Abstract
Motivation: Although gene-expression signature-based biomarkers are often developed for clinical diagnosis, many promising signatures fail to replicate during validation. One major challenge is that biological samples used to generate and validate the signature are often from heterogeneous biological contexts—controlled or in vitro samples may be used to generate the signature, but patient samples may be used for validation. In addition, systematic technical biases from multiple genome-profiling platforms often mask true biological variation. Addressing such challenges will enable us to better elucidate disease mechanisms and provide improved guidance for personalized therapeutics.
Results: Here, we present a pathway profiling toolkit, Adaptive Signature Selection and InteGratioN (ASSIGN), which enables robust and context-specific pathway analyses by efficiently capturing pathway activity in heterogeneous sets of samples and across profiling technologies. The ASSIGN framework is based on a flexible Bayesian factor analysis approach that allows for simultaneous profiling of multiple correlated pathways and for the adaptation of pathway signatures into specific disease. We demonstrate the robustness and versatility of ASSIGN in estimating pathway activity in simulated data, cell lines perturbed pathways and in primary tissues samples including The Cancer Genome Atlas breast carcinoma samples and liver samples exposed to genotoxic carcinogens.
Availability and implementation: Software for our approach is available for download at: http://www.bioconductor.org/packages/release/bioc/html/ASSIGN.html and https://github.com/wevanjohnson/ASSIGN.
Contact: andreab@genetics.utah.edu or wej@bu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Since the advent of high-throughput genomic profiling technologies such as gene expression microarrays and RNA-Seq, many computational and statistical methods have been developed to derive gene expression signatures for disease diagnosis, prognosis and treatment decisions (Golub et al., 1999; Saeys et al., 2007; van de Vijver et al., 2002). Gene expression signatures are often used as surrogate representations of pathway activation or deactivation. The use of expression signatures to quantify pathway activation level has been particularly important for dissecting the complexity of diseases and providing guidelines of targeted therapeutics. To date, gene expression-based pathway analyses mainly face two sources of challenges: (i) limited pathway annotations in curated databases and (ii) ineffective analysis tools.
In reference to the first limitation, many public databases (Ashburner et al., 2000; Kanehisa et al., 2014; Liberzon et al., 2011) provide manually curated pathways that associate genes lists with pathway activity. However, genes in those predefined pathways are not always associated with gene expression changes that differ between disease states. For example, some genes in an annotated pathway might be activated through changes in phosphorylation or protein interaction status. Thus, pathway analysis approaches that use patient gene expression profiles without careful selection for expression-based signature genes with transcriptional change may lead to incorrect results. An alternative way to infer pathway activity is by experimentally perturbing the pathway of interest in controlled settings and projecting the associated molecular signature (e.g. changes in gene expression) onto patient or other target samples to estimate pathway activity levels (Bild et al., 2006; Gustafson et al., 2010; Sweet-Cordero et al., 2005). For example, previous efforts have generated gene expression signatures for growth factor signaling pathways in human primary cells and then used the signatures to predict disease prognosis and drug sensitivity in human cancer cohorts (Bild et al., 2006). Although, these pathway-profiling approaches have been previously shown to generate empirical gene expression-based pathway response signatures, the assumption of homogeneity between in vitro (e.g. perturbation samples) and in vivo (e.g. patient) biological conditions does not always hold due to platform, tissue or disease deregulation status variations.
In effort to address the second concern, factor analysis approaches have been used to identify latent factors (metagenes) associated with pathways and clinical outcome (Bazot et al., 2013; Bhattacharya and Dunson, 2011; West, 2003). However, it is often difficult to interpret the biological meaning of the latent factors identified by these unsupervised approaches or to estimate the absolute activation level for pathways of interest. Supervised classification approaches (Pirooznia et al., 2008) often model pathways one at a time without accounting for pathway correlation or interaction between related pathways. Moreover, supervised classification approaches require expression data from pathway perturbation experiments for building up models, thus often fail to work when only pathway gene lists are available. So far, none of these existing approaches adequately account for tissue, disease or context specificity in assessing gene expression signatures regulated via pathway activation or deactivation. Furthermore, none of them are designed to profile genomic signatures across multiple genomic profiling platforms.
To overcome these limitations, we propose a novel and flexible pathway profiling toolkit called Adaptive Signature Selection and InteGratioN (ASSIGN). ASSIGN relies on a sparse Bayesian factor analysis method to estimate the activation status of pathways under investigation, such as oncogenic pathways, immune response pathways or drug response pathways in individual samples of a genomic dataset for predicting optimal treatment prior to any medication on patients. Here, we use multiple simulated and real datasets to demonstrate the validity and robustness of ASSIGN in estimating pathway activation. In simulated data, the model correctly adapts the pathway signature gene lists in specific biological contexts by excluding irrelevant genes or including relevant genes into signatures. We used five previously published oncogenic signaling pathway signatures to demonstrate the advantages of modeling multiple pathways in concert to account for crosstalk among the pathways. We also used the tumor samples from The Cancer Genome Atlas (TCGA) to show that ASSIGN can robustly combine in vitro signatures generated using one profiling platform with tumor samples profiled using a different platform. Finally, we used profiling data generated from liver tissues exposed to genotoxic hepatocarcinogens to demonstrate the versatility of ASSIGN in identifying and adapting signatures from pre-curated pathway gene lists. Overall, ASSIGN uses a semi-supervised approach that results in more biologically interpretable pathway activation profiles that are adapted to specific tissues or disease contexts, as opposed to more rigid and less interpretable profiles generated by previous approaches. Although, ASSIGN was initially designed for pathway-based analysis from gene expression data, it can easily be extended to other profiling data types such as DNA variation or methylation data.
2 Approach
We define a ‘signature’ as a set of representative genes whose expression changes due to differences in disease status, exposure to a chemical compound/drug or differential regulation of key pathway genes. The signature can also optionally contain the absolute direction changes or expression magnitude changes due to an experimental perturbation. ASSIGN is a pathway analysis toolkit with the flexibility to accommodate profiling analysis needs for a large number of pathways or perturbation profiling scenarios. ASSIGN allows the user the option of choosing either Bayesian regression (signatures known) or factor analysis (signatures unknown) and accommodates multiple signatures simultaneously within a set of samples. Key innovations in ASSIGN allow for broad applicability of the method (Table 1), whereas other existing approaches lack one or more of these critical features. The specific advantages of ASSIGN are described below.
