Abstract
Mobile elements are DNA sequences that can change their position (retrotranspose) within the genome. Although its biological function is largely unappreciated, DNA derived from mobile elements comprises nearly half of the human genome. It has long been thought that neuronal genomes are invariable; however, recent studies have demonstrated that mobile elements actively retrotranspose during neurogenesis, thereby creating genomic diversity between neurons. In addition, mounting data demonstrate that mobile elements are misregulated in certain neurological disorders, including Rett syndrome and schizophrenia.
Proper functioning of the nervous system depends on the establishment of a diverse repertoire of neuronal subtypes and the integration of individual cells into unique neuronal circuits. Within a neuronal subtype, cells display diverse phenotypes and differ in their molecular characteristics, firing patterns and connections. A combination of several molecular mechanisms, including epigenetic regulation, alternative splicing and post-translational modification, contribute to the generation of this neuronal diversity. Somatic mosaicism — the presence of somatic cells with distinct genotypes within one individual — adds an additional level of complexity by generating genomic diversity between neurons. Mobile elements, which are also known as transposable elements, are responsible for the generation of somatic mosaicism, and it has been shown that mobile elements increase their activity specifically during the differentiation of a neural precursor into a neuron1. In this Review, we suggest that mobile element-driven diversity provides a stochastic mechanism by which the coding potential of a single genome can be expanded. Although the full functional impact of mobile DNA in the nervous system remains unknown, we review the literature that supports a role for mobile elements in the development of the nervous system and their potential contribution to neurological diseases.
Introduction to mobile elements
In the 1940s, mobile elements were discovered in maize2. It is now known that as the human genome evolved, DNA sequences capable of mobilizing and inserting themselves (or a copy) into new genomic positions accumulated. This DNA now comprises approximately 45% of our current genome3. Although only a small percentage of these mobile elements are still capable of mobilization, mobile element-derived DNA is abundant in the genome of many organisms4.
Mobile elements fall into two major classes: retrotransposons, which mobilize through an RNA intermediate (see below), and DNA transposons, which mobilize through a process in which the DNA sequence encoding the transposon is cut out of its normal position and ligated into an alternative position within the genome. DNA transposons, such as those first discovered in maize, are inactive in humans and mice and are not discussed in detail in this Review. Retrotransposons, however, remain active in humans and mice, and mobilize through a ‘copy-and-paste’ mechanism that results in their insertion into new locations in the genome as well as replication of a portion of their sequence. During this process, the retrotransposon is transcribed and the RNA intermediate functions as a template for the synthesis of cDNA by an RNA-dependent DNA polymerase. This DNA can integrate back into the genome, resulting in a full or partial copy of the retrotransposon. Retrotransposons are further classified into long terminal repeat (LTR) or non-LTR classes. Herein, we focus on the non-LTR class of retrotransposons, as this is the class that is still active in human genomes (FIG. 1a).
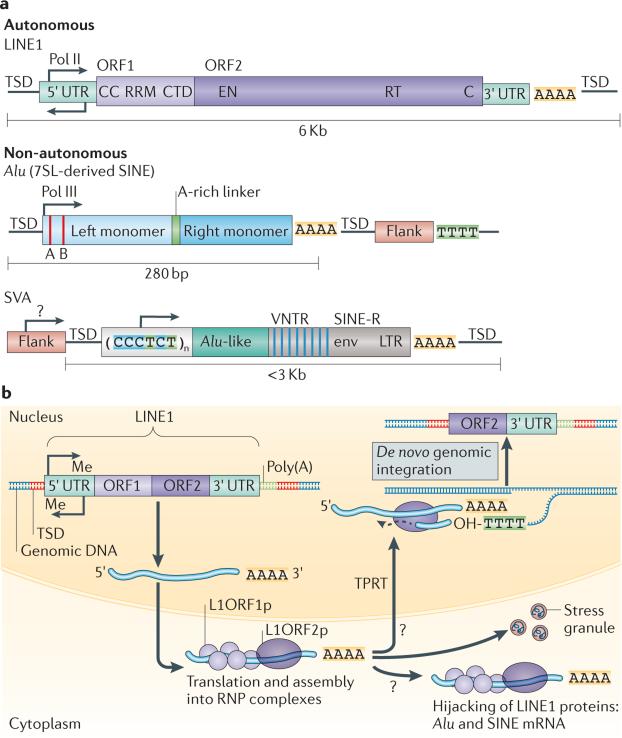
Figure 1. Retrotransposons in humans.
a | The retrotransposons of the long interspersed nuclear element 1 (LINE1) class are the only autonomous mobile elements that are active in humans. Full-length LINE1 elements are 6 kb long and comprise a 5′ untranslated region (UTR) sequence that contains an internal bidirectional DNA polymerase II (Pol II) promoter, followed by open reading frame 1 (ORF1) and ORF2, a 3′ UTR and a poly(A) tail. ORF1 encodes an RNA-binding protein (L1ORF1p) and the protein encoded by ORF2 (L1ORF2p) has an endonuclease (EN) domain and reverse transcriptase (RT) domain. The insertion of LINE1 into DNA during retrotransposition results in target site duplication (TSD), which flanks the new insertion. The non-autonomous elements, Alu and SINE–VNTR–Alu (SVA), are non-coding RNAs that co-opt LINE1 machinery in order to retrotranspose. Alu elements are short interspersed nuclear elements (SINEs) derived from the signal recognition particle RNA 7SL and are 280 bp long. They are transcribed by Pol III and contain A and B box sequences, left and right monomers separated by an A-rich linker and a poly(A) tail. Alu elements do not contain a Pol III termination site, so transcription continues into the flanking sequence until a Pol III termination signal (TTTT) is reached. SVA elements are a composite of other repeats, containing a CCCTCT repeat, two Alu-like sequences, a VNTR and a SINE-R region, which is homologous to the envelope (env) and long terminal repeat (LTR) sequences of human endogenous retrovirus. This is followed by a poly(A) sequence. b | The LINE1 retrotransposition cycle. LINE1 sequences are endogenously encoded in the genome. They are transcribed in the nucleus and assemble a ribonucleoprotein (RNP) complex containing LINE1 RNA, L1ORF1p and L1ORF2p in the cytoplasm. ORF1 and ORF2 sequences can also be co-opted by Alu or SVA sequences, which would result in retrotransposition of Alu or SVA. The RNP complexes can be sequestered as stress granules. The RNP complex accesses the nucleus through nuclear membrane breakdown during cell division or through an unknown import mechanism. L1ORF2p nicks the DNA at a TTAAA sequence and reverse transcribes the RNA through target-primed reverse transcription (TPRT), which results in a 3′ truncated or full-length copy of the LINE1 sequence flanked by TSDs. CC, coiled coil domain; CTD, carboxy-terminal domain; Me, methylation; OH, hydroxyl; RRM, RNA recognition motif. Part b is adapted with permission from REF. 54, © Elsevier (2010).
