Abstract
Purpose
The peroxisome proliferator-activated receptor beta/delta (PPARβ/δ) is a transcription factor with roles in metabolism, angiogenesis, and inflammation. It has yet undefined roles in retinal inflammation and diabetic retinopathy (DR). We used RNA-seq to better understand the role of the antagonist and inverse agonist of PPARβ/δ, GSK0660, in TNFα-induced inflammation. Understanding the underlying mechanisms of vascular inflammation could lead to new treatments for DR.
Methods
RNA was isolated from human retinal microvascular endothelial cells treated with a vehicle, TNFα, or TNFα plus GSK0660. RNA-seq was performed with a 50 bp single read protocol. The differential expression was determined using edgeR and gene ontology, and a pathway analysis was performed using DAVID. RNA-seq validation was performed using qRT-PCR using the primers for ANGPTL4, CCL8, NOV, CXCL10, and PDPK1.
Results
TNFα differentially regulated 1,830 transcripts, many of which are involved in the cytokine–cytokine receptor interaction, chemokine signaling, and inflammatory response. Additionally, TNFα highly upregulated genes involved in leukocyte recruitment, including CCL5, CX3CL1, and CXCL10. GSK0660 differentially regulated 273 transcripts in TNFα-treated cells compared to TNFα alone. A pathway analysis revealed the enrichment of cytokine–cytokine receptor signaling. In particular, GSK0660 blocks the TNFα-induced upregulation of CCL8, a chemokine involved in leukocyte recruitment.
Conclusions
TNFα regulates several genes related to retinal leukostasis in retinal endothelial cells. GSK0660 blocks the effect of TNFα on the expressions of cytokines involved in leukocyte recruitment, including CCL8, CCL17, and CXCL10 and it may therefore block TNFα-induced retinal leukostasis.
Introduction
Diabetic retinopathy (DR) is currently one of the leading causes of irreversible blindness in working-age adults in the United States, and cases are expected to increase threefold within the next 35 years [1]. The pathology of DR consists of two main stages: non-proliferative diabetic retinopathy (NPDR) and proliferative diabetic retinopathy (PDR). NPDR, the early stage, is characterized by vascular dysfunction, including microaneurysms, lipid exudation, and vascular hyperpermeability. PDR, the later and more severe stage, is characterized by abnormal neovascularization. Retinal inflammation plays a critical role in the pathogenesis of both NPDR and PDR.
Retinal inflammation is a result of several inflammatory mediators, including TNFα, IL-8, IL-6, IL-1β, and NF-κB, which are upregulated in the retina or vitreous of patients with DR [2,3]. TNFα, in particular, is important to disease onset and progression. Increased TNFα levels in the sera of children are predictive of NPDR and are observed in the vitreous of patients with PDR [4,5]. Furthermore, TNFα drives leukostasis, endothelial cell apoptosis, and blood-retinal barrier breakdown in DR [6,7]. Leukostasis is a critical pathologic feature of DR because adhered leukocytes can occlude capillaries, leading to focal areas of ischemia and eventual progression to PDR. Additionally, leukocytes may extravasate and promote local cytokine upregulation, increasing retinal inflammation [8]. TNFα contributes to leukostasis through the upregulation of adhesion proteins, including VCAM1, ICAM1, and E-selectin on the lumenal wall of the vascular endothelium, as well as leukocyte recruiting chemokines, such as IL-6 and IL-8 [9,10]. While clearly important in DR, the mechanism of TNFα-induced inflammation in retinal endothelial cells has not been fully characterized.
The peroxisome proliferator-activated receptors (PPARs) are a group of nuclear hormone receptors that have roles in metabolism, cell differentiation, cell proliferation, and inflammation [11]. Of the three isoforms, PPARβ/δ is the least well studied and understood, particularly in terms of its effect on proliferation and inflammation. To date, most reports in the literature on PPARβ/δ and inflammation have focused on the roles of its agonists. Activation of PPARβ/δ inhibits TNFα-induced expression of adhesion molecules and leukocyte adhesion to umbilical vein endothelial cells [12,13]. However, few studies have been done using the specific antagonist of PPARβ/δ, GSK0660, which also has inverse agonist effects when used alone [14]. We have previously shown GSK0660 to be antiangiogenic in the context of oxygen-induced retinopathy [15]. As inflammation, as well as angiogenesis, is an important component of DR, we chose to examine the effect of GSK0660 on TNFα-induced inflammation. In this study, we used RNA-sequencing (RNA-seq) to determine the effect of GSK0660 on TNFα-induced inflammation in human retinal microvascular endothelial cells (HRMECs). RNA-seq has advantages over microarrays in that it is more sensitive, has a broader dynamic range, and allows for the identification of novel transcripts [16].