Table 1.
Comparison of ASSIGN with existing pathway-profiling methods
| GSEA | ssGSEA | BFRM (Binary regression) | BFRM (Factor analysis) | FacPad | ASSIGN | |
|---|---|---|---|---|---|---|
| Software input | ||||||
| Predefined gene list | x | x | x | x | ||
| Magnitude changes | x | x | ||||
| Perturbation expression profiling data | x | x | ||||
| Advanced model features | ||||||
| Multiple signatures | x | x | x | |||
| Context-specific background | x | |||||
| Context-specific signature | x | x | x | |||
| Pathway activity regularization | x | x | ||||
| Method output | ||||||
| Biologically interpretable pathways | x | x | x | x | x | |
| Pathway activity estimates | x | x | x | x | x | |
| Pathway significance estimates | x | x | x | |||
ASSIGN offers a more comprehensive set of features compared with other existing approaches.
2.1 Simultaneous profiling of multiple pathways
ASSIGN can account for pathways simultaneously, compared with other approaches that only consider a single pathway at a time [GSEA (Subramanian et al., 2005), ssGSEA (Barbie et al., 2009), BFRM (West, 2003)]. This feature accounts for ‘cross-talk’ between pathway components by directly modeling correlations and interactions in the pathway signature components that might reduce detection sensitivity and specificity.
2.2 Context specificity in baseline gene expression
Baseline gene expression levels (i.e. expression level when a pathway is inactive) may vary widely due to differences in tissue types or disease status, or across different measurement platforms and can contribute to heterogeneity between in vitro perturbation samples and patient samples. ASSIGN can adaptively estimate background gene expression levels across a set of samples, giving it the unique ability to estimate absolute pathway activity levels or drug efficacy in clinical samples before the samples have received a treatment, even when the signature was generated using a different profiling platform.
2.3 Context specific signature estimation
Many existing signature-based profiling approaches require input signatures in the form of a gene list [GSEA, FacPad (Ma and Zhao, 2012)] or a gene list with static expression magnitude changes (BFRM). While BFRM provides a direct and supervised approach for pathway profiling, it requires the signature to be generated in the same biological context as the patient samples. FacPad allows for the adaptation of signature profiles, but cannot integrate magnitude change information. In addition, FacPad is highly impacted by outliers in the dataset and often suffers from the lack of identifiability of the direction of the signature magnitude. ASSIGN provides the flexibility to use either a signature-based or gene list-based approach and can also use input magnitudes as prior information, thus providing a compromise that allows for adaptive signature refinement while reducing signature over-fitting and direction ambiguity.
2.4 Regularization of signature strength estimates
ASSIGN regularizes signature strength estimates using Bayesian ridge regression (Hsaing, 1975), which ‘shrinks’ signature strength estimates toward zero, especially for signatures with a weak presence or anecdotal correlations in the sample. In addition, ridge regression has well-established benefits in handling correlated covariates (Hsiang, 1975), thus making it advantageous for the simultaneous modeling of correlated signatures.
3 Methods
3.1 Formal definition of ASSIGN model
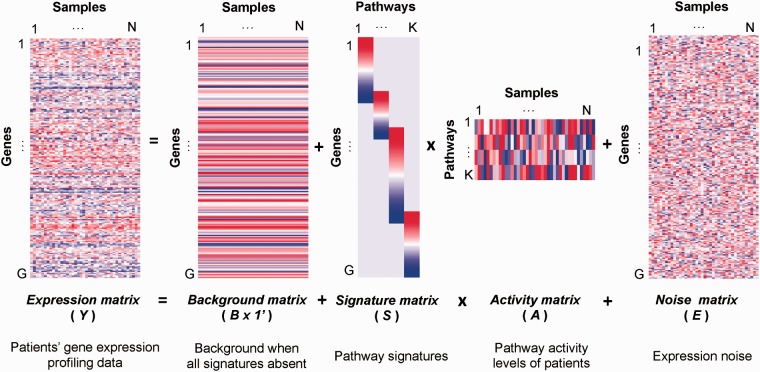
To define the model formally, suppose a gene expression assay profiles G genes on N patient samples of a certain disease type, and let Y be a G × N matrix of observed expression values. Each entry in Y is a gene expression value after data normalization. We apply a Bayesian sparse factor model to decompose the Y matrix as:
| (1) |
Each column of Y represents all the genes for one patient sample. We model the measured expression values of each patient sample in a vector form:where for . Figure 1 contains a visual representation of the ASSIGN model.
Fig. 1.
Visual representation of ASSIGN model
B is a G-vector of the baseline gene expression levels for all genes. We define the prior distribution of B as. The prior parameters and can be set as non-informative or informative from control samples in a pathway perturbation experiment.
Matrix S is the G × K factor loading matrix, with each column representing the gene expression signature of a specific biological pathway. In whole-genome expression profiling, we expect that the majority of genes will not show differential expression in association with any particular factor, and each individual factor will be associated with only a few genes. Thus, the columns k of S will be sparse. The hierarchical spike-and-slab prior distribution of S is: where . is a Bernoulli-distributed binary indicator for ( = 0: the gene is excluded from the signature; = 1: the gene is included in the signature). is sampled with probability . has a diffuse prior ( = 1) when = 1, and a highly precise prior ( = 0.1) when = 0. The choice of prior depends on the prior information of pathway signatures (see Section 3.2 for details).
Matrix A is the K × N factor score (pathway activity) matrix, with each column A.,j representing activation scores of the K pathways for each individual patient sample. Since tumors often rely on the activation of one or two pathways, such as via an ‘oncogene addiction’ (Weinstein, 2002), not all of the K pathways will necessarily be activated in all the individual patient samples. Therefore, any column of A will likely be sparse. Thus, we model the matrix A using a hierarchical spike-and-slab prior similar to the formulation for S. To overcome the ‘sign-flipping’ phenomenon (e.g. non-identifiability) that commonly occurs in factor analysis, we used a truncated normal distribution (0, 1 range) in a modified slab normal prior:
leading to better interpretability of absolute pathway activation levels. In this prior, Φ is the cumulative function of the standard normal distribution, is a binary variable, indicating whether is zero. The binary indicator is assumed to follow a Bernoulli distribution, with, where is a useful parameter that estimates the probability that is assigned to the slab distribution (i.e. the pathway is activated).