Within the non-LTR class of retrotransposons, long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs) remain active in both human and mouse genomes: approximately 80–100 and 3,000 LINE1 elements are retrotransposition-competent in humans and mice, respectively4,5. SINE–VNTR–Alu (SVA) elements are a family of non-autonomous retrotransposons that are specific to the primate lineage, and there are approximately 3,000 SVA elements in the human genome4. LINE1 elements are ~6-kb autonomous elements that encode open reading frame 1 protein (L1ORF1p), an RNA-binding protein6, and L1ORF2p, a protein with endonuclease7 and reverse transcriptase8 activity (FIG. 1b). Whereas L1ORF1p and L1ORF2p function in cis to retrotranspose LINE1 RNA, SINE and composite SVA elements are non-coding and must co-opt LINE proteins in order to mobilize9,10. Therefore, all non-LTR-derived retrotransposition events in mammals depend on LINE1 expression for mobilization. LINE1, SINE and SVA elements can also be divided into subfamilies of related elements based on sequence similarity.
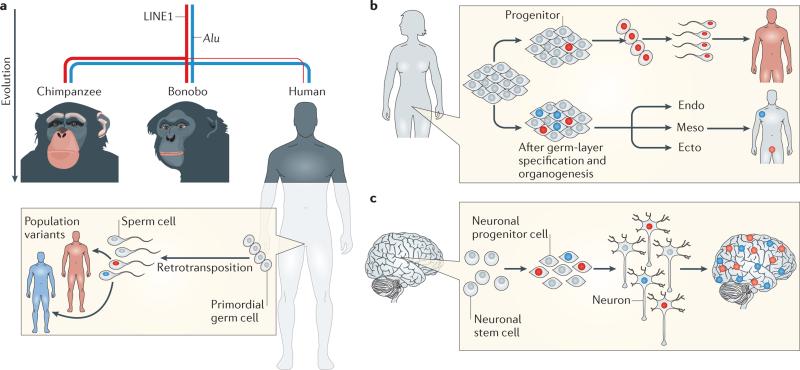
This Review focuses on somatic retrotransposition — that is, retrotransposition events that occur in cells that cannot transmit genetic information to the next generation. Germline retrotransposition, conversely, occurs in germ or pluripotent cells and can be inherited by the subsequent generation (FIG. 2a,b). Many features of somatic retrotransposition are shared by germline retrotransposition. As common mammalian ancestors evolved, species-specific subfamilies of LINE1 and SINE elements also co-evolved, and different subfamilies of these elements remained active within different mammalian germ lines4. For example, during chimpanzee, bonobo and human speciation, lower rates of LINE1 germline retrotransposition occurred in the human lineage (BOX 1). Whether the somatic retrotransposition rates are lower in humans than in chimpanzees and bonobos is unknown.
Figure 2. Consequences of germline and somatic retrotransposition events.
a | As humans, chimpanzees and bonobos evolved from a common ancestor, retrotransposons actively mobilized in the ancestral germ lines, which resulted in the generation of genomic variation that natural selection then acted upon. Alu retrotransposition rates (represented by the thickness of the blue line) remained relatively similar between the three species; however, long interspersed nuclear element 1 (LINE1) retrotransposition rates (represented by the thickness of the red line) were suppressed in the human lineage. Retrotransposition of LINE, Alu and SINE–VNTR–Alu (SVA) elements continues to occur in the human germ line, which creates population variants that are present in every cell of an individual’s body and are also passed on to future generations. Whether LINE1 or Alu somatic retrotransposition rates differ between human and non-human primates is unknown. b | Somatic retrotransposition can happen at any time during embryogenesis. Retrotransposition events that occur in early pluripotent progenitor cells will result in somatic mosaicism: these unique cells will contribute to all tissues of the body of the individual, including the germ line. Somatic retrotransposition that happens after germ-layer specification and organogenesis, however, results in germ-layer- or tissue-specific insertions. These will not contribute to the germ line. c | Somatic retrotransposition increases as neural stem cells differentiate into neurons and results in neurons with unique genomes. Variability exists between the rates of retrotransposition and regions in which it occurs between individuals. High rates of retrotransposition events seem to occur in the hippocampus in some individuals22,28. Figure is adapted from Muotri, A. R., Marchetto, M. C., Coufal, N. G. & Gage, F. H. The necessary junk: new functions for transposable elements. Hum. Mol. Genet. 16, R159–R167 (2007)80, by permission of Oxford University Press.
Germline retrotransposons are a major source of structural variants, deletions and sequence insertions within the human population11–15. The vast majority of these germline variants have unknown functional effects. However, some variants are likely to have functional consequences for the individual. For example, although polymorphic insertions of retrotransposon sequences are abundant in the healthy human population, specific de novo retrotransposon insertions can cause haemophilia16, neurofibromatosis17 and other diseases. In addition to the insertion of the retrotransposon sequence, retrotransposition can mediate the deletion of the host DNA sequence18. Furthermore, retrotransposon events can result in the presence of highly homologous sequences in different genomic locations. These sequences can then recombine, through nonallelic homologous recombination, to cause deletions, duplications, inversions and translocations.
