Abstract
Many signalling proteins involved in diverse functions such as cell growth and differentiation can act as oncogenes and cause cellular transformation. These molecules represent attractive targets for cancer diagnosis or therapy and therefore are subject to intensive investigation.
Aptamers are small, highly structured nucleic acid molecules, isolated from combinatorial libraries by a procedure termed SELEX. Aptamers bind to a target molecule by providing a limited number of specific contact points imbedded in a larger, defined three-dimensional structure. Recently, aptamers have been selected against whole living cells, opening a new path which presents three major advantages: (1) direct selection without prior purification of membrane-bound targets, (2) access to membrane proteins in their native conformation similar to the in vivo conditions and (3) identification of (new) targets related to a specific phenotype. The ability to raise aptamers against living cells opens some attractive possibilities for new therapeutic and delivery approaches. In this chapter, the most recent advances in the field will be reviewed together with detailed descriptions of the relevant experimental approaches.
Keywords: Aptamer, SELEX, ret, delivery, siRNA
1. Introduction
1.1. Intact Cells as Targets
With the first description of SELEX in 1990 (1, 2) the therapeutic potential of aptamers was apparent. In particular, the potential application of RNA ligands as antagonists of clinically relevant protein targets and their advantages over other macromolecular technologies as antibodies or peptides was clear (3, 4). The fact that the entire aptamer identification process is performed in vitro permits the researcher to raise aptamers against virtually any soluble protein. Indeed, several aptamers have been raised that target extracellular soluble proteins with potential therapeutic value. These targets include growth factors, cytokines and coagulation factors, proteins that can be easily produced in huge amounts, and more importantly have the common advantage of being readily accessible to drug ligands that do not cross cell membranes. In fact, some of these aptamers have entered clinical trials, as for example, pegaptinib (Macugen) that targets one isoform of the vascular endothelial growth factor (VEGF165) now approved for treatment of age-related macular degeneration (5).
In addition, aptamers targeting intracellular proteins that act as signalling mediators have also been generated. However, a major obstacle to their use as therapeutics is the development of intracellular delivery approaches, as described for intramers (6).
Aptamer selection approaches that target the cell surface have been developed more recently. In the first paper describing such an approach, Morris et al. demonstrated that SELEX could be used to simultaneously isolate RNA ligands to multiple targets bound to a biological membrane (7). Purified red blood cell ghosts were used as target, with essentially the same SELEX protocol that is used with individual purified proteins. After 25 rounds of SELEX, they have isolated a set of different ligands each targeting distinct red blood cell proteins, thus demonstrating that the procedure could be considered as the resultant of multiple simultaneous and independent experiments. Furthermore, it was evident that increasing the stringency by reducing the target concentrations enriched the pool for the highest affinity ligands.
Homann and Goringer (8) confirmed that SELEX protocol can be performed with live cells and even without the knowledge of all elements of a target’s surface. They were the first to apply the SELEX technology to a parasite system by addressing the question whether aptamers can be selected to recognize the surface of live parasite cells. By using African trypanosomes as a model system, an extracellular blood parasite with a very specific surface architecture, they identified an aptamer family that recognized an invariant surface component of bloodstream stage trypanosomes.
More recently we took advantage of these results to develop a strategy that allowed us to inhibit a transmembrane receptor tyrosine kinase (RTK) by targeting its extracellular region with a high-affinity ligand aptamer (9). RTKs are privileged targets for cancer therapy, which is underscored by the promising outcome of clinical trials with small molecules or antibody inhibitors (10). Indeed, these receptors are large molecules heavily modified by post-translational changes, as glycosylation and phosphorylation and thus purification of even a portion of these receptors may require a long and wasteful procedure. Furthermore these proteins are functional in their membrane-bound conformation therefore, using the extracellular portion as target may frequently lead to isolate ligands that are unable to recognize the functional native receptor (11).
We validated a general strategy to isolate aptamers for an activated mutated transmembrane receptor tyrosine kinase, Ret. Germline mutations in the RET gene are responsible for constitutive activation of the receptor and for inheritance of multiple endocrine neoplasia (MEN) type 2A and 2B syndromes, and of familial medullary thyroid carcinoma (12–16). The RET receptor constitutes a model system of choice (16) in that the transforming mutations, of MEN2A type, located in the extracellular domain simplify the issue of intracellular accessibility for a targeting molecule (14, 15).
We based our experimental approach on the notion that targeting a complex target constituted of multiple proteins, as is the membrane surface of a cell, permits to isolate ligands for individual proteins provided that they are highly represented (7, 9).
Therefore, we used as target of the selection procedure living mammalian cells growing in culture dishes engineered in order to express high levels of the RET mutant protein. These conditions are expected to expose a native protein to the selection procedure, thus best mimicking in vivo conditions. In order to deplete the pool expressing two different forms of the receptor tyrosine kinase Ret: one with a transforming mutation located in the extracellular domain leading to constitutively dimeric Ret, i.e. the target (14), and one in which the receptor remains monomeric, with a transforming mutation located in the intracellular domain, i.e. the sham (15, 17).
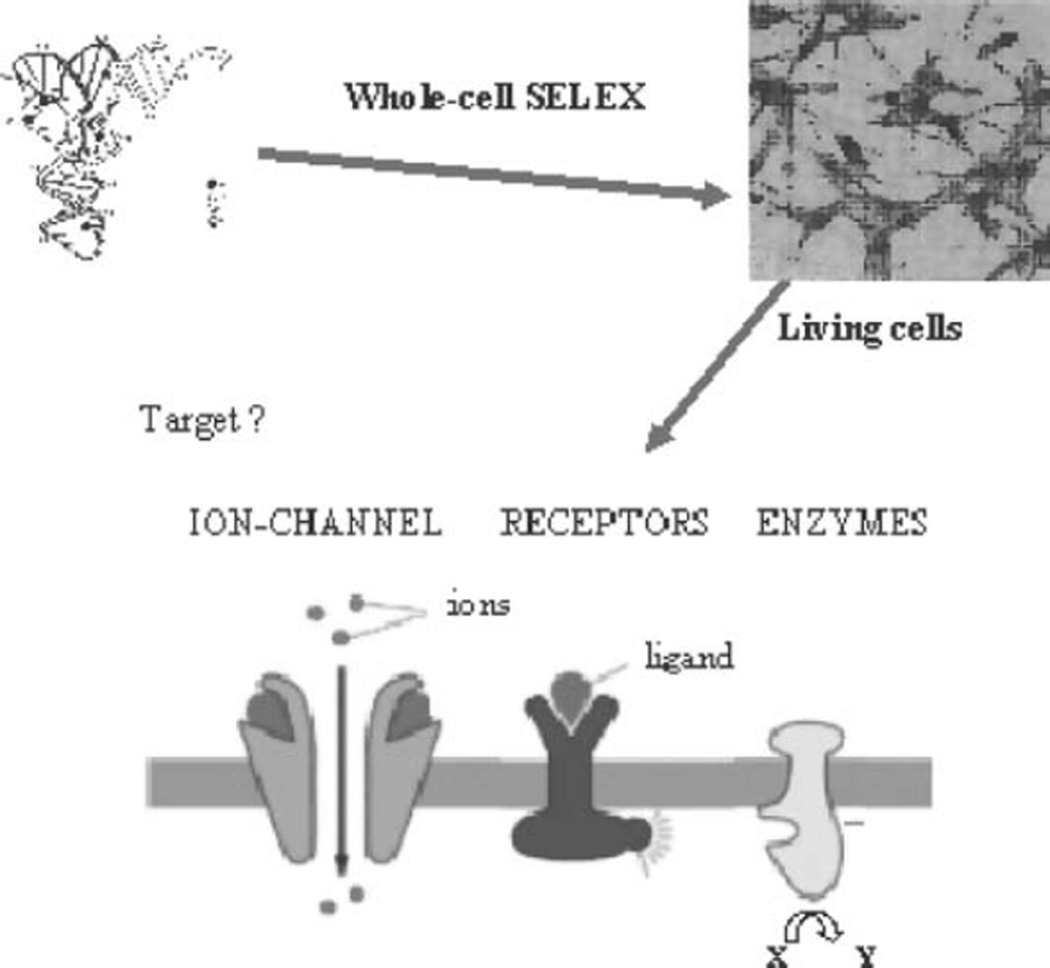
Molecular-level differentiation of neoplastic cells is essential for accurate and early diagnosis, but effective molecular probes for molecular analysis and profiling of neoplastic cells are not yet available. The intact cell-based SELEX strategy is generally applicable to different cell types and holds a great promise in developing specific molecular probes for cancer biomarker discovery and for cancer diagnostic and therapeutic applications (see Fig. 5.1).
