Abstract
Estrogen-signaling pathways are implicated in the development of breast cancer and prostate cancer. Various studies have focused on additional signaling pathways, mediated by estrogen-related receptors (ERRs). ERRs are constitutively active receptors that share a high degree of homology with the classical estrogen receptors (ERs). However, they do not bind to estrogen, while ERs do. ERRs are involved in the development of alternative pathways that lead to the development of cancer and are regarded as potential therapeutic targets for the treatment of breast cancer and prostate cancer that do not respond to conventional therapies. In this review, we first present general structural features of ERRs. Then, we focus on breast cancer and prostate cancer, which are primarily hormone-dependent cancers, and summarizes recent progress in elucidating the involvement of each ERR in these two types of malignancies.
Keywords: estrogen-related receptors, estrogen receptors, estrogen, androgen, breast cancer, prostate cancer
Introduction
Among the 48 members of the nuclear receptor superfamily, the estrogen receptor (ER)-like subfamily (NR3) is one of the seven subfamilies and is composed of three groups: ERs(NR3A), estrogen-related receptors (ERRs or NR3B), and 3-ketosteroid receptors (NR3C), which include the androgen receptor (AR), progesterone receptor (PR), glucocorticoid receptor (GR), and mineralocorticoid receptor (MR). ERRs were initially thought to share a common biological function with ERs, but unexpectedly, they do not bind to estrogen or endogenous ER ligands and are considered orphan nuclear receptors (1).
Estrogens are natural hormones considered to play a major role in promoting the proliferation of both the normal and neoplastic breast epithelium. Growing evidences suggest that estrogen-signaling pathways are implicated in not only in the development of breast cancer but also in that of prostate cancer, which are both hormone-dependent cancers (2). ER-positive breast cancers are preferentially treated with antiestrogens such as selective ER modulators known as SERMs (including tamoxifen which is the most commonly used) or selective ER down-regulators known as SERDs (ICI 182,780) (3). Aromatase inhibitors are used in postmenopausal women to block the enzyme aromatase, which is involved in the final step of estrogen synthesis from circulating androgens (4). Androgen deprivation therapy (ADT) is used for the treatment of locally advanced, biochemically recurrent, and metastatic prostate cancer. However, after the initial response, these therapies for breast and prostate cancer fail and cancer will inevitably recur (5–7). The mechanisms underlying recurrence have not been fully clarified, but the acquisition of alternative intracellular ER signaling may be involved in hormonal therapy resistance.
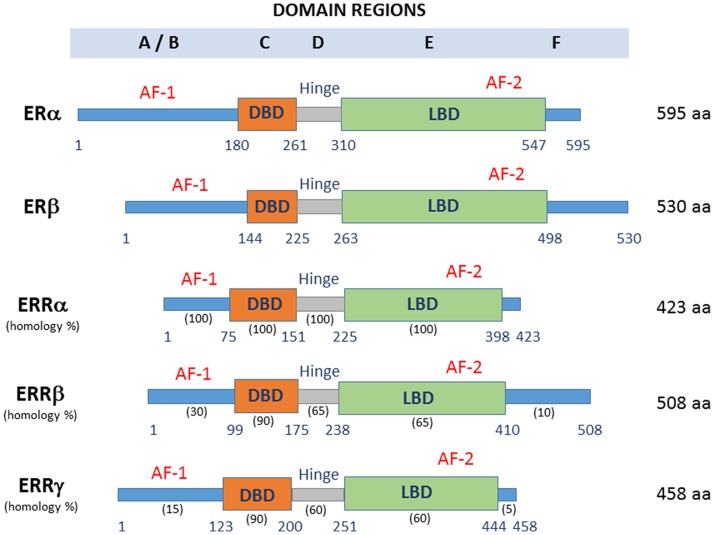
Various studies have focused on additional estrogen-related signaling pathways, mediated by ERRs (8–10). Sequence analysis reveals that ERRs and the classical ERs share a high degree of homology within their DNA and ligand-binding domains (LBDs) (11). The ERR family consists of three closely related members: ERRα, ERRβ, and ERRγ (Figure 1). ERRs, like other nuclear receptors, consist of six conserved regions (A–F): A/B domain containing the N-terminal domain (NTD), C domain with a DNA binding domain (DBD), D domain, which corresponds to the hinge region, E domain containing a putative LBD, and the F domain or C-terminal region. The transcriptional activation function-1 (AF-1) site is located in the N-terminal A/B domain, and its activity does not depend on the presence of activating ligands. The ligand-regulated activation function-2 (AF-2) site is located at E domain and it is known to synergize with AF-1 to regulate gene expression (12, 13). These receptors are constitutively active due to the structure of the LBD that leaves the AF-2 exposed and capable of binding coregulators in the absence of ligand binding.
Figure 1.
Domain structure and protein length of ERRs and ERs. DBD, DNA binding domain; LBD, ligand-binding domain; AF-1, activating function-1; AF-2, activating function-2. The percentage conservation between ERRα and the other two ERRs is shown for each of the five domains.