Retrotransposition in the nervous system
Until recently, mammalian retrotransposition was believed to occur only in germ cells, pluripotent cells and cancer tissues. As a consequence, it was long believed that every cell of the body except immune, cancer and germ cells contains the same DNA sequence. However, in 2005, mobilization of LINE1 retrotransposons that generate neuronal somatic mosaicism within the mammalian brain was described1 (FIG. 2c).
In the past decade, several lines of evidence have proven that LINE1 elements are not only expressed in the mouse, human and Drosophila melanogaster brain but are also actively retrotransposed in these species1,19–22. A combination of three complementary approaches — retrotransposition reporter assays, quantitative PCR (qPCR) to detect an increase in LINE1 copy number and next-generation genome sequencing — has demonstrated that somatic retrotransposition is higher in neurons than in other neural cell types1,22,23. However, many fundamental features of neural retrotransposition remain to be determined. These include the rate of retrotransposition in different regions of the brain, which cell types are more or less prone to retrotransposition, the variability of retrotransposition rates between individuals, the mechanisms that regulate the targeting of insertions and the extent to which neuron-to-neuron genomic differences affect the properties of brain networks.
Evidence of somatic mosaicism in the brain
LINE1-retrotransposition reporter assays indicate whether successful completion of the full retrotransposition cycle, including genomic integration of a newly copied element, can take place within a cell24. In these assays, an active full-length LINE1 element is engineered to contain an enhanced green fluorescent protein (EGFP) or neomycin resistance marker that is only functionally expressed after the full retrotransposition cycle of transcription, splicing and integration of the marker back into the genome is completed24. These assays have demonstrated that neuron-specific retrotransposition takes place in the mouse brain1, that there are increased rates of retrotransposition in mouse and human models of Rett syndrome25 and ataxia telangiectasia26, and that LINE1 retrotransposition is activated in human tissue culture models of neural development22. However, how accurately such reporters reflect endogenous retrotransposition activity remains to be determined. At a minimum, the data demonstrate that neural precursors can support retrotransposition and that retrotransposition and LINE1 expression are detected at higher levels in neural precursor cells than in other somatic cell types. LINE1 does not tend to integrate into particular hotspots in the genome. However, gypsy transposons — LTR retrotransposons that are active in D. melanogaster — often insert into the ovo locus. Endogenous gypsy mobilization can thus be visualized when an endogenous gypsy transposon inserts into engineered ovo sequences, which results in the activation of a GFP reporter21,27. This approach has revealed that somatic gypsy insertions occur in aged D. melanogaster neurons21.
In human neural precursor cells (unlike many other non-transformed cell types), endogenous L1ORF1p is detectable in ribonucleoprotein particles22. A qPCR assay that enables the detection of an increase in the copy number of the most active LINE1 elements in the human genome has also been developed22. This assay revealed that there are higher levels of active LINE1 elements in brain tissue than in non-neuronal tissues22. The magnitude of this increased copy number varies between individuals and also varies between brain regions. For example, in some individuals, the hippocampus may be enriched in LINE1 elements22,28 (FIG. 2c).
A critical limitation of the qPCR assay is that the genomic location of the new insertion cannot be identified. Recent next-generation sequencing approaches have reported endogenous somatic insertions in the human post-mortem brain and D. melanogaster brain19,20,23,28. Investigators have used targeted sequencing approaches to enrich for elements that are capable of retrotransposition, single-cell-targeted sequencing or whole-genome sequencing approaches to identify brain-specific insertions19,20,23,28. Many of these approaches are confounded by several factors, including the rarity of each individual insertion event (such events are likely to be hemizygous and present in a single cell or a subset of cells) and the inherent false discovery rate in next-generation sequencing efforts. Nevertheless, several studies with varying levels of stringency have reported the identification of somatic insertions in the brain19,20,23,28.
Compared with bulk sequencing efforts, the emerging technology of single-cell genomic analysis enables more confident detection of the unique insertions that are present in single cells, although its throughput is limited. Targeted single-cell sequencing in 300 neurons taken from the cortex and caudate of three non-diseased individuals was performed19. This is the only report so far that has successfully identified and validated a brain-specific LINE1 insertion containing all of the known characteristics of LINE1-mediated retrotransposition, including target site duplication. A previous bulk sequencing study was the first to report the identification of endogenous brain-specific insertions and was able to identify two somatic LINE1-mediated insertions with target site duplications, although these were not validated by PCR28.
From the single-cell sequencing data, it was estimated that the rate of unique somatic insertions is <0.6 insertions per neuron in the cortex or caudate19. Using the qPCR copy number assay, a previous study had estimated that 80 to 800 new insertions occur in each hippocampal neuron22. Several possibilities may account for these differing estimates. Perhaps owing to the impact of environmental factors on retrotransposition rates29, variable rates of new somatic insertions probably exist between individuals. Furthermore, in some individuals, the rate of retrotransposition is higher in the hippocampus than in the frontal cortex22,28. In addition, the study that arrived at the larger estimate used exogenous LINE1 DNA copies that were mixed into the PCR reactions at different concentrations to estimate the rate of insertions in hippocampal neurons, which may have limited the accuracy of the assay. Although single-cell sequencing enables the identification and re-validation of insertions, it will not detect insertions if the wrong population of cells is sequenced. Furthermore, owing to the smaller number of insertions identified, the genomic distribution of insertional preference cannot be characterized. Only when additional data from large sample sizes of different brain regions, more individuals and additional tools are available will we know the true rate and distribution of insertions in somatic brain cells. However, even using a conservative estimate of 1 insertion per 300 cells, the human brain would contain more than 100 million unique somatic insertions30.