Fig. 5.1.
The whole-cell SELEX technology allows identifying unique molecular features of cancer cells by selecting aptamers in a physiological context, and, most importantly, it can be done without prior knowledge of the target molecules.
1.2. Cell Internalizing Aptamers as Therapeutic Reagents
The development of aptamers as therapeutics has primarily involved aptamers that bind and inhibit the activity of their protein targets. Another promising application of aptamers is to use them to deliver a variety of secondary reagents specifically to a targeted cell population. Once delivered, the secondary reagents would then impart their therapeutic effect to this subset of cells within the treated individual. Because non-targeted cells would not be exposed to the secondary reagent, the potential for unwanted side-effects such as death of normal cells as occurs with the use of many cancer therapeutics is substantially reduced.
This approach utilizes the cell-type specific expression of cell surface proteins on cell populations of therapeutic value. The idea here is to develop an aptamer to the extracellular portion of such a protein and to then use the aptamer to deliver the secondary reagent to the targeted cell population via binding the targeted protein on the surface of the targeted cell type. Because this binding in some cases also results in the endocytosis of the aptamer/secondary reagent complex, this approach can be used to deliver reagents such as siRNAs that depend on delivery to intracellular compartments for their proper function.
Antibodies and other protein-based reagents have previously been developed to serve comparable roles in targeting therapeutics to specific cell types (18, 19). However, aptamers have a number of important advantages over proteins as therapeutic reagents. A number of these advantages stem from the fact that proteins must be produced in cell culture while aptamers can be chemically synthesized. The production of proteins is thus expensive and complicated by batch-to-batch variability in activity, resulting in a more complicated regulatory approval process. In addition, aptamers can be readily chemically modified to enhance their bioavailability and pharmacokinetics. Another important advantage of RNA aptamers over proteins is the fact that RNA is much less immunogenic than proteins. Therapeutics made from RNA are thus likely to be safer when repeated administrations are necessary. RNA made with pyrimidines modified at the 2’-position, which renders them resistant to extracellular nucleases are even less immunogenic than natural RNA (20).
Aptamers targeting the prostate-specific membrane antigen (PSMA) have been used to deliver both nanoparticles and siRNAs to prostate cancer cells in therapeutic proof of concept studies (21–23). Nanoparticles containing docetaxel or siRNAs targeting cancer cell survival genes, when targeted with PSMA-binding aptamers were internalized by PSMA-expressing prostate cancer cells and resulted in cancer cell death in vitro and retarded tumour growth in vivo. The ability of aptamers to specifically deliver secondary therapeutic reagents has thus been demonstrated for both nanoparticles and siRNAs. It seems likely that aptamers targeting membrane proteins of other therapeutic target cell populations will also prove to be useful reagents in other clinically relevant contexts.
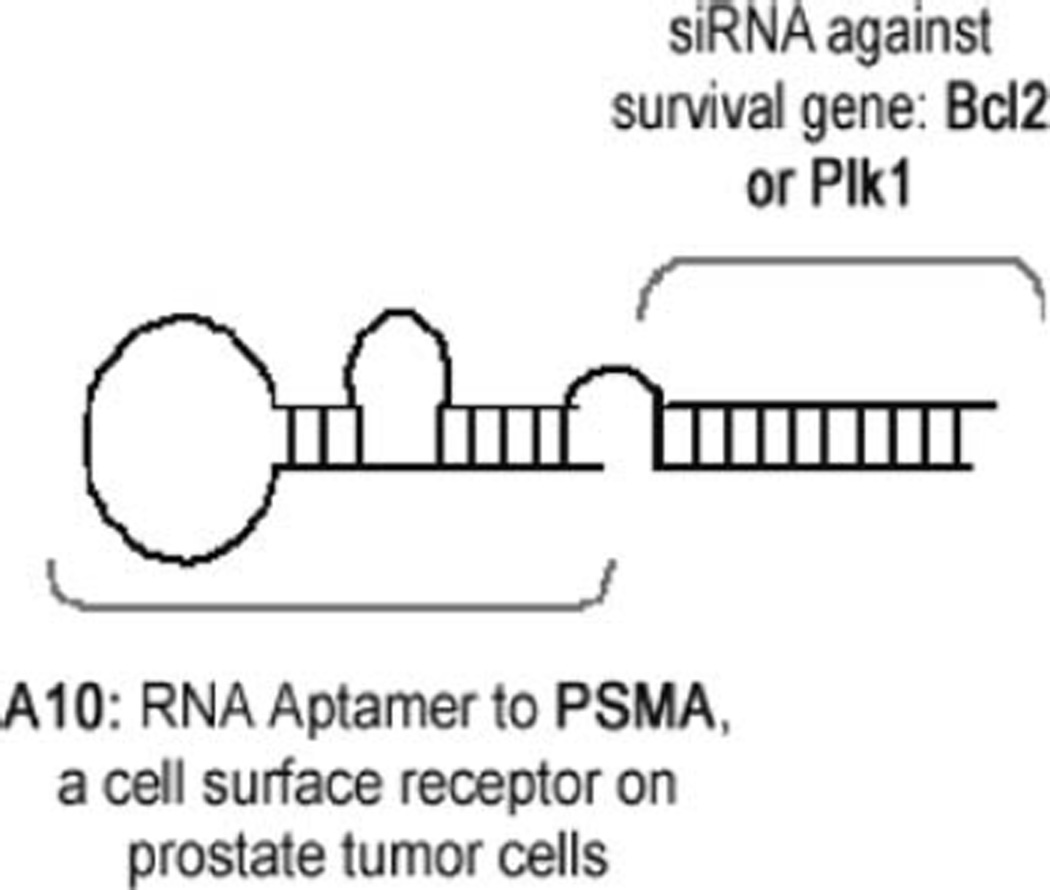
Here, we provide protocols for the approach we used (23) to deliver secondary therapeutic siRNAs specifically to PSMA expressing cells. This approach entails the annealing of two distinct strands of RNA, one strand that consists of the aptamer with an extended tail that makes up the upper strand of the siRNA and a second strand that consists of the lower strand of the siRNA (see Fig. 5.2).