ERR Action Mechanisms
Like other nuclear receptors, the transcriptional activity of ERRs is dependent on the presence of coregulatory proteins, either corepressors or coactivators. Steroid receptor coactivator (SRC)-1, -2, and -3, which regulate hepatic metabolism, fat storage, and energy balance, have been shown to interact with one or more ERR isoform stimulating their transcriptional activity (14, 15). PPARγ coactivator (PGC)-1α and PGC-1β, which play essential roles in metabolic programs, have been shown to directly interact with ERRs, positively regulating the expression and activity or these nuclear receptors (16). The activity of the ERRs as transcription factors is generally inhibited through physical interaction with receptor interacting protein 140, RIP140, also known as nuclear receptor interacting protein 1, NRIP1, a corepressor that competes for interaction with PGC-1α for ERRs binding negatively regulating gene expression (17).
Estrogen-related receptors can bind to DNA on ERR response elements (ERREs) designated by the sequence 5′-TNAAGGTCA-3′ as well as the classical ER response elements (EREs) 5′-AGGTCANNNTGACCT-3′, where N represents any nucleotide (18–20). These orphan nuclear receptors play a central role in regulating cellular metabolism through the regulation of genes involved in glycolysis, oxidative phosphorylation, and tricarboxylic acid cycle (21). ERRα is present in tissues with high metabolism, such as the heart, kidney, intestinal tract, skeletal muscle, and brown adipose tissue (22). The expression of ERRβ and ERRγ is more restricted, being mainly expressed in the heart and kidney (22, 23). Both ERRα and ERRγ are upregulated in preadipocytes and pluripotent mesenchymal cells under adipogenic conditions, positively regulating lipid accumulation (24, 25). ERRβ and ERRγ are also expressed during the early embryonic development and in the central nervous system and spinal cord. Although ERRs have structural and functional similarities, mice deficient for each ERR exhibit different phenotypes, suggesting that these ERRs have specific and unique functions. ERRα-deficient mice are viable, but exhibit a phenotype characterized by reduced body weight, peripheral fat deposits, and resistance to high-fat diet-induced obesity (26). They also exhibit a loss of normal mitochondrial biogenesis (27). On the other hand, ERRβ-deficient mice are lethal due to impaired placenta formation (28). Interestingly, an important role of this orphan receptor in embryonic stem (ES) cells maintenance has been suggested, as it is able to regulate self-renewal in ES cells in cooperation with octamer-binding protein 4, Oct4, and sex determining region Y-box 2, Sox2, replacing kruppel-like factor 4, Klf4, and consequently enhancing the transcription of reprograming factors (29). ERRγ-deficient mice also die during the early postnatal period due to abnormal heart function (30). It has been reported that ERR overexpression increases glycolysis, while depletion results in the opposite effect, reducing aerobic glycolysis (31). Furthermore, ERRα and ERRγ were found to form heterodimers and bind to the promoters of the same target genes to regulate genes controlling both the glycolytic and the oxidative mitochondrial respiration phenotypes (32). These findings suggest that ERRs might contribute to a shift in cellular metabolism from normal mitochondrial oxidative phosphorylation to an aerobic glycolysis typical of cancer cells, known as the Warburg effect, increasing glucose consumption, and ATP production (33). However, a number of evidence suggest that ERRα and ERRγ have opposite functions in regulating cellular metabolism. ERRα could assist in setting a glycolytic profile required for the proliferation of rapidly dividing cells in normal tissues with high-energy demand and tumors consequently promoting cellular proliferation (34–39). By contrast, ERRγ high expression has been detected in tumors with better prognosis (36), and inhibition of this orphan receptor by a microRNA (miR-378*) induces a metabolic switch by promoting a Warburg-like phenotype (40). These findings illustrates the complexity of metabolic reprograming by ERRs in cancer cells, where ERRα and ERRγ could cooperate or play an opposite role in the metabolic transcriptional pathway depending on the cellular environment and the expression of distinct coregulators (41) (Table 1).
Table 1.
Expression of ERR isoforms and implication in breast cancer and prostate cancer.