Other sources of neuronal somatic mosaicism
In addition to the insertion of new mobile element sequences, retrotransposition may contribute to other types of genomic mosaicism. Neuronal genomes are characterized by increased levels of aneuploidy31,32 and large copy number variants (CNVs)33. Using single-cell genomic analysis, it was shown that 13–41% of non-diseased human frontal cortex neurons contain at least one megabase-scale de novo CNV33. Current single-cell genomic analysis has limited ability to detect smaller CNVs; therefore, it is likely that neurons contain even greater numbers of smaller CNVs.
It is unknown how large neuronal CNVs are generated, but it is well established that retrotransposons can generate other genomic insults in addition to the insertion of mobile element sequences. L1ORF2p is an endonuclease that induces double-strand DNA breaks34,35. Non-homologous end joining repair resolves retrotransposon insertions, which occasionally results in deletions and rearrangements in human cells36. Also, repetitive sequences are a major source of non-allelic homologous recombination, as outlined above37. Considering the increased activity of retrotransposons in neuronal development, it is possible that somatic retrotransposition also influences CNV generation in neuronal genomes.
Control of mobile element activity
Regulation of retrotransposon expression
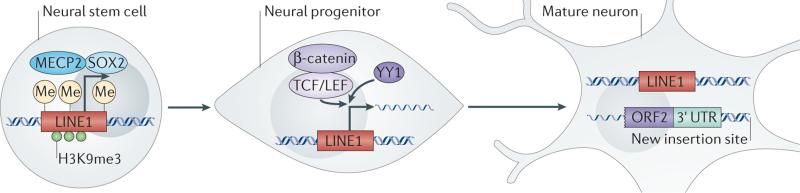
Unregulated LINE1 expression and mobilization can cause genomic instability; therefore, mobile elements are highly suppressed in most cell types. Because retrotransposons mobilize by first being transcribed into an RNA intermediate, the regulation of LINE1 expression is one of several mechanisms by which this process is controlled. Indeed, the first clues that LINE1 retrotransposition might be important in neurogenesis came from observations of LINE1 expression (FIG. 3). In rats, mice and humans, the LINE1 promoter becomes transcriptionally activated as neural precursors differentiate into neurons and glia1,22. The canonical LINE1 promoter contains binding sites for SOX2 (REF. 38), YY1 (REF. 39), RUNX3 (runt-related transcription factor 3)40, and TCF and LEF (WNT signalling pathway transcription factors)41, which are transcription factors that are known to be involved in neurogenesis. SOX2 maintains neural stem cell proliferation and potency and is downregulated as neural stem cells differentiate. In neural stem cells, SOX2, repressive chromatin and methylated promoter DNA bound by methyl-CpG-binding protein 2 (MECP2) collaborate to repress the LINE1 promoter1,25. Upon differentiation, this repression is decreased. Both SOX2 downregulation and promoter-specific DNA demethylation cause a decrease in MECP2 binding1,22,25. At the same time, activation of WNT3A stimulates LINE1 expression through the canonical WNT pathway41. In combination, these events lead to the activation of LINE1 expression during the first few cell divisions as neural progenitors differentiate into neurons.
Figure 3. Regulation of retrotransposition in neural progenitors.
In neural stem cells, the long interspersed nuclear element 1 (LINE1) promoter is repressed by DNA methylation, H3K9me3 modifications, methyl-CpG-binding protein 2 (MECP2; which binds to the methylated (Me) DNA) and SOX2. As neural stem cells transition to progenitors, SOX2 is no longer present. The LINE1 promoter assumes an open chromatin state and becomes de-methylated. MECP2 can no longer bind. The WNT transcription factors, β-catenin and members of the TCF/LEF family activate transcription, perhaps with the cooperation of another transcription factor, YY1. This results in an increase in LINE1 transcription in the progenitor and active retrotransposition. Whether new retrotransposon insertions can occur in postmitotic mature neurons is unknown; however, the de novo insertions that occurred in the progenitor create neurons with unique LINE1 insertion sites. ORF2, open reading frame 2; UTR, untranslated region.
Small RNA-mediated regulation
Small RNA-mediated epigenetic regulation also controls the expression and RNA stability of mobile elements42,43. Research on the role of small RNA-mediated regulation of transposition has largely focused on the germ line; however, small RNA-mediated regulation of transposons in somatic cell types is well documented. In human cells, the 5′ untranslated region (UTR) of LINE1 transcribes both sense and anti-sense RNA, which can function as substrates for small RNA pathways and inhibit LINE1 (REFS 44–46). MOV10, a putative RNA helicase and component of the small RNA-induced silencing complex, was shown to inhibit LINE1 retrotransposition in cancer cell lines47. In the fly brain, depletion of the small RNA pathway proteins Argonaute 2 (Ago2), Dicer 2, Aubergine (Aub), Ago3 and Armitage results in increased expression of LTR elements, LINE-like elements and DNA transposons20,21. Aub and Ago3 are normally depleted from specific mushroom body neurons, and this depletion correlates with increased expression of a number of LTR, LINE-like and DNA transposable elements in these neurons20. This suggests that small RNA-mediated suppression of transposons is lower in certain brain regions, which may result in increased retrotransposition rates in specific mushroom body neurons20. Interestingly, the D. melanogaster mushroom body and mammalian hippocampus are critical regions for learning and memory. It remains to be determined whether the PIWI-interacting RNA (piRNA) pathway also regulates retrotransposition in the mammalian brain. However, there is an intriguing possibility that an evolutionarily conserved mechanism could contribute to increased retrotransposition rates in brain structures that are involved in learning and memory.
