Abstract
Background
Long noncoding RNAs (lncRNAs) have emerged as critical epigenetic regulators with important functions in development and disease. Here, we sought to identify and functionally characterize novel lncRNAs critical for vertebrate development.
Methods and Results
By relying on human pluripotent stem cell differentiation models, we investigated lncRNAs differentially regulated at key steps during human cardiovascular development with a special focus on vascular endothelial cells. RNA sequencing led to the generation of large data sets that serve as a gene expression roadmap highlighting gene expression changes during human pluripotent cell differentiation. Stage-specific analyses led to the identification of 3 previously uncharacterized lncRNAs, TERMINATOR, ALIEN, and PUNISHER, specifically expressed in undifferentiated pluripotent stem cells, cardiovascular progenitors, and differentiated endothelial cells, respectively. Functional characterization, including localization studies, dynamic expression analyses, epigenetic modification monitoring, and knockdown experiments in lower vertebrates, as well as murine embryos and human cells, confirmed a critical role for each lncRNA specific for each analyzed developmental stage.
Conclusions
We have identified and functionally characterized 3 novel lncRNAs involved in vertebrate and human cardiovascular development, and we provide a comprehensive transcriptomic roadmap that sheds new light on the molecular mechanisms underlying human embryonic development, mesodermal commitment, and cardiovascular specification.
Keywords: cardiovascular system, growth and development, RNA, long noncoding, transcriptome, vertebrates
The advent of novel sequencing technologies has revealed that <2% of the human genome encodes for proteins.1–3 Interestingly, despite not being translated into proteins, a large fraction of the mammalian genome is transcribed into what is known as noncoding RNAs (ncRNAs).1–3 The consistent observation of pervasive transcription originating from these noncoding sites suggests that ncRNAs might play a key role in fundamental biological functions.1–3 To date, thousands of ncRNAs have been putatively described. However, the precise functional roles of the vast majority remain unclear.4–6
Among the different classes of ncRNAs, long ncRNAs (lncRNAs), broadly defined as those noncoding transcripts larger than 200 nucleotides,1 represent one of the largest and least understood nucleic acid molecules in vertebrates. Overall, >9000 genomic loci are predicted to code for these transcript subclasses in the human genome.5,7,8 Reports describing important developmental roles for certain lncRNAs in various vertebrates have appeared over the last few years.9–11 Low expression levels and poor sequence conservation have limited the identification and functional characterization of novel lncRNAs.12 With the combination of novel sequencing and analytic technologies and the powerful platforms that pluripotent stem cell models provide for the study of human embryonic development, these limitations can now be circumvented.
Using a methodology recently developed in our laboratory that allows the highly efficient generation of human cardiovascular progenitor cells of mesodermal origin and terminally differentiated vascular endothelial cells,13 we report here the identification and functional characterization of 3 novel human lncRNAs indispensable for the development of the cardiovascular system in different vertebrate species.
Methods
Cell Culture
H1 (WA1, WiCell; passage 25–40) human embryonic stem cells (hESCs) were used for differentiation studies. Terminally differentiated primary human umbilical vein endothelial cells (HUVECs) were used to investigate the transcriptome and functional roles of lncRNA candidates in unmodified vascular endothelial cells. Briefly, hESCs were cultured in chemically defined growth media, mTeSR (Stemcell Technologies), on growth factor–reduced Matrigel (BD Biosciences)-coated plates. After 70% to 80% confluent hESCs were treated with dispase (Invitrogen) for 7 minutes at 37°C, the colonies were dispersed to small clusters and lifted carefully with a 5-mL glass pipette at a ratio of ≈1:6. HUVECs, purchased from Promocell, were cultured in endothelial basal medium supplemented with endothelial growth medium SingleQuots (Lonza). hESC-derived endothelial cells were cultivated in endothelial growth medium-2 bullet kit media (Lonza). Endothelial cells were grown on collagen I–coated plates (BD Biosciences). All cell lines were maintained in an incubator (37°C and 5% CO2) with media changes every day (hESCs) or every second day (HUVECs).
Cell Lineages Analyzed
Pluripotent stem cells were differentiated into early mesoderm-derived cells including c-Kit+ and KDR+ cardiovascular progenitors (day 2–4 of differentiation) and committed CD34+CD31+ bipotent vascular progenitor cells (day 4–8 of differentiation) as previously described.13 Kurian et al13 provides detailed characterization of the differentiation methodologies and cell types generated. Terminally differentiated primary HUVECs were also included in our analyses as positive controls, and no significant differences between differentiated endothelial cells and HUVECs were found.13 Briefly, for the generation of early cardiovascular progenitors (day 2–4) and committed vascular progenitor cells (day 4–8), undifferentiated hESCs were freshly split onto Matrigel-coated plates, making sure that the subcolonies were of small size (≈300–500 cells per colony). Cells were differentiated to different progenitor stages with the use of a chemically defined mesodermal induction media as previously described13 (Dulbecco modified Eagle medium:F12, 15 mg/mL stem cell–grade BSA [MP Biomedicals], 17.5 μg/mL human insulin [SAFC Bioscience], 275 μg/mL human holo transferrin [Sigma Aldrich], 20 ng/mL basic fibroblast growth factor [Humanzyme], 50 ng/mL human vascular endothelial growth factor-165 aa [Humanzyme], 25 ng/mL human bone morphogenetic protein 4 [Stemgent], 450 μmol/L monothioglycerol [Sigma Aldrich], and 2.25 mmol/L each l-glutamine and nonessential amino acids [Invitrogen]). Switching day 8 mesodermal induction media–differentiated cells to endothelial growth medium-2 media as described13 induced endothelial cell differentiation. HUVECs are of primary origin and were therefore not modified during our studies.