| Receptor | Breast cancer |
Prostate cancer |
||||
|---|---|---|---|---|---|---|
| Expression | Implication | Reference | Expression | Implication | Reference | |
| ERRα (NR3B1) | High | Transcriptional activator or repressor | (42) | High | Promotion of cell growth | (39) |
| Promotion of cell growth and metastasis | (34, 39, 43, 44) | |||||
| ERRβ (NR3B2) | Uncertain | Uncertain | (36, 45) | Low | Inhibition of cell growth | (46) |
| ERRγ (NR3B3) | High/low | Promotion or inhibition of cell growth | (36, 47, 48) | Low | Inhibition of cell growth | (49) |
ERRs in Breast Cancer
Breast cancer is the leading cause of cancer-related death in women worldwide (50). It is primarily a hormone-dependent disease that can be regulated by the status of steroid hormones such as estrogen and progesterone. Unfortunately, the emergence of hormone-resistant tumor cells after years of treatment is a major issue affecting patients with breast cancer (7). Breast cancers can be divided into five subtypes that vary in their treatment options and survival outcomes based on gene expression profiles (51–54). ERα positive (ERα+) and progesterone receptor positive (PR+) tumors account for approximately 70% of all cases (55, 56). These ERα+/PR+ tumors can be further classified into HER2+ and HER2− subtypes depending on epidermal growth factor receptor 2, ErbB-2, or HER2 gene expression. Blocking the estrogen production or estrogen binding to the receptor by tamoxifen or aromatase inhibitors is the standard treatment for both early and advanced ERα+ breast cancer (57, 58). For ERα negative (ERα−), progesterone receptor negative (PR−), and HER2+ (ERα−/PR−/HER2+) breast tumors, a combination of pertuzumab, trastuzumab, and docetaxel has been effective (59). However, there are still no approved targeted therapies for triple negative ERα−/PR−/HER2−breast tumors (10–17% of all breast cancer cases) (60, 61), or the normal breast-like or basal-like cancer subtype (15% of the cases) (62, 63), which are mostly triple negative and frequently have TP53 mutations (64).
Much attention has been paid to the role of ERRs in breast cancer, as they are orphan nuclear receptors closely related to ERs. ERRα expression in breast tumors is often high, and it is expressed in tumors with poor prognosis (36). In samples from various cohorts of patients with breast cancer, ERRα mRNA positively correlates with the expression of the oncogene ERBB2 and inversely correlates to that of ERα and PR, which are considered as good prognostic markers for patients with breast cancer (36). The expression of ERRα mRNA and protein positively correlates with the coactivator amplified in breast cancer 1 (AIB1), also known as SRC-3 (65). However, ERRα is able to act as both a transcriptional activator and repressor depending on the cellular context, promoting or inhibiting tumor growth in breast cancer (42). In ER− breast cancer cells, ERRα functions as a transcriptional activator constitutively interacting with coactivators and binding to EREs independently of any ligands. Consequently, ERRα competes with ER in the regulation of estrogen-responsive genes such as the estrogen-regulated trefoil factor 1 (TFF1) (65, 66) and vascular endothelial growth factor (VEGF) (34, 43). In ER+ breast cancer cells, ERRα functions as a transcriptional repressor, interacting with corepressors and binding to negative EREs (42).
ERRα also plays a role in bone metastasis, which occurs in up to 70% of patients with advanced breast cancer (44). In a mouse xenograft model of metastatic human breast cancer, overexpression of wild-type ERRα-reduced metastasis and breast cancer cell growth in the bone, likely by upregulating the osteoclastogenesis inhibitor, osteoprotegerin (OPG). By contrast, ERRα overexpression increases breast cancer cell growth in the mammary gland and the expression of VEGF. Thus, ERRα plays dual roles, promoting the progression and invasion of primary tumors by decreasing osteolytic lesions in the bone (44).
It has been suggested that the induction of c-myc expression by estrogen occurs through the “non-classical” pathway – without binding of ERα to its promoter (67). Another study demonstrated a positive correlation between ERRα, c-myc, and aromatase (37), an enzyme that may enhance estrogen production and stimulate breast cancer progression (38). These studies propose that ERR could play an important role in alternative pathway to classical ERs-dependent pathway in cell signaling through aromatase and c-myc.
ERRα participates in the enhancement of estrogen production by the activation of steroid sulfotransferase (SULT2A1), which maintains high level of peripheral dehydroepiandrosterone sulfate (DHEAS), an important dehydroepiandrosterone (DHEA) metabolite in estrogen synthesis in certain tissues (68). It has also been reported that SULT2A1 inactivates tamoxifen and raloxifene (69). Thus, SULT2A1 activation by ERRα can partly explain the resistance of breast cancer cells expressing ERRα to SERM therapy.
A clinical study analyzing 102 breast cancer samples revealed that the expression of ERRα in more than 10% of malignant cells was associated with a 20% decrease in overall disease survival at 13 years (70). In this work, association between ERα and ERE-containing estrogen-responsive genes was markedly altered according to ERRα status in breast cancer tissues, suggesting that ERRα possibly modulates ERα-mediated and ERE-dependent transcription, and changes the expression of estrogen-responsive genes in breast cancer cells. In a previous clinical study, high levels of ERRα mRNA correlated with ERα-tumor status in 38 tumor specimens (36). These two studies suggest that ERRα mRNA and protein expression are associated with an unfavorable prognosis, and increase the risk of recurrence of breast cancer.
ERRγ also acts differently depending on ER expression. In breast tumors co-expressing ER and PR, ERRγ induces E-cadherin expression and promotes the mesenchymal-to-epithelial (MET) transition, resulting in the inhibition of tumor growth (36, 47). It has been shown that an AAAG tetranucleotide polymorphism in the untranslated region of the ERRγ gene is associated with breast cancer predisposition (71), and that ERRγ mediates tamoxifen resistance in invasive lobular breast cancer. In human breast cancer specimens, both ERRγ mRNA and protein expression are upregulated compared with normal samples, and exogenously transfected ERRγ increased breast cancer cell proliferation (36, 48). Tumors overexpressing ERRγ are also frequently steroid receptor positive, which may reflect hormonal sensitivity and a preferable clinical outcome. Thus, ERRγ mRNA expression is associated with a favorable prognosis of patients with breast cancer (36).
