Abstract
Previous results from two proficiency panels of intracellular cytokine staining (ICS) from the Cancer Immunotherapy Consortium and panels from the National Institute of Allergy and Infectious Disease and the Association for Cancer Immunotherapy highlight the variability across laboratories in reported % CD8+ or % CD4+ cytokine-positive cells. One of the main causes of interassay variability in flow cytometry-based assays is due to differences in gating strategies between laboratories, which may prohibit the generation of robust results within single centers and across institutions. To study how gating strategies affect the variation in reported results, a gating panel was organized where all participants analyzed the same set of Flow Cytometry Standard (FCS) files from a four-color ICS assay using their own gating protocol (Phase I) and a gating protocol drafted by consensus from the organizers of the panel (Phase II). Focusing on analysis removed donor, assay, and instrument variation, enabling us to quantify the variability caused by gating alone. One hundred ten participating laboratories applied 110 different gating approaches. This led to high variability in the reported percentage of cytokine-positive cells and consequently in response detection in Phase I. However, variability was dramatically reduced when all laboratories used the same gating strategy (Phase II). Proximity of the cytokine gate to the negative population most impacted true-positive and false-positive response detection. Recommendations are provided for the (1) placement of the cytokine-positive gate, (2) identification of CD4+ CD8+ double-positive T cells, (3) placement of lymphocyte gate, (4) inclusion of dim cells, (5) gate uniformity, and 6) proper adjustment of the biexponential scaling.
Key terms: ICS, assay harmonization, proficiency panel, immune monitoring, gating
Intracellular cytokine staining (ICS) is a common flow cytometry-based method for rapid quantitation of cytokine-producing antigen-specific T cells. This method is more comprehensive than ELISA or ELISPOT as it allows for simultaneous analysis of cell phenotype and cytokine production on a single cell level. ICS is an important monitoring tool for clinical trials to assess the immune response to vaccination, transplantation, and other preventive or therapeutic immune interventions (1,2). However, ICS is a biological assay that can lead to variable results within and across institutions that cannot easily be controlled due to the lack of a “gold reference standard” to confirm an accurate measurement of the true expected value. As such, proficiency panels have been designed as an external validation tool and to identify sources of variability among participants with the aim to introduce assay harmonization guidelines. In initial proficiency panels, variables that impact assay performance are identified, and in subsequent panels, iterative testing leads to the development of harmonization guidelines that can be implemented by the scientific community. Assay harmonization can improve the quality of data and allow for greater ease in comparing and interpreting data across multiple laboratories (3). The high degree of complexity in an ICS assay can introduce multiple sources of variation, starting with (i) sample preparation (freezing, thawing, and stimulation of T cells) and continuing through (ii) the staining procedure (antibody/fluorochrome choice, secretion inhibitor, and fixation/permeabilization reagent selection), (iii) data acquisition that is influenced by instrument setup (voltages and compensation) and type of flow cytometer, and finally, (iv) data analysis (4,5). ICS proficiency panels have been conducted by the National Institute of Allergy and Infectious Disease, the Division of Acquired Immunodeficiency Syndrome (DAIDS), the Cancer Immunotherapy Immunoguiding Program, and the Cancer Immunotherapy Consortium (CIC), and all came to the same general conclusion that a large source of variation in reported results is due to different gating strategies (4,6–9).
To our knowledge, there is no standardized method of gating for ICS experiments. Currently, the decision about precisely where to draw a gate is highly subjective and gate placement is largely based on the operator’s visual assessment. Furthermore, gating strategy, or the sequence of plots to identify a final population, is also very subjective and based on the operator’s preference. For instance, defining how to draw the cytokine-positive gate is of particular importance in ICS gating as it can potentially change a positive response into a negative response or vice versa. Choice of gating hierarchy can also confound the assay outcome, for example, not gating on CD3 T cells could allow for CD4 dim-expressing macrophages to be included in the T-cell analysis or not using a singlet gate may create a false-positive that is really a cytokine-producing monocyte bound to a noncytokine-producing T cell. Hence, training, guidance, and experience are crucial for successful analysis of ICS-based flow cytometry experiments. Unfortunately, very little organized flow cytometry training is available, leaving the decision how to set gates mainly to locally established standards and personal preferences of operators. Automatic gating algorithms are in development, but require a combination of flow cytometry, bioinformatics, and programming knowledge; as such they are only available at certain institutions as a research tool (10,11). These also tend to be panel-specific and/or require algorithm training to work effectively. There are very few papers, courses, or instruction in how to properly gate flow cytometry data (12–14). Coexistence of various gating strategies is not necessarily a problem but should at least be accompanied by full reported transparency on how gates were set in a given experiment, making it easier for third-party investigators to judge reported data sets and to compare results generated across institutions. Still, full gating strategies are rarely published in scientific papers, although several initiatives including MyFlowCyt and MIATA are attempting to improve the reporting of flow cytometry data (15,16). Therefore, we designed a gating proficiency panel to study how gating strategies affect the variation in reported percentage of cytokine-positive CD4+ and CD8+ T lymphocytes from an ICS experiment. In this in silico proficiency panel, we eliminated all other sources of experimental variability with the exception of gating strategy and gate placement.