Matrix E is the G × N noise matrix. Each column of E follows a multivariate normal distribution with mean 0 and a G × G diagonal covariance matrix Σ. The precision of the gth gene is assumed to follow a Gamma prior with shape parameter u and rate parameter v. In practice, we use non-informative priors for u and v.
Based on the settings of prior distributions above and prior parameters (Supplementary Table S1), we computed the full conditional posterior distribution for each parameter in the model. Since the prior distributions we use are conjugate for the likelihood functions; the parameters in B, A, S and E were jointly approximated using Gibbs sampling. In practice, we observed that 2000 iterations are sufficient for convergence. To calculate the posterior mean of the parameters, we ran 2000 iterations until the MCMC chain converged and discarded the first half of values in the chain. Typical computational time is 80 seconds for a dataset with 22 000 genes profiled on 100 patient samples.
3.2 Signature gene selection
For gene selection, to set the priors of and for the matrix S, we apply a Bayesian variable selection approach (George and Mcculloch, 1997) to compute prior weights and signal strengths of genes from the gene expression profiles and use the genes with the highest signal strengths and weights when estimating pathway activation status. Briefly, we define to be the expression measurement for gene g of pathway k, and sample j. We assume + , where is an indicator variable denoting perturbation status for sample j (control = 0; treatment = 1), is the gene-specific background expression level of pathway k, is the change in expression due to treatment status, and is a Gaussian error term. We place a mixture prior on , i.e. | ∼ (1−)N(0, ) +N(0, ), where has a diffuse prior ( = 1) when = 1, and a highly precise prior ( = 0.1) when = 0, where is an indicator variable that is equal to 1 when the gene is differentially expressed in the perturbation experiment and 0 otherwise. We assume a priori that it follows a Bernoulli(π) distribution, where π = 0.01. Genes are ranked by the posterior mean of (expression change) and the posterior probability of (statistical significance), and the top 50–200 genes are selected for the signature (number specified by user), where is set to ; is set to . ASSIGN concatenates a set of genes involved in at least one pathway signature for further profiling.
3.3 Gene expression profiling data
3.3.1 Novel pathway data
To experimentally validate the method, we transfected human primary mammary epithelial cells in vitro using a recombinant adenovirus that expresses EGFR, MEK and EGFR+MEK (both pathways active) as described previously (Bild et al., 2006). After transfection, total RNA was extracted and purified using a Qiagen RNeasy kit and sequencing libraries were generated using Illumina TruSeq stranded mRNA sample preparation kits. The Illumina Hi-Seq 2000 platform was used to produce single-end reads (50 bp in length). These data have been deposited in the GEO database under accession number GSE59765. The EGFR and MEK pathway RNA-Seq data were combined with PI3K microarray data (described below) for multi-pathway profiling to validate the cross-platform feature of ASSIGN.
3.3.2 Existing pathway data
We used data for five oncogenic pathways (β-catenin, E2F3, MYC, RAS, SRC) (GEO accession: GSE3151) profiled on Affymetrix HG-U133 Plus 2.0 arrays and PI3K pathway perturbation data (GSE12815) profiled on HG-U133A microarrays. We also obtained mRNA expression data for liver tissues from the DrugMatrix dataset (Ganter et al., 2005; GSE57822) profiled on Affymetrix Rat 230 2.0 arrays. Each sample corresponds to rat-liver tissue treated with a different carcinogenic compound, annotated using the Carcinogenic Potency Database (CPDB) (Fitzpatrick, 2008). RNA-sequencing counts summarized at the gene level for TCGA breast carcinomas and tumor adjacent normal tissues were also downloaded from the TCGA data portal. Details of all the datasets used in this study are listed in Supplementary Table S2.
3.4 Data preprocessing and normalization
For the cell line perturbation experiments, the microarray mRNA expression profiles were preprocessed and normalized using SCAN.UPC (Piccolo et al., 2012, 2013). The rat liver data were normalized using fRMA (McCall et al., 2010). The RNA-Seq cell-line data were aligned to the reference genome using TopHat v2.0.6 (Trapnell et al., 2009), then quantified to FPKM for each gene/transcript using Cufflinks v2.0.2 (Trapnell et al., 2010). For the RNA-seq data from TCGA, Level 3 summarized values were used (Cancer Genome Atlas Network, 2012); reads were mapped to the reference genome using MapSplice v12_07 (Wang et al., 2010), read counts were estimated using RSEM v1.1.13 (Li and Dewey, 2011) and RSEM read counts are normalized using upper-quantile normalization. In these analyses, we intensionally used data that were preprocessed using different normalization procedures to demonstrate the robustness of ASSIGN’s adaptive profiling approach. PCA was conducted on the expression profiles to validate that there were no batch effects. Finally, to evaluate whether batch/platform effects existed in the TCGA data, we plotted the first two principal components and observed no obvious clustering by batch/platform (Supplementary Fig. S1A). As expected, a clear separation of tumor and normal tissues were observed on the PCA plot (Supplementary Fig. S1B). To integrate gene expression data across experiments and platforms, we applied ComBat (Johnson et al., 2007) in two steps, first to adjust for batch effects within the perturbation experiments and second to adjust for batch effects across the signature and patient (e.g. TCGA) datasets.
3.5 Simulation data
We used simulation to generate expression data for four pathways (25 samples with one of the four pathway perturbed) and 1000 genes (250 genes per pathway). The values of elements in the B vector were independently simulated from a Uniform (0, 1) distribution. The values for significant genes in each pathway of S matrix were simulated independently from a Normal (0, 1) distribution with a constraint on absolute values greater than 1, and simulated insignificant genes in the S matrix independently from Normal (0, 0.01). The noise term, E, for each gene was simulated from Normal (0, 0.5). Y is the summation of B, SA and E. We used this simulation to evaluate ASSIGN under three scenarios: (i) an ideal scenario where the predetermined pathway signature exactly represents the pathway signature in a disease environment, (ii) signature perturbation experiments that do not fully represent the pathway signatures in a disease environment and (iii) when one or more of the pathways are deregulated, thus requiring significant adaptation of the gene list, signature magnitudes and background expression profile. Detailed descriptions of data generation and the results are given in the Supplementary Materials.