Methods
Culture of human retinal endothelial cells and RNA isolation
Primary HRMECs were purchased from Cell Systems (Kirkland, WA) and grown in an endothelial basal medium (Lonza; Walkersville, MD) with 10% fetal bovine serum (FBS) and endothelial growth supplements (EGM SingleQuots; Lonza). Cultures were kept in a humidified cell culture incubator at 37 °C with 5% CO2. Cells were plated in six-well dishes coated with attachment factor (Cell Signaling; Danvers, MA) and grown to 70% subconfluency. The medium was changed to 2% FBS with one of the following treatment schemes: a vehicle (0.1% DMSO) for 24 h then a vehicle for 4 h, a vehicle for 24 h then 1 ng/ml TNFα (Sigma-Aldrich; St. Louis, MO) + a vehicle for 4 h, or 10 µM GSK0660 (Tocris; Minneapolis, MN) for 24 h followed by TNFα + GSK0660 for 4 h. The work area was cleaned using RNaseZap® (Life Technologies; Grand Island, NY) and then total RNA was isolated from cell lysates using an RNeasy kit (Qiagen; Valencia, CA) according to the manufacturer’s directions.
Library preparation and RNA-sequencing
RNA samples were submitted to the Vanderbilt VANTAGE core for RNA-seq. RNA quality was determined using the 2100 Bioanalyzer (Agilent Technologies; Santa Clara, CA). The RNA integrity number (RIN) of each sample was 10. Libraries were prepared using the TruSeq RNA Sample Prep Kit (Illumina; San Diego, CA) to enrich for poly(A)-containing mRNA and generate cDNA. Library quality was also confirmed using the 2100 Bioanalyzer. The libraries were sequenced using a 50 bp single read protocol on the Illumina HiSeq 2500 (Illumina). Sequence data were deposited at the NCBI Short Read Archive under the accession number SRP053124.
Sequence alignment and differential expression
Sequence alignment and differential expression were performed by the Vanderbilt VANGARD core. TopHat v2.0.9 was used to align sequences to the UCSC human reference genome hg19 using default parameters [17]. Raw counts of mapped reads were generated and then used by the MultiRankSeq program, which utilizes the edgeR algorithm to determine differential expression [18]. Comparisons were made between vehicle- and TNFα-treated HRMECs, as well as between TNFα-treated HRMECs and TNFα-treated HRMECs with GSK0660. Transcripts were considered significant with an adjusted p value of <0.05. The list was further reduced to transcripts with a fold change greater than or equal to 2. The Euler diagram was generated using the R package utility, VennDiagram [19].
Gene ontology and pathway analysis
The lists of differentially expressed genes were submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 for gene ontology (GO) and pathway analysis [20,21]. GO was determined using the GOTERM_BP_FAT data set, which includes the lower levels of the biologic process ontology. GO terms were considered enriched with an EASE score of <0.05. Pathway enrichment was determined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway annotation. Pathways were considered enriched with an EASE score of <0.05.
qRT-PCR validation
RNA was reverse transcribed to cDNA using the High-Capacity cDNA Archive Kit (Applied Biosystems; Carlsbad, CA) according to the manufacturer’s directions. Quantitative real-time PCR (qRT-PCR) was performed by amplification of the gene of interest (ANGPTL4, CCL8, NOV, CXCL10, or PDPK1) versus β-actin using gene-specific TaqMan Gene Expression Assays (Applied Biosystems). Data were analyzed using the comparative Ct method and Ct values were normalized to β-actin levels. Statistical significance was determined using the statistical software JMP (SAS Institute; Cary, NC) and an ANOVA with a student’s t post-hoc analysis. Data were considered significant at p<0.05.
Results
RNA-seq quality and alignment
To determine the effect of GSK0660 on the TNFα-dependent gene expression in HRMECs, we treated HRMECs with a vehicle, TNFα plus a vehicle, or TNFα plus GSK0660 and performed a transcriptomic analysis using RNA-seq with three samples per treatment. The total number of reads ranged between 28,275,640 and 33,252,277, which covered 32,009 different transcripts (Appendix 1). There was no difference in the total number of reads across the nine samples (ANOVA, p = 0.07). On average, 96.5% of the reads mapped to the UCSC human genome hg19 and between 311 and 1,084 reads were removed due to low quality.