Additional host mechanisms
Additional mechanisms within the cell, termed host mechanisms, suppress retrotransposition through post-transcriptional mechanisms. RNA- and DNA-editing proteins, such as AID, APOBECs, and ADAR proteins, act on retrotransposons and restrict their mobilization48,49. DNA repair processes and non-homologous end joining proteins inhibit LINE1 retrotransposition or lead to 5′ truncation of new LINE1 insertions, which restricts the duplication of full-length retrotransposition-competent LINE1 sequences50,51. Some host suppression mechanisms, such as DNA repair processes, may act equally in both neuronal and nonneuronal tissues. Other processes, such as ADAR editing, may occur more frequently in brain tissue, where ADAR-specific A-to-I RNA editing is abundant in transcriptome data sets, compared with other tissues52.
Benefits of somatic retrotransposition
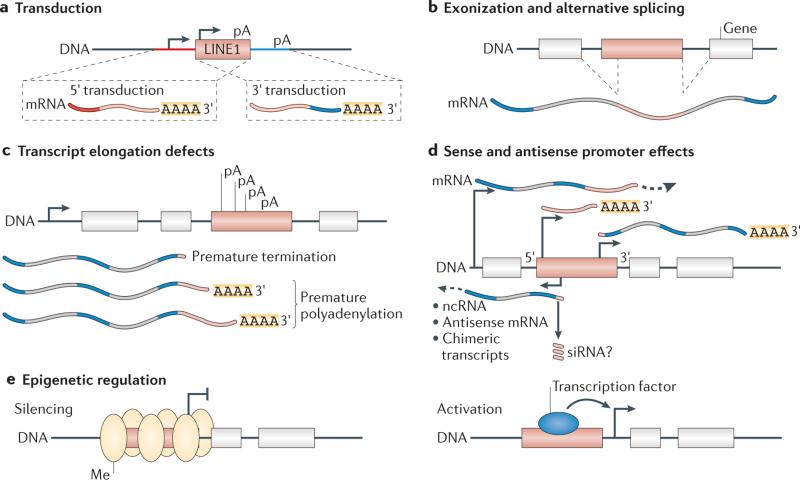
Mobile element insertions generate cells with unique genomes, leading to distinct transcriptomes. Depending on when the somatic transposon insertion occurs, it could be present in as many as half of the cells in an individual or may be restricted to as few as one unique copy in one single cell. Both somatic and germline insertions probably have the same potential to affect cellular function by altering proximal gene expression and RNA splicing, and by causing premature polyadenylation of RNA (FIG. 4).
Figure 4. Impact of mobile element insertions on the transcriptome.
Retrotransposon insertions can affect genes near the site of insertion. a | Transduction is a process through which sequences flanking the 3′ and 5′ regions of long interspersed nuclear element 1 (LINE1) elements (red and blue) can be carried by LINE1 into new locations in the genome. Transduced sequences can bring novel exons, regulatory sequences or poly(A) sequences to a new genomic location. b | LINE1 and Alu elements contain cryptic splice signals. This can cause alternative splicing of mRNA transcripts that results in LINE1 or Alu being spliced into the transcript. c | LINE1 elements also contain internal polyadenylation (pA) signals that can cause aberrant premature polyadenylation or termination of transcription. d | The 5′ untranslated region (UTR) of LINE1 encodes sense and antisense promoters. Transcripts that originate from these promoters can generate novel antisense, non-coding or chimeric transcripts. Similarly, the 3′ UTR also encodes a promoter that can initiate transcription of novel sequences. e | Mobile elements are targeted by many epigenetic mechanisms. Insertion of a mobile element can bring novel epigenetic regulation to the region, including altered levels of DNA methylation (Me) or heterochromatin formation that can initiate at the element and spread to the flanking sequence. Insertion of a mobile element can also provide novel transcription factor-binding sequences, which results in binding and activation of proximal promoters. ncRNA, non-coding RNA; siRNA, small interfering RNA. Figure is adapted with permission from REF. 54, © Elsevier (2010).
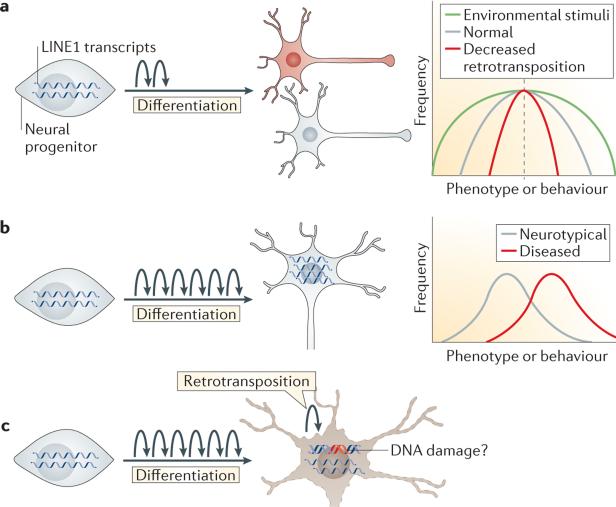
The evidence of increased somatic retrotransposition in neurons suggests that it may have a unique role within the brain (FIG. 5a). Insertions that are targeted into specific regions of the genome could have specific functional consequences, whereas random positioning of insertions could have a more expansive role in somatic diversification. Owing to the large size of many genes that are specifically expressed in the brain and also the open chromatin state that is a feature of these genes in neurons, brain-specific somatic retrotransposition probably affects neuronal genes more frequently than it affects other genes20,28,53. Brain-specific insertions into genes that are important for neural function, including those encoding dopamine receptors and neurotransmitters, have been identified in both humans and D. melanogaster20,28. These insertions would probably affect the expression or function of these genes in individual neurons. However, highly constrained targeting of retrotransposon insertions that would result in a specific functional impact has not been observed. Thus, we suggest that somatic retrotransposition functions as a more stochastic generator of neuronal diversity and broadens the variance of cellular and organismal phenotypes54,55.
Figure 5. Effects of somatic mosaicism in neurons.
