Summary
Hsp70 family proteins are highly conserved chaperones involved in protein folding, degradation, targeting and translocation, and protein complex remodeling. They are comprised of an N-terminal nucleotide binding domain (NBD) and a C-terminal protein substrate binding domain (SBD). ATP binding to the NBD alters SBD conformation and substrate binding kinetics, but an understanding of the mechanism of interdomain communication has been hampered by the lack of a crystal structure of an intact chaperone. Were-port here the 2.6 Å structure of a functionally intact bovine Hsc70 (bHsc70) and a mutational analysis of the observed interdomain interface and the immediately adjacent interdomain linker. This analysis identifies interdomain interactions critical for chaperone function and supports an allosteric mechanism in which the interdomain linker invades and disrupts the interdomain interface when ATP binds.
Introduction
Hsp70s are ubiquitous chaperones (in yeast, there are 14 representatives [James et al., 1997]) whose many cellular functions (Young et al., 2003; Bukau and Horwich, 1998; Rothman and Schmid, 1986; Glick, 1995) are specified through compartmentalization and interaction with cochaperones (Cyr et al., 1994). In their ATP bound forms, Hsp70s bind and release substrates quickly, while, in the ADP bound state, substrate binding and release are slow (Schmid et al., 1994; Greene et al., 1995). Structures of isolated SBDs reveal substrate peptides bound in a hydrophobic pocket covered by an α-helical lid, which would have to open to allow substrate binding or release (Zhu et al., 1996). The fast substrate binding/ release state is therefore presumed to correspond to the ATP bound and lid open conformation, while the slow substrate binding/release form may represent the conformation with ADP bound and the lid closed. Cycles of ATP hydrolysis in the NBD may not only alter SBD conformation and substrate binding kinetics but may also drive movement of the SBD relative to the NBD to provide a power stroke and allow Hsp70s to serve as motors in processes such as protein translocation through transport channels (Voisine et al., 1999). A large ATP-driven conformational change in these chaperones is consistent with small-angle X-ray scattering (SAXS), indicating that ATP binding to bHsc70 or E. coli Hsp70 (DnaK) changes their radii of gyration (Wilbanks et al., 1995; Shi et al., 1996). Crystal structures of isolated NBDs from bovine and human Hsc70 (in ATP and ADP bound forms), and of isolated SBDs (with and without bound peptide substrates) from rat Hsc70 and DnaK have been determined (Zhu et al., 1996; Flaherty et al., 1990, 1994; Sriram et al., 1997; Wang et al., 1998; Morshauer et al., 1999). However, the crystal structure of a functionally intact chaperone containing both the NBD and SBD has not been described. This has hindered our understanding of the mechanism of action of these proteins. We report here the crystal structure of a functionally intact bHsc70 molecule. Together with analysis of mutants in the interdomain linker and interface, this structure provides insight into the allosteric mechanism of the Hsp70 chaperones.
Results
Crystallization and Structure Determination
A truncated bHsc70 (bHs70ΔCterm, aa 1–554) that contains both the NBD and SBD, but lacks the 10 kDa C-terminal oligomerization domain, is functional but aggregates less than full-length protein (Ungewickell et al., 1997; Wilbanks et al., 1995). Crystals of this truncated protein could be grown but diffracted poorly. In an effort to reduce surface entropy and generate new crystal contacts, we prepared eight mutants in which clusters of exposed, charged residues were changed to alanines (Derewenda, 2004). One of these (E213A/D214A) gave improved crystals, and both its kinetics and stochiometry of action in a clathrin cage disassembly assay, which recapitulates in vivo uncoating of endocytic vesicles (Barouch et al., 1994), were identical to wt (Figure S1). A 2.6 Å data set was collected from a single crystal using a rotating anode X-ray source (Table S1). A molecular replacement solution for the NBD was obtained with coordinates for residues 3–384 of bHsc70 (3HSC), and a solution for the SBD was identified with residues 395–516 of DnaK (1DKZ). Density for the linker between these domains and 35 C-terminal residues was apparent, and, after building these regions and rebuilding of the NBD and SBD, a model of residues 1–554 and 118 waters was refined to an R/Rfree of 22.0/29.7 (Table S1).
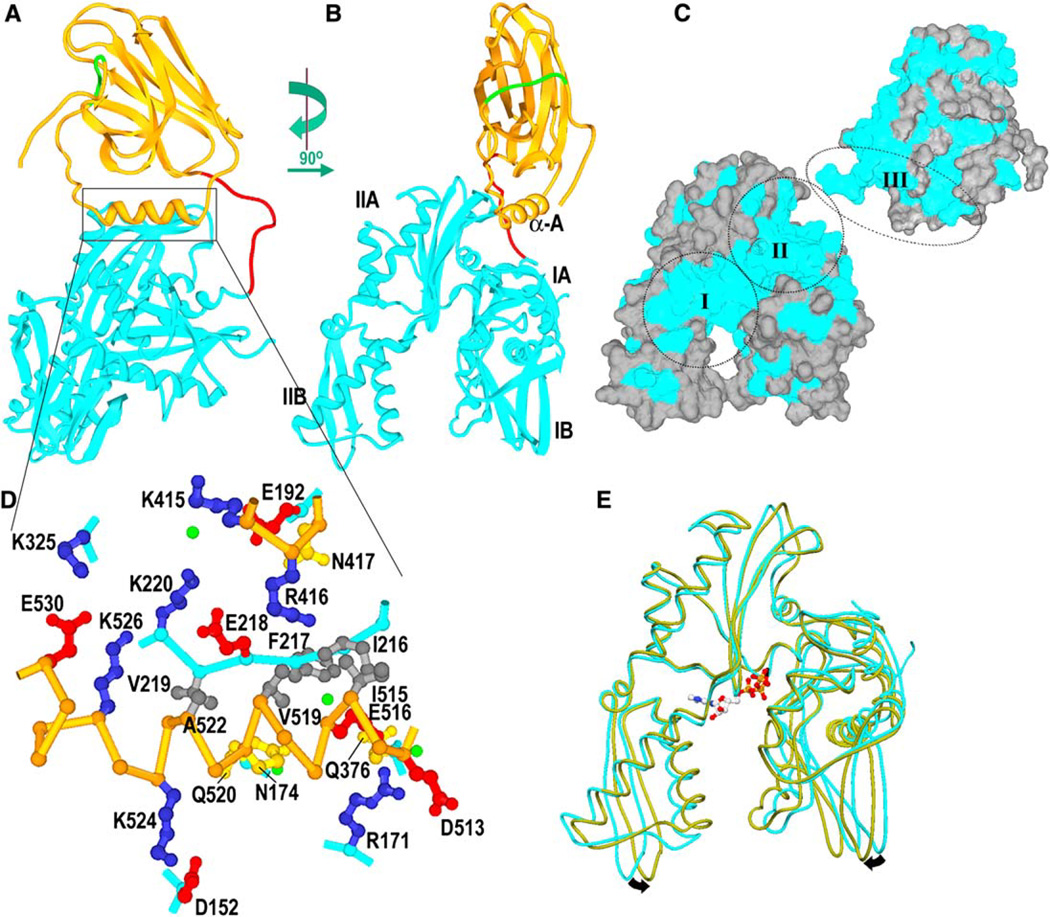
Overall Structure and the Interdomain Interface
The structure (Figures 1A and 1B) reveals the SBD (aa 395–554, orange) connected by an exposed linker (aa 384–394, red) to the NBD (aa 1–383, cyan), with residues 415–417 and helix A of the SBD (aa 513–524; helix lettering as in Zhu et al. [1996]) resting in a groove between lobes IA and IIA of the NBD. Helix B of the SBD (aa 527–554) is partially unwound, and residues 539–544 (in green; Figures 1A and 1B) occupy the substrate binding pocket. Such intramolecular binding has been observed in the NMR structure of an isolated SBD (Wang et al., 1998) and probably reflects destabilization of helix B due to truncation of the helical lid subdomain (aa ~507–650).
Figure 1. bHsc70ΔCterm Structure.
(A) Ribbons representation of bHsc70 with the NBD (aa 1–383) in cyan, the SBD (aa 395–554) in orange, the linker (aa 384–394) in red, and aa 539– 544, which occupy the substrate binding site, in green.
(B) As in (A), but with the molecule rotated by 90° as indicated.
(C) Solvent-accessible surface of bHsc70 with residues that are identical in bHsc70 and the DnaK NBD and SBD colored blue and other residues in gray. The SBD has been broken away from the NBD and rotated to expose its interface with the NBD. The two largest regions of conserved surface in the NBD are around the ATP binding site (“I”) and the interface with the SBD (“II”). The interdomain linker and the interface with the NBD on the SBD (“III”) are also well conserved.