Differential expression
The differential expression of transcripts was determined using the MultiRankSeq program, which determines the differential expression from raw read counts using the edgeR algorithm. Comparisons were made between the TNFα- and vehicle-treated cells, as well as between the TNFα- and TNFα plus GSK0660-treated cells. Transcripts were considered significant with an adjusted p value of <0.05 and a fold change of at least 2.0. Using these parameters, 1,830 transcripts were differentially expressed in the TNFα-treated cells compared to vehicle-treated cells. TNFα plus GSK0660 treatment altered the expression of 273 transcripts compared to TNFα alone (Table 1).
Table 1. Summary of RNA-seq differential expression analysis.
| Treatment comparison | Transcripts with adj p<0.05 | Upregulated Transcripts (>=2) | Downregulated Transcripts (<=-2) |
|---|---|---|---|
|
TNFα versus Vehicle |
1830 |
746 |
1084 |
| TNFα + GSK0660 versus TNFα | 273 | 169 | 104 |
Differential expression was determined using edgeR and transcripts were considered significantly changed with adjusted p<0.05 and fold change of at least 2.0.
Effect of TNFα on HRMECs
Stimulation of HRMECs with TNFα resulted in large changes to the gene expression (Appendix 2), with the top 10 upregulated and downregulated transcripts of protein-coding genes summarized in Table 2. Notably, TNFα increased the expressions of CCL5, CX3CL1, and CXCL10, all of which play roles in leukocyte recruitment. Additionally, TNFα increased its own transcription.
Table 2. Top 10 upregulated and downregulated protein-coding genes by TNFα in HRMEC.
| Gene Symbol | Log2 Fold Change | Adjusted P Value | Ensembl ID |
|---|---|---|---|
| Upregulated Genes | |||
| LAD1 |
6.421452313 |
1.10E-07 |
ENSG00000159166 |
| CSF2 |
6.39995987 |
<0.000001 |
ENSG00000164400 |
| TNFAIP6 |
6.358926206 |
2.18E-154 |
ENSG00000123610 |
| CX3CL1 |
6.121874962 |
<0.000001 |
ENSG00000006210 |
| CXCL10 |
5.706389982 |
6.14E-222 |
ENSG00000169245 |
| HLA-DOB |
5.667011352 |
5.01E-13 |
ENSG00000241106 |
| ETV3L |
5.616009893 |
1.07E-05 |
ENSG00000253831 |
| CCL5 |
5.487915198 |
5.76E-257 |
ENSG00000161570 |
| TNF |
5.486418289 |
4.71E-52 |
ENSG00000232810 |
| GBP7 |
5.455106479 |
1.78E-05 |
ENSG00000213512 |
| Downregulated Genes | |||
| RBM20 |
−4.29030613 |
0.009588413 |
ENSG00000203867 |
| PAK6 |
−4.29073104 |
0.026177447 |
ENSG00000137843 |
| OR1F1 |
−4.291371288 |
0.00855682 |
ENSG00000168124 |
| MYO18B |
−4.474679753 |
0.004370114 |
ENSG00000133454 |
| CSRNP3 |
−4.474760229 |
0.004370114 |
ENSG00000178662 |
| CR1 |
−4.634084683 |
0.003662894 |
ENSG00000203710 |
| ARL14 |
−4.783629776 |
0.001145234 |
ENSG00000179674 |
| STOX2 |
−4.915666474 |
0.000890805 |
ENSG00000173320 |
| FAM151A |
−4.916414121 |
0.000585614 |
ENSG00000162391 |
| MUC20 | −5.040180048 | 0.000485122 | ENSG00000176945 |
Transcript fold change and adjusted p value of transcripts affected by TNFα-treated HRMEC compared to vehicle-treated were determined by the edgeR algorithm.
To determine the function of the transcripts differentially expressed by TNFα, we used the DAVID tool for functional annotation. In terms of gene ontology, 344 GO biologic pathway terms were significantly enriched, with the top 20 summarized in Figure 1. Significant terms included immune response, response to wounding, and inflammatory response. Another common theme included terms for regulating endothelial behavior, such as regulation of cell proliferation, vasculature development, cell adhesion, and chemotaxis.
Figure 1.
Top 20 biologic pathway GO terms enriched in TNFα-treated HRMECs. Biologic Pathway GO term enrichment in TNFα-treated HRMECs compared to vehicle-treated HRMECs was determined using DAVID. Terms were considered significant at p<0.05.