ERRβ expression has also been detected in breast tumors. It has been shown that ERRβ expression is associated with that of ERβ, and that ERRβ levels inversely correlate with the S-phase fraction, suggesting that this orphan receptor inhibits cellular proliferation, or possibly promotes cellular differentiation (36). However, another report found that it acts as a proliferative gene (45). Thus, the potential role of ERRβ in breast cancer remains unclear.
ERRs in Prostate Cancer
Prostate cancer, which is dependent on androgens for proliferation and survival, is the second-leading cause of cancer-related mortality, after lung cancer, in men from developed countries (72). With early diagnosis, radical prostatectomy and/or radiation therapy are potentially curative. For advanced or metastatic prostate cancer, hormonal therapies, reducing androgen levels by surgical or chemical castration or inhibiting the AR protein by small molecules, are used (57). However, after an initial response, the cancer eventually recurs in an incurable, castration-resistant form (5, 6), as a result of amplification of AR protein, mutations of AR gene, and elevated production of AR variants (73).
In addition to androgen-signaling pathways, estrogen-signaling pathways are implicated in the development of prostate cancer (1), and estrogen has been used for the treatment of advanced prostate cancer (74). The direct effect of estrogens on normal and malignant prostate tissues is assumed to be mediated through ERα and ERβ (75, 76).
ERRα mRNA has been detected in prostate cancer cell lines and human prostate cancer tissue (77). Although a heterogeneous ERRα staining was found in immunohistochemical analysis using prostate cancer tissues with low Gleason score (GS), increased ERRα protein expression was detected in human prostate tissue from 106 surgical resected prostate samples in a study that showed a positive correlation between ERRα expression and GS (78). The enhanced expression of ERRα might play a role in the development of human prostate cancer and serve as a significant prognostic factor for the disease.
By contrast, reduced ERRβ and ERRγ expression in some prostatic carcinomas has been reported (77), and overexpression of ERRβ or ERRγ results in the suppression of cell proliferation in both androgen-sensitive and insensitive prostate cancer cells, suggesting that these receptors present antiproliferative or tumor-suppressing functions in prostate cancer (46, 49). Furthermore, it has been shown that ERRβ directly transactivates a promoter upstream of the cyclin-dependent kinase inhibitor, p21 gene, resulting in the inhibition of the cell cycle progression (46). Clinicopathological studies for the expression of both orphan receptors in human prostate tissues have been performed using immunohistochemical analysis (79). Cancerous lesions and benign foci obtained from radical prostatectomy were stained and immunoreactivity scores (IR scores) were evaluated from the proportion of immunoreactive cells and staining intensity, revealing that ERRβ and ERRγ IR scores are significantly lower in cancerous tissues. In the same study, patients with high ERRα IR score, but low ERRγ IR score, presented a poorer cancer-specific survival when compared to the other group, suggesting that the expression of ERRβ and ERRγ could be a useful prognostic indicator of prostate cancer (79).
ERRs Agonists and Antagonists
Although no endogenous ligands have been identified for these receptors several natural phytoestrogens (three isoflavones: genistein, daidzein, and biochanin A, and one flavone: 6, 3, 4-trihydroxyflavone) have been identified as potential ligands of these receptors with agonistic activities, by structure-based virtual screening and biological functional assays (80, 81). Phytoestrogens are produced by plants, and represent the major natural exogenous sources of estrogenic compounds. DY131 is another ERR agonist, specific to ERRβ and ERRγ that was shown to enhance growth inhibition, which was caused by overexpression of these nuclear receptors (46, 49). Inhibition of ERRα with the inverse agonist XCT790 reduces cell proliferation of various cancer cell lines, including prostate and breast cancer cells (39).
On the other hand, diethylstilbestrol (DB00255) and the tamoxifen metabolite, 4-hydroxytamoxifen, have been shown to interact with ERRβ and ERRγ and act as antagonists (82, 83). SR16388, a novel steroidal antiestrogen, inhibits the interaction between its coactivator peroxisome proliferator-activated receptor γ coactivator-1 α to inhibit ERR α activity (84).