RNA Sequencing
Total RNA from roughly 1×107 cells of each of the groups indicated above was isolated with TRIzol (Invitrogen). Intact total RNA samples were treated with DNase1 and sent to the Beijing Genomics Institute for sequencing. Before sequencing, all samples were subjected to quality control processes to ensure the lack of contaminating DNA and the integrity of the RNA. All the RNA samples met the following RNA quality threshold: optical density (OD)260/280=2 to 2.2; OD260/230≥2.0; 28S:18S>1.0, RNA integrity number>7. Whole-transcriptome sequencing was then performed. Briefly, TruSeq Stranded Total RNA with a Ribo-Zero Human Kit (Illumina) was used to remove ribosomal RNA and to prepare strand-specific paired-end RNA sequence libraries. Ninety million 2× 100-bp paired end reads were sequenced on an Illumina HiSeq2000 instrument for each library. Reads were aligned to the hg19/GRCh37 version of the human genome by the use of STAR (version 2.1.4a). Only reads that aligned to a single unique location were kept for further analysis. Quantification of reads on each strand in 10-kb windows across the entire genome and comparisons with DNA sequencing data were used to further evaluate RNA purity and integrity (Figure I in the online-only Data Supplement).
Primers Used in This Study
Table I in the online-only Data Supplement provides a list of primers and their respective sequences.
Morpholino Embryo Injections
Morpholinos (Gene Tools) were designed against highly conserved regions in the lncRNAs or alternatively to block putative splice sites (Table II in the online-only Data Supplement). A pair of morpholinos was generated and tested per lncRNA. For zebrafish injection, morpholinos were dissolved in water at a 2-mmol/L stock concentration and diluted to a 2-ng/nL working concentration in PBS/phenol red solution. Embryo injections were performed by injecting ≈1 nL morpholino solution at the 1-cell stage with the use of a FemtoJet (Eppendorf). Morphants were evaluated at 24, 48, 72, and 96 hours in a StereoLumar stereoscope (Zeiss). For murine experiments, CD-1 female mice were superovulated by injecting pregnant mare's serum gonadotropin followed by human chorionic gonadotropin 48 hours later. The female mice were housed with males after human chorionic gonadotropin injection. One-cell–stage embryos were collected the next day after 20 hours and were injected with 10 pL of 2-nmol/L stock Terminator morpholino with a FemtoJet. The embryos were cultured until the blastocyst stage in K-modified simplex optimized medium, and RNA was extracted with the use of an RNeasy Mini Kit (Qiagen).
Data Availability
All data sets in this study are available in the National Center for Biotechnology Information Gene Expression Omnibus under the accession number GSE54969.
Statistical Evaluation
Statistical analyses of all end points were performed by statisticians at the Salk Institute and University of California San Diego using Excel, SPSS, or GraphPad software. All data presented a normal distribution. Statistical significance was evaluated with a standard unpaired Student t test (2 tailed; P<0.05) when appropriate. For multiple-comparison analysis, 1-way ANOVA with the Dunnett posttest correction was applied when appropriate (P<0.05). Comparisons of groups with small sample sizes (n<6) were performed as follows: Mann-Whitney test (2 sided; 95% confidence level; P<0.05) was used, and a Kruskal-Wallis test with the Dunn posttest correction (P<0.05) was applied for multiple comparisons. All data are presented as mean±SD and represent a minimum of 3 independent experiments with at least 3 technical replicates unless otherwise stated.
Results
Noncoding RNAs Account for Half of the Transcriptome During Human Vascular Differentiation
With the goal of identifying and functionally validating novel genetic elements underlying cardiovascular vertebrate development, we first decided to perform an RNA sequencing–based screening of transcripts differentially regulated during stem cell differentiation. We focused on our recently reported methodology for the efficient differentiation of human pluripotent cells to early c-Kit+ cardiovascular progenitors, committed CD34+CD31+ vascular progenitors, and terminally differentiated functional vascular cells.13 We selected 5 different stages: undifferentiated hESCs, mesoderm committed cells (day 2; KDR+), early cardiovascular progenitor cells (day 4; c-Kit+), committed vascular progenitor cells (day 8; CD34+CD31+), and vascular endothelial cells (VE-cadherin+endoglin+). Kurian et al13 provide detailed characterization of the generated cell lineages (Figure IA in the online-only Data Supplement). After rRNA depletion and size removal of species whose length was smaller than 150 nucleotides, we generated data sets with a sequencing depth of 90 million paired-end reads of 100 bp per sample (Figure 1A and 1B and Figure IIB and IIC and Table III in the online-only Data Supplement). To provide an overall estimation of the transcriptome changes occurring during the differentiation of pluripotent cells to vascular progenitor cells, we first decided to collectively assess global expression changes in all cell lineages, including early c-Kit+ cardiovascular cells appearing from day 2 to 4 during the course of differentiation and committed vascular progenitor cells (day 4–8).13 To describe the global transcriptomic changes across the genome, we investigated the genomic coverage of RNA sequencing data (at least 1 read). All different cell types indicated that ≈56% of the genome was transcriptionally activated during vascular progenitor cell differentiation14–17 (Figure IIB in the online-only Data Supplement). More than 25 000 transcripts were expressed on differentiation (reads per kilobase per million >0.1), with 13 796 transcripts exhibiting at least 3-fold changes in expression. Among these 13 796 transcripts, a total of 44% represented ncRNAs (Figure 1A and Figure IIB in the online-only Data Supplement). After the exclusion of repetitive sequences, we identified a total of 406 novel transcripts that were not annotated previously in any database (83 were promoter antisense, 188 were intergenic, and 135 were anti-sense gene sequences).14–17 By considering both annotated and nonannotated transcripts, we observed that one of the largest groups differentially regulated during differentiation was lncRNAs (≈1924). These observations therefore suggest that lncRNAs have a functional involvement during mesoderm development and cell fate specification.5,9,10,18
Figure 1.