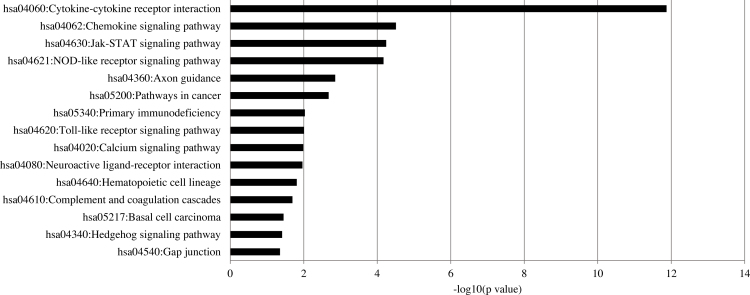
To annotate further the transcripts, we used the KEGG database to determine enriched pathways. The pathways most highly enriched included the cytokine–cytokine receptor interaction, chemokine signaling pathway, and Jak-STAT signaling pathway (Figure 2). Additional pathways through which TNFα may affect HRMECs included calcium signaling, Toll-like receptor signaling, hedgehog signaling, and the complement cascade.
Figure 2.
KEGG pathways enriched in TNFα-treated HRMECs. Pathway enrichment was determined for differentially expressed transcripts by TNFα compared to the vehicle using DAVID. Pathways were considered enriched at p<0.05.
The effect of GSK0660 on TNFα-treated HRMEC
The effect of GSK0660 on the TNFα-regulated gene expression in HRMECs was determined; the top upregulated and downregulated protein-coding transcripts are summarized in Table 3, while the full data set can be found in Appendix 3. Among those most highly downregulated by GSK0660 in TNFα-treated cells, CCL8 is of interest due to its role in leukocyte recruitment. Also of note, GSK0660 prevented the TNFα-induced downregulation of FAM151A, MUC20, STOX2, and ARL14. These four transcripts were among those most highly downregulated by TNFα compared to the vehicle. Additionally, GSK0660 affected the transcription of one (CXCL10) of the top 10 genes upregulated by TNFα.
Table 3. Top 10 protein-encoding genes that were upregulated or downregulated by GSK0660 in TNFα-treated HRMEC.
| Gene Symbol | Log2 Fold Change | Adj P Value | Ensembl ID |
|---|---|---|---|
| Upregulated Genes | |||
| CBFA2T3 |
4.970269827 |
0.001038075 |
ENSG00000129993 |
| FAM151A |
4.837332922 |
0.002008674 |
ENSG00000162391 |
| MUC20 |
4.702950855 |
0.011947675 |
ENSG00000176945 |
| STOX2 |
4.133641693 |
0.027737782 |
ENSG00000173320 |
| ARL14 |
4.133638802 |
0.027737782 |
ENSG00000179674 |
| BEAN1 |
3.364047225 |
0.004043429 |
ENSG00000166546 |
| WISP2 |
3.130060871 |
0.012964106 |
ENSG00000064205 |
| GPLD1 |
3.129996475 |
0.012964106 |
ENSG00000112293 |
| UBASH3A |
2.850826781 |
0.04000081 |
ENSG00000160185 |
| GOLGA8R |
2.850824538 |
0.04000081 |
ENSG00000186399 |
| Downregulated Genes | |||
| KIT |
−1.992297751 |
3.64E-30 |
ENSG00000157404 |
| PKD2L2 |
−2.016705824 |
0.026490386 |
ENSG00000078795 |
| LCP2 |
−2.553894739 |
0.000489648 |
ENSG00000043462 |
| TMPRSS9 |
−2.784724078 |
0.006155424 |
ENSG00000178297 |
| FOXG1 |
−2.834209583 |
0.039176409 |
ENSG00000176165 |
| CH25H |
−3.087262332 |
0.014984916 |
ENSG00000138135 |
| CCL8 |
−3.445451904 |
0.000244835 |
ENSG00000108700 |
| HIST1H3J |
−4.043009187 |
0.039661319 |
ENSG00000197153 |
| CCDC73 |
−4.252616175 |
0.022003857 |
ENSG00000186714 |
| SERPING1 | −4.25281999 | 0.022002453 | ENSG00000149131 |
Transcript fold change and adjusted p value were determined using the edgeR algorithm.
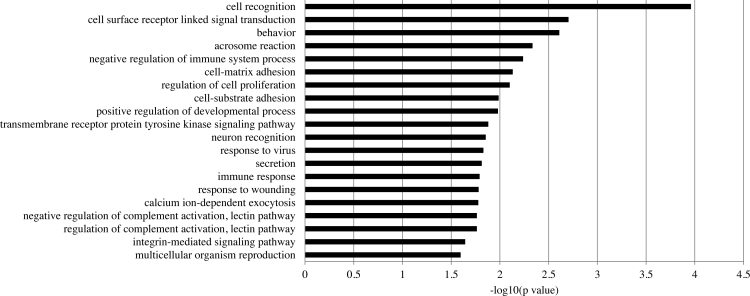
GO revealed 33 GO terms that were significantly enriched. Similar to TNFα treatment alone, TNFα treatment plus GSK0660 affected the regulation of cell proliferation and response to wounding (Figure 3). The terms also included reproduction, integrin-mediated signaling pathway, regulation of complement activation, and cell recognition. There was only one KEGG pathway enriched: the cytokine–cytokine receptor interaction (p = 0.01).