(D) Expanded view of interactions between the SBD (C-α trace in orange) and NBD (C-α trace in cyan; side chains in blue, red, yellow, or dark gray for +, −, polar, or nonpolar, respectively; waters in green). Interdomain interactions include the following: salt bridges between K325 and E530 and between K524 and D152; longer-range ionic interactions between E218 and both R426 and K526 as well as between R171 and D513; a cluster of six hydrophobes (I216, F217, V219, I515, V519, A522); one direct (N417:E192) and five water-mediated H bonds (between Q520 and N174, K220 and K415, E516 and Q376, and R376 and the F217 amide).
(E) Superposition of the NBD from the bHsc70 structure (cyan), with structure of an isolated bHsc70 NBD (1kax) crystallized with bound ATP (olive). ATP binding may induce the closing suggested by the arrows.
The molecule’s shape and dimensions are consistent with SAXS studies (Wilbanks et al., 1995; Shi et al., 1996) that have shown DnaK and bHsc70 to be bilobed, elongated, prolate molecules with dmax values of 110– 130 Å (dmax for a truncated Hsc70 like that described here is 5 Å smaller, and dmax for this structure is 105 Å). The interacting regions of the SBD and NBD show shape and charge complementarity, bury 900 Å of protein surface on each domain, and include six direct or water-mediated H bonds, five ionic interactions, and a clustering of hydrophobes (Figure 1D). The interdomain interface encompasses the most well-conserved region on the NBD surface (region II in Figure 1C) and a highly conserved region on the SBD surface (region III, Figure 1C).
Conformation of the NBD
No density for bound nucleotide is visible in the electron density, and the NBD is found to be 7.4° more open than the structure of an isolated NBD complexed with ATP or ADP (Figure 1E). Such “open” NBD conformations have been observed in complexes of isolated NBDs with nucleotide exchange factors (NEFs; Harrison et al., 1997; Sondermann et al., 2001). The observation that the nucleotide-free enzyme assumes the open conformation in the absence of NEFs suggests that nucleotide release induces the open conformation of the NBD.
Preparation of Mutants within the Interdomain Interface and Linker
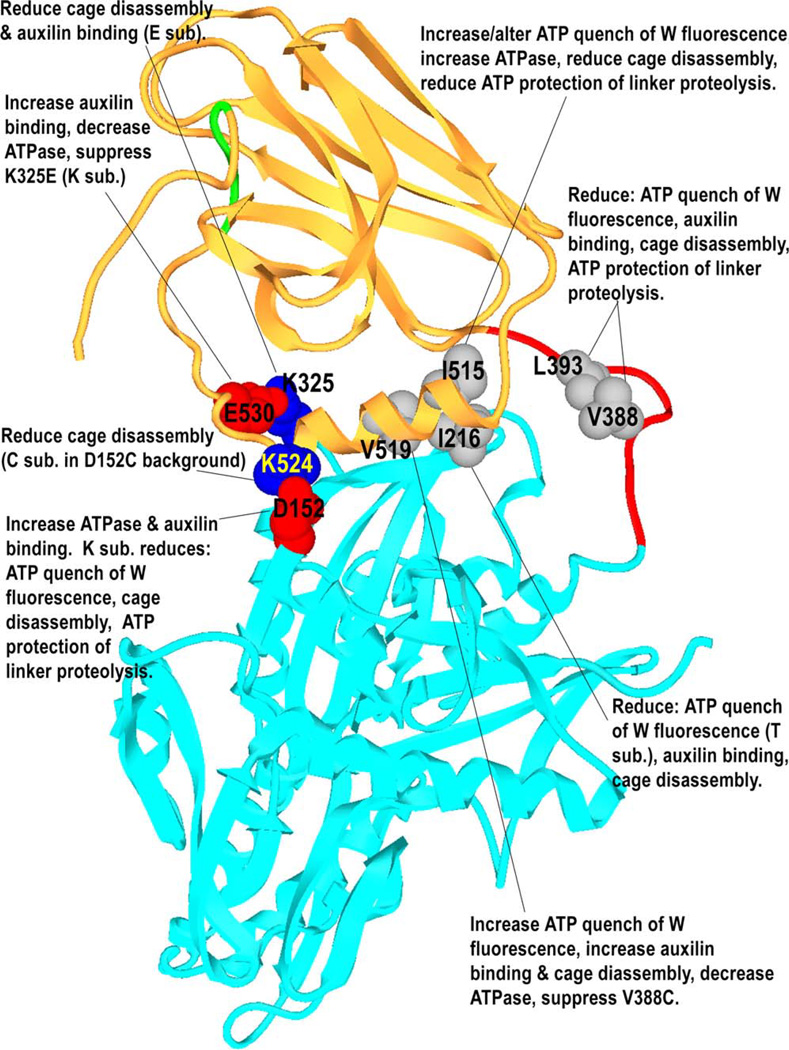
Mutants D152C, D152K, K325E, K325C, K524C, K524D, E530C, and E530K were constructed to assess the effects of disrupting ionic interactions between the NBD and the C-terminal region of SBD helix A (Figure 1D). Mutants I216T, I216C, I216H, I515C, and V519C were constructed to disturb hydrophobic interactions between the NBD and the N-terminal region of SBD helix A (Figure 1D). We also constructed V388C, L393C, and a quadruple arginine substitution for residues 391–394 in the interdomain linker. The multiply arginine-substituted enzyme was proteolyzed in vivo and could not be purified. However, V388C, L393C, and all the interface mutants could be purified. Subsequently, a set of double mutants (D152K/K524D, D152C/K524C, K325E/E530K, K325C/E530C, I216C/V388C, V388C/ L393C, V388C/I515C, and V388C/V519C) was constructed to detect interactions between mutations or test proximity relationships by disulfide crosslinking (Figure 7 illustrates the positions of the mutated residues in the structure). Mutations were constructed in the bHsc70ΔCterm background (referred to here as wt) because full-length recombinant bHsc70 has been reported to be inactive in vesicle uncoating, suggesting that it is not properly folded (Rajapandi et al. [1998]; note that even preparation of properly folded bHsc70ΔCterm required expression at 30°C), and because oligomerization of the full-length protein complicates binding studies.
Figure 7.
Effects of Mutations of Different Amino Acids in the Interdomain Interface or Linker on Cage Disassembly, ATP Quenching of W Fluorescence, ATP Protection of the Linker from Proteolysis, ATPase Rates, and Auxilin Binding
Mutant Enzyme Clathrin Cage Disassembly Activity
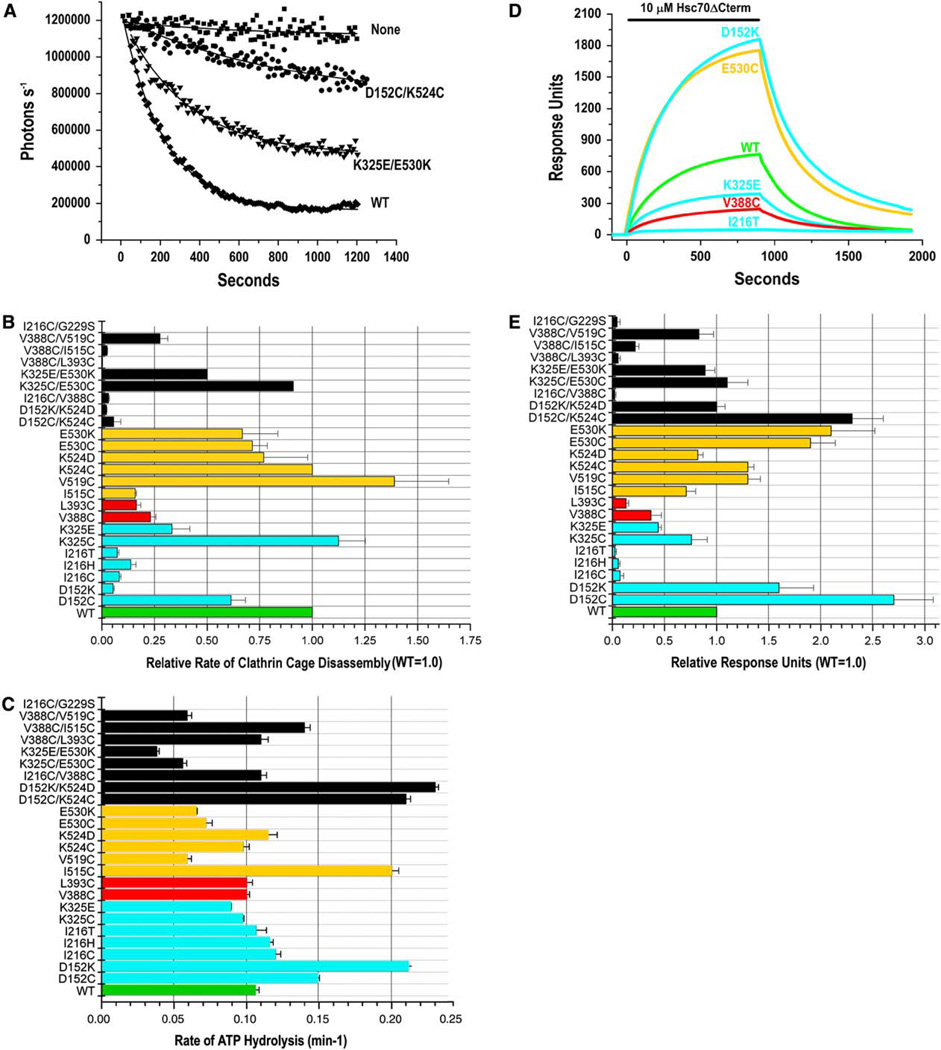
Hsc70 uncoats clathrin-coated vesicles in vivo (Morgan et al., 2001; Newmyer and Schmid, 2001). In vitro, disassembly of clathrin cages requires ATP, the J cochaper-one auxilin, and an Hsc70 with both an NBD and SBD (Rothman and Schmid, 1986; Barouch et al., 1994; Ungewickell et al., 1995, 1997; Mayer et al., 1999; Suh et al., 1998). Mutations that affect interdomain communication maytherefore affect clathrin cage disassembly. To quantitatively compare mutant enzyme disassembly activity, the decrease in light scattering that occurs as cages are disassembled was monitored continuously following initiation of disassembly by addition of Hsc70ΔCterm (preincubated with 1 mM ATP) to reactions containing auxilin and cages. Data were fit to single exponential decays. A representative experiment is shown in Figure 2A: with a clathrin cage concentration corresponding to 0.2 µM triskelia and with 0.6 µM wt Hsc70ΔCterm, 50% of the cages are disassembled in ~130 s, while the K325E/E530K and D152C/K524C double mutants are ~2- and ~20- fold slower (Figure 2A). Data for all mutants is presented in Figure 2B.