Transcriptome kinetics during human embryonic stem cell differentiation to endothelial cells. A, Annotation of transcripts expressed >0.1 reads per kilobase per million (RPKM) during differentiation of H1 human embryonic stem cells into endothelial cells. B, Total genes regulated >3-fold between key stages of vascular differentiation. Fractions attributed to lncRNA regulation are shown in dark gray and black. C, Dynamics of key lineage-restricted markers during vascular differentiation from human embryonic stem (ES) cells to endothelial cells. D, Gene ontology enrichment for biological processes regulated at each specific stage of differentiation. E, Contour plots depicting the relative fraction of genes presenting bivalent methylation marks (H3K4me3 and H3K27me3) at promoters of each gene (ChIP-Seq reads [log2] from –2kb, +2kb from the GENCODE TSS). Bivalent genes are defined as those with >5 (log2) normalized ChIP-Seq reads. Pie chart represents the summary of genes activated during vascular differentiation that are driven by bivalent promoters, including lncRNAs. F, Regulatory motif enrichment in the promoters (–300 bp, +50 bp) of critical genes at each stage of differentiation. HUVEC indicates human umbilical vein endothelial cell.
Noncoding RNA Regulation Accounts for the Majority of Transcriptomic Changes Regulating Lineage Specification
Next, we focused our attention on the analyses of each respective cell lineage generated during differentiation. Similar to recent reports,19,20 we validated the robustness of our system by focusing our attention on genes characteristic of each of the different stages analyzed: pluripotent cells (day 0; POU5F1 [Oct4], NANOG, SOX2, ZFP42),13,19 mesoderm-committed cardiovascular progenitor cells (day 2–4; T [Brachyury], MSX1, GSC, EOMES, WNT3A, SNAI2, EVX1),9,10,13 committed vascular progenitor cells (day 4–8; GATA1, GATA2, LMO2, ETS1, HOXB4),13,21 and endothelial cells (CDH5, VWF, EPHB2, NRF2F2, ENG)13 (Figure 1C). As expected, all genes appeared upregulated at the corresponding developmental stages (Figure 1C). Gene ontology analysis highlighted the expression of key genetic circuitries driving cardiovascular commitment (upregulated at day 2) and blood vessel and heart development (upregulated from day 2–8; Figure 1D). These results are in agreement with previous reports and further demonstrate that early cardiac and vascular developmental programs share common genetic pathways before further cell-type specification in humans.21,22 Chromatin modification analysis identified an increasing number of bivalent sites as differentiation progressed, for both protein-coding and lncRNA sites (Figure 1E and Figure IID in the online-only Data Supplement). Differentiation to endothelial cells resulted in the upregulation of >1000 genes and the downregulation of ≈2500 genes (Figure 1B). Genome-wide methylation profiling highlighted significant hypomethylation at lncRNA sites (19%; Figure IIE in the online-only Data Supplement). Together, these results suggested an important regulatory role for lncRNAs during lineage specification (eg, compared with 5% for microRNAs; Figure IIE in the online-only Data Supplement).
Identification of ncRNAs Differentially Regulated During Human Pluripotent Stem Cell Differentiation to Vascular Endothelial Cells
To identify enriched DNA motifs acting as regulatory elements, we used HOMER, an analytic tool suitable for promoter enrichment analyses.15 Promoter motif enrichment (–300, +50 bp from the transcriptional start site) revealed that transcripts differentially expressed during differentiation harbor binding sites for the major transcriptional networks regulating vascular development compared with nonregulated promoters (Figure 1F). Key developmental drivers differentially regulated in a time-dependent manner included members of the homeobox gene family, mesodermal regulators (EOMES, GATA, SMAD), and vascular transcription factors (ETS, HIF; Figure 1F). Interestingly, lncRNAs made up the largest class of noncoding transcripts subjected to strict temporal regulation patterns during differentiation. These results are in line with the notion that lncRNAs are more tightly controlled in terms of timing and cell-type specificity than protein-coding transcripts.18,23 We then selected 4 distinct groups of lncRNAs, corresponding to the 4 top-level clusters, highlighted by hierarchical clustering (Figure IIIA in the online-only Data Supplement). These 4 distinct groups had the highest average dissimilarity between one another and correlated with the different developmental stages analyzed, with ≈300 lncRNAs expressed specifically in pluripo-tent stem cells, ≈100 in mesodermal progenitors (day 0–2), ≈250 in early cardiovascular progenitor cells (day 2–4), ≈550 in late vascular progenitors (day 4–8), and finally, ≈200 in differentiated endothelial cells (Figure IIIA in the online-only Data Supplement). Furthermore, antisense lncRNA expression demonstrated a positive correlation with the respective sense protein-coding counterparts in many cases (Figure IIIB in the online-only Data Supplement). We next used a stringent 5-step filtering process at each indicated differentiation stage to identify novel lncRNAs, the sequence and functionality of which might be conserved across different vertebrate species (Figure IIIA in the online-only Data Supplement). To do this, we focused on the following parameters: (1) cell stage–specific patterns of expression distinguishing pluripotent cells, cardiovascular progenitor cells, and terminally differentiated endothelial cells in multiple biological replicates (at least 3); (2) significant sequence conservation across different vertebrate species; (3) exon-intron structure data; (4) availability of expressed sequenced tags in human, mouse, and zebrafish to evaluate coding potential and facilitate expression analyses; and (5) physical location in close proximity to known cardiovascular regulatory elements. Our analyses highlighted a total of 75 lncRNAs that successfully fulfilled the first 3 criteria (ie, specific expression, exon-intron structure, partial sequence conservation across vertebrates) and at least 1 of the latter (eg, expressed sequenced tag data in lower vertebrates). Next, we randomly selected 3 previously uncharacterized lncRNAs specifically expressed in pluripotent stem cells (TERMINATOR), vascular progenitors (ALIEN), and mature endothelial cells (PUNISHER) for further characterization (names were retrospectively assigned on the basis of the phenotypes elicited in zebrafish). ncRNA identification relies, to a great extent, on known sequence-defining gene characteristics (eg, well-defined promoters, exon-intron structure, codon conservation, and ribosomal footprints), as well as a lack of coding potential.6,24 None of these transcripts possessed coding potential, as determined by GeneID analysis24 (Figure IIIC in the online-only Data Supplement), as well as a lack of identifiable amino acid domains and lack of codon conservation across evolution or ribosomal footprints (data not shown).