The specific goals of this gating panel were (1) to obtain an overview of the gating strategies currently used in the immune monitoring field for ICS, (2) to identify gating strategies which produce results that reflect the underlying rate of cytokine-positive cells, and 3) to summarize the findings into general recommendations for harmonization of gating strategies, as an overall consensus document. Considering the importance of reliable and harmonized immune monitoring and the similarity of assays across fields, the CIC conducted this ICS Gating panel as a collaborative effort together with the Association for Cancer Immunotherapy, the Cancer Vaccine Collaborative of the Cancer Research Institute (CVC/CRI), DAIDS, the External Quality Assurance Program Oversight Laboratory, the Federation of Clinical Immunology Societies, the International AIDS Vaccine Initiative, the Immunology of Diabetes Society, the Immune Tolerance Network, and the Society for Immunotherapy of Cancer.
Materials and Methods
Participants
One hundred ten laboratories participated in the ICS Gating panel from different fields and organizations, comprising laboratories working in academia, industry, and government (see Supporting Information Table S1 for complete list of participants). Three requirements had to be met for participation: (1) the laboratory must have experience with analyzing data from ICS experiments, (2) laboratories needed to participate in both phases of the Gating panel, and (3) the same operator should perform the analysis during both phases of the panel. Each laboratory received an individual laboratory ID number. One laboratory contributed two data sets (two different protocols for gating) for Phase I, and one laboratory did not complete Phase II. Therefore, the analysis is based on 111 data sets for Phase I and 109 data sets for Phase II.
Panel Setup
The participating laboratories were required to download nine Flow Cytometry Standard (FCS) files from the CIC proficiency panel website. The laboratories then analyzed these files using their laboratory-specific gating protocol (Phase I). After completing the analysis for Phase I and uploading all of the results to the CIC website, the laboratories were then able to download a gating protocol (Supporting Information Fig. S1) drafted by consensus from the organizers of the panel. The laboratories analyzed the nine FCS files for a second time, this time using the consensus instructions (Phase II) and uploaded their Phase II results to the website. The data reporting requirements for each phase were a survey, an Excel file with the results of the analysis, and a PowerPoint graphical display of their gating strategy for each FCS file. The Excel file captured (1) the percentage of lymphocytes in the “lymphocyte gate,” (2) the percentage of CD3+ CD8+ cells, (3) the percentage of CD3+ D4+ cells, (4) the percentage of cells in the CD8+ cytokine+ gate, and (5) the percentage of cells in the CD4+ cytokine+ gate. The survey for Phase I asked participating laboratories about their gating protocol. The survey for Phase II asked laboratories to compare their results from Phase I and Phase II.
FCS File Description
There were nine FCS data files that were FCS 3.0 format. The FCS files used in this panel were a subset of files created by a laboratory that detected all of the positive responses during the second CIC ICS proficiency panel conducted in 2009. The files were from peripheral blood mononuclear cells (PBMC) from three healthy human donors (Donors 1, 2, and 3) obtained with IRB approval from the Immunology Quality Assurance Center Laboratory of the Duke Human Vaccine Institute, a division of the Duke University Medical Center in Durham, NC, as described previously (17). PBMCs were each stimulated with three conditions: unstimulated (negative control), CEF peptide pool (HLA class I restricted T cell epitopes of cytomegalovirus, Epstein-Barr virus, and flu virus), and CMV pp65 peptide pool. The cells were stained with five antibodies: a dump channel (CD14 and CD19 Pacific Blue), CD3-FITC, CD4-PE-Cy7, CD8-Alexa 700, and IL-2/IFNγ-APC. No Fluoresence Minus One (FMO) controls were provided, and the original laboratory did not do doublet exclusion or use a dead cell marker. Each FCS file contained a minimum of 100,000 cells.