Figure 2. Effects of Interdomain Interface and Linker Mutations on Clathrin Cage Disassembly, ATPase Rates, and Auxilin Binding.
(A) Clathrin cage disassembly measured by number of photons scattered per second versus time for the wt and two mutant bHsc70Δcterm enzymes. “None” is a reaction with no chaperone. Data are fit to single exponential decays; error bars show ranges from two experiments.
(B) Relative rates of clathrin cage disassembly (wt = 1.0) for all enzymes. Data bars in all panels are colored green for wt, cyan for single mutants in the NBD, red for single mutants in the linker, orange for single mutants in the SBD, and black for all double mutants. Error bars are ±SE for at least two experiments.
(C) Single-turnover ATPase rates for all enzymes; error bars show ranges from two experiments.
(D) Representative SPR experiment showing wt and mutant enzyme binding to immobilized auxilin with 1 mM ATP and an ATP-regenerating system (no binding is observed in the absence of ATP or coupled auxilin).
(E) Summary of binding for all enzymes. Binding is relative to wt controls, which were run in parallel for every experiment. Error bars are ±SE for two to four determinations.
Substitutions at I216 and I515, which make the most extensive interdomain hydrophobic contact (Figure 1D), reduce disassembly rates 6- to 10-fold. However, a mutation at the adjacent V519 results in a modest but reproducible increase in disassembly activity. Substitutions at V388 and L393 in the linker reduce disassembly rates 4- to 6-fold. Mutations of residues involved in ionic interactions are generally less disruptive than mutations of the hydrophobic amino acids: substitutions at E530 or K524 do not markedly affect disassembly activity, while, at D152 and K325, the effect on disassembly depends on the nature of the introduced amino acid, with Cys substitutions being neutral while introductions of oppositely charged residues reduce disassembly rates by 3- and 20-fold for K325E and D152K, respectively (Figure 2B).
Many of the double mutants had more severe effects than the single mutants. V338C/L393C, for example, displayed a 60-fold lower disassembly rate than either V388C or L393C, indicating that the disruptive effects of mutations of hydrophobic side chains in the linker or interface are additive. Observation of intragenic suppression in the D152K/K524D and K325E/E530K double mutants would be consistent with a role for the D152:K524 and K325:E530 salt bridges in chaperone function, though the absence of such suppression could be ambiguous since steric considerations can also affect whether functional salt bridges are recovered in such double mutants, and individual side chains can have functions distinct from their ability to participate in an ionic interaction with a specific partner. In fact, K524D did not suppress the disassembly defect of D152K (though the proximity of these two residues is supported by efficient crosslinking in a D152C/K524C mutant, see below). However, partial suppression of K325E was observed in K352E/E530K (Figure 2B). V519C also partially suppressed disassembly by V388C, though this effect was modest—V519C/V338C was reproducibly 25% more active than V388C—and not allele-specific, since V519C was also more active than the wt enzyme.
Effects on ATP Binding and Hydrolysis
Interactions with the SBD modulate the ATPase activity of the NBD: peptide binding to the SBD, for example, increases ATPase rates by 2- to 6-fold (Ha et al., 1997). Mutations that affect NBD:SBD interactions could therefore alter ATP hydrolysis by the NBD. To detect such effects, we measured ATP binding and single-turnover ATPase rates. With the exception of I216C/G229S, which was inadvertently generated during construction of I216C, all the enzymes had ATP binding activity like wt (data not shown). Gly229 is near the ATP binding site, and the I216C/G229S enzyme was inactive in cage disassembly and ATP hydrolysis (Figures 2B and 2C) and was used as a negative control in these and subsequent experiments. Many mutants displayed ATPase rates similar to wt. However, D152C, D152K, and I515C rates were increased ~1.5- to 2-fold, and rates for V519C, E530K, and E530C were reduced by ~40% (Figure 2C). Though these effects were modest, their significance is underscored by these observations: (1) rate determinations were highly reproducible from assay to assay, with variations typically 5%–6% (error bars in Figure 2B give the range from two experiments); (2) substitution of different residues at the same position has similar effects on ATPase rates (i.e., C and K at 152 or 530); and (3) similar effects are seen in double mutants that carry one of the single mutations that affect ATP hydrolysis; the D152C/K524C, D152K/K524D and V388C/ I515C mutants display increased ATPase rates, while the K325C/E530C, K325E/E530K, and V388C/V519C mutants display reduced rates (Figure 2C). The magnitude of these effects is comparable to the stimulation in ATPase activity due to substrate binding to the SBD and, for SBD mutations at 515/519/530, we may conclude that these effects are due to changes in interdomain coupling. The D152 mutations could, however, be affecting ATPase rates via a direct effect on the NBD. To test this, we generated the 44 kDa NBD fragment from wt and D152K by proteolysis and measured their ATPase rates to be 0.30–0.32 min−1 and 0.31–0.32 min−1, respectively (ranges from two experiments), indicating that D152 mutants are also affecting ATPase rates via changes in interdomain coupling.
We also measured the effect of a substrate peptide (ALLLSAPRRGAGKKC) on the ATPase rates of these enzymes but did not observe significant ATPase stimulation even at very high (1024 M) peptide concentrations for any of the enzymes (data not shown). Notably, the ATPase rate of bHsc70ΔCterm is 3- to 6-fold higher than for full-length bHsc70 (Ha et al., 1997; Johnson and McKay, 1999). This is comparable to the stimulation conferred by peptide and suggests that the C-terminally truncated enzyme is autostimulated via intramolecular binding of the unwound B helix, so that little additional stimulation is observed upon peptide addition.
Effects on Auxilin Binding
Hsc70 binding to J cochaperones requires ATP and both the NBD and SBD (Ungewickell et al., 1997; Mayer et al., 1999; Suh et al., 1998). Hsc70 mutations can therefore affect J cochaperone binding directly, if they target residues that contact the cochaperone, or indirectly, if they interfere with the ATP-driven conformational transition that allows binding to the J cochaperone. We measured binding of the mutant enzymes to immobilized auxilin using surface plasmon resonance (SPR). A representative experiment is shown in Figure 2D, and data for all mutants is summarized in Figure 2E. All I216 mutations show large reductions in binding (20-fold or more), while K325E, V388C, and L393C exhibit more modest (2.5- to 7-fold) reductions. Despite its poor disassembly activity, I515C shows wt-like binding, and several mutations, including one (D152K) that exhibits very poor disassembly activity, display increased binding. Though the increases in binding are modest (1.6- to 2.7-fold), they are reproducible, and confidence that these effects are due to the mutations is increased by the observation that different substitutions at the same residue have similar effects (K and C at 152 or 530) and that D152C/K524C and D152K/K524D display increased auxilin binding like the D152 single mutants. The K325E/E530K and K325C/ E530C mutants, on the other hand, do not exhibit the enhanced binding seen in the E530 single mutants, indicating that mutations at K325 suppress the increased binding of the E530 mutations. However, this suppression may not be allele specific, since the K325 single mutants show reduced binding. Another example of such suppression is seen with V388C/V519C, which corrects the poor binding of V388C.