Fig. 5.2.
Schematic of PSMA aptamer–siRNA chimera.
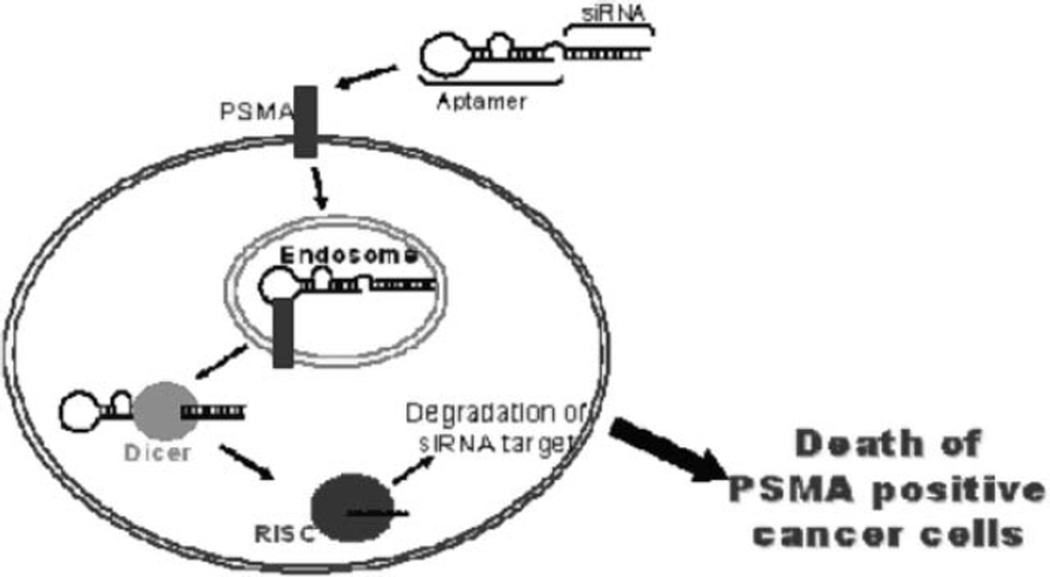
Using this approach, we showed that when added to cells expressing the aptamer target receptor on the surface, the aptamer– siRNA chimeras are rapidly internalized. Importantly, we showed that internalization of the chimera results in silencing of the siRNA target, by an RNAi-mediated mechanism, resulting in the death of the targeted cancer cell (see Fig. 5.3). Advantages of this approach include the facts that this reagent (an aptamer– siRNA chimera) consists only of RNA, which has important advantages (see above) as a therapeutic material, and that it can easily be carried out in labs that have the reagents and equipment to carry out basic molecular biology procedures.
Fig. 5.3.
Mechanism of aptamer–siRNA chimera-mediated targeting and silencing. Target specificity by the RNA chimera can be achieved both at level of the aptamer (PMSA-specific) as well as at the level of the siRNA (by silencing cancer cell-specific survival factors). This approach leads to selective killing of cancer cells that express both the cell surface receptor PSMA and prostate cancer-specific survival factors (e.g. Plk1 and Bcl2).
2. Materials
2.1. RNA Transcription and Purification
Transcription buffer (5×): 0.2 M Tris–HCl (pH 7.5), 30 mM MgCl2, 50 mM NaCl and 10 mM spermidine.
Transcription mix: Transcription buffer (1×) with 1 mM 2’F-Py (2’F-2’-dCTP and 2’F-2’-dUTP, TriLink Biotech, San Diego, CA), 1 mM ATP, 1 mM GTP (Amersham Pharmacia Biotech), Uppsala Sweden, 10 mM dithiothreitol (DTT) (Sigma, St. Louis, MO), 0.5 u/µl RNAse inhibitors (Amersham Pharmacia Biotech), 5 µg/ml inorganic pyrophosphatase (Roche, Germany).
Loading solution: prepare a solution of 480 µl of formamide, 10 µl water, 10 µl EDTA and Bromophenol Blue (BBF, Bio-Rad, Hercules, CA).
Denaturing polyacrylamide gel (8% final concentration): 40% acrylamide/bis solution (37.5:1) (Bio-Rad) dissolved in Tris– borate–ethylenediamine tetraacetic acid (EDTA) buffer (TBE) containing 7 M urea and N,N,N’,N’ tetramethyl-ethylendiamine (TEMED, Bio-Rad) (see Note 1) and ammoniumpersulfate (Sigma).
Ammoniumpersulfate: Prepare 10% solution in water and immediately freeze in aliquots at –20°C.
Elution buffer: 300 mM NaOAc with 200 mM EDTA.
32P-αUTP (3,000 Ci/mmol, Amersham Pharmacia Biotech); T7 RNA/DNA polymerase (the mutant T7Y639F RNA polymerase) (Epicentre), DNase I (Amersham Pharmacia Biotech).
2.2. Cell Culture
PC12/MEN2A and PC12/MEN2B are PC12 cells stably expressing Ret9C634Y and Ret9M918T proteins, respectively.
Growth medium for PC12 cells: RPMI 1640 (Gibco/BRL, Bethesda, MD) with 10% heat-inactivated horse serum (HS, Gibco/BRL), 5% heat-inactivated fetal bovine serum (FBS, Gibco/BRL), 2 mM glutamine.
Growth medium for PC12/MEN2A and PC12/MEN2B: the same medium for PC12 cells but supplemented with HAT medium supplement 50× (Sigma), 250 µg/ml xantine (Sigma) and 25 µg/ml micophenolic acid (Sigma).
Xantine is dissolved in 0.1NNaOH at 5 mg/ml and adjust pH of 10.8 with 3 N HCl and pH 10.5 with 1 N HCl, filtered and stored at dark and at room temperature (see Note 2).
Micophenolic acid (Sigma) is dissolved at 25 mg/ml in EtOH and stored at room temperature.
Solution of trypsin and EDTA from Gibco/BRL.
2.3. Counter-Selection and Selection Steps
Buffer of incubation for the RNAs: RPMI 1640 without serum
Washing buffer: RPMI 1640 without serum.
Total yeast RNA from Sigma.
Total RNA extraction kit from Ambion Inc. (Texas, USA).