3.6 Software implementation and application
ASSIGN is available as a Bioconductor package, written in the R programming language and is freely available for download at http://www.bioconductor.org/packages/release/bioc/html/ASSIGN.html. As input, ASSIGN requires gene expression data from patient/test samples, and a signature perturbation dataset or signature gene list. When perturbation data are given, ASSIGN automatically generates pathway signatures based on the raw gene expression data from one or more perturbations. When signature perturbation datasets are unavailable, the user can provide predetermined signature gene lists (e.g. from public databases, prior differential expression experiments). ASSIGN outputs a matrix of signature strengths for each sample and the prior/posterior signature gene lists and magnitude changes. The software also provides the user with output from a complete internal cross-validation on the perturbation data, MCMC posterior convergence diagnostics and an evaluation of classification accuracy when patient labels are provided by the user. The user can specify model parameters/features such as background adaptation, signature adaptation and regularization of signature strength. The model specification options for the analyses in this study are listed in Supplementary Table S3.
4 Results
To overcome challenges from pathway ‘cross-talk’ and heterogeneity from biological and technical sources, we developed the ASSIGN toolkit that allows for flexible profiling of multiple correlated signatures into specific disease, tissue and patient contexts. Here, we demonstrate the features of ASSIGN using simulation, cross validation and several publicly available genomic datasets. In Section 4.1, we use three simulated scenarios to evaluate the model’s abilities to estimate pathway-activation status and filter irrelevant genes. In Sections 4.2 and 4.3, we illustrate ASSIGN’s ability to account for context-specific background levels and to crosstalk among multiple pathways. In Section 4.4, we evaluate the effectiveness of ASSIGN to overcome cross-tissue and cross-platform obstacles to estimates pathway activity in a large breast carcinoma dataset. In Section 4.5, we adapt curated signatures of DNA damage response pathways to estimate pathway signature strength in liver profiling samples. In these sections we compare ASSIGN in multiple contexts with existing methods such as GSEA, ssGSEA, BFRM and FacPad and demonstrate a general advantage of ASSIGN over these existing approaches.
4.1 Simulation studies
We conducted a simulation study to evaluate the performance of ASSIGN under three scenarios to test the ability of ASSIGN to effectively estimate background, signature and activity profiles. Details regarding data generation for each scenario are given in Supplementary Materials. In the first simulation scenario, we evaluated ASSIGN’s ability to estimate a pathway’s activity when pathway signatures are known a priori. ASSIGN accurately estimated the activation level of the pathways (Supplementary Table S4A). In the second simulation scenario, we attempted to estimate signatures obtained from pathway perturbation experiments that require context-specific adaptation. ASSIGN was able to closely estimate the posterior mean of the activation levels and accurately estimate the correct posterior means of the background and the signature (Supplementary Table S4B). Here, we observed that 91% of the insignificant genes and 98% of the significant genes were respectively dropped from or added to the posterior (Supplementary Fig. S2). In the third simulated scenario, we showed that ASSIGN was capable of detecting more than one activated pathway (Supplementary Table S4C). Furthermore, we discovered that knowledge of the regulation status of only 10 genes out of 250 total significant genes was sufficient to overcome the sign-flipping issue and correctly estimate a pathway activation status.
4.2 Profiling of interconnected oncogenic pathways
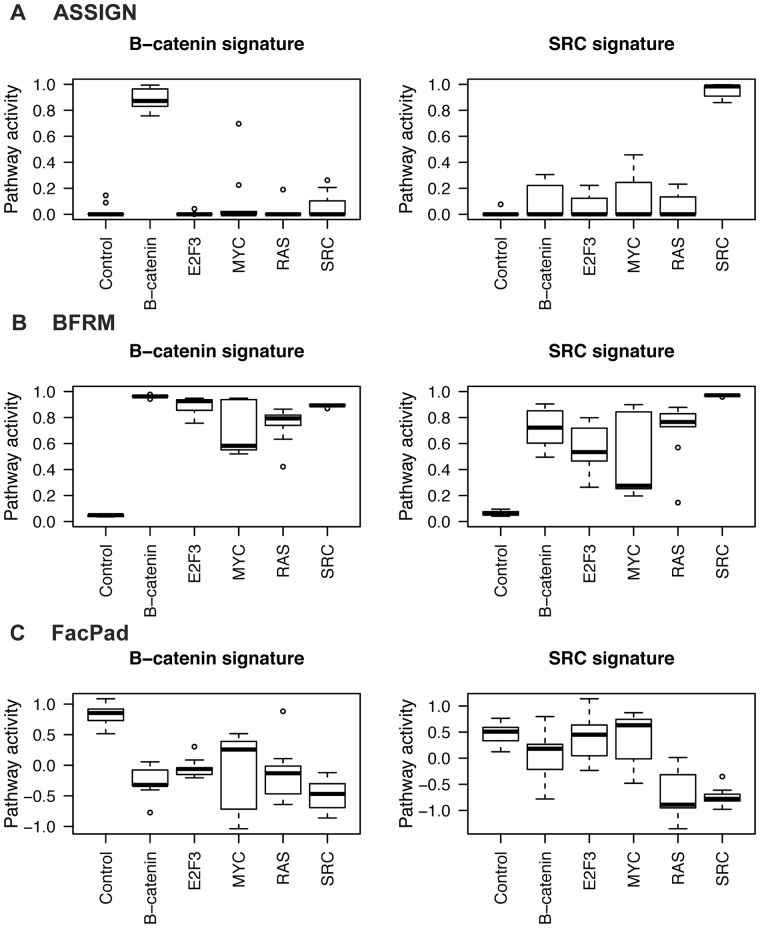
Many pathway analysis methods use a single-pathway approach where the pathways are profiled independently. However, because pathways interact with each other as part of complex biological systems, analyzing multiple pathways simultaneously provides better insight into pathway function and activity. We validated our multiple-pathway-based model by predicting activity of five previously published oncogenic pathways (β-catenin, E2F3, MYC, RAS, SRC) in human cell lines (Bild et al., 2006). In these signatures, about 17% of the genes exhibit significant expression changes in more than one pathway and also exhibit high correlation across the pathway gene expression signatures (Supplementary Table S5). We used ASSIGN to estimate pathway activity profiles for all five pathway sets via cross validation. ASSIGN consistently predicted pathway activity profiles accurately in all of these samples (Fig. 2A). In contrast, the single-pathway BFRM approach (West, 2003) and FacPad incorrectly estimated pathway activity profiles for four of the five pathways (Fig. 2B, C). Consequently, the false-positive pathway activation profiles from these approaches could interfere with clinical decisions for selecting the appropriate targeted therapies for cancer patients.
Fig. 2.