Effects on ATP Quenching of Tryptophan Fluorescence
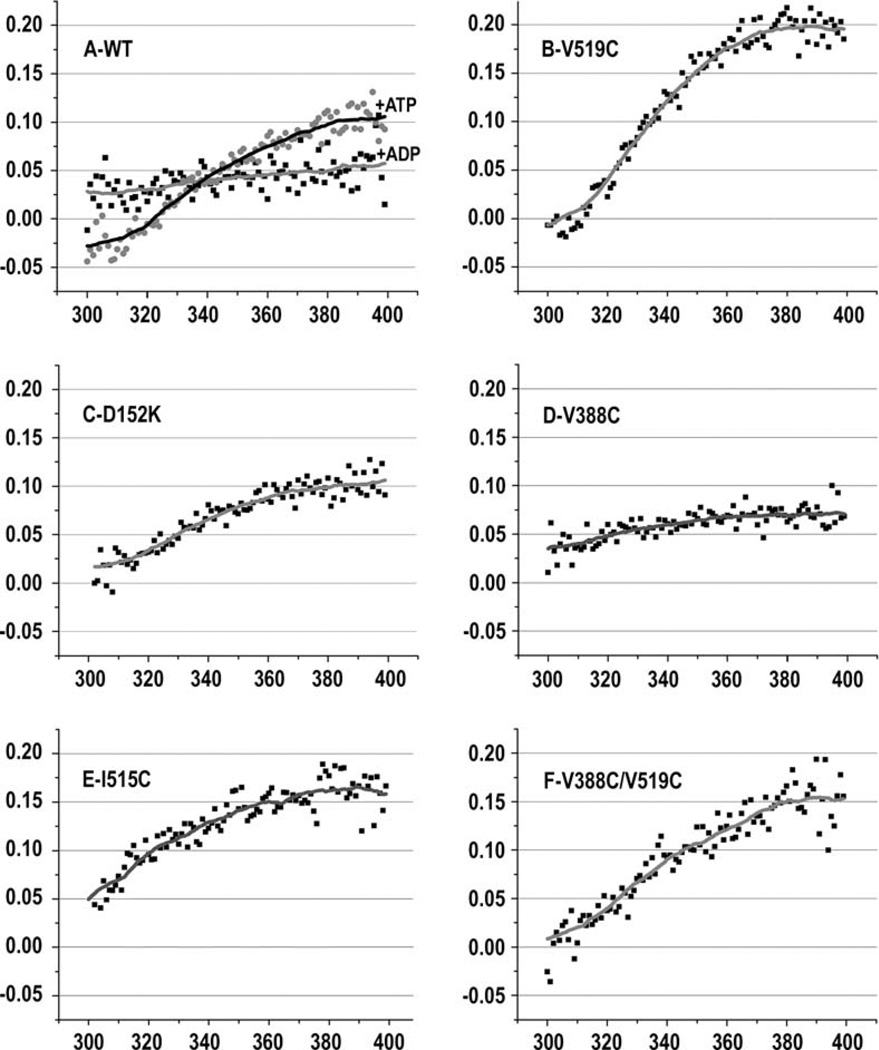
DnaK and bHsc70 contain a single tryptophan in the NBD (W90 in bHsc70). ATP binding quenches and blue shifts the fluorescence emission from this tryptophan (Moro et al., 2003), indicating that ATP binding alters NBD conformation. The extent of the blue shift and quench differs in an intact Hsp70 versus an isolated NBD, indicating that interactions with the SBD modulate the ATP-driven conformational change in the NBD (Montgomery et al., 1999). Mutations that affect interdomain communication may therefore affect ATP quenching of fluorescence emission. To identify such effects, we measured fluorescence for the mutant Hsc70s. The fractional differences in emission with and without ATP for all mutants are presented in Figure S2, and representative data are plotted in Figure 3. In the wt enzyme (Figure 3A), ATP quenches long wavelength emission (370–400 nm) by ~10%. The magnitude of the ATP quench is smaller at shorter wavelengths, and, below 320 nm, fluorescence is increased by ATP (ATP has a similar effect on fluorescence from full-length Hsc70 purified from bovine brain, Figure S3). ADP binding causes a smaller (~5%) quench that is similar at high and low wavelengths (Figure 3A). Fluorescence quenching by ATP in mutants that had at least 25% of wt cage disassembly activity (D152C, K325C/E, K524C/D, E530C/K, K325C/E530C, and K325E/E530K) is similar to that seen with wt (Figure S2). However, V519C—which is more active than wt (Figure 2B)—stood out in that the magnitude of the ATP quench is approximately two times larger than wt (Figure 3B).
Figure 3. Effects of Interdomain Interface and Linker Mutations on ATP-Induced Fluorescence Changes.
Fractional differences in W fluorescence spectra with and without ATP ([fluorescence - ATP] - [fluorescence + ATP])/(fluorescence -ATP]) are plotted. In (A), spectral differences due to ADP are also plotted for comparison.
The fractional spectral difference plots for most of the less-active enzymes differ from those of the wt or more active enzymes (Figure S2). They exhibit either a reduction in the magnitude of the quench at long wavelengths (I216T, V388C [Figure 3D], L393C, D152K/K524D, and V388C/I216C) and/or an increase in the quench at short wavelengths (D152K [Figure 3C], I216T, V388C [Figure 3D], L393C, I515C, D152K/K524D, V388C/I216C, V388C/L393C, V388C/I216C, V388C/L393C, and V388C/ I515C). The plot for I515C is unique in that this poorly active enzyme displays a quench that is larger than wt at high wavelengths, but the quenching does not drop at lower wavelengths and remains above 5% over the entire wavelength range (Figure 3E). There were also three reduced-activity mutants—I216C, I216H (panel U), and D152C/K524C (Figure S2)—that exhibit fractional spectral differences similar to the active enzymes, indicating that the presence of wt-like ATP-dependent changes in fluorescence emission is not sufficient to ensure wt levels of chaperone activity.
Examples of intragenic suppression were also observed. Both I515C (Figure S2) and V519C partially suppress the loss of ATP quenching of fluorescence seen in V388C, with the stronger suppression occurring in the V388C/V519C mutant (compare Figures 3B, 3D, and 3F).
