: 10.1136/gut.47.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Ou J.H.J. Mechanisms of liver injury. III. Oxidative stress in the pathogenesis of hepatitis C virus. American Journal of Physiology-Gastrointestinal And Liver Physiology. 2006;290(5):G847-G51. doi: 10.1152/ajpgi.00522.2005. [DOI] [PubMed] [Google Scholar]
- Dao T., Ohashi K., Kayano T., Kurimoto M., Okamura H. Interferon-gamma-inducing factor, a novel cytokine, enhances Fas ligand-mediated cytotoxicity of murine T helper 1 cells. Cell. Immunol. 1996;173(2):230–235. doi: 10.1006/cimm.1996.0272. [DOI] [PubMed] [Google Scholar]
- Davis K.L., Mitra D., Medjedovic J., Beam C., Rustgi V. Direct economic burden of chronic hepatitis C virus in a United States managed care population. J. Clin. Gastroenterol. 2011;45(2):E17–E24. doi: 10.1097/MCG.0b013e3181e12c09. [DOI] [PubMed] [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials. Control Clin. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Dinarello C.A. Interleukin-18, a proinflammatory cytokine. Eur. Cytokine Netw. 2000;11(3):483–486. [PubMed] [Google Scholar]
- Fassio E. Hepatitis C and hepatocellular carcinoma. Ann. Hepatol. 2010;9:S119–S122. [PubMed] [Google Scholar]
- Haas S.L., Weiss C., Bugert P. Interleukin 18 promoter variants (− 137G > C and − 607C > A) in patients with chronic hepatitis C: association with treatment response. J. Clin. Immunol. 2009;29(5):620–628. doi: 10.1007/s10875-009-9302-z. [DOI] [PubMed] [Google Scholar]
- Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Hindorff L.A., Sethupathy P., Junkins H.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran M., Manzoor S., Parvaiz F. Predictive potential of IL-18 − 607 and osteopontin − 442 polymorphism in interferon-based therapy of HCV infection in the Pakistani population. Viral Immunol. 2014;27(8):404–411. doi: 10.1089/vim.2014.0044. [DOI] [PubMed] [Google Scholar]
- Ioannidis J.P. Genetic associations: false or true? Trends Mol. Med. 2003;9(4):135–138. doi: 10.1016/s1471-4914(03)00030-3. [DOI] [PubMed] [Google Scholar]
- Ioannidis J.P., Trikalinos T.A., Khoury M.J. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am. J. Epidemiol. 2006;164(7):609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- Jackson D., White I.R., Riley R.D. Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat. Med. 2012;31:3805–3820. doi: 10.1002/sim.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri R., Grant W., Kauf T.L., McHutchison J. The burden of hepatitis C virus infection in children: estimated direct medical costs over a 10-year period. J. Pediatr. 2006;148(3):353–358. doi: 10.1016/j.jpeds.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Lauer G.M., Walker B.D. Medical progress: hepatitis C virus infection. N. Engl. J. Med. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- Mandour M., Nemr N., Kishk R., Ahmed E. Impact of the IL-18 gene polymorphism in response to antiviral therapy in chronic HCV genotype 4 patients. Rev. Soc. Bras. Med. Trop. 2014;47(2):137–142. doi: 10.1590/0037-8682-0024-2014. [DOI] [PubMed] [Google Scholar]
- Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- McGuinness P.H., Painter D., Davies S., McCaughan G.W. Increases in intrahepatic CD68 positive cells, MAC387 positive cells, and proinflammatory cytokines (particularly interleukin 18) in chronic hepatitis C infection. Gut. 2000;46(2):260–269. doi: 10.1136/gut.46.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHutchison J.G., Bacon B.R. Chronic hepatitis C: an age wave of disease burden. Am. J. Manag. Care. 2005;11(10):S286–S295. [PubMed] [Google Scholar]
- Niederau C., Lange S., Heintges T. Prognosis of chronic hepatitis C: results of a large, prospective cohort study. Hepatology (Baltimore, Md) 1998;28(6):1687–1695. doi: 10.1002/hep.510280632. [DOI] [PubMed] [Google Scholar]
- Peano G., Menardi G., Ponzetto A., Fenoglio L.M. HLA-DR5 antigen. A genetic factor influencing the outcome of hepatitis C virus infection? Arch. Intern. Med. 1994;154(23):2733–2736. doi: 10.1001/archinte.1994.00420230126015. [DOI] [PubMed] [Google Scholar]
- Peters J.L.A.J. Sutton, Jones D.R., Abrams K.R., Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295(6):676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- Poynard T., Bedossa P., Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. Lancet. 1997;349(9055):825–832. doi: 10.1016/s0140-6736(96)07642-8. [DOI] [PubMed] [Google Scholar]
- Rauch A., Kutalik Z., Descombes P. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138(4):1338-U173. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J. Clin. Investig. 2009;119(7):1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Chakraborti A., Das A., Dhiman R.K., Chawla Y. Elevation of interleukin-18 in chronic hepatitis C: implications for hepatitis C virus pathogenesis. Immunology. 2009;128(1):e514–e522. doi: 10.1111/j.1365-2567.2008.03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard C.W., Finelli L., Alter M. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005;5(9):558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Stadhouders P., Cooreman M.P. Chronic hepatitis C virus disease: an evaluation of procedures for diagnosis and treatment. Neth. J. Med. 1997;51(6):213–224. doi: 10.1016/s0300-2977(97)00045-4. [DOI] [PubMed] [Google Scholar]
- Thomas D.L., Astemborski J., Rai R.M. The natural history of hepatitis C virus infection — host, viral, and environmental factors. JAMA. 2000;284(4):450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- Thursz M., Fontanet A. HCV transmission in industrialized countries and resource-constrained areas. Nat. Rev. Gastroenterol. Hepatol. 2014;11(1):28–35. doi: 10.1038/nrgastro.2013.179. [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Matsui K., Kawada N. IL-18 accounts for both TNF-alpha- and Fas ligand-mediated hepatotoxic pathways in endotoxin-induced liver injury in mice. J. Immunol. 1997;159(8):3961–3967. [PubMed] [Google Scholar]
- WHO Consulatation Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat. 1999;6:35–47. [PubMed] [Google Scholar]
- Wong J.B., McQuillan G.M., McHutchison J.G., Poynard T. Estimating future hepatitis C morbidity, mortality, and costs in the United States. Am. J. Public Health. 2000;90(10):1562–1569. doi: 10.2105/ajph.90.10.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeremski M., Petrovic L.M., Talal A.H. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J. Viral Hepat. 2007;14(10):675–687. doi: 10.1111/j.1365-2893.2006.00838.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
Original Data.