Figure 3.
Top 20 GO terms enriched in GSK0660-treated samples. The list of differentially expressed transcripts in TNFα plus GSK0660-treated HRMECs compared to TNFα alone was submitted to DAVID. Terms were considered significant at p<0.05.
Of the 273 genes differentially expressed by TNFα co-treatment with GSK0660, 91 were also differentially expressed by treatment with TNFα compared to the vehicle (Figure 4). GSK0660 exacerbated the TNFα-induced upregulation of eight genes (ATP2C2, GATA6, GCKR, LIF, SEMA3A, SERPINB2, TNFRSF18, and TSLP) and TNFα-induced the downregulation of 11 transcripts. GSK0660 counteracted the effects of TNFα on the remaining 72 transcripts. A KEGG pathway analysis of these 91 transcripts included the enrichment of the cytokine–cytokine receptor interaction (CCL8, CCL17, CXCL10, LIF, TSLP, and TNFRSF18) and chemokine signaling (CCL8, CCL17, CXCL10, and SHC3).
Figure 4.
Euler diagram of transcripts differentially expressed by TNFα and GSK0660. TNFα treatment resulted in the differential expression of 1,830 transcripts compared to the vehicle. The addition of GSK0660 regulated 91 of these transcripts. In addition, co-treatment of GSK0660 and TNFα resulted in the differential expression of 182 transcripts that were not affected by TNFα alone.
qRT-PCR validation
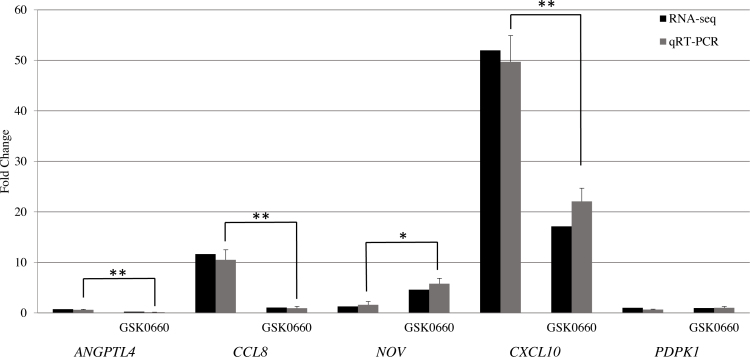
For validation of the RNA-seq, we confirmed the expressions of ANGPTL4, CCL8, NOV, CXCL10, and PDPK1 by qRT-PCR. ANGPTL4 is a well-known PPARβ/δ target and is known to be downregulated by GSK0660. CCL8 and CXCL10 are chemokines and their TNFα-induced expressions were blocked by GSK0660 in the RNA-seq. NOV was upregulated by GSK0660 in TNFα-treated HRMECs, but not by TNFα alone in the RNA-seq. In HRMECs, the TNFα increased the expressions of ANGPTL4, CCL8, and CXCL10, but it had no effect on NOV. GSK0660 reduced the expressions of ANGPTL4, CCL8, and CXCL10 in TNFα-treated cells and increased the expression of NOV. The PDPK1 expression was not affected by either treatment using RNA-seq or RT–PCR (Figure 5). Taken together, these data confirm gene expression changes seen in RNA-seq.
Figure 5.
qRT-PCR validation of RNA-seq targets. HRMECs were pre-treated with the vehicle or GSK0660 for 24 h, followed by stimulation with 1 ng/ml TNFα for 4 h. The mRNA expression was evaluated by RNA-seq and qRT-PCR. The fold change for RNA-seq was determined by the edgeR algorithm, while the fold change for qRT-PCR was determined by the comparative Ct method and it is relative to the β-actin expression levels. All fold changes are relative to HRMECs treated with the vehicle alone. Error bars indicate standard deviation for three samples in each group. *p = 0.0003, **p<0.0001.