Oncogenic pathway activity prediction via cross-validation. Predicted pathway activity for (A) ASSIGN, (B) BFRM and (C) FacPad. Activation levels of two oncogenic pathways (Bcat, Src) were estimated for cell lines with one of five pathways activated (β-catenin, E2F3, MYC, RAS, SRC). The ASSIGN and BFRM values range between zero (inactive pathway) and one (active pathway). FacPad was designed for relative pathway activation comparisons and activation levels can range from negative infinity to infinity
4.3 Adapting background levels across heterogeneous samples
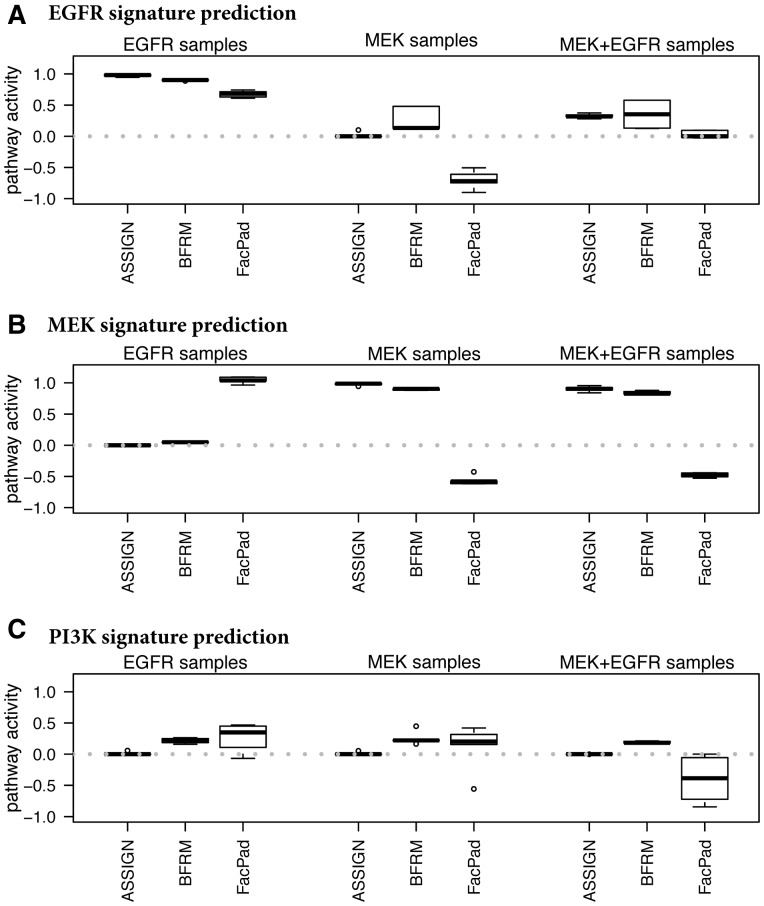
To further evaluate the importance of correcting for context-specific baseline expression levels, we estimated pathway activity for the EGFR and MEK co-activated RNA-Seq samples using the EGFR and MEK pathway signatures profiled using RNA-Seq. We also included a previously published PI3K signature that was generated in a different cell type (lung epithelial cells compared with mammary epithelial cells) using a microarray profiling technology. To validate the adaptive background feature of ASSIGN, we compared three ASSIGN model settings: (i) background (i.e. expression levels when no pathways are active) fixed to the observed values in the control samples of the EGFR/MEK pathway coactivated experiment; (ii) background fixed to the value in the control samples of the PI3K activation experiment; (iii) background fixed as in (ii) but allowing for ASSIGN background estimation. We observed that the pathway activation level was correctly estimated in model (i), which included the correct background and (iii) with the ASSIGN adapted background, but not in (ii) with a non-adaptive incorrect background (Supplementary Fig. S3). The posterior mean of B estimated in model (iii) converged almost exactly to the true values (Cor. = 0.99), whereas the background values used in model (ii) deviate from the true values (Cor. = 0.60). Thus, the ASSIGN model (iii) with adaptive background correctly estimates EGFR and MEK pathway activity in EGFR and MEK co-activated samples even when the background is unknown (Fig. 3). In these samples, we observed that the EGFR signature is strong in the EGFR-only samples and the MEK signature is strong in the MEK-only samples. Both EGFR and MEK are upregulated in the EGFR+MEK samples, with EGFR signal being overall lower, potentially due to stronger negative feedback on the pathway with concurrent activation of EGFR and MEK (Avraham and Yarden, 2011; Klinger et al., 2013). For the sake of comparison across methods, we applied the FacPad and BFRM methods to these scenarios. FacPad requires a baseline level for each sample and takes the ratio of treated samples and control samples as input. When true baseline information of the EGFR and MEK coactivated samples was not available, FacPad failed to estimate the correct pathway activation level (Fig. 3). BFRM correctly estimated the EGFR and MEK pathways in the EGFR and MEK coactivated samples when the background in the patient samples perfectly matched the training samples, albeit slightly less significantly than ASSIGN. However, BFRM does not adjust for the background expression level across platforms, and thus estimated elevated PI3K levels in the EGFR and MEK samples (Fig. 3).
Fig. 3.
Pathway activity prediction using cross-platform generated pathway signatures. Comparison of ASSIGN, BFRM and FacPad predicted EGFR (A), MEK (B) and PI3K (C) pathway activity in EGFR, MEK and EGFR+MEK activated RNA-Seq samples. EGFR and MEK pathway signatures were profiled via RNA-Seq, whereas PI3K pathway signature was profiled via microarray. ASSIGN detected two pathways (EGFR and MEK) activated at the same time in the EGFR+MEK samples and correctly predicted that the PI3K pathway was inactive, whereas BFRM and FacPad estimated PI3K pathway activation incorrectly. FacPad also estimated active pathways as inactive and inactive pathways as active (so called ‘sign-flipping’. See Section 3)
4.4 Cross-platform and cross-tissue pathway profiling
We examined activity levels for our RNA-seq based EGFR and MEK pathways combined with a previously published PI3K signature generated on a different cell type and on a microarray profiling technology. We used ASSIGN to estimate pathway activation status in RNA-seq data from breast carcinomas and matched adjacent normal breast samples from TCGA. In addition, we compared pathway activation in the breast carcinomas based on four molecular subtypes: basal-like, luminal A, luminal B and Her2 (Supplementary Fig. S4). For all three pathways, ASSIGN consistently found known pathway-molecular subtype relationships confirmed by other studies and outperformed BFRM and FacPad (Cheang et al., 2008; Hoeflich et al., 2009; López-Knowles et al., 2010; Moestue et al., 2013). All approaches estimated significantly higher EGFR activity in tumor samples in general as well as in all four subtypes of breast cancer compared with normal tissue (Table 2A). ASSIGN correctly predicted MEK activity to be higher in the basal-like subtype and PI3K activity to be higher both in basal-like and Her2 subtype than normal tissues (Table 2B, C). BFRM failed to recognize higher MEK activity and higher PI3K activity in basal-like subtypes (Table 2B, C). FacPad incorrectly predicted MEK activities to be significantly lower than normal tissue (Table 2B; Supplementary Fig. S4).