Effects on ATP Suppression of Linker Proteolysis
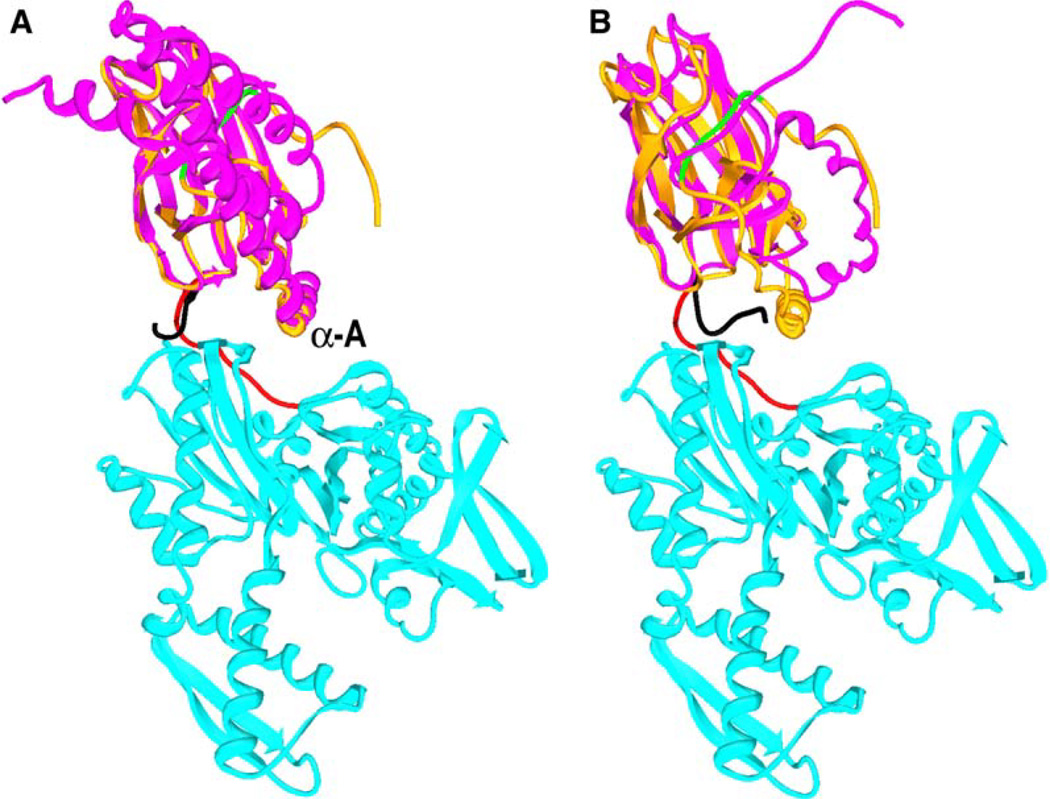
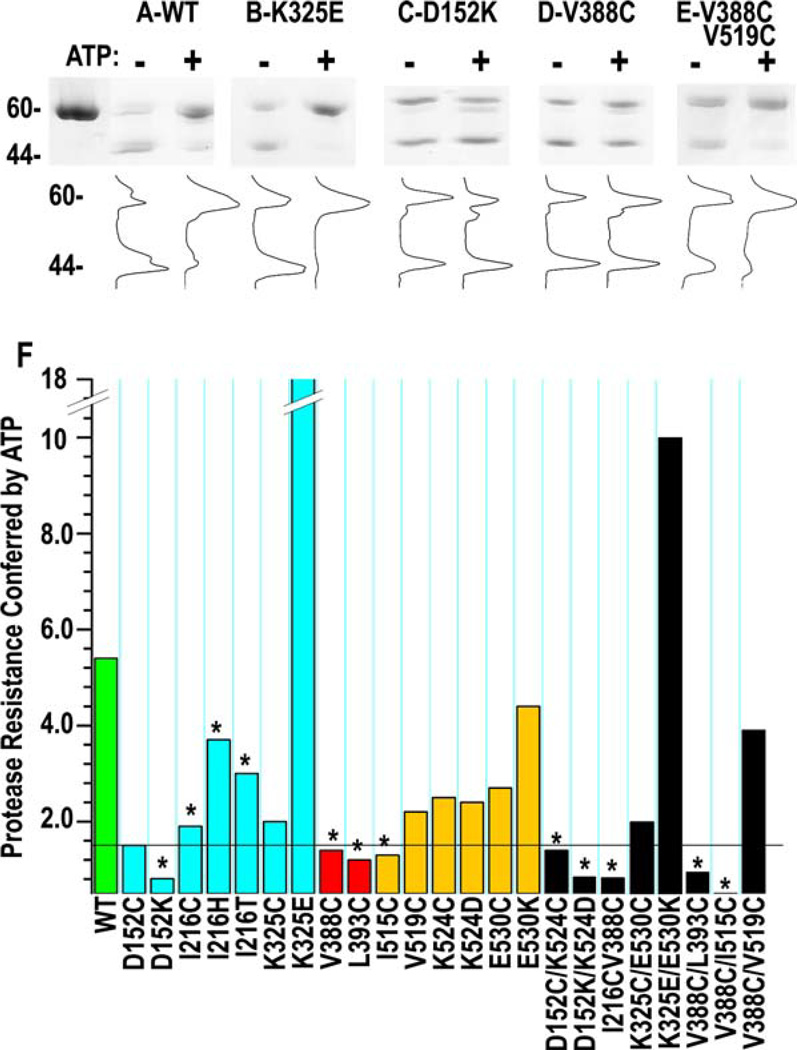
Crystal and NMR structures of isolated SBDs reveal the interdomain linker in two conformations: a linker “out” form in which the linker projects into solvent and assumes a conformation like that seen in the structure reported here, and a linker “in” form in which the linker is packed onto a hydrophobic patch on the SBD (Zhuet al., 1996; Wang et al., 1998). Figure 4 shows these alternate conformations in superpositions of the bHsc70 structure with DnaK SBD in the linker out conformation (Figure 4A) and the rat Hsc70 SBD in the linker in conformation (Figure 4B). The idea that the linker can alternate between in and out is supported by studies of DnaK and BiP (an Hsp70 homolog), which show that ATP protects the linker from proteolysis (Buchberger et al., 1995; Wei et al., 1995), suggesting that ATP binding causes the linker to move from an exposed to a buried conformation. To extend these observations to bHsc70 and to evaluate the effects of the mutations on this transition, we measured protease K sensitivity in the absence and presence of ATP. Representative SDS gels together with densitometric traces are shown in Figures 5A–5E. Digestion of wt enzyme in the absence of ATP leads to cleavage in the linker and rapid generation of the 44 kDa NBD as the major fragment. The fraction of 44 kDa fragment released from the wt enzyme is reduced 5- to 6-fold by ATP (Figures 5A and 5F), indicating that the linker becomes less protease accessible upon ATP binding.
Figure 4. Alternate Conformations of the Interdomain Linker.
(A) bHsc70ΔCterm structure colored as in Figure 1A with DnaK SBD (pdb 1dkx in magenta with the interdomain linker in black) superimposed.
(B) bHsc70ΔCterm with the rat Hsc70 SBD (pdb 1bpr in magenta with the interdomain linker in black) superimposed.
Figure 5. Effects of Interdomain Interface and Linker Mutations on ATP-Induced Protection of the Linker from Proteolysis.
(A–E) Proteolysis of indicated bHsc70ΔCterm enzymes in the absence (−) or presence (+) of ATP. Cleavage in the linker generates the 44 kDa NBD fragment. Densitometric traces are shown below their respective lanes.
(F) Degree of protease resistance conferred by ATP for all mutants is presented as the ratio of the fraction of 44 kDa fragment in the absence of ATP to the fraction in the presence of ATP ([%44 kDa(−ATP)]/[%44 kDa(+ATP)]). The horizontal line corresponds to a 1.5-fold decrease in linker protease sensitivity.
Quantitative data for all mutants are presented in Figure 5F, with mutants displaying less than 25% of wt cage disassembly activity highlighted by asterisks. With the exception of the I216 mutants, the enzymes with reduced disassembly activity all exhibit little or no ATP suppression of proteolysis in the linker (<1.5-fold protection; Figures 5C, 5D, and 5F), while all the active enzymes exhibit ATP protection of the linker, ranging from 1.5-fold for D152C up to 18-fold for K325E (Figures 5B and 5F). The significance of the enhanced ATP protection of the linker in K325E is corroborated by the observation of similar enhancement in the K325E/ E530K mutant. The observation of ATP protection of the linker in the I216 mutants correlates with the observation that these poorly active mutants also display ATP-dependent changes in fluorescence similar to wt (Figure S2). Further, just as V519C suppresses loss of activity and ATP-dependent fluorescence quenching in V388C (Figures 2B and 3), it also suppresses loss of ATP suppression of proteolysis in this mutant (Figures 5E and 5F).
We also characterized nucleotide-dependent changes in linker protease sensitivity in full-length Hsc70 purified from bovine brain and detected a similar ATP-dependent decrease in linker protease sensitivity (Figure S4), indicating that this conformational change is not due to truncation of the protein. ADP did not protect the linker from proteolysis in either bHsc70 or bHsc70ΔCterm (Figure S4), consistent with protease sensitivity and NMR studies that the nucleotide-free and ADP states of the enzyme are similar in conformation and distinct from the ATP form (Buchberger et al., 1995; Wei et al., 1995; Revington et al., 2004).
In summary, this characterization reveals that mutations in the interdomain interface and linker functionally affect interdomain communication, as assessed by effects on cage disassembly, ATPase rates, and auxilin binding. These functional effects correlate with structural measures of interdomain communication, with less-active enzymes revealing changes in ATP quenching of fluorescence emission and loss of ATP suppression of linker proteolysis.