Data Analysis and Interpretation
Central analysis of the gating panel was performed using the survey responses, analysis results data, and gating graphical displays. A central review of all the dot plots/histograms from Phase I and II was conducted on the submitted Power-Point files to assess gating strategies. The central review evaluated (1) if uniform gates were used (both within donor and across donors), (2) if biexponential scaling was properly adjusted, (3) if placement of dump, lymphocyte, CD3, CD4, CD8, and cytokine gates were adequate (did not contain negative cells and included all positive cells), and (4) if double-positive (DP) cells were included in a gate. The reviews were performed by two independent reviewers experienced in the field. There is no gold-standard method of gating, and thus the review is necessarily subjective. Therefore, to standardize the review, prior to conducting the review, all the reviewers discussed what factors were important to examine and how to assess each of the factors of interest. Furthermore, the findings of the reviewers were compared, and where there were differences, a consensus was reached. A note about the plots used for Figures 3–5: all of the images shown in these figures are the actual graphs provided by the participating laboratories. Each laboratory used their own preferred software, scaling, axis labeling, fonts, color scheme, and type of plot, as such the plots look very different from each other. This further emphasizes the differences in gating between laboratories.
Figure 3.
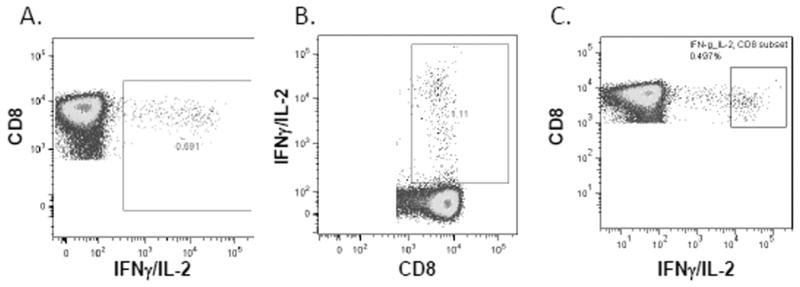
Different approaches to the placement of cytokine-positive gate. Actual graphs from different participating laboratories, all of the examples are from the same FCS file, Donor 2, CEF stimulated. (A) An example of a cytokine gate with adequate proximity. (B) A cytokine gate that is too close to the negative population. (C) A cytokine gate that is drawn too far from the negative population, missing many cytokine-positive cells. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 5.
Uniform gates and biexponential scaling. Actual graphs from different participating laboratories, all of the examples are from the same FCS file, Donor 2, CEF stimulated. (A) The cytokine gate for Donor 2 is displayed with unstimulated on the left and CEF stimulated on the right. The cytokine gates are not uniform and are much larger in the CEF-stimulated sample. (B) Three dot plots are shown with incorrectly applied biexponential scaling. The dot plot on the left has a trimodal CD4 population, the middle dot plot has a trimodal CD8 population, and the dot plot on the right displays under-scaled populations, where some of the cells are pushed against the axis. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Statistical Analysis
The percentages of cytokine-positive cells reported by each laboratory are graphically presented for both phases of the panel and are summarized using descriptive statistics. The predefined response for each donor and reagent was based on repeated pretesting results, which were confirmed by the results of the 2009 ICS proficiency panel. Those donor–reagent combinations that were expected to be negative based on the pretesting results were categorized as a false-positive if the laboratory had greater than two times background for that combination. Similarly, laboratories that had less than two times background for donor–reagent combinations that were expected to be positive based on pretesting results were assumed to have missed a response. To examine the association between response detection and laboratory protocol, laboratories were grouped into three groups: (1) laboratories that detected five or six responses and no false-positive responses, (2) laboratories that detected five or six responses and had false-positive responses, and (3) laboratories that detected fewer than five responses. The gating protocols and other laboratory characteristics were compared for these three groups of laboratories. The variables considered originated from the responses to the questions from the surveys about the laboratories’ gating protocol and the central review of the Power-Point files.