2.4. Restriction Fragment Length Polymorphism (RFLP) Analysis
Polymerase chain reaction (PCR) buffer (10×): 100 mM Tris–HCl (pH 8.3), 15 mM MgCl2, 500 mM KCl.
PCR mix: PCR buffer (1×) with 200 µM dATP, 200 µM dGTP, 200 µM dCTP, 200 µM dTTP (Amersham Pharmacia Biotech), 2 µM primers, DNA of each cycle of selection and Taq polymerase (0.02 U/µl) (Roche, New Jersey, USA).
[γ-32P]ATP (3,000 Ci/mmol, Amersham Pharmacia Biotech).
REact 1 (10×) for digestion: 500 mM Tris–HCl (pH 8.0), 100 mM MgCl2, 500 mM NaCl.
RsaI, AluI, HaeIII, HhaI enzymes from Invitrogen.
Denaturing polyacrylamide gel (6% final concentration): 40% acrylamide/bis solution (37.5:1) dissolved in TBE buffer containing 7 M urea.
2.5. Binding Analysis
Dephosphorylation buffer (10×): 500 mM Tris–HCl (pH 8.5), 1 mM EDTA.
Buffer for phosphatase alkaline (PA) inactivation: 200 mM EGTA.
Phosphorylation buffer (10×): 500 mM Tris–HCl (pH 8.2), 100 mM MgCl2, 1 mM EDTA, 50 mM DTT, 1 mM spermidine.
Buffer of incubation of RNAs: RPMI 1640 without serum.
Washing buffer: RPMI 1640 without serum.
Recovering buffer: 0.6% sodium dodecyl sulphate (SDS).
PA for dephosphorylation from Boehringer Mannheim; T4 Polynucleotide Kinase for phosphorylation from Roche; [γ-32P]ATP (6,000 Ci/mmol, Amersham Pharmacia Biotech).
2.6. Cell Lysis and Western Blotting for Functional Analysis of Selected Aptamers
Lysis solution: 50 mM Tris–HCl (pH 8.0) with 150 mM NaCl, 1% Nonidet P-40, 2 µg/ml aprotin, 1 µg/ml pepstatin, 2 µg/ml leupeptin (Roche) and 1 mM Na2VO4(Sigma).
Separating buffer (4×): 1.5MTris–HCl (pH 8.7), 0.4% SDS.
Stacking buffer (4×): 0.5% Tris–HCl (pH 6.8), 0.4% SDS.
Denaturing polyacrylamide gel (10% – final concentration): 40% acrylamide/bis solution (37.5:1).
Running buffer (5×): 125 mM Tris, 960 mM glycine, 0.5% SDS.
Laemmli buffer: 2% SDS, 5% β-mercaptoethanol, 0.001% bromophenol blue, 10% glycerol.
Pre stained molecular weight marker: Kaleidoscope markers from Bio-Rad.
Supported polyvinylidenedifluoride (PVDF) membrane from Millipore, Bedford, MA, and 3 MM chromatography paper from Whatman, Maidstone, UK.
Transfer buffer: 25 mM Tris, 190 mM glycine, 20% methanol, 0.05% SDS (see Note 3).
Tris-buffered saline with Tween (T-TBS): prepare 10× stock with 1.37 M NaCl, 27 mM KCl, 250 mM Tris–HCl (pH 7.4); dilute 100 ml of TBS 10× with 900 ml water and add Tween at the concentration required for use. 1
Blocking buffer: 5% nonfat dry milk in the T-TBS required.
Primary antibody dilution buffer: 5% nonfat dry milk in the TTBS required.
Enhanced chemiluminescent (ECL) reagent from Amersham Pharmacia Biotech and Bio-Max ML film (Kodak).
Stripping buffer: 62.5 mM Tris–HCl (pH 6.8), 2% SDS, 100 mM β-mercaptoethanol.
2.7. Generation of Transcription Template
High quality DNA oligonucleotides can be obtained desalted, from many sources such as Integrated DNA Technologies, Oligos, etc., and Promega. Longer oligos (>50 nucleotides) should be ordered PAGE purified.
2.8. In Vitro Transcription
10× NTP mix: 30 mM 2’F-CTP, 30 mM 2’F-UTP, 10 mM 2’OH-ATP, 10 mM 2’OH-GTP, dissolved in water. 2’-fluoro modified NTPs can be obtained from Trilink Inc. Unmodified NTPs can be obtained from a number of sources including New England Biolabs and Roche.
Mutant (Y639F) T7 RNA Polymerase (Epicentre), Inorganic pyrophosphatase (Roche),
5× T7 RNA polymerase buffer: 20% w/v PEG 8000, 200 mM Tris–HCl (pH 8.0), 60 mM MgCl2, 5 mM spermidine HCl, 0.01% w/v triton X-100, 25 mM DTT.
5’-FAM-G can be custom-synthesized by Trilink Inc.
2.9. Gel Purification
Urea (Sigma), acrylamide (Bio-Rad), 10× TBE (Sigma), formamide (Sigma), xylene cyanol (Sigma), bromophenol blue (Sigma), Centrex spin filters, Centricon YM-30 filtration units (Millipore), Dulbecco’s phosphate-buffered saline RNAse-free DNAse (NEB).
For 2× formamide gel loading buffer, combine: 0.01 g xylene cyanol, 0.02 g bromophenol blue, 500 µl 10× TBE, 9.5 ml formamide.
For 10% acrylamide/urea gel solution, combine: 115 g urea, 62.5 ml 40% acrylamide/bis (29:1), 12.5 ml 10× TBE and water for a final volume of 250 ml. Heat to dissolve urea, filter-sterilize and store at 4°C, protected from light (see Note 6).
2.10. RNA Oligonucleotides
High quality RNA oligonucleotides can be obtained from a number of commercial vendors including Dharmacon and Promega.
The following RNA oligos will anneal to the upper sense strands of the chimeric RNAs that can be produced with the protocol included below: A10-Plk1 Antisense siRNA: 5’GCACUUGGCAAAGCCGCCCdTdT3’. A10-CON Antisense siRNA: 5’ACGUGACACGUUCGGAGAAdTdT3’.
2.11. Buffers for Cell Surface Binding Assays
Binding buffer: 20 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM CaCl2, 0.02% BSA. Fix solution: DPBS plus 1% formaldehyde.
2.12. Reagents for In Vitro siRNA Activity Assays
RIPA buffer
An antibody for Plk1 is available from Zymed. An antibody for Bcl-1 is available from DykoCytomation.
PERM/FIX and PERM/WASH buffers are available from Pharmacia.
3. Methods
3.1. The Rationale of the SELEX Methodology
The SELEX method includes steps of: (i) incubating the library with the target molecule; (ii) partitioning unbound nucleic acids from those bound specifically to the selector cell type; (iii) dissociating the nucleic acid–target complexes; and (iv) amplifying of the nucleic acids pool enriched for specific ligands. Usually the positive selection is preceded by a negative or counter-selection step against the capture system in the absence of the desired target. This ensures the selection of aptamers directed against the target but not the other elements of the aptamer capturing moiety. To select aptamers against a transmembrane protein it is useful to perform a counter-selection step with the non-expressing cell line or otherwise with a cell line expressing the target protein in a different conformation state (as for example, monomeric vs. dimeric for a tyrosine kinase transmembrane receptor).