Discussion
Using RNA-seq, this study confirms further the effect TNFα has on HRMECs, as well as the role the PPARβ/δ inverse agonist GSK0660 plays in TNFα-induced inflammation. Using RNA-seq over microarray allows for greater sensitivity and a broader range of fold changes. Additionally, the data generated by the RNA-seq can be probed for the discovery of novel transcripts or can be queried in the future after novel transcripts have been characterized. In our study, TNFα had an effect on several signaling pathways including the Jak-STAT pathway, Toll-like receptor pathway, and the complement cascade, replicating findings in other cell types [22-24]. TNFα also differentially expressed several genes involved in cytokine–cytokine signaling and chemokine signaling, suggesting the role of TNFα in retinal inflammation. Further evidence suggests this role may be tied to retinal leukostasis, as TNFα induced the upregulation of several leukocyte adhesion genes, such as VCAM1, ICAM1, and SELE, the gene encoding E-selectin. Besides adhesion protein genes, TNFα also upregulated several leukocyte-recruiting genes, including CCL8, CXCL10, CX3CL1, and CCL5. Taken together, these data suggest a role for TNFα-induced inflammation in retinal endothelial cells that is likely to contribute to leukostasis.
PPARβ/δ has only recently been studied in inflammation, with most work suggesting that PPARβ/δ agonism is anti-inflammatory. According to the studies published, the activation of PPARβ/δ prevented TNFα-induced inflammation in adipocytes, human umbilical vein endothelial cells, and proximal tubular cells [12,13,25,26]. In addition, PPARβ/δ agonists have been shown to be protective against inflammation in both hyperoxia-induced lung injuries and spinal cord injuries in rodents [27,28]. There has been little work done, however, on the PPARβ/δ antagonist and inverse agonist GSK0660, and those studies typically co-administer it with the agonist. Studies that have used GSK0660 by itself have showed its role thus far in reversing ginseng-enhanced cardiac contractility, worsening spinal cord injury in diabetic rats, reducing retinal vascular permeability, and inhibiting psoriasis-like skin disease [29-32]. As well, our laboratory has shown that GSK0660 inhibits retinal endothelial cell proliferation and oxygen-induced retinopathy [15]. While the anti-angiogenic properties of GSK0660 make it a possible therapeutic for DR, it was not known whether GSK0660 would exacerbate or protect against inflammation, another component of this disease.
The RNA-seq analysis revealed GSK0660 differentially regulated several transcripts in TNFα-treated HRMECs. These transcripts have possible diverse roles in cell proliferation, wound response, cell recognition, and calcium ion-dependent exocytosis. Importantly, the only pathway significantly enriched was the cytokine–cytokine receptor interaction. This finding becomes even more significant when the list of transcripts is limited to those both differentially expressed by TNFα compared to the vehicle and by TNFα plus GSK0660 treatment compared to TNFα treatment alone. GSK0660 prevents the TNFα-induced upregulation of CCL8, CCL17, and CXCL10. CCL8, which is also known as MCP-2, CCL17, and CXCL10 are chemokines that attract and activate leukocytes. Interestingly, these data suggest a possible role for GSK0660 in the prevention of TNFα-induced leukostasis, particularly related to chemokine recruitment. This result is unexpected, as the agonists of PPARβ/δ have been shown to inhibit TNFα-induced cell adhesion through the inhibition of the VCAM1 and ICAM1 expressions [12,13]. These results suggest that PPARβ/δ may have a contradictory effect on TNFα-induced leukostasis in that its activation inhibits cell adhesion through inhibition of the adhesion molecule expression, while the inhibition of PPARβ/δ prevents leukocyte recruitment.
TNFα is a potent inflammatory factor in HRMECs, with a role in leukocyte recruitment and adhesion to the vascular endothelium. Our RNA-seq revealed a novel role for GSK0660 in the regulation of the TNFα-dependent expression of cytokines known to be involved in leukostasis. GSK0660 may block some of the TNFα-dependent changes in expressions that facilitate leukocyte recruitment. Future studies will be aimed at investigating GSK0660 in this context.
Acknowledgments
This project was funded by grants from the National Eye Institute: R01EY07533 and P30EY08126, an unrestricted grant from Research to Prevent Blindness, and the Phyllis G. and William B. Snyder Endowed Chair.
Appendix 1. Summary of reads mapping to the human genome (UCSC hg19) using TopHat v2.0.9.
To access these data, click or select the words “Appendix 1.” Total reads for 3 samples each of vehicle, TNFα, and TNFα + GSK0660 treated HRMEC were generated using RNA-seq and then mapped to the human genome UCSC hg19.
Appendix 2. Transcripts differentially regulated by TNFα treatment in HRMEC.
To access these data, click or select the words “Appendix 2.” Transcript fold change and adjusted p value were determined using the edgeR algorithm and were considered to be significant with the adjusted p value <0.05 and a fold change ≥2.
Appendix 3. Transcripts differentially regulated by co-treatment of GSK0660 and TNFα treatment in HRMEC compared to TNFα treatment alone.