Conclusion
Although the role of orphan nuclear receptors in cancer is becoming clearer as a result of advances in previous studies, their function still remains to be elucidated. A better understanding of ERRs in breast and prostate cancer will provide new insights into cancer biology as well as the discovery of novel small molecules that bind to these orphan receptors. This knowledge will be helpful for the identification of novel hormonal therapeutic strategies and cancer treatments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. K. Ikeda (Saitama Medical University) and Dr. K. Horie (Saitama Medical University) for critical reading of this manuscript and helpful suggestions. This work was supported by grants of the Cell Innovation Program and the P-DIRECT from the MEXT, Japan (SI), grants from JSPS, Japan (SI), and a grant of the Program for Promotion of Fundamental Studies in Health Sciences from NIBIO, Japan (SI). This work was supported by grants of the Cell Innovation Program (http://www.cell-innovation.org/) and the P-DIRECT (http://p-direct.mext.go.jp/) from the MEXT, Japan (S.I.), grants from JSPS KAKENHI (Grant Numbers 23249040 and 15K15353), Japan (S.I.), and a grant of the Program for Promotion of Fundamental Studies in Health Sciences from NIBIO (09-12), Japan (S.I.).
References
- 1.Roshan-Moniri M, Hsing M, Butler MS, Cherkasov A, Rennie PS. Orphan nuclear receptors as drug targets for the treatment of prostate and breast cancers. Cancer Treat Rev (2014) 40:1137–52. 10.1016/j.ctrv.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 2.Bonkhoff H, Berges R. The evolving role of oestrogens and their receptors in the development and progression of prostate cancer. Eur Urol (2009) 55:533–42. 10.1016/j.eururo.2008.10.035 [DOI] [PubMed] [Google Scholar]
- 3.Howell SJ, Johnston SR, Howell A. The use of selective estrogen receptor modulators and selective estrogen receptor down-regulators in breast cancer. Best Pract Res Clin Endocrinol Metab (2004) 18:47–66. 10.1016/j.beem.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 4.Smollich M, Gotte M, Fischgrabe J, Radke I, Kiesel L, Wulfing P. Differential effects of aromatase inhibitors and antiestrogens on estrogen receptor expression in breast cancer cells. Anticancer Res (2009) 29:2167–71. [PubMed] [Google Scholar]
- 5.Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: adaptive responses in the androgen axis. Cancer Treat Rev (2014) 40:426–33. 10.1016/j.ctrv.2013.09.011 [DOI] [PubMed] [Google Scholar]
- 6.Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog (2011) 10:20. 10.4103/1477-3163.83937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer (2004) 11:643–58. 10.1677/erc.1.00776 [DOI] [PubMed] [Google Scholar]
- 8.Giguere V. To ERR in the estrogen pathway. Trends Endocrinol Metab (2002) 13:220–5. 10.1016/S1043-2760(02)00592-1 [DOI] [PubMed] [Google Scholar]
- 9.Horard B, Vanacker JM. Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J Mol Endocrinol (2003) 31:349–57. 10.1677/jme.0.0310349 [DOI] [PubMed] [Google Scholar]
- 10.Yang N, Shigeta H, Shi H, Teng CT. Estrogen-related receptor, hERR1, modulates estrogen receptor-mediated response of human lactoferrin gene promoter. J Biol Chem (1996) 271:5795–804. 10.1074/jbc.271.10.5795 [DOI] [PubMed] [Google Scholar]
- 11.Laudet V, Hanni C, Coll J, Catzeflis F, Stehelin D. Evolution of the nuclear receptor gene superfamily. EMBO J (1992) 11:1003–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tora L, Gronemeyer H, Turcotte B, Gaub MP, Chambon P. The N-terminal region of the chicken progesterone receptor specifies target gene activation. Nature (1988) 333:185–8. 10.1038/333185a0 [DOI] [PubMed] [Google Scholar]
- 13.Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol (2003) 17:1901–9. 10.1210/me.2002-0384 [DOI] [PubMed] [Google Scholar]
- 14.Karmakar S, Foster EA, Smith CL. Unique roles of p160 coactivators for regulation of breast cancer cell proliferation and estrogen receptor-alpha transcriptional activity. Endocrinology (2009) 150:1588–96. 10.1210/en.2008-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Benlhabib H, Mendelson CR. cAMP enhances estrogen-related receptor alpha (ERRalpha) transcriptional activity at the SP-A promoter by increasing its interaction with protein kinase A and steroid receptor coactivator 2 (SRC-2). Mol Endocrinol (2009) 23:772–83. 10.1210/me.2008-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skrzypczak M, Kapka-Skrzypczak L, Cyranka M, Treeck O, Wrobel A, Matosiuk D. Nuclear estrogen receptors co-activation mechanisms. Curr Med Chem (2013) 20:3317–38. 10.2174/09298673113209990006 [DOI] [PubMed] [Google Scholar]