Somatic retrotransposition in neurons might have cellular and organismal phenotypic effects. a | Under healthy conditions, a moderate level of retrotransposition (represented by curved arrows) during the differentiation of neural progenitors could expand the coding potential of the genome and create more diversity among neuronal subtypes. On an organismal level, we speculate that retrotransposition expands the variation of phenotypes and behaviours, creating an intangible variance54,55. This is illustrated on the graph on the right by the grey line showing an intermediate level of variance in a normally distributed population of behavioural phenotypes. With decreased levels of retrotransposition, we speculate that phenotypes would be more similar and have a narrow distribution but unchanged mean (illustrated by the red line). Environmental stimuli may enhance variation even further (illustrated by the green line). b | Increased retrotransposition in neurodevelopmental disorders such as Rett syndrome and schizophrenia may lead to diseased neurons, with perhaps less connectivity or complexity. On an organismal level, we speculate that this may occur because phenotypes or behaviours are shifted to a more extreme mean, resulting in a disease phenotype. It is possible that targeting of insertions to certain loci or the cellular response to increased retrotransposition results in a more extreme behavioural phenotype. c | Increased long interspersed nuclear element 1 (LINE1) activity is also implicated in neurodegeneration. LINE1 may act as a DNA-damaging agent or retrotransposon transcripts may function as toxic RNAs, thereby increasing vulnerability to apoptosis.
At the individual-neuron level, somatic retrotransposition could alter synaptic activity, the response of a neuron to stimuli or the competitive innervation of neuronal circuitry, depending on which genes are affected. On the organismal level, somatic retrotransposition has the potential to contribute to the ‘intangible variance’ observed between genetically identical individuals54,55. The mechanisms of somatic retrotransposition may remain conserved to enable individuals to respond to unforeseen environmental triggers. Although natural selection allows for positive and negative selection of genomic variants that are favourable to historical evolutionary pressures, a static somatic genome is unable to change or respond when presented with a new, unanticipated environmental signal. If somatic retrotransposition can be triggered by environmental factors, it could enable genomic changes that may benefit the organism. For example, voluntary exercise increased LINE1 insertions in the hippocampus when a mouse containing a reporter for LINE1 retrotransposition was used29. In the absence of somatic retrotransposition, we predict that genetically identical individuals would have decreased phenotypic variance.
Mobile elements and disease
Although a controlled level of retrotransposition may be beneficial for neuronal genomes, upregulated mobile elements may also have deleterious effects on cognitive function (FIG. 5b,c). Evidence from studies of patients and/or animal models suggests that transposon mis-regulation may occur in various neurological disorders, including post-traumatic stress disorder (PTSD), alcoholism, neurodegeneration, ageing and neurodevelopmental disorders21,23,25,26,56,57. Two forms of mobile element misregulation have been observed: altered levels of mobile element expression and increased numbers of somatic insertions. However, it is unknown how or even if mobile element misregulation directly causes neurological disorders.
Neurodevelopmental disorders
Rett syndrome was the first neurodevelopmental disorder in which an increase in somatic retrotransposon insertions was discovered25. Rett syndrome is an X-linked autism spectrum disorder that primarily affects females and is caused by mutations in MECP2 (REF. 58). MECP2 is thought to act as a transcriptional repressor59 or activator60 and can repress LINE1 transcription61. MECP2 binds to methylated DNA at the LINE1 promoter and represses LINE1 transcription in neural progenitor cells25. Mecp2-knockout mice display an increase in LINE1 and other repetitive element expression25. In addition, both the brains of patients with Rett syndrome and Mecp2-knockout mice have higher levels of somatic LINE1 retrotransposition than non-diseased controls25. However, an increased retrotransposon insertion number is probably not the sole cause of all Rett syndrome phenotypes, as re-expression of MECP2 in postmitotic neurons or microglia can restore normal lifespan and activity in Rett syndrome mouse models62–64, which presumably retain cells with high rates of somatic retrotransposon insertions. Although some rescue of cognitive defects is observed in these mice, the impact of increased retrotransposition on the full spectrum of cognitive or behavioural phenotypes remains unknown. In addition, LINE1 transcript levels in these mice probably return to basal levels, which may be important in the phenotypic rescue.
Recently, it was shown that individuals with schizophrenia also have increased levels of somatic LINE1 retrotransposition copy numbers in neurons23. Using the qPCR copy number assay22,25, the authors showed that LINE1 copy number is increased in the prefrontal cortex of patients with schizophrenia, in an induced pluripotent stem cell model of 22q11 deletion and in mouse and macaque immune activation models of schizophrenia23. Patients with schizophrenia seem to have a variable increase in LINE1 copy number. It is possible that this reflects assay noise; however, inter-individual and intra-individual cell-to-cell differences may also contribute to the variability. Finally, whole-genome sequencing was performed on bulk brain and liver tissues to identify tissue-specific insertions in post-mortem patient and control tissues. Similar levels of tissue-specific insertions are found in both the brains and livers of patients and control individuals, but no validation was performed to verify these tissue-specific insertions. However, putative tissue-specific insertions are found more frequently in genes associated with synaptic function and schizophrenia relative to control genes. This raises the possibility that an increased frequency of insertions and a higher percentage of neurons with detrimental mutations in synaptic genes contribute to schizophrenia pathology.
Neurodegeneration and ageing
Another disorder associated with increased somatic retrotransposition, ataxia telangiectasia, is a neurodegenerative disorder caused by mutations in the gene encoding ataxia telangiectasia mutated (ATM)26. ATM is a serine/threonine kinase involved in DNA damage signalling65. DNA repair pathways, including those mediated by ATM, are known to inhibit or cause 5′ truncation of LINE1 insertions,26,51,66. Similar to Rett syndrome, ATM deficiency corresponds to an increase in LINE1 retrotransposition activity and copy number26. Considering the ability of LINE1 to cause DNA damage34, LINE1-related genomic instability may cause or increase neurodegeneration.