Characterization of 3 Novel lncRNAs Differentially Expressed During Endothelial Cell Differentiation
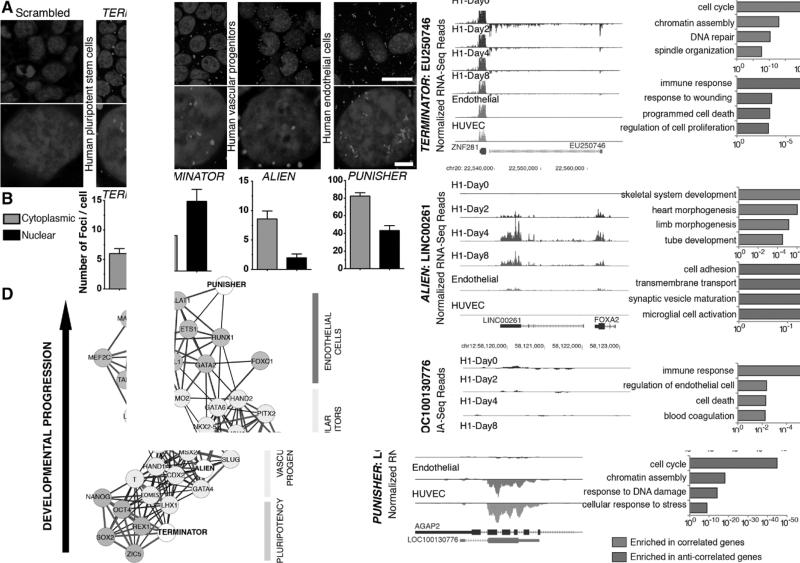
Next, we sought to determine the functionality of the 3 different lncRNAs selected. To do so, we first focused on localization studies in the different cell lineages in which the 3 lncRNAs were identified as differentially regulated (pluripotent cells, cardiovascular progenitors, and endothelial cells). Fluorescence in situ hybridization experiments18 revealed preferential expression of TERMINATOR in the nucleus of pluripotent stem cells, whereas nuclear, perinuclear, and cytosolic localization of ALIEN and PUNISHER was found in cardiovascular progenitors and endothelial cells, respectively (Figure 2A and 2B). lncRNAs have been demonstrated to play important roles in gene regulation during cell fate specification and development.3,9,10,25,26 To comprehensively characterize the gene networks associated with the different lncRNAs, we performed Pearson correlation analyses in which protein-coding mRNA expression was systematically evaluated and associated with each of the 3 different lncRNAs. We next focused exclusively on those transcripts with a correlation coefficient >0.85 for further gene ontology analyses. Expression of TERMINATOR correlated positively with genes involved in cell cycle, DNA repair, and chromatin assembly and negatively with genes involved in cell death and regulation of proliferation (Figure 2C and Figure IVA and IVB in the online-only Data Supplement). ALIEN expression correlated positively with transcripts involved in skeletal muscle development, heart morphogenesis, and tube formation and correlated inversely with cell adhesion, membrane transport, and neural function related genes (Figure 2C and Figure IVA and IVB in the online-only Data Supplement), suggesting that ALIEN might possess a functional role during early cardiovascular development before vascular specialization. Finally, PUNISHER demonstrated a positive correlation with genes participating in definitive vascular development while negatively correlating with cell-cycle regulators, chromatin modifiers, and DNA damage response genes (Figure 2C and Figure IVA and IVB in the online-only Data Supplement). Next, we performed RNA immunoprecipitation coupled to mass spectrometry analysis (Table IV in the online-only Data Supplement). Protein complex analysis on pulldown led to the identification of proteins involved in RNA binding,27 posttranscriptional control, and epigenetic remodeling28 (Figure 2C and Table IV in the online-only Data Supplement). Spring-embedded algorithms (Figure 2D) revealed hits closely correlating with each lncRNA. TERMINATOR expression was associated with POU5F1 (Oct4), SOX2, ZIC5, and REX1, all of them known regulators of pluripotency. ALIEN demonstrated a high degree of correlation with pivotal drivers of mesoderm and cardiovascular commitment, including T (Brachyury), EOMES, MIXL1, and GATA4 (Figure 2D). Finally, expression of transcription factors essential for endothelial cells such as TAL1 and FOXC1 correlated with PUNISHER (Figure 2D). Together, these results indicate a stage-specific function for each of the different lncRNAs in regulating gene expression. Additionally, none of the identified lncRNAs were physically associated with polypeptides of the ribosome translational machinery. Thus, this confirms the noncoding nature of the selected transcripts.
Figure 2.