Table 2.
Comparison of predicted pathway activity in breast carcinoma and adjacent normal tissue by ASSIGN, BFRM and FacPad
| A. EGFR pathway | |||
|---|---|---|---|
| ASSIGN Mean diff (P-values) | BFRM Mean diff (P-values) | FacPad Mean diff (P-values) | |
| Tumor versus Normal | 0.20 (<0.001) | 0.13 (<0.001) | 1.81 (<0.001) |
| Basal versus Normal | 0.28 (<0.001) | 0.13 (<0.001) | 1.68 (<0.001) |
| Her2 versus Normal | 0.23 (<0.001) | 0.11 (<0.001) | 1.54 (<0.001) |
| Luminal A versus Normal | 0.09 (<0.001) | 0.07 (<0.001) | 0.98 (<0.001) |
| Luminal B versus Normal | 0.20 (<0.001) | 0.22 (<0.001) | 1.81 (< 0.001) |
|
B. MEK pathway | |||
|---|---|---|---|
| ASSIGN Mean diff (P-values) | BFRM Mean diff (P-values) | FacPad Mean diff (P-values) | |
| Tumor versus Normal | −0.02 (0.695) | −0.00 (0.426) | 0.92 (<0.001) |
| Basal versus Normal | 0.04 (0.009) | 0.00 (0.308) | −0.64 (<0.001) |
| Her2 versus Normal | 0.03 (0.069) | −0.00 (0.518) | 0.53 (<0.001) |
| Luminal A versus Normal | −0.01 (0.703) | −0.00 (0.613) | 0.91 (<0.001) |
| Luminal B versus Normal | −0.02 (0.089) | −0.00 (0.103) | 0.92 (<0.001) |
|
C. PI3K pathway | |||
|---|---|---|---|
| ASSIGN Mean diff (P-values) | BFRM Mean diff (P-values) | FacPad Mean diff (P-values) | |
| Tumor versus Normal | 0.02 (0.013) | −0.02 (0.219) | 0.23 (<0.001) |
| Basal versus Normal | 0.12 (<0.001) | 0.01 (0.178) | 1.06 (<0.001) |
| Her2 versus Normal | 0.06 (<0.001) | −0.03 (0.028) | 0.85 (<0.001) |
| Luminal A versus Normal | 0.00 (0.763) | −0.02 (0.101) | 0.25 (0.049) |
| Luminal B versus Normal | 0.02 (0.094) | −0.02 (0.115) | 0.30 (0.033) |
Pathway activity comparison between breast carcinoma and normal tissues, and breast carcinoma subtypes (Basal, Her2, Luminal A, Luminal B) and normal tissues using two-sample t-test. P-values of t-tests are listed in the table.
4.5 Context-specific signature predictions in individual samples
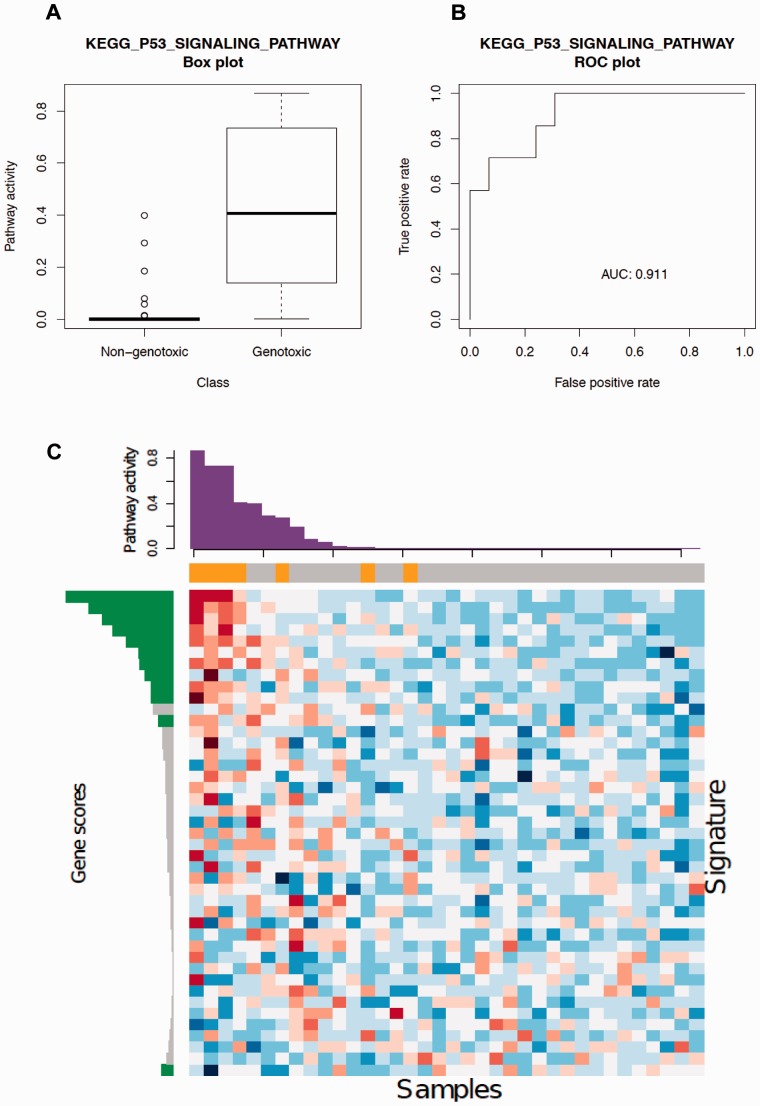
To evaluate ASSIGN’s signature adaptation features and single sample prediction abilities, we investigated pathway activation status in liver samples from Rattus norvegicus exposed to genotoxic or non-genotoxic carcinogens. We estimated how well we could use curated pathway signatures from existing databases to predict genotoxicity of the carcinogenic compounds. For validation purpose, we used the outcome of an Ames Salmonella test as a proxy for genotoxicity (Mortelmans and Zeiger, 2000) available through CPDB (Fitzpatrick, 2008) for the carcinogenic compounds under consideration. In this study, we focused on the association of the activity level of DNA damage response/repair pathways with genotoxic carcinogen exposures. Among DNA damage response/repair pathways from the MSigDB database, 9 pathways were identified as differentially activated between the two groups (genotoxic versus non-genotoxic) by at least two of four approaches: ASSIGN, GSEA, ssGSEA and FacPad (Table 3). BFRM was not included in this analysis because it requires gene expression profiling data from pathway perturbation experiments to train its model (these are not available here). We applied GSEA, ssGSEA and FacPad to test the enrichment of DNA damage/repair pathways in genotoxic group and to validate ASSIGN predictions. FacPad yielded results largely inconsistent with the other methods; FacPad often produced mean differences between two groups that were in opposite directions than the other approaches. Although GSEA and ssGSEA approaches yielded results similar to ASSIGN, we note that ASSIGN did not require genotoxic status to estimate the pathway activation level. Furthermore, in contrast to GSEA, ssGSEA and FacPad, ASSIGN is able to estimate absolute pathway activity for each individual sample (Supplementary Table S6). ssGSEA outputs an enrichment score for each sample, but this score is on a relative (not absolute) scale. Therefore, pathway enrichment/activation can only be determined in contexts containing multiple control samples (Supplementary Table S7). The ASSIGN predictions of genotoxic carcinogen exposure using the KEGG P53 signaling pathway in rat samples closely matched the genotoxicity labels from the bacterial assays with AUC = 0.91 (Figure 4-A and 4-B).