Results
Overall Variability
Phase I
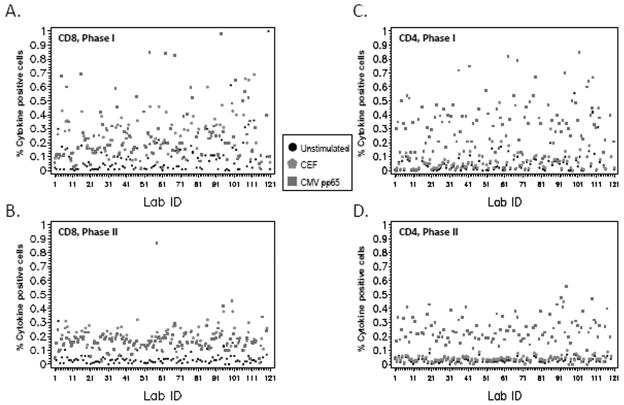
In the first phase, participants used their laboratory-specific gating protocol to gate the nine FCS files. Supporting Information Table S2 provides an overview of the responses to the survey about laboratory-specific gating protocols. Of the 110 laboratories, there were 110 different approaches to gating. Figures 1A and 1C show the CD8+ and CD4+ T-cell cytokine-positive percentages, respectively, for Donor 1 from each participating laboratory for each of the three reagents (unstimulated, CEF, and CMV) when laboratories used their own gating protocols (Phase I). The CEF and CMV responses are widely variable among the participating laboratories, ranging from 0 to 1%. There was also large variability in the reported percentage of cytokine-positive cells when laboratories used their own gating protocols for the other two donors (Supporting Information Table S3). The background responses (unstimulated group) were much higher in Phase I, with some laboratories having a background greater than 0.5% (Fig. 2A).
Figure 1.
Decreased variability if same gating strategy is used. Graph describes the percentage of cytokine-positive CD8+ (A and B) and CD4+ T cells (C and D) for Donor 1 with each of the three stimulants (unstimulated, CEF, and CMV) for all 110 participating laboratories. Unstimulated results are indicated by a black dot, CEF results are indicated by a red pentagon, and CMV results are indicated by a green square. The results from (A and C) Phase I using laboratory-specific gating protocol and (B and D) Phase II using consensus gating protocol are shown. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
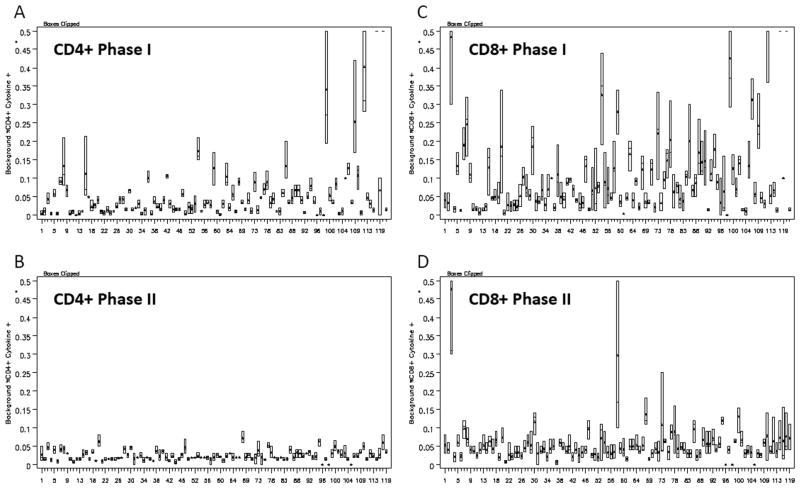
Figure 2.
Background is decreased when laboratories use the consensus gating strategy. Box plot of (A) CD4+ background cytokine-positive levels (all three cytokine-positive percentages reported for the unstimulated samples of the three donors for each laboratory in Phase I, (B) all CD4+ background cytokine-positive levels for each laboratory in Phase II, (C) all CD8+ background cytokine-positive levels for each laboratory in Phase I, and (D) of all CD8+ background cytokine-positive levels for each laboratory in Phase II.
Phase II
Figures 1B and 1D illustrate the CD8+ and CD4+ T cell cytokine-positive percentages for Donor 1 from each participating laboratory for each of the three reagents (unstimulated, CEF, and CMV) when laboratories used the gating protocol drafted by consensus from the organizers of the panel (Phase II). It is evident when comparing Figures 1A and 1B (CD8+) and Figures 1C and 1D (CD4+) that the CEF and CMV responses are less variable across laboratories when the same consensus gating SOP is used compared with the reported results where laboratories used their own gating SOP. In fact, the standard deviation was between 2.5 and 25 times larger in Phase I than Phase II with half of the donor/reagent CD4+ or CD8+ results in Phase I having a standard deviation of at least nine times larger than the standard deviation of the same donor/reagent/CD+ combinations in Phase II (Supporting Information Table S3). The mean background response in Phase I for CD4+ T cells was 0.07% and for CD8+ T cells was 0.13%, whereas the CD4+ and CD8+ background cytokine responses were 0.02% and 0.05%, respectively, in Phase II. This is highlighted in Figures 2A and 2B where the background responses seen when laboratories use the same consensus gating protocol (Phase II) are markedly lower than the background responses illustrated in Phase I.