Characterization of TERMINATOR, ALIEN, and PUNISHER, 3 novel developmentally regulated lncRNAs. A, Representative images of subcellular localization of TERMINATOR in human embryonic stem (ES) cells, ALIEN in vascular progenitors, and PUNISHER in primary endothelial cells as determined by RNA in situ hybridization using specific locked nucleic acid (LNA) probes. A scrambled control LNA probe has been tested in all 3 different cell types with similar results (representative pictures on the left). B, Quantification of nuclear and cytosolic lncRNA foci (n ≥5). C, Left, RNA-Seq read density coverage along lncRNA loci. Right, Gene ontology functional enrichment analysis from all genes exhibiting expression profiles similar to TERMINATOR (top), ALIEN (middle), or PUNISHER (bottom). D, Network depicting correlated gene expression profiles of the uncharacterized lncRNAs and key developmental transcription factors. A Pearson correlation threshold of 0.85 was used to define edges in the network. Thick gray lines indicate higher correlation relative to black lines. Data are represented as mean±SD. Scale bars: 25 μm (A, top) and 5 μm (A, bottom). HUVEC indicates human umbilical vein endothelial cell.
Novel lncRNAs Functionally Control Pluripotency, Cardiovascular Commitment, and Endothelial Cell Identity
To gain insights into the physiological relevance of the identified lncRNAs, we first evaluated their expression pattern during mouse and zebrafish embryonic development. In accordance with their expression during human vascular differentiation, Terminator expression was maximal at the blastocyst stage in mouse and at 6 hours after fertilization in zebrafish, closely correlating with the expression of Pou5f1 (Oct4) and Nanog (Figure 3A and Figures IIC and V in the online-only Data Supplement). Terminator levels were undetectable in 1-cell–stage zebrafish embryos immediately after fertilization, suggesting that Terminator was not already present in the oocyte before fertilization (Figure 3A). Alien was expressed in the allantois and lateral plate mesoderm of E8.5 mouse embryos and zebrafish embryos 12 hours after fertilization (Figure 3B), correlating with the expression of T (Brachyury), Meox1, and Mixl1 (Figure V in the online-only Data Supplement). The endothelial cell–specific lncRNA Punisher showed the highest levels of expression in mouse embryos at embryonic day 12.5 and in zebrafish at 72 hours after fertilization, once the vasculature was formed (Figure 3C and Figure V in the online-only Data Supplement). PhyloP analysis for detailed evolutionary conservation revealed relatively short, highly conserved sequence regions of 250 to 500 bp (>90%–95%) across vertebrates29 (Figure 3D and Table V in the online-only Data Supplement). Additionally, as previously mentioned, there were described expressed sequenced tags transcribed from the conserved loci in mouse and in zebrafish. TERMINATOR was found to be an intergenic lncRNA located next to ZNF281 in human and mouse (in both cases in chromosome 1). It presents H3K27Ac sites in start and end regions (marking actively transcribed chromatin), and its genomic location is conserved in sense. ALIEN represents a subclass of lncRNAs, a lincRNA (long interspersed noncoding RNA). It was found located in an intergenic region with low gene density. It is encoded in sense with exons, and its location is conserved next to FOXA2 in mouse, humans, and zebrafish. Finally, PUNISHER was found to be an antisense lncRNA covering exonic and intronic sequences of the AGAP2 gene. Its location is conserved in mouse and humans. It is transcribed as an antisense to AGAP2, and it presents H3K27Ac sites. Taken together, our results indicate that these 3 lncRNAs are conserved from zebrafish to humans and show similar stage-specific expression during development.
Figure 3.
lncRNA sequence conservation and expression profiles through vertebrate evolution and development. A through C, Dynamic expression pattern in developing mouse and zebrafish embryos for Terminator (A), Alien (B), and Punisher (C) as determined by quantitative reverse transcription–polymerase chain reaction (n=5 per group). D, Vertebrate genomic alignments and PhyloP conservation scores for each uncharacterized lncRNA across 44 species. See Table V in the online-only Data Supplement for the full list of animals used for PhyloP conservation analysis.1–7 Data are represented as mean±SD. E indicates embryonic day; hpf, hours postfertilization; and MEF, mouse embryonic fibroblast.
To further characterize the functional roles of the different lncRNAs during vertebrate development, we designed a series of experiments in different vertebrate systems, including zebrafish and mammalian murine embryos. Antisense morpholino oligonucleotides, synthetic molecules used to sterically block RNA binding sites in the absence of degradation, targeting highly conserved regions or putative splice sites in the identified lncRNAs were designed to induce specific knockdown or functional blockade.30,31 Loss-of-function experiments were first performed in the transgenic zebrafish strains fli1:GFP (vascular reporter) and cmlc2:GFP (cardiac reporter; Figure 4A) with specific antisense morpholino and compared with the corresponding nontargeting sequences used as negative controls. Morpholino injections targeting terminator compromised development at the gastrulation stage and resulted in >70% lethality, whereas the surviving embryos exhibited developmental arrest and severe cardiovascular defects (Figure 4B–4D and Tables VI and VII in the online-only Data Supplement). In addition, delayed epiboly stages and detachment of the cell mass from the yolk at 5 to 7 hours after fertilization could be observed early during development (Figure VI in the online-only Data Supplement), in line with peak expression of terminator at 6 hours after fertilization. Morpholino-mediated loss of function of alien led to severe impairment in multiple anatomic structures, highlighting the specific role of alien in mesodermal specification. Among other mesoderm-related defects, alien inhibition resulted in defective vascular patterning, with pronounced branching defects abrogating the correct formation of dorsal and intersegmental blood vessels and defective cardiac chamber formation, demonstrating that alien specifically functions in an early developmental progenitor stage common to both the vascular and cardiac lineages (Figure 4B–4D and Tables VI and VII in the online-only Data Supplement). Inhibition of punisher also resulted in severe defects in the vasculature, including defective branching and compromised vessel formation (Figure 4B–4D and Tables VI and VII in the online-only Data Supplement). Abrogation of Punisher activities demonstrated significant changes in vessel tube number and length and severely impaired cardiac development and function (Figure 4E–4G and Tables VI and VII in the online-only Data Supplement). To confirm the in vivo role of terminator and punisher, we performed rescue experiments. Coinjection of mature human lncRNA sequences alongside the respective morpholinos sufficed for rescuing the observed phenotypes with a significant effect in embryo survival, animal morphology, and branching and development of the cardiovascular system (Figure VII and Table VIII in the online-only Data Supplement). It should be noted that zebrafish morpholinos could not efficiently target the human lncRNA counterparts because of sequence divergence. Therefore, these rescue experiments further highlighted a qualitatively specific functional role for each of the different lncRNAs. Together, our results demonstrated a specific function for the identified lncRNAs in the differentiation of pluripotent cells to mesoderm and further specification of cardiovascular followed by vascular and endothelial cell specification in vertebrates.