- 17.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson JA. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol (1998) 12:864–81. 10.1210/mend.12.6.0123 [DOI] [PubMed] [Google Scholar]
- 18.Pettersson K, Svensson K, Mattsson R, Carlsson B, Ohlsson R, Berkenstam A. Expression of a novel member of estrogen response element-binding nuclear receptors is restricted to the early stages of chorion formation during mouse embryogenesis. Mech Dev (1996) 54:211–23. 10.1016/0925-4773(95)00479-3 [DOI] [PubMed] [Google Scholar]
- 19.Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol (1997) 17:5400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barry JB, Laganiere J, Giguere V. A single nucleotide in an estrogen-related receptor alpha site can dictate mode of binding and peroxisome proliferator-activated receptor gamma coactivator 1alpha activation of target promoters. Mol Endocrinol (2006) 20:302–10. 10.1210/me.2005-0313 [DOI] [PubMed] [Google Scholar]
- 21.Deblois G, Giguere V. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer (2013) 13:27–36. 10.1038/nrc3396 [DOI] [PubMed] [Google Scholar]
- 22.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell (2006) 126:789–99. 10.1016/j.cell.2006.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev (2008) 29:677–96. 10.1210/er.2008-0017 [DOI] [PubMed] [Google Scholar]
- 24.Ijichi N, Ikeda K, Horie-Inoue K, Yagi K, Okazaki Y, Inoue S. Estrogen-related receptor alpha modulates the expression of adipogenesis-related genes during adipocyte differentiation. Biochem Biophys Res Commun (2007) 358:813–8. 10.1016/j.bbrc.2007.04.209 [DOI] [PubMed] [Google Scholar]
- 25.Kubo M, Ijichi N, Ikeda K, Horie-Inoue K, Takeda S, Inoue S. Modulation of adipogenesis-related gene expression by estrogen-related receptor gamma during adipocytic differentiation. Biochim Biophys Acta (2009) 1789:71–7. 10.1016/j.bbagrm.2008.08.012 [DOI] [PubMed] [Google Scholar]
- 26.Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol Cell Biol (2003) 23:7947–56. 10.1128/MCB.23.22.7947-7956.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab (2008) 19:269–76. 10.1016/j.tem.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J, Sladek R, Bader JA, Matthyssen A, Rossant J, Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature (1997) 388:778–82. 10.1038/42022 [DOI] [PubMed] [Google Scholar]
- 29.Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol (2009) 11:197–203. 10.1038/ncb1827 [DOI] [PubMed] [Google Scholar]
- 30.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab (2007) 6:13–24. 10.1016/j.cmet.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 31.Cai Q, Lin T, Kamarajugadda S, Lu J. Regulation of glycolysis and the Warburg effect by estrogen-related receptors. Oncogene (2013) 32:2079–86. 10.1038/onc.2012.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab (2007) 5:345–56. 10.1016/j.cmet.2007.03.007 [DOI] [PubMed] [Google Scholar]
- 33.Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci U S A (2008) 105:7821–6. 10.1073/pnas.0711677105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein RA, Chang CY, Kazmin DA, Way J, Schroeder T, Wergin M, et al. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res (2008) 68:8805–12. 10.1158/0008-5472.CAN-08-1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CY, Kazmin D, Jasper JS, Kunder R, Zuercher WJ, McDonnell DP. The metabolic regulator ERRalpha, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell (2011) 20:500–10. 10.1016/j.ccr.2011.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res (2002) 62:6510–8. [PubMed] [Google Scholar]
- 37.Jarzabek K, Koda M, Kozlowski L, Sulkowski S, Kottler ML, Wolczynski S. The significance of the expression of ERRalpha as a potential biomarker in breast cancer. J Steroid Biochem Mol Biol (2009) 113:127–33. 10.1016/j.jsbmb.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 38.Yang C, Zhou D, Chen S. Modulation of aromatase expression in the breast tissue by ERR alpha-1 orphan receptor. Cancer Res (1998) 58:5695–700. [PubMed] [Google Scholar]
- 39.Bianco S, Lanvin O, Tribollet V, Macari C, North S, Vanacker JM. Modulating estrogen receptor-related receptor-alpha activity inhibits cell proliferation. J Biol Chem (2009) 284:23286–92. 10.1074/jbc.M109.028191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, et al. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab (2010) 12:352–61. 10.1016/j.cmet.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 41.Deblois G, St-Pierre J, Giguere V. The PGC-1/ERR signaling axis in cancer. Oncogene (2013) 32:3483–90. 10.1038/onc.2012.529 [DOI] [PubMed] [Google Scholar]
- 42.Kraus RJ, Ariazi EA, Farrell ML, Mertz JE. Estrogen-related receptor alpha 1 actively antagonizes estrogen receptor-regulated transcription in MCF-7 mammary cells. J Biol Chem (2002) 277:24826–34. 10.1074/jbc.M202952200 [DOI] [PubMed] [Google Scholar]