Table 3.
Comparison of pathway activity between genotoxic and non-genotoxic groups reported in P-values
| Pathways |
ASSIGN | GSEA | ssGSEA | FacPad |
|---|---|---|---|---|
| AMUNDSON_DNA_DAMAGE_RESPONSE_TP53 | <0.001 | 0.027 | <0.001 | 0.279 |
| AMUNDSON_GENOTOXIC_SIGNATURE | 0.032 | 0.074 | 0.002 | 0.290 |
| KEGG_P53_SIGNALING_PATHWAY | <0.001 | 0.077 | <0.001 | 0.464 |
| KYNG_DNA_DAMAGE_BY_4NQO | 0.041 | 0.198 | 0.027 | 0.159 |
| KYNG_DNA_DAMAGE_BY_4NQO_OR_GAMMA_RADIATION | 0.695 | 0.054 | 0.014 | 0.001 |
| KYNG_DNA_DAMAGE_BY_GAMMA_AND_UV_RADIATION | 0.002 | 0.042 | 0.001 | 0.024 |
| KYNG_DNA_DAMAGE_BY_UV | 0.024 | 0.117 | 0.023 | 0.320 |
| KYNG_DNA_DAMAGE_DN | 0.014 | 0.002 | <0.001 | 0.221 |
| KYNG_DNA_DAMAGE_UP | 0.009 | 0.058 | 0.038 | 0.703 |
Pathway activity compared between genotoxic and non-genotoxic groups (two sample t-test for ASSIGN, FacPad and ssGSEA; Kolomogorov–Smirnov test for GSEA). The results were mostly consistent among the ASSIGN, GSEA and ssGSEA approaches, but mostly inconsistent with FacPac approach. DNA damage response/repair pathways were significantly differentially activated (P-value) between two groups for at least two approaches.
Fig. 4.
KEGG P53 signaling pathway signature in tissues exposed to carcinogens. (A) ASSIGN predicted pathway activity in rat liver tissues exposed to non-genotoxic or genotoxic carcinogens. The boxplot exhibits an association between genotoxic carcinogen exposure and P53 signaling pathway activation. (B) ROC curve for ASSIGN predicted signature strengths of the KEGG P53 signaling pathway. The corresponding area under the curve (AUC) is 0.911, suggesting an excellent model predictive ability. (C) Heatmap of 43 predefined P53 signaling pathway genes in 36 rat liver samples. Each row represents a gene and each column represents a sample. The color bar above the heatmap represents the treatment labels for each corresponding sample (orange: genotoxic; grey: non-genotoxic). The bar plot above the heatmap is the ASSIGN predicted signature strength for each corresponding sample. The bar plot on the left is the ASSIGN predicted posterior signature (green: gene included in the posterior signature; grey: gene not included)
4.5.1 Context-specific signatures
We further examined the adaptive pathway KEGG P53 signature estimated by ASSIGN. The predefined signature of the KEGG P53 signaling pathway from MSigDB is a curated gene set for Homo sapiens. ASSIGN adapts this signature to R. norvegicus when predicting the pathway activity level in rat samples. For the adaptive signature of this pathway, we observed that 65% of the genes in the KEGG P53 signaling pathway were dropped out from the significant gene list (posterior probability <0.90) (Supplementary Table S7). In addition, for the genes retained in the list, although the magnitude of gene expression level is not provided in the predefined signature, it was estimated and adapted to the rat samples (Supplementary Table S7). We plotted a heatmap, ordering the samples by the activity level of the context-specific R. norvegicus KEGG P53 signaling pathway. The gene expression profiles of those 36 rat samples were naturally clustered by pathway activity predicted by ASSIGN (Fig. 4C).
5 Conclusions and Discussion
We have developed the ASSIGN approach for simultaneously determining the strengths of multiple molecular signatures in patient samples. Our ASSIGN framework is specifically designed for cases where the signatures or relevant signature gene lists are known a priori. ASSIGN does not accommodate situations where signatures are completely unknown. ASSIGN uses sparse Bayesian regression and factor analysis approaches to simultaneously profile multiple pathway signatures. ASSIGN is a flexible toolkit that allows for signatures in the form of gene sets, gene sets with direction and magnitudes or signatures extracted directly from profiling data. ASSIGN also allows for adapting the background and the signatures to better accommodate specific tissues, biological systems or disease contexts.