Response Detection
Ideally, based on extensive pretesting, a laboratory should have detected six positive responses, one low-positive response (<0.15%) and five negative responses. A laboratory was considered to have detected a positive response if the antigen stimulation (CMV or CEF) was at least twice the background of the unstimulated sample from the same donor. About 73% of laboratories detected five or six responses in Phase I versus 92% in Phase II (Table 1). Furthermore, 23% of laboratories in Phase I detected two or more false-positive responses (greater than two times background in a negative donor stimulation) when compared with only 9% of laboratories in Phase II.
Table 1.
Total number of positive, low-positive, and false-positive responses detected within a laboratory
| NUMBER OF DETECTED RESPONSES WITHIN A LABORATORYa | PHASE I (LABORATORY SOP): NUMBER OF LABORATORIES | PHASE II (GATING STRATEGY SOP): NUMBER OF LABORATORIES | |
|---|---|---|---|
| True-positive responses | 0 | 0 | 0 |
| 1 | 3 (3%) | 2 (2%) | |
| 2 | 3 (3%) | 1 (1%) | |
| 3 | 8 (7%) | 1 (1%) | |
| 4 | 16 (14%) | 5 (5%) | |
| 5 | 20 (18%) | 17 (16%) | |
| 6 | 61 (55%) | 83 (76%) | |
| True low-positive responses | 0 | 67 (60%) | 56 (51%) |
| 1 | 44 (40%) | 53 (49%) | |
| False-positive responses | 0 | 58 (52%) | 66 (61%) |
| 1 | 28 (25%) | 33 (30%) | |
| 2 | 22 (20%) | 10 (9%) | |
| 3 | 2 (2%) | 0 | |
| 4 | 1 (1%) | 0 |
Response is positive if >2 times background (unstimulated).
Gating Process
To come to a consensus on harmonizing gating protocols, it is important to examine each step of the gating process, including gate sequence and gate placement, and to determine which approach leads to detection of all true-positive responses without false-positive results. Laboratories were grouped into three groups according to the number of true-positive and false-positive responses from Phase I where laboratories used their own gating protocol: (1) laboratories that detected five or six responses and no false-positive responses (n =37), (2) laboratories that detected five or six responses and had false-positive responses (n =44), and (3) laboratories that detected fewer than five responses (n =28). An analysis was performed to examine the association between response detection using these three groups of laboratories and laboratory protocol variables to understand which variables influence gating results. Importantly, even laboratories that followed the same gating strategy had differences in their results that were found to be associated with gate placement. The overall gating recommendations based on this analysis can be found in Table 2.
Table 2.
Summary of current gating recommendations for assay harmonization
| RECOMMENDATION | |
|---|---|
| Cytokine gate | Cytokine gate should not include negative cells and should include all of the cytokine-positive cells (both high- and low-positive cells). |
| Double-positive cells | Gate on CD4 and CD8 to allow clear identification of the double-positive cells. Include as a separate population or with either CD4 or CD8 cells but not with both. |
| Dim cells | Draw gates around the main populations of CD3, CD4, and CD8 that include dim events. |
| Lymphocyte gate | Lymphocyte gate should be large enough to include all lymphocytes. It should not include RBC or debris. |
| Uniform gates | Uniform gates should be used for all samples within a donor. |
| Biexponential scaling | Proper adjustment of biexponential scaling is important to clearly and completely visualize all populations. |
Cytokine Gate
During the central review of all gating strategies, three main approaches to gate on the cytokine-positive cells were observed: (1) placing the gate so that no negative cells were included and all of the cytokine-positive cells, both low and high positives were included (adequate proximity; Fig. 3A), (2) placing the cytokine gate too close to the cytokine-negative population and often including some of the cytokine-negative cells in the gate (Fig. 3B), and (3) placing the gate too far from the cytokine-negative cell population (Fig. 3C). Table 3 describes the association of the central review analysis for the cytokine gates and response detection (5/6 responses and NO false positives, 5/6 responses and false positives, and <5 responses). About 89–92% of laboratories that detected five or six responses and no false positives had a gate that included all cytokine-positive cells (high and low positives), whereas 64% of laboratories with five or six positive responses and false positives had a cytokine gate that did not include cytokine low cells. Laboratories that detected fewer than five responses had the highest percentage (57%) of gating too close to the negative population. Laboratories that detected five or six responses but had false positives had the largest percentage (64–66%) of placing their cytokine gates too far from the negative population. Laboratories that set the CD4 cytokine gate too far from the negative population had a median background of 0.01. Laboratories that had a CD4 cytokine gate of adequate proximity had a median background of 0.03, whereas laboratories that set their gates too close to the negative population had a median background of 0.08, a much higher background.
Table 3.