After reiterating these steps (the number of rounds of selection necessary is determined by both the type of library used as well as by the specific enrichment achieved per selection cycle), the resulting oligonucleotides are subjected to DNA sequencing. The sequences corresponding to the variable region of the library are screened for conserved sequences and structural elements indicative of potential binding sites and subsequently tested for their ability to bind specifically to the target cell.
3.2. Preparation of RNA Libraries
The starting RNA library pool is a library of 2’F-Py RNA molecules containing a 50 nt random sequence flanked by two fixed regions for the amplification reaction. The PCR amplifications of this library are performed using the following set of primers:
P20: 5’TCCTGTTGTGAGCCTCCTGTCGAA3’
P10: 5’TAATACGACTCACTATAGGGAGACAAGAATAAACGCTCAA3’
The complexity of the starting pool was roughly 1014 2’F-Py RNAs (1–5 nmol).
The transcription reactions are performed at 37°C for 12 h in the transcription mix with 10 µCi/µl 32P-αUTP, 1 pmol/µl DNA and 2.5 u/µl of the mutant form of T7, T7Y639F RNA polymerase (see Notes 4 and 5).
Following transcription, the RNA is treated with DNase I to remove contamination of ssDNA. Ten units of DNase I are added at end of transcription and incubated at 37°C for 20 min. Large volume RNA transcriptions are concentrated and desalted with phenol/chloroform/isoamyl alcohol (25:24:1), ethanol precipitated in the presence of 0.1 mg/ml of linear acrylamide (Ambion) and centrifuged at 14,000 rpm for 30 min at 4°C. The pellet is suspended in 20 µl of loading solution and purified by 8% denaturing polyacrylamide gel. Gel purification of full-length RNA is important to prevent artefacts. RNA is passively eluted from the gel at 42°C in elution buffer and the concentration is determined spectrophotometrically assuming that one A260 unit is equal to 40 µg/ml of RNA.
3.3. Selection Strategies
In each cycle two counter-selection steps were performed.
Denaturation/renaturation step: 2’F-Py RNAs (1–5 nmol) are heated at 85°C for 5 min in 3 ml of RPMI 1640 serum free, snap-cooled on ice for 2 min, and allowed to warm up to 37°C, before incubation with the cells.
Following the denaturation/renaturation step, the pool of 2’F-Py RNAs (resuspended in 3 ml of RPMI 1640) is first incubated for 30 min at 37 °C with 107 PC12 cells in order to eliminate non-specific binders of the PC12 cell surface.
The unbound sequences are recovered by centrifugation and incubated for 30 min at 37°C with 107 adherent PC12/ MEN2B cells that express an allele of RET (RET/M918T) mutated in the intracellular domain.
For the selection step, the unbound sequences from the second counter-selection are recovered and incubated with 107 adherent PC12/MEN2A cells expressing the RETC634Y mutated in the extracellular domain, for 30 min at 37°C in the presence of total yeast RNA as non-specific competitor RNA. Finally, the bound sequences are recovered after several washings with 5 ml of RPMI by total RNA extraction.
During the selection process, the selective pressure is progressively increased by increasing the number of washings (from one for the first cycle up to five for the last three cycles) and the amount of non-specific RNA competitor (100 µg/ml in the last three cycles), and by decreasing the incubation time (from 30 to 15 min from round 5) and the number of cells exposed to the aptamers (5 × 106 in the last three cycles).
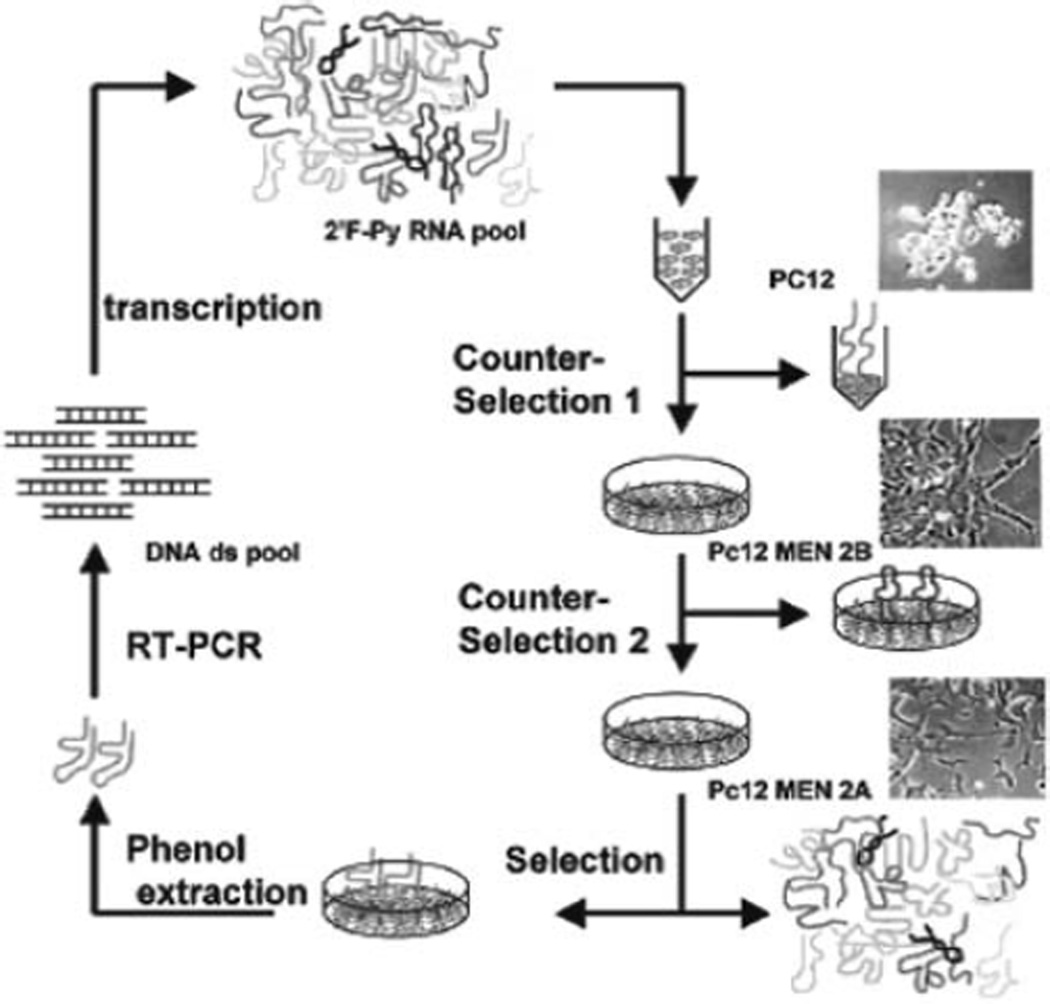
The approach used for cell-SELEX is reported in Fig. 5.4 (adapted from (9)).