- 43.Stein RA, Gaillard S, McDonnell DP. Estrogen-related receptor alpha induces the expression of vascular endothelial growth factor in breast cancer cells. J Steroid Biochem Mol Biol (2009) 114:106–12. 10.1016/j.jsbmb.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fradet A, Sorel H, Bouazza L, Goehrig D, Depalle B, Bellahcene A, et al. Dual function of ERRalpha in breast cancer and bone metastasis formation: implication of VEGF and osteoprotegerin. Cancer Res (2011) 71:5728–38. 10.1158/0008-5472.CAN-11-1431 [DOI] [PubMed] [Google Scholar]
- 45.Lu D, Kiriyama Y, Lee KY, Giguere V. Transcriptional regulation of the estrogen-inducible pS2 breast cancer marker gene by the ERR family of orphan nuclear receptors. Cancer Res (2001) 61:6755–61. [PubMed] [Google Scholar]
- 46.Yu S, Wong YC, Wang XH, Ling MT, Ng CF, Chen S, et al. Orphan nuclear receptor estrogen-related receptor-beta suppresses in vitro and in vivo growth of prostate cancer cells via p21(WAF1/CIP1) induction and as a potential therapeutic target in prostate cancer. Oncogene (2008) 27:3313–28. 10.1038/sj.onc.1210986 [DOI] [PubMed] [Google Scholar]
- 47.Tiraby C, Hazen BC, Gantner ML, Kralli A. Estrogen-related receptor gamma promotes mesenchymal-to-epithelial transition and suppresses breast tumor growth. Cancer Res (2011) 71:2518–28. 10.1158/0008-5472.CAN-10-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ijichi N, Shigekawa T, Ikeda K, Horie-Inoue K, Fujimura T, Tsuda H, et al. Estrogen-related receptor gamma modulates cell proliferation and estrogen signaling in breast cancer. J Steroid Biochem Mol Biol (2011) 123:1–7. 10.1016/j.jsbmb.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 49.Yu S, Wang X, Ng CF, Chen S, Chan FL. ERRgamma suppresses cell proliferation and tumor growth of androgen-sensitive and androgen-insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res (2007) 67:4904–14. 10.1158/0008-5472.CAN-06-3855 [DOI] [PubMed] [Google Scholar]
- 50.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin (2011) 61:69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 51.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature (2000) 406:747–52. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 52.Reis-Filho JS, Westbury C, Pierga JY. The impact of expression profiling on prognostic and predictive testing in breast cancer. J Clin Pathol (2006) 59:225–31. 10.1136/jcp.2005.028324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouzier R, Wagner P, Morandi P, Pusztai L. Gene expression profiling of primary breast cancer. Curr Oncol Rep (2005) 7:38–44. 10.1007/s11912-005-0024-y [DOI] [PubMed] [Google Scholar]
- 54.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A (2001) 98:10869–74. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson WF, Chatterjee N, Ershler WB, Brawley OW. Estrogen receptor breast cancer phenotypes in the surveillance, epidemiology, and end results database. Breast Cancer Res Treat (2002) 76:27–36. 10.1023/A:1020299707510 [DOI] [PubMed] [Google Scholar]
- 56.Niemeier LA, Dabbs DJ, Beriwal S, Striebel JM, Bhargava R. Androgen receptor in breast cancer: expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol (2010) 23:205–12. 10.1038/modpathol.2009.159 [DOI] [PubMed] [Google Scholar]
- 57.Damber JE, Aus G. Prostate cancer. Lancet (2008) 371:1710–21. 10.1016/S0140-6736(08)60729-1 [DOI] [PubMed] [Google Scholar]
- 58.Mohamed A, Krajewski K, Cakar B, Ma CX. Targeted therapy for breast cancer. Am J Pathol (2013) 183:1096–112. 10.1016/j.ajpath.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 59.Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med (2015) 372:724–34. 10.1056/NEJMoa1413513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California Cancer Registry. Cancer (2007) 109:1721–8. 10.1002/cncr.22618 [DOI] [PubMed] [Google Scholar]
- 61.Criscitiello C, Azim HA, Jr, Schouten PC, Linn SC, Sotiriou C. Understanding the biology of triple-negative breast cancer. Ann Oncol (2012) 23(Suppl 6):vi13–8. 10.1093/annonc/mds188 [DOI] [PubMed] [Google Scholar]
- 62.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology (2008) 52:108–18. 10.1111/j.1365-2559.2007.02889.x [DOI] [PubMed] [Google Scholar]
- 63.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature (2012) 490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist (2011) 16(Suppl 1):61–70. 10.1634/theoncologist.2011-S1-61 [DOI] [PubMed] [Google Scholar]
- 65.Heck S, Rom J, Thewes V, Becker N, Blume B, Sinn HP, et al. Estrogen-related receptor alpha expression and function is associated with the transcriptional coregulator AIB1 in breast carcinoma. Cancer Res (2009) 69:5186–93. 10.1158/0008-5472.CAN-08-3062 [DOI] [PubMed] [Google Scholar]