Analysis of Phase I cytokine gates
| GATE | TYPE OF GATE | N | 5/6 OR 6/6 RESPONSES DETECTED AND NO FALSE-POSITIVE RESPONSES (N =37) | 5/6 OR 6/6 RESPONSES DETECTED AND FALSE- POSITIVE RESPONSES (N =44) | <5/6 POSITIVE RESPONSES (N =28) |
|---|---|---|---|---|---|
| CD4+ Cytokine+ Gate: Positive cells |
Gate includes all cytokine-positive cells | 71 | 34 (92%) | 13 (30%) | 24 (86%) |
| Gate does not include cytokine low cells | 33 | 3 (8%) | 28 (64%) | 2 (7%) | |
| Gate does not include cytokine high cells | 2 | 0 (0%) | 1 (2%) | 1 (4%) | |
| Other | 3 | 0 (0%) | 2 (5%) | 1 (4%) | |
|
| |||||
| CD4+ Cytokine+ Gate: Gate proximity |
Gate does not include negative cells and includes all cytokine-positive cells (high and low positives) | 42 | 20 (54%) | 13 (30%) | 9 (32%) |
| Gate too far from the cytokine-negative population (>1 log from negative population) | 33 | 3 (8%) | 28 (64%) | 2 (7%) | |
| Gate too close to the cytokine-negative population (includes cytokine-negative cells) | 31 | 14 (38%) | 1 (2%) | 16 (57%) | |
| Other | 3 | 0 (0%) | 2 (5%) | 1 (4%) | |
|
| |||||
| CD8+ Cytokine+ Gate: Positive cells |
Gate includes all cytokine-positive cells | 71 | 33 (89%) | 15 (34%) | 23 (82%) |
| Gate does not include cytokine low cells | 35 | 4 (11%) | 28 (64%) | 3 (11%) | |
| Gate does not include cytokine high cells | 1 | 0 (0%) | 0 (0%) | 1 (4%) | |
| Other | 2 | 0 (0%) | 1 (2%) | 1 (4%) | |
|
| |||||
| CD8+ Cytokine+ Gate: Gate proximity |
Gate does not include negative cells and includes all cytokine-positive cells (high and low positives) | 41 | 19 (51%) | 13 (30%) | 9 (32%) |
| Gate too far from the cytokine-negative population (>1 log from negative population) | 35 | 4 (11%) | 29 (66%) | 2 (7%) | |
| Gate too close to the cytokine-negative population (includes cytokine-negative cells) | 31 | 14 (38%) | 1 (2%) | 16 (57%) | |
| Other | 2 | 0 (0%) | 1 (2%) | 1 (4%) | |
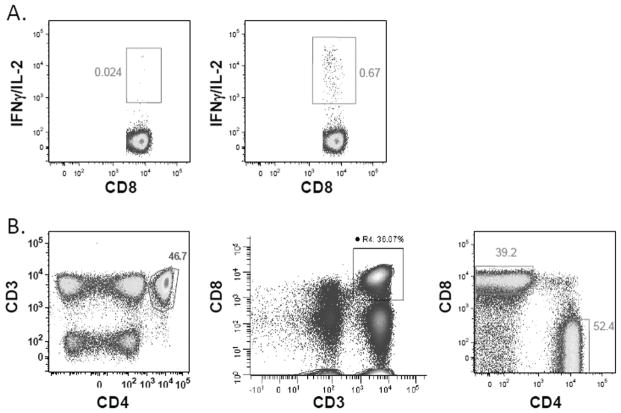
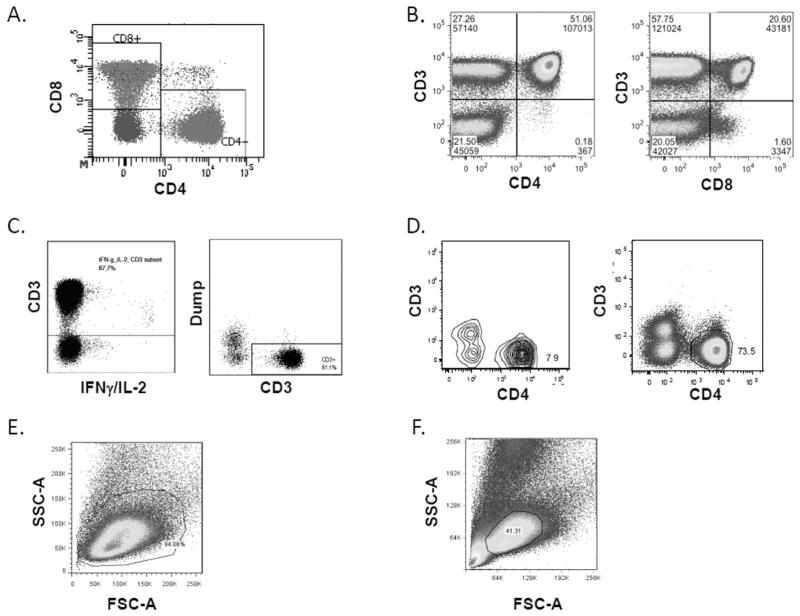
CD4+ CD8+ Double-Positive T Cells
Most laboratories (81%) indicated in the survey that they excluded CD4+ CD8+ DP T cells or gated them separately, and 10% indicated that they included DP cells in both the CD4 and CD8 gates. In the central review of the Phase I PowerPoint files, 63% of laboratories excluded DP cells and 34% included DP cells in both the CD4 and CD8 gates. Therefore, even though more than 80% of the laboratories indicated that they excluded DP cells in the survey, many of the laboratories do not apply that in practice to their gating strategy. As shown in Figure 4A, when CD4 and CD8 are gated in the same dot plot, it is easy to identify the CD4+ CD8+ DP cells. However, when CD4 and CD8 are gated versus CD3, the DP population is not differentiated, and often the DP cells are included with both CD4 and CD8 gates (Fig. 4B). It is important to note that CD4+ CD8 dim T cells are a separate T-cell population from CD4+ CD8+ DP T cells and have been shown to be MHC Class II dependent, respond to antigens such as CMV, and should be included in the CD4+ T cell gate (18).
Figure 4.