Fig. 5.4.
Schematic protocol for the selection of PC12/MEN2A cell-specific aptamers.
To monitor the evolution of the pool the appearance of four base restriction sites in the population (by RFLP analysis) is analysed, which reveals the emergence of distinct families in the library. The PCR product of each cycle (about 500 ng) are end-labelled with [γ-32P] ATP and digested with a mix of four restriction enzyme. The endonuclease used are: RsaI, AluI, HaeIII, HhaI (10 units/enzyme) in the buffer REactI (Invitrogen Life Technologies) for 1 h at 37°C.
Following ethanol precipitation, the digested samples are loaded onto 6% denaturing polyacrylamide gel. The gel is wrapped and an autoradiography film is exposed.
3.4. Cloning and Sequencing
After 15 rounds of selection, sequences are cloned with TOPOTA cloning kit (Invitrogen, Carlsbad, CA, United States) and analysed. About 100 clones are usually analysed in a SELEX protocol by using bioinformatics alignment tools. In general, SELEX-derived sequences contain regions of strong sequence conservation separated by regions of high variability. This hampers the usage of global alignment tools and resulted in the application of modified alignment protocols specifically tailored to identify and score sequence patterns in aptamers. This usually hallows the identification of conserved and variable nucleotide positions and permits the grouping of the various sequences into quasi-phylogenetic families. Conserved motifs within a sequence family are frequently candidates for specific target recognition elements.
3.5. Binding Analysis and Kd-Determination
To determine the binding of individual aptamers (or the starting pool as a control) to PC12 cells and derivatives, the aptamers are dephosphorylated with PA at 37°C for 1 h and end-labelled with T4 polynucleotide kinase in presence of [γ-32P] ATP at 37°C for 30 min.
Binding of individual 5’-32P-labelled RNAs is performed in 24-well plates in triplicate. 105 cells per well are incubated with various concentrations of individual aptamers for 10 min at 37°C in the presence of 100 µg/ml polyinosine as a nonspecific competitor. After extensive washings (5 × 500 µl of RPMI 1640), bound sequences are recovered in 350 µl of 0.6% SDS, and the amount of radioactivity recovered is normalized to the number of cells by measuring the protein content of each well.
- Apparent Kd values for each aptamers are determined by Scatchard analysis according to the equation:
where [T]tot represents the total target concentration. The aptamers exhibit an affinity for the target in the low nanomolar range.
3.6. In Vitro and In Vivo Functional Analysis
Once isolated the sequences with the best binding properties, they are tested for the ability to block RET dependent intracellular signaling pathways.
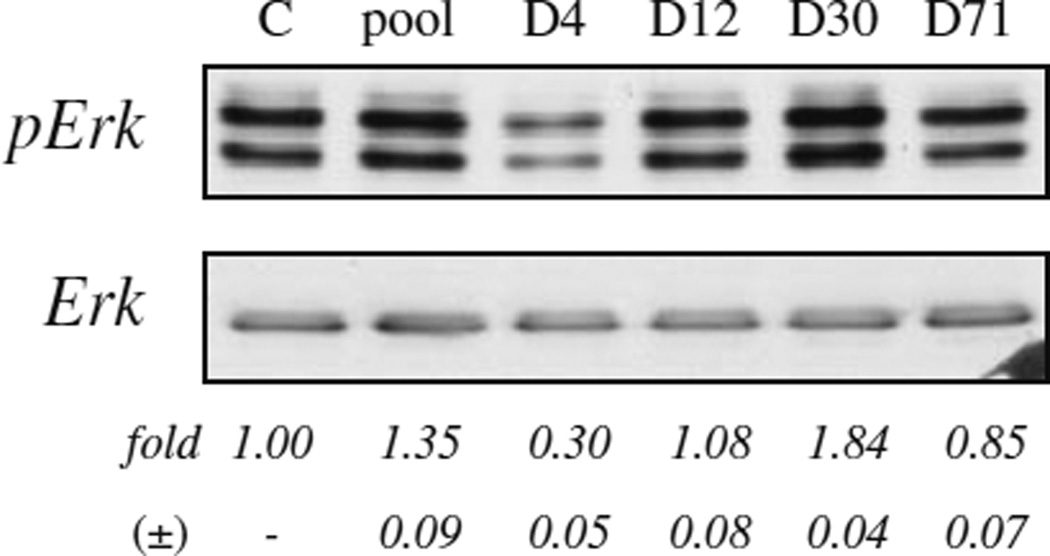
To assess the effects of aptamers on RET activity, PC12/MEN2A cells (160,000 cells per 3.5-cm plate) are serum starved for 2 h and then treated for 16 h with 150 nM RNA aptamer, or the starting RNA pool after a short denaturation–renaturation step. Cell lysates are analysed by immunoblotting. The primary antibodies used were: anti-Ret (H-300), anti-Erk1 (C-16) (Santa Cruz Biotechnology Inc., Santa Cruz, CA); anti-(Tyrphosphorylated) Ret, anti-phospho-p44/42 MAP Kinase, also indicated as pERK (E10; Cell Signaling, Beverly, MA). An example of the results produced is shown in Fig. 5.5 (adapted from (9)).
Fig. 5.5.
PC12/MEN2A cells were either left untreated or treated with the indicated RNA aptamer, or the starting RNA pool (pool). Cell lysates were immunoblotted with anti-pErk antibody, then stripped and reprobed with anti-ERK antibody to confirm equal loading. Values below the blots indicate signal levels relative to untreated controls.
A clear example of the importance to choose the best selection procedure for generating aptamers specifically binding a transmembrane protein is illustrated by the comparison of the different strategies carried out to select RNA aptamers against the RETC634Y receptor (9, 11). While several aptamers selected against the recombinant extracellular domain of transmembrane proteins recognize their targets on the cell surface (24–27), in the case of RET, however, a SELEX protocol performed on the recombinant extracellular domain of RETC634Y did not provide aptamers able to recognize the protein in a cell surface environment (Cerchia et al. (9)), which most likely implies a different mode of recognition for the native and recombinant proteins. Furthermore, a different SELEX against whole-living cells without counter-selection against PC12/MEN2B, and a crossover SELEX alternating RETC634Y expressing cells and the recombinant purified extracellular domain of the RETC634Y protein as targets, were performed. The crossover SELEX leads to a higher enrichment of aptamers against RET. However, the selected aptamers using pure whole-living cells SELEX display a better apparent Kd. This study brings new insights into the respective advantages of each of these different methods for the selection of aptamers targeting membrane proteins.
3.7. Generation of Transcription Template
-
Transcription templates for use with the T7 RNA polymerase are produced by generating double-stranded DNA that encodes a T7 promoter sequence in the 5’-end. The aptamer sequence followed by the sequence of the upper strand of the siRNA are then encoded in the 3’-end of the template. In the case of the PSMA aptamer siRNA chimera targeting Plk1, the following DNA oligos can be used to generate such a template with PCR.