Figure 4.
In vivo functional evaluation of conserved lncRNAs during zebrafish development by morpholino (MO)-mediated loss of function. A, Morpholinos were designed to block specific highly conserved regions or putative splice sites and were injected into cardiovascular reporter zebrafish embryos. B, Representative phenotypes observed for the different knockdowns during vascular (fli1:GFP) and cardiac (cmlc2:GFP) development at 48 hours postfertilization (hpf). C and D, Quantification of vascular (C) and cardiac (D) phenotypes obtained for the different morpholinos (n>70). E and F, Analysis of defects in blood vessel formation by quantification of tube number (E) and tube length (F; n >70). G, Heart function measured as heartbeats per minute for the different morphants (n >10). Data are represented as mean±SD. Scale bars, 5 μm (B). Wt indicates wild-type. *P<0.05.
Identified lncRNAs Demonstrate a Critical Role at Specific Stages During Mammalian Development
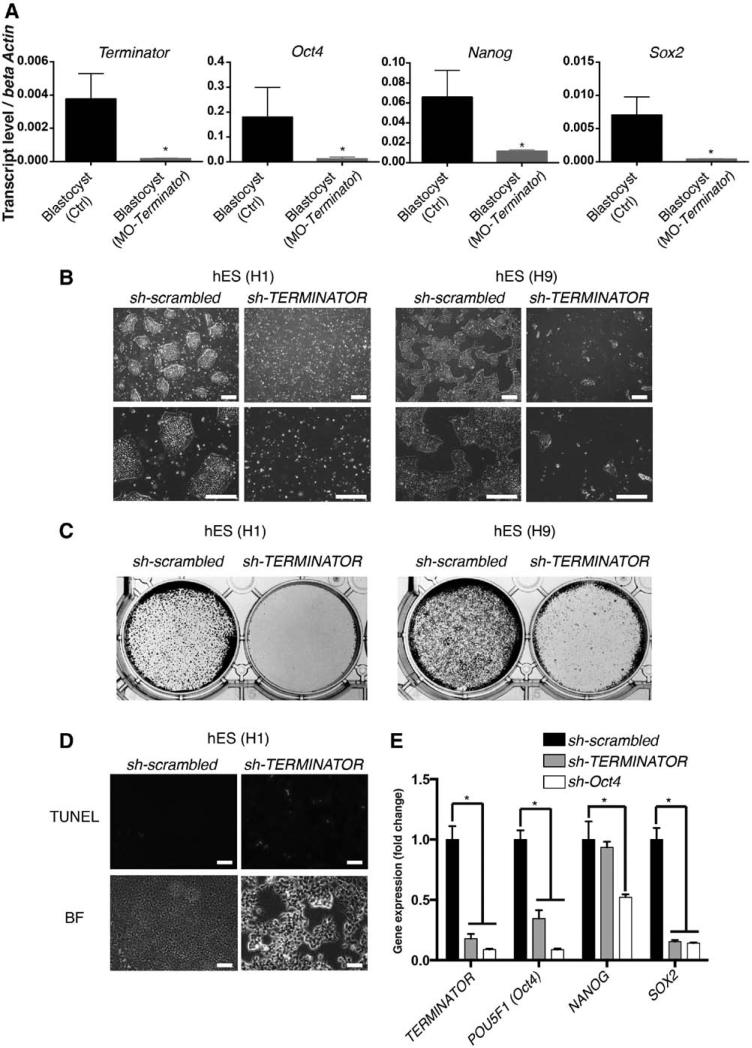
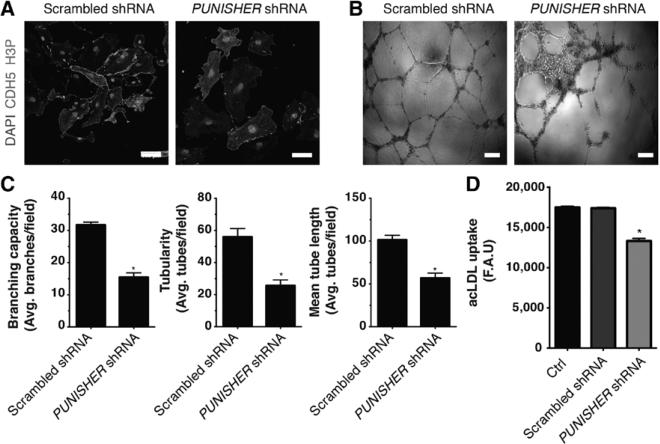
Finally, to provide an overview of the role that the identified lncRNAs play in mammalian development, we evaluated the role of all 3 lncRNAs in mammals by loss-of-function experiments in mouse embryos and human cells. To do so, 3 independent biological replicates were pooled prior to microarray hybridization. Further analysis indicated that TERMINATOR blockade in mouse embryos and human ESCs resulted in downregulation of the pluripotency factors POU5F1 (Oct4), SOX2, and NANOG at the blastocyst stage and led to significant cell death (Figure 5A–5E). Downregulation of TERMINATOR resulted in the upregulation of 506 genes and the downregulation of 185 different genes involved in cell-cell interactions and chromatin remodeling necessary for the maintenance of a pluripotent state (Figure 6). ALIEN knockdown resulted in the significant upregulation of 738 genes involved in cell adhesion and extracellular matrix remodeling, whereas downregulation of 503 genes related to angiogenesis and blood vessel development could be observed (Figure 6). Finally, PUNISHER knockdown resulted in profound gene expression changes in endothelial cells, with 802 genes involved in mitosis and cell division being downregulated and 831 genes involved in cell adhesion and extracellular interactions being upregulated (Figure 6). PUNISHER knockdown resulted in decreased histone H3 phosphorylation (Figure 7A), a marker indicative of mitosis, and impaired human vessel maturation (Figure 7B and 7C). PUNISHER knockdown also impaired acetyl–low-density lipoprotein uptake, a hallmark of endothelial cell functionality, therefore indicating severely impaired endothelial cell function compared with scrambled shRNA controls and unmodified endothelial cells (Figure 7D).