DP T cell gate, dim gates, and lymphocyte gate. Actual graphs from different participating laboratories, all of the examples are from the same FCS file, Donor 2, CEF stimulated. (A) CD4 and CD8 are gated in the same dot plot, making it very easy to identify the DP cells. (B) CD4 and CD8 are gated versus CD3. The DP cell population is not differentiated and thus is included with both CD4 and CD8 gates. (C) CD3 dim or low-positive cells are included in the CD3 gate. (D) CD3 gate is drawn too tightly and the dim cells are not included in the gate. (E) Lymphocyte gate is large enough to include all lymphocytes. (F) Lymphocyte gate is too narrow and excludes many lymphocytes. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Inclusion of Dim Cells
Dim cells are cells that have lower expression of CD3, CD4, or CD8. As stimulated T cells can downmodulate their surface receptors, it is often thought that these dim cells are activated T cells that could secrete cytokines (18,19). Most laboratories (74%) indicated in the survey that they included CD3 dim cells, and 76% indicated that they included CD4/CD8 dim cells. Among those that stated they included dim cells, the main reasons given were due to downregulation of coreceptor (55%) and that some dim cells indicate a positive response (23%). Among those that indicated that they did not include dim cells, the main reasons given were that dim cells can be myeloid cells (41%), dim cells are DP cells (15%), and that there were no dim cells present (11%). As a CD14 and CD19 dump gate was included, the dim cells in these files are not myeloid cells; otherwise they would have been excluded with the dump gate.
In the central review of the Phase I dot plots, 44% of laboratories included CD3 dim cells, 47% included CD4 dim cells, and 51% included CD8 dim cells. Therefore, even though 75% of the laboratories indicated that they include the dim cells in the survey, less than half of the laboratories apply that in practice. Examples of including dim cells in the CD3 gate are shown in Figure 4C, and Figure 4D shows an example of drawing the CD3 gate too tight and thereby excluding CD3 dim cells. Similar graphs were seen with CD4 and CD8 dim cells (data not shown).
Lymphocyte Gate
In the review of the Phase I PowerPoint files, 45% of laboratories had a lymphocyte gate that included all lymphocytes and had no debris or red blood cells, 22% had a gate that excluded lymphocytes, 17% had a gate that was too large as it included nonlymphocyte cells, and 13% had a gate that included red blood cells or debris. Figure 4E shows an example of a lymphocyte gate that is large enough to include all lymphocytes, and Figure 4F shows a gate that is too tight and excludes many lymphocytes.
Uniform Gates
Uniform gating is the practice of applying the same set of gates to all applicable samples (Fig. 5A). Most laboratories (73%) indicated in the survey that they used the same gates for all samples within a donor for consistency and comparability of results. In the review of the Phase I PowerPoint files, 88% used uniform gates within a donor and 73% used uniform gates across donors. In the group of laboratories that detected five or six positive responses and no false-positive responses, only 3% did not use uniform gates within a donor, whereas in laboratories that had false-positive responses, 16% did not use uniform gates within a donor.