A10 template primer: 5’GGGAGGACGATGCGGATCAGCCATGTTTACGTCACTCCTTGTCAATCCTCATCGGCAGACGACTCGCCCGA-3’
Plk1 siRNA 3’-primer: 5’AAGCACTTGGCAAAGCCGCCCTTTCGGGCGAGTCGTCTG3’
A105’-primer: 5’TAATACGACTCACTATAGGGAGGACGATGCGG3’
In this case, the template oligo is used as the template (in trace amounts) in a PCR with the A10 5’-primer and Plk1 siRNA 3’- primer as amplification oligos. The choice of the 3’-primer determines the sense siRNA sequence that is appended to the aptamer sequence. Substitution of the Plk1 siRNA 3’-primer with the following control primer will produce the A10 aptamer with a control siRNA sequence in place of that targeting Plk1: Control siRNA 3’-primer: 5’AAACGTGACACGTTCGGAGAATTTC GGGCGAGTCGTCTG3’
The generation of the correct-sized PCR product should be confirmed by running a sample on a 2% agarose gel with a DNA size ladder. This PCR product can then be purified (for instance, with the Qiagen PCR cleanup kit) in preparation for transcription (see Note 7).
3.8. In Vitro Transcription
For a 250 µl reaction, combine: 50 µl 5× T7 RNA polymerase buffer, 25 µl 10× NTP mix, 2 µl IPPI, 125 pmol DNA transcription template, 3 µl T7 (Y639F) polymerase and water for a final volume of 250 µl. Incubate at 37°C for 4–6 h. 5’-FAM-G can be added for a final concentration of 4 mM to produce RNA labelled with FAM only at the 5’ position (see Section 2.8.).
3.9. Gel Purification
Add 1 µl RNAse-free DNAse (Roche) to the transcription reaction and incubate at 37°C for 10 min.
Extract reaction twice with an equal volume of chloroform. Then concentrate with a YM-30 Centricon spin filtration unit until volume is less than 100 µl. Add an equal volume of 2× formamide gel loading buffer and heat to 65°C for 5 min.
Transfer to ice and then load on a pre-run 10% acrylamide/urea gel. Gel is prepared by adding 75 µl 10% ammonium persulfate and 25 µl TEMED to 25 ml of 10% acrylamide/ urea gel solution (see Note 8).
Separate plates and transfer gel to a piece of plastic wrap over a UV-shadowing screen. Use short wavelength handheld UV-light to visualize RNA (see Note 9).
Excise the piece of gel with the RNA and transfer to a 15 ml centrifuge tube with 2.5 ml of DPBS. Incubate on a rotator at 37°C for 4 h. Spin liquid with eluted RNA through a Centrex spin filter to remove any small pieces of gel that may be have been carried over in the liquid and then concentrate with a YM-30 Centricon spin filter unit. Wash twice by adding 2.5 ml DPBS and repeating spin.
Quantify RNA by measuring OD260 of a 1:50 dilution of the recovered material.
3.10. Annealing Reaction
The in vitro transcribed aptamer/siRNA upper strand is diluted to 10 µM and the complementary siRNA lower strand is diluted to 20 µMin DPBS (with calcium and magnesium) and this mixture is heated to 65°C for 5 min to denature the oligos. The reaction is then cooled to 37°C for 10 min to allow the two RNAs to anneal (see Note 10).
3.11. Cell Surface Binding Assay
Binding of the aptamer–siRNA chimeras to prostate cancer cells expressing PSMA can be assessed with flow cytometry. First, trypsinize PC-3 or LNCaP cells, wash twice with 500 µL PBS, and fix in 400 µL of fix solution for 20min at room temperature. Wash cells with PBS to remove formaldehyde, resuspend in binding buffer and incubate at 37°C for 20 min. Then pellet cells and resuspend in 100 µL 37°C binding buffer containing 400 nM FAM-labelled A10 aptamer or FAM-labelled aptamer– siRNA chimera (see Note 11 regarding the volume and RNA concentration of the labelling reaction). Incubate cells at 37°C for 30–40 min, wash three times with 500 µl 37°Cbinding buffer and resuspend in 400 µL 37°C fix solution. Incubate at 37°C for 5–10 min and then measure cellular fluorescence with flow cytometry.
3.12. Cell Culture (for siRNA Activity Assays)
Plate LNCaP or PC-3 cells in 6-well dishes at a density of 60% confluency. Incubate with aptamer siRNA chimeras or transfect with corresponding siRNAs (without aptamers). For siRNA transfections, transfect with 400 nM siRNA with Superfect (Qiagen) following manufacturer’s instructions 1 and 3 days after plating cells. Add aptamer siRNA chimeras at the same concentration directly to the culture media of appropriate wells 1 day after plating cells. Remove media from these wells and replace with fresh media also supplemented with 400 nM of the appropriate chimera 3 days after plaling cells. Grow cells for an additional 2 days and then process as detailed below for either immunoblotting or flow cytometry.
3.13. siRNA Activity Assay (Immunoblotting)
Trypsinize cells transfected with siRNAs or treated with aptamer–siRNA chimeras as described above and then wash with PBS. Pellet cells and resuspend in RIPA buffer. Incubate on ice for 20 min. Pellet cells again and transfer supernatants to fresh tubes. Quantify protein concentration in supernatants with a Bradford assay. Run 50 µg of each on an SDS-PAGE gel (8.5% acrylamide for Plk1, 15% acrylamide for Bcl-2). Transfer protein to a PVDF membrane via electrophoresis. Block membranes with 5% milk in PBS. Then incubate membranes in block plus 1:1,000 of either anti-Plk1 or anti-Bcl2 antibodies diluted in 5% milk in PBS.
3.14. siRNA Activity Assay (Flow Cytometry)
Trypsinize cells transfected with siRNAs or treated with aptamer– siRNA chimeras, wash three times with 500 µl PBS and then count with a hemacytometer. Resuspend 400,000 cells in 400 µl FIX/PERM buffer for a final concentration of 5 × 105 cells/ml and incubate at room temperature for 20 min. Pellet cells, resuspend in 500 µl PERM/WASH buffer, wash three times with 500 µl PERM/WASH buffer and then resuspend in 50 µl PERM/WASH buffer plus 20 µg/ml anti-human Plk1, antihuman Bcl2 or the appropriate isotype-matched control antibody. Incubate cells at room temperature for 40 min, wash three times with 500 µl PERM/WASH buffer and then incubate for 30 min at room temperature in 50 µl PERM/WASH buffer with 1:500 diluted anti-mouse IgG-APC. Wash cells three times with PERM/WASH buffer and then resuspend in PBS. Measure cellular fluorescence with flow cytometry.