To access these data, click or select the words “Appendix 3.” Transcript fold change and adjusted p value were determined using the edgeR algorithm and were considered to be significant with the adjusted p value <0.05 and a fold change ≥2.
References
- 1.Saaddine JB, Honeycutt AA, Narayan KM, Zhang X, Klein R, Boyle JP. Projection of diabetic retinopathy and other major eye diseases among people with diabetes mellitus: United States, 2005–2050. Arch Ophthalmol. 2008;126:1740–7. doi: 10.1001/archopht.126.12.1740. [DOI] [PubMed] [Google Scholar]
- 2.Yuuki T, Kanda T, Kimura Y, Kotajima N, Tamura J, Kobayashi I, Kishi S. Inflammatory cytokines in vitreous fluid and serum of patients with diabetic vitreoretinopathy. J Diabetes Complications. 2001;15:257–9. doi: 10.1016/s1056-8727(01)00155-6. [DOI] [PubMed] [Google Scholar]
- 3.Tang J, Kern TS. Inflammation in diabetic retinopathy. Prog Retin Eye Res. 2011;30:343–58. doi: 10.1016/j.preteyeres.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zorena K, Myśliwska J, Myśliwiec M, Balcerska A, Hak Ł, Lipowski P, Raczyńska K. Serum TNF-alpha level predicts nonproliferative diabetic retinopathy in children. Mediators Inflamm 2007; 2007:92196. [DOI] [PMC free article] [PubMed]
- 5.Murugeswari P, Shukla D, Rajendran A, Kim R, Namperumalsamy P, Muthukkaruppan V. Proinflammatory cytokines and angiogenic and anti-angiogenic factors in vitreous of patients with proliferative diabetic retinopathy and eales' disease. Retina. 2008;28:817–24. doi: 10.1097/IAE.0b013e31816576d5. [DOI] [PubMed] [Google Scholar]
- 6.Huang H, Gandhi JK, Zhong X, Wei Y, Gong J, Duh EJ, Vinores SA. TNFalpha is required for late BRB breakdown in diabetic retinopathy, and its inhibition prevents leukostasis and protects vessels and neurons from apoptosis. Invest Ophthalmol Vis Sci. 2011;52:1336–44. doi: 10.1167/iovs.10-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joussen AM, Doehmen S, Le ML, Koizumi K, Radetzky S, Krohne TU, Poulaki V, Semkova I, Kociok N. TNF-alpha mediated apoptosis plays an important role in the development of early diabetic retinopathy and long-term histopathological alterations. Mol Vis. 2009;15:1418–28. [PMC free article] [PubMed] [Google Scholar]
- 8.Schröder S, Palinski W, Schmid-Schonbein GW. Activated monocytes and granulocytes, capillary nonperfusion, and neovascularization in diabetic retinopathy. Am J Pathol. 1991;139:81–100. [PMC free article] [PubMed] [Google Scholar]
- 9.Mackay F, Loetscher H, Stueber D, Gehr G, Lesslauer W. Tumor necrosis factor alpha (TNF-alpha)-induced cell adhesion to human endothelial cells is under dominant control of one TNF receptor type, TNF-R55. J Exp Med. 1993;177:1277–86. doi: 10.1084/jem.177.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Chi L, Stechschulte DJ, Dileepan KN. Histamine-induced production of interleukin-6 and interleukin-8 by human coronary artery endothelial cells is enhanced by endotoxin and tumor necrosis factor-alpha. Microvasc Res. 2001;61:253–62. doi: 10.1006/mvre.2001.2304. [DOI] [PubMed] [Google Scholar]
- 11.Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nat Rev Immunol. 2002;2:748–59. doi: 10.1038/nri912. [DOI] [PubMed] [Google Scholar]
- 12.Fan Y, Wang Y, Tang Z, Zhang H, Qin X, Zhu Y, Guan Y, Wang X, Staels B, Chien S, Wang N. Suppression of pro-inflammatory adhesion molecules by PPAR-delta in human vascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:315–21. doi: 10.1161/ATVBAHA.107.149815. [DOI] [PubMed] [Google Scholar]
- 13.Piqueras L, Sanz MJ, Perretti M, Morcillo E, Norling L, Mitchell JA, Li Y, Bishop-Bailey D. Activation of PPARbeta/delta inhibits leukocyte recruitment, cell adhesion molecule expression, and chemokine release. J Leukoc Biol. 2009;86:115–22. doi: 10.1189/jlb.0508284. [DOI] [PubMed] [Google Scholar]