Transposon misregulation may also be involved in TAR DNA-binding protein 43 (TDP43)-related neuro-degenerative disorders, including amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration. TDP43 is an aggregation-prone RNA-binding protein that is mutated in certain familial forms of ALS. TDP43 binds to approximately 30% of the mammalian transcriptome, influences alternative splicing and is involved in the regulation of very long introns67–69. A re-analysis of in vivo TDP43 binding targets revealed that TDP43 binds to many transposon sequences, including LINE, SINE and LTR retrotransposons70. In patients with frontotemporal lobar degeneration, TDP43 binding to transposons is reduced, and depletion or overexpression of TDP43 results in dramatic upregulation of the same families of transposons. It is unknown whether altering the levels of TDP43 results in increased numbers of somatic retrotransposition events. Perhaps misregulation results in genomic instability, DNA damage or toxicity as a result of excess transposon-derived RNA70.
Transposons are also activated during the course of normal ageing in D. melanogaster21. LINE-like elements, named R1 and R2, and gypsy elements are highly expressed in an age-dependent manner in flies. Increasing transposon expression by disrupting Ago2, an endogenous repressor of transposons and other genes, results in an increase in transposon expression that correlates with accelerated, age-dependent memory decline and decreased lifespan21. Blocking DNA damage-induced apoptosis in aged postmitotic neurons is sufficient to extend the animal’s lifespan and delay age-dependent memory decline21. These intriguing findings suggest that transposons are activated during ageing and that the DNA damage response to de novo insertions contributes to age-dependent neuronal decline. However, it remains to be seen whether transposon misregulation and the resulting DNA damage could lead to ageing and neurodegeneration in both flies and humans.
Transposons respond to environmental shocks
Growing evidence implicates transposons as sensors of environmental stress in the brain, reminiscent of an earlier hypothesis that transposons are a mechanism of programmed response to unknown ‘shocks’ (REF. 57). Under control conditions, transposon expression is repressed by epigenetic mechanisms, and these epigenetic mechanisms are altered in a tissue-specific response to stress. As a homeostatic mechanism to respond to environmental stress, somatic retrotransposition may ultimately benefit the organism. However, under pathological conditions of extreme environmental stress, retrotransposition may also contribute to disease. For example, rats subjected to stress-enhanced fear learning, a model for PTSD, upregulated LINE1 transcripts in the amygdala71. Furthermore, acute restraint of rats induces upregulation of the silencing H3K9-specific methyltransferase SUV39H2, specifically in the hippo-campus72, which correlates with silencing of B2 SINE and IAP elements. Human alcoholic brains display upregulation of SINE, LTR and LINE1 transcripts, which correlates with decreased epigenetic silencing73. Also, TRIM28, a critical endogenous retrovirus silencer, is upregulated in the alcoholic brain. Although related, expression of mobile elements is distinct from increased insertion events. Many mobile element sequences are contained within other transcriptional units, such as introns and the UTRs of mRNAs74. Collectively, the evidence of upregulation of mobile element sequences in response to environmental stimuli complements the growing evidence that mobile element regulation may have a profound impact on cognition.
Conclusions and future perspectives
Mounting evidence challenges the notion that mobile elements are merely ‘junk’ DNA sequences that are silent and stable in the developing soma. They are now implicated in healthy and diseased brain function. Whether beneficial or detrimental, the impact of mobile elements remains largely enigmatic. To move beyond correlational studies, future research will need to provide functional evidence of a direct involvement of retrotransposons in the phenotype being studied. How increased retrotransposition rates could contribute to such distinct phenotypes as Rett syndrome, neurodegeneration, ageing and schizophrenia remains unknown. Perhaps increased retrotransposition is a more general phenomenon of cellular stress and disease that has a different impact depending on when, where and how frequently insertions occur. Individuals harbour diverse retrotransposon element insertion positions, and perhaps different elements have altered activation in various circumstances. Although manipulation of repetitive sequences remains a technological challenge, retrotransposon inhibitors, genome editing and emerging genome-wide technologies are likely to contribute to our future understanding of somatic retro transposition. Although nearly a decade has passed since the discovery of somatic retrotransposition, we are just beginning to disentangle the influence of mobile elements on the brain.
Box 1 | Evolutionary impact of mobile elements.
Retrotransposons are prehistoric elements that have shaped genome evolution. The reverse transcriptases present in long interspersed nuclear element 1 (LINE1) elements are related to ancient mobile group II introns, which are found in mitochondria and eubacteria75. Mobile elements have contributed genomic sequences that are crucial for mammalian neurogenesis, including transcription factor-binding sites for PAX6 (REF. 76), an essential regulator of neurogenesis, and for alternative splicing signals74. Retrotransposons remained active at varying rates during human and great ape speciation and contributed species-specific insertions (FIG. 2a). During primate evolution, the rate of Alu retrotransposition was 15-fold higher in human, chimpanzee and bonobo lineages than in gorilla and orangutan lineages77. Interestingly, LINE1 activity rates were suppressed in the human lineage compared with chimpanzee and bonobo lineages77,78, which may have resulted in a less diverse human population compared to non-human primate populations. By comparing human and non-human primate induced pluripotent stem cells (iPSCs), it was demonstrated that human iPSCs suppress LINE1 activity, as they have higher levels of Piwi-like protein 2 and APOBEC3B, which are known suppressors of LINE1 retrotransposition78. The selection for increased LINE1 suppressors in the human lineage may be a consequence of a population bottleneck created by a combination of factors, such as pandemic pathogens or extreme climate change. A speculative hypothesis suggests that less (but not null) genomic variability in humans could have contributed to increased cultural evolution79.
Epigenetic regulation
A process that alters the state of gene expression through changes in chromatin structure (that is, DNA or histone modifications).
Alternative splicing
A process whereby different mRNAs can be produced from a single gene through the differential incorporation of exons into the mature transcript during splicing. Frequently, various mature proteins are generated from a single gene.