Use of Cys-Substituted Enzymes to Assess Amino Acid Accessibility and Proximity
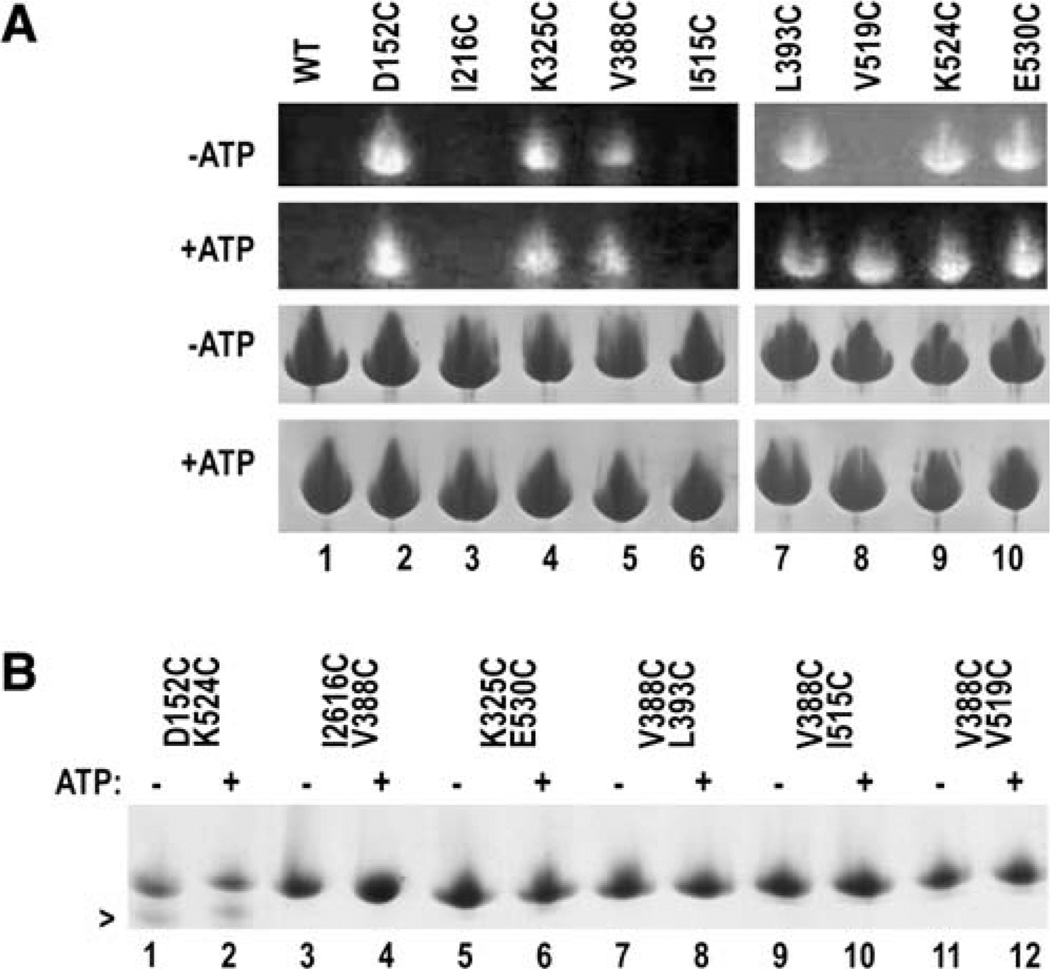
In structures of isolated SBDs or NBDs, residues 152, 216, 388, 393, 515, 519, 524, and 530 are exposed, but in the bHsc70 structure, positions 216, 515, and 519 are buried in the interdomain interface. To corroborate this, we reacted enzymes bearing single Cys substitutions at these positions with a thiol-specific fluorescent dye (monochlorobimane) in either the presence or absence of ATP. Reacted proteins were resolved by SDS-PAGE and visualized by UV illumination (Figure 6A, upper panels). The wt enzyme contains two buried cysteines and is not reactive with the dye (lane 1). In the absence of ATP, enzymes with Cys at 152, 325, 388, 393, 524, and 530 are labeled, while enzymes substituted at 216, 515, and 519 are not (Figure 6A, “−ATP”). In the presence of ATP, V519C also becomes labeled (Figure 6A, “+ATP”), suggesting that this part of the interdo-main interface becomes more solvent accessible upon ATP binding. We draw no conclusion regarding the lack of ATP-dependent labeling changes for I216C and I515C, since these mutants are poorly active.
Figure 6. Accessibility and Side Chain Proximity in Singly and Doubly Cys-Substituted Enzymes.
(A) (Top two panels) UV illumination of SDS-PAGE of singly Cys-substituted enzymes reacted with monochlorobimane in either the absence (“−ATP”) or presence (“+ATP”) of ATP. Lower two panels show Coomassie blue staining of the same gels.
(B) Nonreducing Coomassie-stained SDS-PAGE of enzymes treated with 3 mM oxidized glutathione in either the absence (“−”) or presence (“+”) of ATP.
We also measured disulfide bond formation in doubly Cys-substituted enzymes. The structure predicts that disulfide bond formation should be most efficient for D152C/K524C and L393C/V388C since the α carbons for these pairs are 8.2 and 6.8 Å apart, respectively (though the turn that brings L393 and V388 close together via packing of these aliphatic side chains would probably be disrupted in the double mutant). The α carbons for K325 and E530, however, are 11.5 Å apart (twice the α carbon separation in a disulfide), while the distances between the V388 and I216, I515, and V519 α carbons are 14.1, 17.7, and 21.9 Å, respectively, so minimal disulfide crosslinking is expected for the respective doubly Cys-substituted enzymes. Figure 6B shows nonreducing SDS-PAGE of these enzymes following treatment with 3 mM oxidized glutathione. A species migrating more rapidly than the reduced protein forms in the D152C/K524C reactions. No such species are seen in reducing buffer or in experiments with wt or singly Cys-substituted enzymes (data not shown), indicating that this rapidly migrating species corresponds to a protein with a disulfide between 152 and 524. Barely detectable levels of species migrating faster than the reduced protein and unaffected by addition of ATP also form in the K325C/E530C, V388C/I515C, and V388C/ V519C reactions. The small shifts in migration (relative to the reduced protein) for the latter species compared to the 152–524 disulfide-bonded enzyme are consistent with the differences in sequence separation between the disulfide-bonded amino acids in these enzymes. For this same reason, we do not expect to detect a disulfide-bonded protein in the V388C/L393C reactions, since a disulfide between these two residues would not appreciably affect migration.
Discussion
The phenotypes of the mutations characterized here (summarized in Figure 7) indicate that interdomain communication occurs across the interdomain interface observed in this crystal structure and involves the adjacent interdomain linker. Previous observations of alternate linker conformations in isolated SBDs (Zhu et al., 1996; Wang et al., 1998), and of ATP suppression of linker proteolysis (Buchberger et al., 1995; Wei et al., 1995), are consistent with the linker moving from an exposed to a buried conformation upon ATP binding. Such a role for the linker agrees with its hydrophobic character and conservation: comparison of archaeal, eubacterial, eukaryotic, and organellar Hsp70 sequences identifies the C-terminal half of the linker (bHsc70 388–396, VQDLLLLDV) as the most well-conserved element in the Hsp70s (Karlin and Brocchieri, 1998). We find that mutations that abolish ATP suppression of linker proteolysis also affect ATP quenching of tryptophan fluorescence and reduce clathrin cage disassembly activity. This indicates that these conformational changes are a necessary part of chaperone mechanism.
However, while necessary, these conformational changes are not sufficient for full activity. Mutations at I216, for example, have severe effects on auxilin binding and strong effects on cage disassembly but retain partial ATP-dependent linker protease resistance and, with the exception of I216T, show wt-like W fluorescence quenching by ATP. Thus, these mutations have strong effects on allosteric function but lesser effects on some structural aspects of allosteric communication. These mutations may allow part of the allosteric mechanism—those detected by changes in W fluorescence and linker protease sensitivity—to occur but may disrupt an important aspect of the mechanism that occurs directly through the interface. Mutations at I515 and V519, which sit on the same face of SBD helix A, increase the magnitude of the ATP-driven fluorescence quench but otherwise differ in that V519C shows ATP suppression of linker proteolysis and a wt-like decrease in the quench at short wavelengths, while I515C is defective in both of these structural signals of allosteric communication. I515C also decreases cage disassembly and increases ATPase rates but does not affect auxilin binding, while V519C exhibits enhanced disassembly and reduced ATPase rates. If the magnitude of the fluorescence quench reflects, in part, the magnitude of a shift in the equilibrium from a conformation in which the interdomain interface is intact to one in which this interface is partially disrupted by invasion of the linker, then I515C and V519C may both enhance the shift toward the latter conformation by destabilizing the interface, but I515C may have additional effects on function via direct changes in functionally important interactions with the NBD or the buried linker.
The binding site on Hsp70 for J domain cochaperones is poorly defined, in part because Hsp70 mutations affecting J domain binding may do so directly or by interfering with Hsp70 conformational change. However, mutations at DnaK R169 (bHsc70 R171) suppress a binding defect of a DnaJ with a mutation in the asp of the invariant HPD motif, suggesting that these two residues interact (Suh et al., 1998), and we have found that an auxilin J domain with a Cys substitution at the corresponding D876 crosslinks to an R171C bHsc70 mutant (J.J. and R.S., unpublished data). Modeling the J domain:bHs70 interaction so as to place these two residues in proximity puts positively charged J domain residues important for binding Hsc70 (Jiang et al., 2003) next to negatively charged regions of bHsc70 (Figure S5A). Though such modeling is tentative, it creates no steric clashes and places the J domain near the interdomain interface so that it could contact both the SBD and NBD and influence conformational changes in Hsp70 (Figure S5B).