3.15. In Vivo Testing of PSMA-Targeting Aptamer–siRNA Chimeras
Because the A10 aptamer binds specifically to the human orthologue of PSMA, chimeras made with the A10 aptamer will only target cells of human origin or cells engineered to express human PSMA. One approach for in vivo testing is to grow human prostate cancer tumours in nude mice as described below. The use of a prostate cancer cell line that does not express PSMA (PC-3) can serve as a negative control, while the PSMA-expressing prostate cancer cell line LNCaP can produce the aptamer-targeted tumour. Culture PC-3 cells in Ham’s F12-K medium supplemented with 2 mM l-glutamine, 1.5 g/L sodium bicarbonate, and 10% FBS. LNCaP cells were propagated in RPMI 1640 medium containing l-glutamine supplemented with 1.5 g/L sodium bicarbonate, 4.5 g/l glucose, 10 mM HEPES, 1.0 mM sodium pyruvate, and 10% FBS. Trypsinize cells, wash with DPBS and resuspend in DPBS. Count with a hemacytometer. Pellet cells again and resuspend in DPBS + 50% Matrigel with a cell concentration of 5 × 107 cells per milliliter. Inject 100 µl subcutaneously into the flanks of nude mice. Monitor tumour growth by examining animals and measuring any visible tumours every other day. Allow tumours to reach 0.5–1.0 cm in diameter and then inject with 200 pmol of chimeras diluted in 75 µl DPBS every other day. Continue to monitor tumour growth by measuring tumours every 2–3 days using a caliper and sacrifice animals if tumours grow excessively large (>2.0 cm in diameter).
Acknowledgements
This work was supported by the European Molecular Imaging Laboratory (EMIL) Network (LSHC-2004-503569) and by the MIUR-FIRB Grant (#RBIN04J4J7). We wish to thank C.L. Esposito, B. Tavitian, F. Duconge and D. Libri for fruitful discussions.
Footnotes
TEMED is best stored at room temperature in a desiccator. Buy small bottles as it may decline in quality (gels will take longer to polymerize) after opening.
The solution precipitates when exposed to light, if this occurs you need to prepare a new solution.
Transfer buffer can be used for up to five transfers within 1 week so long as the voltage is maintained constant for each successive run (the current will increase each time). Adequate cooling to keep the buffer no warmer than room temperature is essential in order to prevent heat-induced damage to the apparatus and the experiment.
T7Y639F RNA polymerase is used to improve yields.
2’F-Py RNAs are used because of their increased resistance to degradation by seric nucleases.
If urea precipitates, warm to 37°C to re-dissolve prior to pouring gel.
It is recommended that an aliquot of this DNA duplex be sequenced from both 5’ and 3’ ends to confirm the correct sequence.
Run gel until the first dye front is close to the bottom.
Only observe through UV-blocking eyeglasses because UV light is harmful to eyes.
Because the lower strand is in excess, there should be residual, unpaired lower strand RNA in the mixture following annealing. For many applications, this is not a concern. For instance, if the chimera is to be applied to cells in culture in the presence of serum, this RNA, which does not include 2’-fluoro modified pyrimidines, will be rapidly degraded by serum nucleases. However, if necessary, this RNA can be removed by purifying the aptamer–siRNA chimera on a non-denaturing acrylamide gel.
Because generation of FAM-labelled RNAs is expensive, it is usually desirable to minimize the volumes of these labelling reactions in order to conserve RNA. However, the concentration of RNA used must be rather high because the incorporation efficiency of FAM in the in vitro transcriptions is probably less than 50%. For this reason, it may be necessary to increase the concentration of RNA into the micromolar range to achieve binding saturation.
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Science. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 4.Osborne SE, Ellington AD. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 5.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 6.Famulok M, Mayer G. Intramers and aptamers: applications in protein-function analyses and potential for drug screening. Chem Bio Chem. 2005;6:19–26. doi: 10.1002/cbic.200400299. [DOI] [PubMed] [Google Scholar]
- 7.Morris KN, Jensen KB, Julin CM, Weil M, Gold L. High affinity ligands from in vitro selection: complex targets. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Homann M, Goringer HU. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. 1999;27:2006–2014. doi: 10.1093/nar/27.9.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerchia L, Ducongé F, Pestourie C, Boulay J, Aissouni Y, Gombert K, Tavitian B, de Franciscis V, Libri D. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3:e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 11.Pestourie C, Cerchia L, Gombert K, Aissouni Y, Boulay J, de Franciscis V, Libri D, Tavitian B, Duconge F. Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides. 2006;16:323–335. doi: 10.1089/oli.2006.16.323. [DOI] [PubMed] [Google Scholar]
- 12.Jhiang SM. The RET proto-oncogene in human cancers. Oncogene. 2000;19:5590–5597. doi: 10.1038/sj.onc.1203857. [DOI] [PubMed] [Google Scholar]
- 13.Ichihara M, Murakumo Y, Takahashi M. RET and neuroendocrine tumors. Cancer Lett. 2004;204:197–211. doi: 10.1016/S0304-3835(03)00456-7. [DOI] [PubMed] [Google Scholar]
- 14.Hansford JR, Mulligan LM. Multiple endocrine neoplasia type 2 and RET: from neoplasia to neurogenesis. J. Med. Genet. 2000;37:817–827. doi: 10.1136/jmg.37.11.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi M. The GDNF/Ret signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12:361–373. doi: 10.1016/s1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- 16.Putzer BM, Drosten M. The RET proto-oncogene: a potential target for molecular cancer therapy. Trends Mol. Med. 2004;10:351–357. doi: 10.1016/j.molmed.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Manié S, Santoro M, Fusco A, Billaud M. The RET receptor: function in development and dysfunction in congenital malformation. Trends Genet. 2001;17:580–589. doi: 10.1016/s0168-9525(01)02420-9. [DOI] [PubMed] [Google Scholar]
- 18.Pirollo KF, Zon G, Rait A, Zhou Q, Yu W, Hogrefe R, Chang EH. Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Human Gene Ther. 2006;17:117–124. doi: 10.1089/hum.2006.17.117. [DOI] [PubMed] [Google Scholar]
- 19.Song E, Zhu P, Lee S, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 20.Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: A central role for 2’-hydroxyl uridines in immune responses. Eur. J. Immunol. 2006;36:1222–1230. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- 21.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu TC, Twu KT, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1115. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 24.Chen CH, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 26.Mi J, Zhang X, Giangrande PH, McNamara JO, 2nd, Nimjee SM, Sarraf-Yazdi S, Sullenger BA, Clary BM. Targeted inhibition of alphavbeta3 integrin Cell-Specific Aptamers for Targeted Therapies 77 with an RNA aptamer impairs endothelial cell growth and survival. Biochem. Biophys. Res. Commun. 2005;338:956–963. doi: 10.1016/j.bbrc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 27.Mori T, Oguro A, Ohtsu T, Nakamura Y. RNA aptamers selected against the receptor activator of NF-kappaB acquire general affinity to proteins of the tumor necrosis factor receptor family. Nucleic Acids Res. 2004;32:6120–6128. doi: 10.1093/nar/gkh949. [DOI] [PMC free article] [PubMed] [Google Scholar]