Stochastic mechanism
A mechanism that is governed by random effects.
Autonomous elements
Elements that mobilize by themselves and do not require any other transposable elements for mobilization.
Endonuclease
An enzyme that cleaves a polynucleotide chain.
Reverse transcriptase
An enzyme that generates complementary DNA from an RNA template.
Speciation
The evolutionary process by which new biological species arise.
Polymorphic insertions
Mobile element insertions into specific locations of the genome that are present in some individuals and absent in others.
Next-generation genome sequencing
Sequencing carried out using high-throughput sequencing technologies that are based on massively parallel pyrosequencing technology and that enable the discovery of rare sequences (for example, small RNAs).
Aneuploidy
A condition in which extra or missing chromosomes are present within a cell or organism.
Copy number variants
(CNVs). Changes in the normal number of copies of a given gene or locus. Usually, there are two copies of each locus, but if, for example, duplications or triplications occur the number of copies will increase.
Induced pluripotent stem cell
A cell that is created from differentiated cell types — for example, fibroblasts — and is reprogrammed by a cocktail of transcription factors (or other approaches) back to a pluripotent state. This cell can now be differentiated into cells of distinct lineages: for example, neurons.
Acknowledgements
The authors thank M. L. Gage for editorial comments. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, The Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002, and NIH TR01 MH095741.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [This article describes the first demonstration of somatic retrotransposition in the brain.] [DOI] [PubMed] [Google Scholar]
- 2.McClintock B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 8.Mathias SL, et al. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 9.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 10.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witherspoon DJ, et al. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart C, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing AD, Kazazian HH. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazazian HH, Jr, et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 17.Wallace MR, et al. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 19.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [Along with reference 28, the authors use high-throughput sequencing to identify endogenous brain-specific LINE1 insertions in post-mortem human tissue.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrat PN, et al. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [This work shows that somatic transposition in the brain is conserved in the fly. Also, specific types of mushroom body neurons have decreased levels of known retroelement repressors (Aub and Ago3), which correlates with increased expression of certain retroelements.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nature Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [This study demonstrates that neuronal transposon mobilization accumulates in the ageing fly. Increasing retroelement expression by mutating Ago2 also results in accelerated cognitive decline, suggesting that transposons contribute to age-dependent cognitive decline.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bundo M, et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:306–313. doi: 10.1016/j.neuron.2013.10.053. [The authors use LINE1 copy number assays to demonstrate an increase in the number of LINE1 sequences in neurons from patients with schizophrenia.] [DOI] [PubMed] [Google Scholar]
- 24.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 25.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coufal NG, et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl Acad. Sci. USA. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrador M, Sha K, Li A, Corces VG. Insulator and Ovo proteins determine the frequency and specificity of insertion of the gypsy retrotransposon in Drosophila melanogaster. Genetics. 2008;180:1367–1378. doi: 10.1534/genetics.108.094318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [Along with reference 19, the authors use high-throughput sequencing to identify endogenous brain-specific LINE1 insertions in post-mortem human tissue.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl Acad. Sci. USA. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingsbury MA, et al. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc. Natl Acad. Sci. USA. 2005;102:6143–6147. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell. Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belancio VP, Roy-Engel AM, Deininger PL. All y'all need to know ‘bout retroelements in cancer. Semin. Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang N, Zhang L, Zhang Y, Kazazian HH. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuwabara T, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nature Neurosci. 2009;12:1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Yang N, Kazazian HH. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature Struct. Mol. Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 45.Ciaudo C, et al. RNAi-dependent and independent control of LINE1 accumulation and mobility in mouse embryonic stem cells. PLoS Genet. 2013;9:e1003791. doi: 10.1371/journal.pgen.1003791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Heras SR, et al. The microprocessor controls the activity of mammalian retrotransposons. Nature Struct. Mol. Biol. 2013;20:1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodier JL, Cheung LE, Kazazian HH. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koito A, Ikeda T. Intrinsic restriction activity by AID/APOBEC family of enzymes against the mobility of retroelements. Mob. Genet. Elements. 2011;1:197–202. doi: 10.4161/mge.1.3.17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogerd HP, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 51.Morrish TA, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 52.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 53.Thomas CA, Paquola A, Muotri AR. LINE-1 retrotransposition in the nervous system. Annu. Rev. Cel Dev. Biol. 2012;28:555–573. doi: 10.1146/annurev-cellbio-101011-155822. [DOI] [PubMed] [Google Scholar]
- 54.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 56.Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res. 2013;35:69–76. [PMC free article] [PubMed] [Google Scholar]
- 57.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 58.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 59.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001;29:4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tropea D, et al. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc. Natl Acad. Sci. USA. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc. Natl Acad. Sci. USA. 2004;101:6033–6038. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Derecki NC, et al. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki J, et al. Genetic evidence that the nonhomologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nature Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ayala YM, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE. 2012;7:e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ponomarev I, Rau V, Eger EI, Harris RA, Fanselow MS. Amygdala transcriptome and cellular mechanisms underlying stress-enhanced fear learning in a rat model of posttraumatic stress disorder. Neuropsychopharmacology. 2010;35:1402–1411. doi: 10.1038/npp.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hunter RG, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc. Natl Acad. Sci. USA. 2012;109:17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deininger PL. Alu elements: know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou YH, Zheng JB, Gu X, Saunders GF, Yung WK. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 2002;12:1716–1722. doi: 10.1101/gr.188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hormozdiari F, et al. Rates and patterns of great ape retrotransposition. Proc. Natl Acad. Sci. USA. 2013;110:13457–13462. doi: 10.1073/pnas.1310914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchetto MC, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitehead H, Richerson PJ, Boyd R. Cultural selection and genetic diversity in humans. Selection. 2002;3:115–125. 2002. [Google Scholar]
- 80.Muotri AR, Marchetto MC, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum. Mol. Genet. 2007;16:R159–R167. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]