Our conclusion that the interdomain interface on the NBD is the groove between lobes IA and IIA is consistent with a recent NMR chemical shift and dipolar coupling analysis that identified interdomain interface regions in a truncated T. thermophilus DnaK (Revington et al., 2005), as well as with identification of NBD mutations in E. coli DnaK that affect interdomain communication and are at or near this groove (Gassler et al., 1998). Assignment of part of the interface on the SBD to residues 415–417 (Figure 1D) is supported by identification of a K414 mutant in DnaK (R416 in bHsc70) that eliminates interdomain coupling (Montgomery et al., 1999). Identification of helix A as forming most of the interdomain interface in the SBD is more controversial. A DnaK lacking the helical subdomain of the SBD, including helix A, exhibits ATP stimulation of peptide release (Revington et al., 2005), suggesting that allosteric communication is present in this mutant. However, deletion of SBD helix A in ecDnaK eliminates the ATP quenching of W fluorescence that is considered a signature of allosteric communication (Moro et al., 2003), and our analysis reveals that mutations at residues 515 or 519 near the N terminus and at 530 near the C terminus of helix A affect interdomain communication as measured by changes in ATPase rates, W fluorescence, auxilin binding, cage disassembly, or ATP effects on linker protease sensitivity. Further, disulfide crosslinking in D152C/K524C supports the proximity of the helix A C terminus to an NBD residue involved in allosteric communication, and the lack of thiol reactivity of I515C and V519C mutants is consistent with these residues being buried in the interdomain interface.
Though it is possible that prokaryotic DnaKs and eukaryotic Hsc70s differ in the role played by SBD helix A in interdomain communication, it may be more likely that, in both groups of enzymes, deletion of the SBD helical subdomain eliminates some aspects of interdomain coupling (such as those detected by W fluorescence quenching) but that partial coupling persists through interactions with bHsc70 residues 415–417 (413–415 in DnaK) of the β sandwich portion of the SBD. It is also possible that partial function in such a deletion depends on intragenic suppression due to lack of the helical lid. That is, interdomain interactions with helix A may be important for opening the lidto allow substrate release, but if the lid is deleted, these interactions become partially dispensable. In fact, deletion of the SBD lid facilitates the conformational change involved in peptide release: W fluorescence quenching occurs ~50-fold more rapidly in a lidless DnaK than in a full-length molecule (Slepenkov and Witt, 2002). An analogous example of intragenic suppression may be provided by a bHsc70 E543K mutation that disrupts a salt bridge that forms a “latch” to close the lid. This mutation suppresses ATP binding site mutants that weaken transduction of the ATP-induced conformational change from the NBD to the SBD, presumably because it eliminates an interaction that must be broken during this conformational change (Johnson and McKay, 1999). Similarly, we find that V519C structurally and functionally suppresses V388C, possibly because V519C weakens interdomain interface interactions and facilitates invasion of this interface by a suboptimal linker sequence.
The possibility that the interdomain interface is partially disrupted upon ATP binding is supported by the observation that V519C becomes reactive with a thiol-specific dye upon ATP binding (Figure 6A), by the observation that DnaK K414 (R416 in bHsc70; Figure 1D) becomes trypsin sensitive in the presence of ATP (Buchberger et al., 1995), and by results from chemical shift analyses indicating that the interface is looser in the ATP versus ADP or nucleotide-free states (Revington et al., 2005). Intrusion of the linker into the interdomain interface would, in fact, cause severe steric clashes unless the two domains, or parts of each domain, move to relieve these clashes. Such movements could account for the global shape changes indicated by SAXS studies (Wilbanks et al., 1995; Shi et al., 1996) and could provide for a power stroke in processes such as protein translocation (Glick, 1995). If modeled on the basis of the rat SBD (Figure 4B), the hydrophobic linker would displace the cluster of hydrophobic interactions made by the N terminus of helix A, but interactions made by the C terminus of helix A and the base of the helical lid (Figures 1D and 7) would not be directly disrupted. If this led to differential movement of parts of the SBD upon ATP binding (i.e., if the β sandwich part of the SBD were to move while the helical lid remained anchored through persistent interactions with the NBD), then this could rupture lid:sandwich interactions, allowing the lid to open and release substrate.
How could ATP binding to the NBD trigger movement of the linker into the interdomain interface? The linker emerges from NBD helix K. If the linker is modeled in the bHsc70 structure on the basis of the rat SBD shown in Figure 4B, then V388 is placed 25.5 Å from D383 of helix K. This is too long a distance to be spanned by five residues and might require that helix K move or partially unravel to allow the change in linker conformation. D386 at the N terminus of helix K interacts directly with ATP, and helix K packs tightly against the 1–2 β hairpin of the NBD. T13 at the tip of the 1–2β hairpin interacts with the ATP γ-phosphate, and mutations in this threonine disrupt transduction of the ATP signal to the SBD (Sousa and McKay, 1998). It is therefore possible that direct interactions with ATP induce shifts in the positions of the 1–2 β hairpin and/or helix K and that these shifts alter hairpin:helix interactions to allow the helix to move or partially unravel so that the linker can invade the interdomain interface. The structural and mutational analysis described here provides a basis for formulation of such specific mechanistic hypotheses that may now be tested.
Experimental Procedures
Crystallization and Structure Determination
Seven triple mutants (K108A/E110A/K112A, D186A/K187A/K188A, K250A/K251A/D252A, E283A/D285A/E289A, K325A/D327A/K328A, K357A/E358A/K361A, and D383A/K384A/E386A) were constructed in truncated bHsc70 (bHsc70ΔCterm; aa 1–554) using two-stage PCR (Wang and Malcolm, 1999). E213A/D214A was constructed subsequently based on the observation of high B factors for these residues in a human Hsp70 NBD (Sriram et al., 1997). bHsc70ΔCterm was produced in E. coli BL21/DE3/plysS (Novagen) and purified by anion exchange on Q-Sepharose (pH 8.0), cation exchange on S-Sepharose (pH 6.5), and gel exclusion on Superdex 200 (resins from Amersham Biosciences). For crystallization, 5 µl of protein at 13 mg/ml in 10 mM Tris (pH 8.0), 50% glycerol, 1 mM EDTA, and 1 mM DTT was mixed with 5 µl of 4 M trimethylamine oxide and 10 µl of 18% PEG 8K and incubated under paraffin oil at 16°C. Crystals nucleated in 1–2 days and grew to triangular plates 0.3 mm on each edge and 0.05 mm thick in 3 weeks. Crystals were frozen in liquid nitrogen directly from the mother liquor. Data were collected on a Rigaku Cu rotating anode X-ray source with imaging plate detector and processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997). Molecular replacement and refinement were done with MOLREP (Vagin and Teplyakov, 1997) and CNS (Brünger et al., 1998), and model building with O (Jones et al., 1991).
Clathrin Cage Disassembly Assays
Cages were assembled with purified clathrin and assembly protein fraction (AP180, AP-2, AP-1, and auxilin) from bovine brain clathrin-coated vesicles (Keen et al., 1979) or with recombinant mouse GST-AP180. Disassembly of cages (at a clathrin concentration of 0.42 µM) was performed by incubating cages with the indicated concentrations (Figure S1) of Hsc70ΔCterm and 1 mM ATP in 20 mM imidazole, 25 mM KCl, 10 mM (NH4)2SO4, 2 mM Mg acetate, and 1 mM DTT (pH 7.0) (buffer C) at 25°C for the indicated time, separating released clathrin from cages by ultracentrifugation and quantifying the results by SDS-PAGE (Barouch et al., 1994; Morgan et al., 2001). Measurement of disassembly using dynamic light scattering was done on a Protein Solutions 800 instrument. Cages assembled from clathrin and GST-AP180 at a clathrin concentration of 0.2 µM were mixed with auxilin at 0.1 µM in buffer C. Disassembly was initiated by adding Hsc70ΔCterm (0.3–1 µM) preincubated with 1 mM ATP to the reactions in the light-scattering cuvette and continuously monitoring light scattering for 20 min.
Protease Sensitivity Studies
Analysis of protease sensitivity as a function of the presence of ATP was done as described (Wei et al., 1995).
Tryptophan Fluorescence Measurements
Tryptophan fluorescence was measured with an Aminco SPF-500C with excitation at 295 nm (2 nm bandpass), and emission measured over 300–400 nm (5 nm bandpass) and with 1 mg/ml protein in 10 mM Tris (pH 8.0), 50 mM KCl, and 5 mM MgCl2. Following measurement in the absence of ATP, ATP was added directly to the cuvette to 0.4 mM, and measurement of the spectra was initiated 1 min after mixing. Three scans, 2 min apart, were taken and in all cases were virtually superimposable, indicating that these were steady-state measurements.
Measurement of Auxilin Binding
Binding of wt or mutant Hsc70ΔCterm to immobilized auxilin was measured using SPR (Jiang et al., 2003) with 10 µM bHsc70ΔCterm and inclusion of an ATP-regenerating system (1 mM ATP, 30 units/ mL creatine kinase, 10 mM creatine phosphate).
Assessment of Cysteine Accessibility and Side Chain Proximity in Singly and Doubly Cys-Substituted Enzymes
Assessment of cysteine accessibility and side chain proximity in singly and doubly Cys-substituted enzymes was done by diluting enzymes stored in 10 mM Tris (pH 8.0), 1 mM EDTA, 1 mM DTT, and 50% glycerol into buffer C to give a final protein concentration of 1 mg/ml and a DTT concentration of <0.2 mM. Either monochlorobi-mane (24 mM in methanol) or oxidized glutathione (100 mM in water) was then added to 1 or 3 mM, respectively. Reactions were incubated for 2 hr, resolved by nonreducing SDS-PAGE, and visualized by staining with Coomassie blue or illumination with UV light.