Figure 5.
Terminator is essential for pluripotent stem cells survival. A, Morpholino (MO)-mediated loss of function of Terminator in mouse blastocyst–stage embryos led to significant downregulation of pluripotency factors Oct4, Nanog, and Sox2, indicating a fundamental role during very early development (n >8). B and C, Massive cell death in human embryonic stem (hES) cell lines (H1 and H9) was induced by loss of TERMINATOR, as indicated by representative bright-field pictures of sh-scrambled and sh-TERMINATOR–treated cell culture (B). In C, treated cells were stained by crystal violet. D, TERMINATOR knockdown induced apoptosis in human pluripotent stem cells as indicated by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining. E, Downregulation of essential pluripotency transcription factors Oct4 and Sox2 on TERMINATOR knockdown (n=5). Data are represented as mean±SD. Scale bars, 200 μm (B) and 75 μm (D). *P<0.05. BF indicates bright-field.
Figure 6.
Gene expression profiling of key molecular networks influenced by lncRNA knockdown during human vascular differentiation. Heat map depicting genetic networks differentially regulated on shRNA-mediated knockdown of TERMINATOR (human embryonic stem [ES] cells), ALIEN (vascular progenitors), and PUNISHER (human umbilical vein endothelial cells), respectively. Enriched biological processes are indicated on the right.
Figure 7.
PUNISHER knockdown results in severe functional aberrations in human primary endothelial cells. A, Loss of PUNISHER in endothelial cells led to significant reduction in cell proliferation indicated by loss of phosphorylated histone H3 48 hours after knockdown. B, Representative bright-field pictures showing well-organized capillary-like structures formed by scramble-treated endothelial cells (left) as opposed to PUNISHER knockdown endothelial cells (right). C, Quantitative analysis after Matrigel tube formation assay on knockdown of PUNISHER indicated severe defects in branching (left), tubularity (middle), and mean tube length (right; n ≥5). D, Acetylated low-density lipoprotein (acLDL) functional assay showed significantly reduced uptake on PUNISHER knockdown (n=5). Note that the control group represents primary endothelial cells. Data are represented as mean±SD. Scale bars, 100 μm (A) and 1 mm (B). *P<0.05. F.A.U. indicates fluorescence arbitrary units.
Discussion
Recent discoveries indicating that the mammalian transcriptome is comprised of a large majority of noncoding transcripts (≈60%) as opposed to coding RNAs (≈7%) have brought about the question of to whether noncoding sequences can play a role in controlling cellular fate and functionality, ultimately affecting biological diversity.25,32–34 Despite major efforts, little information is available on the actual cellular functions of ncRNAs in the context of human development, physiology, and disease.9,10,19,35,36 Among the existing information, lncRNAs identified in pluripotent cells have been ascribed mainly to gene-network interactions, the regulation of chromatin, and the control of the pluripotency network.19,33,35 Similarly, other recent reports have focused on the identification of lncRNAs during cardiomyocyte generation and identified Braveheart and Fenderr as critical players during the development of the early cardiovascular system and ultimately the murine heart.9,10
Pluripotent cell differentiation has been demonstrated to be a suitable platform for recapitulating key developmental stages in a dish and for the establishment of cellular disease models.37,38 Therefore, with the goal of comprehensively investigating human cardiovascular development, we analyzed human ESCs during the course of differentiation to mesoderm, cardiovascular progenitor cells, and differentiated vascular endothelial cells. By relying on our recently reported methodologies for the differentiation of human stem cells to cardiovascular progenitors and terminally differentiated vascular endothelial cells, we report here the identification and characterization of 3 novel human lncRNAs (see Table IX in the online-only Data Supplement for a summary of findings for each lncRNA). First, we found a total of 75 novel lncRNAs with different expression patterns depending on the differentiation stage. Among those lncRNAs fulfilling our preselection criteria, we next selected 3 novel noncoding transcripts (TERMINATOR, ALIEN, and PUNISHER) for further characterization in different vertebrate models. We found that TERMINATOR specifically controls pluripotent stem cell identity, ALIEN impairs cardiovascular development, and PUNISHER compromises endothelial cell function. The differentiation platform used, however, presents several limitations. As with most protocols on pluripotent stem cell differentiation, the generated cells generally present a fetal-like signature, thus potentially obviating lncRNAs that are specifically expressed in adult cells. In addition, in vitro differentiation protocols and genetic profiling suffer from the inherent disadvantage of providing snapshots of what is otherwise a continuous process. Thus, we cannot rule out that, despite collecting samples every second day, other more tightly regulated lncRNAs are underrepresented in our data sets. Despite these limitations and considering the role that the identified lncRNAs play during cardiovascular development, it is tempting to speculate that deregulation of lncRNAs affecting early embryonic development might be one of the causes of congenital cardiovascular diseases and malformations. Indeed, our work indicates that deregulation of lncRNAs and the resulting changes in gene expression have profound implication in embryonic development and cardiovascular system formation and function across different vertebrate species. Although the implications of lncRNAs in human disease remain obscure and we are only now starting to unravel the complex role that non-coding genetic elements play in the context of disease, it seems clear that leveraging next-generation sequencing technologies with patient data collection might shed new light on how aberrant lncRNA expression functionality correlates with disease. Understanding the precise role of ncRNAs during development and disease might ultimately open new avenues for the development of human therapeutics.