We have demonstrated the usefulness of our approach in multiple simulated and real-data examples and showed that ASSIGN performs favorably in these datasets compared with other existing approaches. For example, because ASSIGN evaluates multiple pathway signatures simultaneously, it accounts for confounding events between interactive pathways. Here, we applied ASSIGN to five highly correlated oncogenic pathways and compared results with BFRM, a single pathway-based approach. Although, BFRM achieves similar sensitivity to ASSIGN, BFRM has much lower specificity. In addition, ASSIGN can use either curated pathway signature gene lists or perturbation signatures in a flexible way. Most supervised learning methods, such as BFRM, require perturbation datasets as input. GSEA and FacPad can only use curated pathway gene lists. For pathway signature profiling, the selection of multiple pathways is based on the biological knowledge of pathway interaction. However, we recommend a maximum of about a dozen of correlated pathways in ASSIGN to avoid multicollinearity and unidentifiability issues of the model.
The adaptive background feature of ASSIGN allows for the estimation of absolute pathway activity levels in a biologically interpretable manner (ranging between 0 and 1). No existing factor analysis approach or supervised learning approach accommodates this feature, and thus can only achieve relative activation status. The enrichment scores estimated by ssGSEA do not have biological meaning unless compared with control samples for relative pathway strength. GSEA estimates one overall enrichment score, but does not predict for individual samples. Furthermore, ASSIGN allows for the refinement and adaptation of pathway signatures within a dataset, in contrast to other regression-based or supervised learning algorithms in which the predetermined pathway signature is static (Pirooznia et al., 2008; Ringnér et al., 2002). This unique feature not only reduces the bias of pathway strength estimation, but also curates pathway signatures to be cell- or tissue-specific future applications.
In addition to pathway activation level estimation, ASSIGN can be used to predict patients’ drug response, carcinogen exposure, pathogen immune response on the basis of gene expression signature strength. The input data of ASSIGN is assumed to follow a normal distribution. To accommodate to different types of omic data such as methylation microarray data or SNP array data, a more generalized model may need to be developed in the future. In addition, in future work we plan to allow for multiple background profiles in the patient dataset, whereas the current version of ASSIGN only allows for a single baseline expression profile. We also hope to evaluate extensions of ASSIGN to integrate multi-omic data types and to better accommodate the discrete nature of sequencing data. Overall, ASSIGN results in more biologically interpretable pathway activation profiles that are adapted to specific tissues or disease contexts, as opposed to more rigid and less interpretable profiles from previous approaches.
Supplementary Material
Acknowledgements
The authors thank the Linux Clusters for Genetic Analysis and the Shared Computing Cluster at Boston University for computational support for this project. The authors thank Marc E. Lenburg and Paola Sebastiani for critical reading of their manuscript.
Funding
This research was supported by funds from the NIH (U01CA164720) and (T15LM007124).
Conflict of Interest: none declared.
References
- Ashburner M., et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham R., Yarden Y. (2011) Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol., 12, 104–117. [DOI] [PubMed] [Google Scholar]
- Barbie D.A., et al. (2009) Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature, 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazot C., et al. (2013) Unsupervised Bayesian linear unmixing of gene expression microarrays. BMC Bioinformatics, 14, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Dunson D.B. (2011) Sparse Bayesian infinite factor models. Biometrika, 98, 291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild A.H., et al. (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature, 439, 353–357. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheang M.C.U., et al. (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res., 14, 1368–1376. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick R.B. (2008) CPDB: Carcinogenic Potency Database. Med. Ref. Serv. Q., 27, 303–311. [DOI] [PubMed] [Google Scholar]
- Ganter B., et al. (2005) Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J. Biotechnol., 119, 219–244. [DOI] [PubMed] [Google Scholar]
- George E.I., Mcculloch R.E. (1997) Approaches for Bayesian variable selection. Stat. Sin., 339–374. [Google Scholar]
- Golub T.R., et al. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science, 286, 531–537. [DOI] [PubMed] [Google Scholar]
- Gustafson A.M., et al. (2010) Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Transl. Med, 2, 26ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich K.P., et al. (2009) In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res., 15, 4649–4664. [DOI] [PubMed] [Google Scholar]
- Hsaing T. (1975) A Bayesian view on ridge regression. The Statistician, 24, 267–268. [Google Scholar]
- Johnson W.E., et al. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8, 118–127. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., et al. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res., 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger B., et al. (2013) Network quantification of EGFR signaling unveils potential for targeted combination therapy. Mol. Syst. Biol., 9, 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., et al. (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics, 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Knowles E., et al. (2010) PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int. J. Cancer J. Int. Cancer, 126, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Ma H., Zhao H. (2012) FacPad: Bayesian sparse factor modeling for the inference of pathways responsive to drug treatment. Bioinf., 28, 2662–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall M.N., et al. (2010) Frozen robust multiarray analysis (fRMA). Biostatistics, 11, 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moestue S.A., et al. (2013) Metabolic biomarkers for response to PI3K inhibition in basal-like breast cancer. Breast Cancer Res. BCR, 15, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortelmans K., Zeiger E. (2000) The Ames Salmonella/microsome mutagenicity assay. Mutat. Res., 455, 29–60. [DOI] [PubMed] [Google Scholar]
- Piccolo S.R., et al. (2012) A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics, 100, 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S.R., et al. (2013) Multiplatform single-sample estimates of transcriptional activation. Proc. Natl. Acad. Sci., 110, 17778–17783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M., et al. (2008) A comparative study of different machine learning methods on microarray gene expression data. BMC Genomics, 9, S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringnér M., et al. (2002) Analyzing array data using supervised methods. Pharmacogenomics, 3, 403–415. [DOI] [PubMed] [Google Scholar]
- Saeys Y., et al. (2007) A review of feature selection techniques in bioinformatics. Bioinformatics, 23, 2507–2517. [DOI] [PubMed] [Google Scholar]
- Subramanian A., et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet-Cordero A., et al. (2005) An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat. Genet., 37, 48–55. [DOI] [PubMed] [Google Scholar]
- Trapnell C., et al. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinf., 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vijver M.J., et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med., 347, 1999–2009. [DOI] [PubMed] [Google Scholar]
- Wang K., et al. (2010) MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res., 38, e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein I.B. (2002) Cancer. addiction to oncogenes—the Achilles heal of cancer. Science, 297, 63–64. [DOI] [PubMed] [Google Scholar]
- West M. (2003) Bayesian factor regression models in the ‘Large p, Small n’ Paradigm. Bayesian Statistics, 7, 723–732. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.