ATP Binding and ATPase Assays
ATPase rates were measured by adding 20 µl of wt or mutant Hsc70ΔCterm proteins at 2 mg/ml in buffer C to 20 µl buffer C containing 0.5% v/v of α-32P ATP (3000 Ci/mM; 10 mCi/ml). Aliquots (4 µl) were then taken at 1, 2, 4, 8, 16, and 32 min and mixed with 4 µl of 2% SDS, 50 mM EDTA; 2.5 µl aliquots were spotted onto TLC plates and resolved in 0.5 M LiCl, 0.5 M formic acid, followed by quantification with a Molecular Dynamics Phosphorimager. The percent of ATP as a function of time was then fit to a single exponential decay to obtain ATPase rates. Measurement of ATP binding was done by mixing equal volumes of buffer C containing α-32P ATP as in the ATPase reactions with enzymes at concentrations ranging from 0.01 to 2 µM. Two minutes after mixing, reactions were filtered through nitrocellulose that had been pretreated by washing with 0.1 M NaOH, followed by washing with buffer C. Retained radioactivity was quantified with a Molecular Dynamics Phosporimager to determine the protein concentration at which ~50% of input radioactivity was retained. No attempt was made to determine precise binding constants, since the mutants (with the exception of I216C/ G229S) all displayed binding similar to the wt enzyme. A set of experiments done with buffer in which KCl was replaced with NaCl to slow ATP hydrolysis (Ha et al., 1997) gave similar results.
Supplementary Material
Acknowledgments
We thank Dr. D. McKay for an expression plasmid for bHsc70ΔCterm. This work is supported by GM-52522 and AQ-1486 from the NIH and Welch Foundation (to R.S.) and MDA3473 and NS29051 from the MDA and NINDS (to E.M.L.). Support for the X-ray Crystallography Core and the UTHSCSA Center for Macromolecular Interactions, from the UTHSCSA Executive Research Committee and San Antonio Cancer Institute, is acknowledged.
Footnotes
Supplemental Data
Supplemental Data include four figures and one table and can be found with this article online at http://www.molecule.org/cgi/content/full/20/4/513/DC1/.
Accession Numbers
The bHsc70ΔCterm coordinates have been deposited in the Protein Data Bank under ID code 1YUW.
References
- Barouch W, Prasad K, Greene LE, Eisenberg E. ATPase activity associated with the uncoating of clathrin baskets by Hsp70. J. Biol. Chem. 1994;269:28563–28568. [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Theyssen H, Schroder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdo-main communication. J. Biol. Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaper-one machines. Cell. 1998;92:351–359. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Cyr D, Langer T, Douglas M. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, De Luca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, Wilbanks SM, De Luca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. J. Biol. Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS. Can Hsp70 Proteins Act as Force-Generating Motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- Greene LE, Zinner R, Naficy S, Eisenberg E. Effect of nucleotide on the binding of peptides to 70-kDa heat shock protein. J. Biol. Chem. 1995;270:2967–2973. doi: 10.1074/jbc.270.7.2967. [DOI] [PubMed] [Google Scholar]
- Ha J-H, Hellman U, Johnson ER, Li L, McKay DB, Sousa MC, Takeda S, Wernstedt C, Wilbanks SM. Destabi-lization of peptide binding and interdomain communication by an E534K mutation in the bovine 70-kDa heat shock cognate protein, a molecular chaperone. J. Biol. Chem. 1997;272:27796–27803. doi: 10.1074/jbc.272.44.27796. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. Functional Specificity Among Hsp70 Molecular Chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- Jiang J, Taylor AB, Prasad K, Ishikawa-Brush Y, Hart PJ, Lafer EM, Sousa R. Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry. 2003;42:5748–5753. doi: 10.1021/bi034270g. [DOI] [PubMed] [Google Scholar]
- Johnson ER, McKay BB. Mapping the role of active site residues for transducing an ATP-induced conformational change in bovine 70-kDa heat shock cognate protein. Biochemistry. 1999;38:10823–10830. doi: 10.1021/bi990816g. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution. J. Mol. Evol. 1998;47:565–577. doi: 10.1007/pl00006413. [DOI] [PubMed] [Google Scholar]
- Keen JH, Willingham MC, Pastan I. Clathrin-coated vesicles: isolation, dissociation, and factor-dependent reassociation of clathrin baskets. Cell. 1979;16:303–312. doi: 10.1016/0092-8674(79)90007-2. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 1999;289:1131–1144. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70 kda molecular chaperone, DnaK, which alter substrate affinity or interdo-main coupling. J. Mol. Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Jin S, Augustine GJ, Lafer EM. Uncoating of clathrin-coated vesicles in presynaptic terminals: roles for Hsc70 and auxilin. Neuron. 2001;32:289–300. doi: 10.1016/s0896-6273(01)00467-6. [DOI] [PubMed] [Google Scholar]
- Moro F, Ferna´ndez V, Muga A. Interdomain interaction through helices A and B of DnaK peptide binding domain. FEBS Lett. 2003;533:119–123. doi: 10.1016/s0014-5793(02)03752-3. [DOI] [PubMed] [Google Scholar]
- Morshauer RC, Hu W, Wang H, Pang Y, Flynn GC, Zuiderweg ER. High-resolution solution structure of the 18 kDA substrate-binding domain of the mammalian chaperone protein Hsc70. J. Mol. Biol. 1999;289:1387–1403. doi: 10.1006/jmbi.1999.2776. [DOI] [PubMed] [Google Scholar]
- Newmyer SL, Schmid SL. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J. Cell Biol. 2001;152:607–620. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rajapandi T, Wu C, Eisenberg E, Greene L. Characterization of D10S and K71E mutants of human cytosolic hsp70. Biochemistry. 1998;37:7244–7250. doi: 10.1021/bi972252r. [DOI] [PubMed] [Google Scholar]
- Revington M, Holder TM, Zuiderweg ERP. NMR study of nucleotide-induced changes in the nucleotide binding domain of Thermus thermophilus Hsp70 chaperone DnaK: implications for the allosteric mechanism. J. Biol. Chem. 2004;279:33958–33967. doi: 10.1074/jbc.M313967200. [DOI] [PubMed] [Google Scholar]
- Revington M, Zhang Y, Yip GNB, Kurochkin AV, Zuiderweg ERP. NMR investigations of allosteric processin in a two-domain Thermus Thermophilus Hsp70 molecular chaperone. J. Mol. Biol. 2005;27:163–183. doi: 10.1016/j.jmb.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Schmid S. Enzymatic recycling of clathrin from coated vesicles. Cell. 1986;46:5–9. doi: 10.1016/0092-8674(86)90852-4. [DOI] [PubMed] [Google Scholar]
- Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- Shi L, Kataoka M, Fink A. Conformational characterization of DnaK and its complexes by small-angle X-ray scattering. Biochemistry. 1996;35:3297–3308. doi: 10.1021/bi951984l. [DOI] [PubMed] [Google Scholar]
- Slepenkov SV, Witt SN. Kinetic analysis of interdo-main coupling in a lidless variant of the molecular chaperone DnaK: DnaK’s lid inhibits transition to the low affinity state. Biochemistry. 2002;41:12224–12235. doi: 10.1021/bi0263208. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- Sousa MC, McKay DB. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry. 1998;37:15391–15399. doi: 10.1021/bi981510x. [DOI] [PubMed] [Google Scholar]
- Sriram M, Osipuk J, Freeman C, Morimoto RI, Joachimiak A. Human Hsp70 binds two calcium ions within the ATPase domain. Structure. 1997;5:403–414. doi: 10.1016/s0969-2126(97)00197-4. [DOI] [PubMed] [Google Scholar]
- Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interactions of the Hsp70 molecular chaper-one, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, Holstein SE, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, Holstein SEH. Functional interaction of the auxilin J domain with the nucleotide- and substrate-binding modules of Hsc70. J. Biol. Chem. 1997;272:19594–19600. doi: 10.1074/jbc.272.31.19594. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Wang H, Kurochkin AV, Pang Y, Hu W, Flynn GC, Zuiderweg ER. NMR solution structure of the 21 kDA chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interactions. Biochemistry. 1998;37:7929–7940. doi: 10.1021/bi9800855. [DOI] [PubMed] [Google Scholar]
- Wei J, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- Wilbanks SM, Chen L, Tsuruta H, Hodgson KO, McKay DB. Solution small-angle X-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry. 1995;34:12095–12105. doi: 10.1021/bi00038a002. [DOI] [PubMed] [Google Scholar]
- Young JC, Barral JM, Hartl FU. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 2003;28:541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.