- 66.Surowiak P, Materna V, Gyorffy B, Matkowski R, Wojnar A, Maciejczyk A, et al. Multivariate analysis of oestrogen receptor alpha, pS2, metallothionein and CD24 expression in invasive breast cancers. Br J Cancer (2006) 95:339–46. 10.1038/sj.bjc.6603254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeNardo DG, Cuba VL, Kim H, Wu K, Lee AV, Brown PH. Estrogen receptor DNA binding is not required for estrogen-induced breast cell growth. Mol Cell Endocrinol (2007) 277:13–25. 10.1016/j.mce.2007.07.006 [DOI] [PubMed] [Google Scholar]
- 68.Seely J, Amigh KS, Suzuki T, Mayhew B, Sasano H, Giguere V, et al. Transcriptional regulation of dehydroepiandrosterone sulfotransferase (SULT2A1) by estrogen-related receptor alpha. Endocrinology (2005) 146:3605–13. 10.1210/en.2004-1619 [DOI] [PubMed] [Google Scholar]
- 69.Apak TI, Duffel MW. Interactions of the stereoisomers of alpha-hydroxytamoxifen with human hydroxysteroid sulfotransferase SULT2A1 and rat hydroxysteroid sulfotransferase STa. Drug Metab Dispos (2004) 32:1501–8. 10.1124/dmd.104.000919 [DOI] [PubMed] [Google Scholar]
- 70.Suzuki T, Miki Y, Moriya T, Shimada N, Ishida T, Hirakawa H, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res (2004) 64:4670–6. 10.1158/0008-5472.CAN-04-0250 [DOI] [PubMed] [Google Scholar]
- 71.Galindo CL, McCormick JF, Bubb VJ, Abid Alkadem DH, Li LS, McIver LJ, et al. A long AAAG repeat allele in the 5’ UTR of the ERR-gamma gene is correlated with breast cancer predisposition and drives promoter activity in MCF-7 breast cancer cells. Breast Cancer Res Treat (2011) 130:41–8. 10.1007/s10549-010-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin (2013) 63:11–30. 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 73.Nyquist MD, Dehm SM. Interplay between genomic alterations and androgen receptor signaling during prostate cancer development and progression. Horm Cancer (2013) 4:61–9. 10.1007/s12672-013-0131-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin (1972) 22:232–40. [DOI] [PubMed] [Google Scholar]
- 75.Fujimura T, Takahashi S, Urano T, Ogawa S, Ouchi Y, Kitamura T, et al. Differential expression of estrogen receptor beta (ERbeta) and its C-terminal truncated splice variant ERbetacx as prognostic predictors in human prostatic cancer. Biochem Biophys Res Commun (2001) 289:692–9. 10.1006/bbrc.2001.6038 [DOI] [PubMed] [Google Scholar]
- 76.Ho SM. Estrogens and anti-estrogens: key mediators of prostate carcinogenesis and new therapeutic candidates. J Cell Biochem (2004) 91:491–503. 10.1002/jcb.10759 [DOI] [PubMed] [Google Scholar]
- 77.Cheung CP, Yu S, Wong KB, Chan LW, Lai FM, Wang X, et al. Expression and functional study of estrogen receptor-related receptors in human prostatic cells and tissues. J Clin Endocrinol Metab (2005) 90:1830–44. 10.1210/jc.2004-1421 [DOI] [PubMed] [Google Scholar]
- 78.Fujimura T, Takahashi S, Urano T, Kumagai J, Ogushi T, Horie-Inoue K, et al. Increased expression of estrogen-related receptor alpha (ERRalpha) is a negative prognostic predictor in human prostate cancer. Int J Cancer (2007) 120:2325–30. 10.1002/ijc.22363 [DOI] [PubMed] [Google Scholar]
- 79.Fujimura T, Takahashi S, Urano T, Ijichi N, Ikeda K, Kumagai J, et al. Differential expression of estrogen-related receptors beta and gamma (ERRbeta and ERRgamma) and their clinical significance in human prostate cancer. Cancer Sci (2010) 101:646–51. 10.1111/j.1349-7006.2009.01451.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suetsugi M, Su L, Karlsberg K, Yuan YC, Chen S. Flavone and isoflavone phytoestrogens are agonists of estrogen-related receptors. Mol Cancer Res (2003) 1:981–91. [PubMed] [Google Scholar]
- 81.Tremblay GB, Kunath T, Bergeron D, Lapointe L, Champigny C, Bader JA, et al. Diethylstilbestrol regulates trophoblast stem cell differentiation as a ligand of orphan nuclear receptor ERR beta. Genes Dev (2001) 15:833–8. 10.1101/gad.873401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coward P, Lee D, Hull MV, Lehmann JM. 4-Hydroxytamoxifen binds to and deactivates the estrogen-related receptor gamma. Proc Natl Acad Sci U S A (2001) 98(15):8880–4. 10.1073/pnas.151244398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tremblay GB, Bergeron D, Giguere V. 4-Hydroxytamoxifen is an isoform-specific inhibitor of orphan estrogen-receptor-related (ERR) nuclear receptors beta and gamma. Endocrinology (2001) 142(10):4572–5. 10.1210/en.142.10.4572 [DOI] [PubMed] [Google Scholar]
- 84.Chao WR, Amin K, Shi Y, Hobbs P, Tanabe M, Tanga M, et al. SR16388: a steroidal antiangiogenic agent with potent inhibitory effect on tumor growth in vivo. Angiogenesis (2011) 14(1):1–16. 10.1007/s10456-010-9191-z [DOI] [PubMed] [Google Scholar]