- 14.Shearer BG Steger DJ, Way JM, Stanley TB, Lobe DC, Grillot DA, Iannone MA, Lazar MA, Willson TM, Billin AN. Identification and characterization of a selective peroxisome proliferator-activated receptor beta/delta (NR1C2) antagonist. Mol Endocrinol. 2008;22:523–9. doi: 10.1210/me.2007-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capozzi ME, McCollum GW, Savage SR, Penn JS. Peroxisome proliferator-activated receptor-beta/delta regulates angiogenic cell behaviors and oxygen-induced retinopathy. Invest Ophthalmol Vis Sci. 2013;54:4197–207. doi: 10.1167/iovs.13-11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Zhao S, Ye F, Sheng Q, Shyr Y. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. BioMed Research International. 2014;2014:248090. doi: 10.1155/2014/248090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang W Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miscia S, Marchisio M, Grilli A, Di Valerio V, Centurione L, Sabatino G, Garaci F, Zauli G, Bonvini E, Di Baldassarre A. Tumor necrosis factor alpha (TNF-alpha) activates Jak1/Stat3-Stat5B signaling through TNFR-1 in human B cells. Cell Growth Differ. 2002;13:13–8. [PubMed] [Google Scholar]
- 23.Syed MM, Phulwani NK, Kielian T. Tumor necrosis factor-alpha (TNF-alpha) regulates Toll-like receptor 2 (TLR2) expression in microglia. J Neurochem. 2007;103:1461–71. doi: 10.1111/j.1471-4159.2007.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheerin NS, Zhou W, Adler S, Sacks SH. TNF-alpha regulation of C3 gene expression and protein biosynthesis in rat glomerular endothelial cells. Kidney Int. 1997;51:703–10. doi: 10.1038/ki.1997.101. [DOI] [PubMed] [Google Scholar]
- 25.Serrano-Marco L, Chacón MR, Maymó-Masip E, Barroso E, Salvadó L, Wabitsch M, Garrido-Sánchez L, Tinahones FJ, Palomer X, Vendrell J, Vázquez-Carrera M. TNF-alpha inhibits PPARbeta/delta activity and SIRT1 expression through NF-kappaB in human adipocytes. Biochim Biophys Acta. 2012;1821:1177–85. doi: 10.1016/j.bbalip.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Kume S, Tanaka Y, Isshiki K, Araki S, Chin-Kanasaki M, Sugimoto T, Koya D, Haneda M, Sugaya T, Li D, Han P, Nishio Y, Kashiwagi A, Maegawa H, Uzu T. GW501516, a PPARdelta agonist, ameliorates tubulointerstitial inflammation in proteinuric kidney disease via inhibition of TAK1-NFkappaB pathway in mice. PLoS ONE. 2011;6:e25271. doi: 10.1371/journal.pone.0025271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao XC, Fang YQ, You P, Zhang S, Ma J. Protective role of peroxisome proliferator-activated receptor beta/delta in acute lung injury induced by prolonged hyperbaric hyperoxia in rats. Respir Physiol Neurobiol. 2014;199:9–18. doi: 10.1016/j.resp.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Paterniti I, Esposito E, Mazzon E, Galuppo M, Di Paola R, Bramanti P, Kapoor A, Thiemermann C, Cuzzocrea S. Evidence for the role of peroxisome proliferator-activated receptor-beta/delta in the development of spinal cord injury. J Pharmacol Exp Ther. 2010;333:465–77. doi: 10.1124/jpet.110.165605. [DOI] [PubMed] [Google Scholar]
- 29.Lin JW, Cherng YG, Chen LJ, Niu HS, Chang CK, Niu CS. Ginseng is useful to enhance cardiac contractility in animals. BioMed Research International. 2014;2014:723084. doi: 10.1155/2014/723084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai CC, Lee KS, Chen SH, Chen LJ, Liu KF, Cheng JT. Decrease of PPARdelta in Type-1-Like Diabetic Rat for Higher Mortality after Spinal Cord Injury. PPAR Res. 2014;2014:456386. doi: 10.1155/2014/456386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suarez S, McCollum GW, Bretz CA, Yang R, Capozzi ME, Penn JS. Modulation of VEGF-Induced Retinal Vascular Permeability by Peroxisome Proliferator-Activated Receptor-beta/delta. Invest Ophthalmol Vis Sci. 2014;55:8232–40. doi: 10.1167/iovs.14-14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hack K, Reilly L, Palmer C, Read KD, Norval S, Kime R, Booth K, Foerster J. Skin-targeted inhibition of PPAR beta/delta by selective antagonists to treat PPAR beta/delta-mediated psoriasis-like skin disease in vivo. PLoS ONE. 2012;7:e37097. doi: 10.1371/journal.pone.0037097. [DOI] [PMC free article] [PubMed] [Google Scholar]