Our work establishes an analytic pipeline for the systematic study of lncRNAs in cardiovascular development and demonstrates that appropriate in vitro systems can be used to identify novel players controlling lineage commitment and human development. Ultimately, broad-scale application of genomic strategies based on the use of pluripotent cells might shed new light on the fundamental mechanisms underlying human development and disease.
Supplementary Material
CLINICAL PERSPECTIVE.
Long noncoding RNAs (lncRNAs) are a new class of regulatory RNA molecules able to modulate diverse processes such as development, pluripotency, and disease by altering posttranscriptional and posttranslational regulation, recombination, protein complex formation, and cell-cell signaling. Accordingly, it is clear that identification and characterization of novel lncRNAs regulating human development are of major interest. Human pluripotent stem cells have the capacity to generate any given somatic cell lineage on differentiation. In vitro differentiation recapitulates key developmental stages, thus allowing comprehensive studies on early human development and disease. Leveraging methodologies for the differentiation of cardiovascular lineages from human pluripotent stem cells in combination with next-generation sequencing, we provide here the transcriptomic changes underlying vertebrate cardiovascular development. In addition, we provide an analytic framework for the identification of novel genetic elements underlying the development of the cardiovascular system in multiple vertebrate species. Our work indicates that deregulation of specific lncRNAs and the resulting changes in gene expression may have profound implications in embryonic development and cardiovascular system formation and function across different vertebrate species. The combination of next-generation sequencing technologies with patient data collection might shed new light on how aberrant lncRNA expression correlates with disease. A better understanding of the precise role of noncoding RNAs during development and disease might ultimately open new avenues for the development of human therapeutics specifically targeting this previously overlooked family of genes.
Acknowledgments
We thank M. Schwarz, P. Schwarz, I. Dubova, M. Marti, C. Fabregat, and D. Okamura for administrative and technical support. We also thank Ruoping Chen and the Beijing Genomics Institute for sequencing services and Nasun Hah for valuable advice.
Sources of Funding
Work in Dr Yeo's laboratory was supported by grants from the National Institutes of Health (U54 HG007005, R01 HG004659, and R01 NS075449) and the California Institute for Regenerative Medicine (RB3-05009). Work in Dr Izpisua Belmonte's laboratory was supported by grants from the National Institutes of Health (5U01HL107442), CIRM, the G. Harold and Leila Y. Mathers Charitable Foundation, and the Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002). Dr Kurian was partially supported by the California Institute for Regenerative Medicine (CIRM). Dr Nivet was partially supported by an F.M. Kirby Foundation postdoctoral fellowship. Dr Sancho-Martinez was partially supported by a Nomis Foundation postdoctoral fellowship.
Footnotes
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/lookup/suppl/doi:10.1161/CIRCULATIONAHA.114.013303/-/DC1.
Disclosures
None.
References
- 1.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. doi: 10.1073/ pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson SR, Marguerat S, Bähler J. Exploring long non-coding RNAs through sequencing. Semin Cell Dev Biol. 2012;23:200–205. doi: 10.1016/j.semcdb.2011.12.003. doi: 10.1016/j.semcdb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T, Guigó R, Johnson R. The long non-coding RNAs: a new (p)layer in the “dark matter.”. Front Genet. 2011;2:107. doi: 10.3389/fgene.2011.00107. doi: 10.3389/fgene.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, Dunagin M, Pimkin M, Gore M, Sun D, Konuthula N, Raj A, An X, Mohandas N, Bodine DM, Hardison RC, Weiss MJ. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurian L, Sancho-Martinez I, Nivet E, Aguirre A, Moon K, Pendaries C, Volle-Challier C, Bono F, Herbert JM, Pulecio J, Xia Y, Li M, Montserrat N, Ruiz S, Dubova I, Rodriguez C, Denli AM, Boscolo FS, Thiagarajan RD, Gage FH, Loring JF, Laurent LC, Izpisua Belmonte JC. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat Methods. 2013;10:77–83. doi: 10.1038/nmeth.2255. doi: 10.1038/nmeth.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. doi: 10.1016/j. molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saldanha AJ. Java Treeview: extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 19.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, Zhang X, Coyne M, Fostel JL, Holmes L, Meldrim J, Guttman M, Epstein C, Park H, Kohlbacher O, Rinn J, Gnirke A, Lander ES, Bernstein BE, Meissner A. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen CA, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung WY, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Bondue A, Tännler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 26.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryo-genesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong C, Popp MW, Maquat LE. Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods. 2012;58:88–93. doi: 10.1016/j.ymeth.2012.06.020. doi: 10.1016/j.ymeth.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. doi: 10.1016/j. cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–D63. doi: 10.1093/nar/gks1172. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNARoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerød A, Børresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daley GQ. The promise and perils of stem cell therapeutics. Cell Stem Cell. 2012;10:740–749. doi: 10.1016/j.stem.2012.05.010. doi: 10.1016/j.stem.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets in this study are available in the National Center for Biotechnology Information Gene Expression Omnibus under the accession number GSE54969.