Biexponential Scaling
Proper adjustment of biexponential scaling is important because difficulty visualizing all populations due to the cells being pushed against the axis may result in detecting fewer responses. In addition, applying too much biexponential transformation can result in “valley artifacts” where the negative population has merged into the positive population, making it difficult to discern the demarcation between positive and negative cells (20). “Trimodal” populations also make it difficult to assess where to place the gate (Fig. 5B). About 32% of laboratories that detected less than five responses had populations pushed against the axis, whereas only 14% of the laboratories that detected all of the responses had populations pushed against the axis.
Discussion
Overall Variability
The variability in Phase I results confirms the findings from previous proficiency panels that gating approach is a significant source of the variability in reported results. Unfortunately, there is no gold standard for ICS experiments and in general for flow cytometry assays. As this assay is measuring a biological response in live cells, there is inherently more variability than assays measuring a soluble analyte. Therefore, it is important that the analysis of these complex immune assays is precise, reproducible, and as accurate as possible. The results from this proficiency panel show that the variation in detecting a positive response is decreased if all laboratories follow the same consensus gating protocol and that the background responses are decreased and much less variable. Therefore, these results demonstrate the urgent need for harmonization of gating protocols to ensure comparability of results across laboratories. Gating recommendations based on the results from this gating panel are summarized in Table 2, and these recommendations are expanded on in the following sections. These recommendations are based on consensus from the 110 participating laboratories.
Cytokine Gate
The placement of the cytokine gate relative to the negative population is the step in the gating protocol that most clearly impacted false-positive and true-positive response detection in this panel. It is important to note that a positive response was defined in this panel as at least two times above background. These findings indicate that laboratories that set the cytokine gate too far (over a log distant) from the negative population and did not include cytokine low cells were more likely to get false-positive responses, 64% versus 11% (Table 3). This is probably due to the low background seen when the gates are set far from the negative population, leading to negative responses being incorrectly determined to be a positive response. Laboratories that set the cytokine gate too close to the negative population (actually including some negative cells in the gate) were more likely to detect fewer responses (Table 3). This is probably due to the large background seen when the gates are set too close which would only allow a large response to be determined as a positive response. Laboratories might assume that the larger the cytokine gate, the less the likelihood of missing a cytokine-positive cell and thus the higher the chances of detecting a positive response. However, the opposite is true as placing the gate so close to the negative/positive boundary artificially increases the background and results in underestimation of the cytokine response.
To decrease the likelihood of detecting false positives and to increase the magnitude of the response, cytokine gates should not include negative cells and should include all of the cytokine-positive cells (both high- and low-positive cells). A dot plot of CD4 versus cytokine or CD8 versus cytokine should be used to draw a gate starting a short distance from the negative population, as shown in Figure 3A. The gate should be positioned to not include any of the negative cells. The negative cells usually consist of the main population and a “halo” of individual cells just next to the main cells. Draw the cytokine gate next to the boundary of the halo cells. The gate should be sufficiently large to include all cytokine high cells. Backgating strategies, for example, gating on cytokine low cells and establishing where they fall in earlier gates, can be applied to help determine the optimal location to place the cytokine gate and to ensure that all cytokine-positive cells are included in the gate.
CD4+ CD8+ Double-Positive T Cells
The discrepancy in the number of laboratories that reported that they excluded DP cells and laboratories that actually excluded DP cells in practice could be due to the fact that many laboratories plotted CD4 and CD8 on separate plots and therefore inadvertently included the DP cells in both the CD4 and CD8 plots.
Therefore, it is advisable to gate on CD4 versus CD8 expression to allow clear identification of the subset of CD4+ CD8+ DP T cells. If it is of interest to include DP cells, it may be best to include them as a separate population or with only one of the T-cell populations (CD4 or CD8 cells) but not with both CD4 and CD8 cells.
Inclusion of Dim Cells
As stimulated T cells can downmodulate their surface receptors, it is suggested that the CD3, CD4, and CD8 gates be drawn large enough to include the main population and all dim events (21,22). Not collecting dim cells may result in detecting fewer positive responses. Backgating on the cytokine-positive population should be performed to determine if the CD3, CD4, and CD8 dim cells contain cytokine-positive cells.
Lymphocyte Gate
The lymphocyte gate should be large enough to include all lymphocytes. However, the gate should not include red blood cells or debris, which can cause autofluorescence or nonspecific binding of antibodies that could lead to false-positive responses and/or increased background.
Uniform Gates
Applying uniform gating within a donor can affect the detection of positive responses. If a laboratory was to change its cytokine-positive gate between the unstimulated sample and antigen-stimulated sample, it could decrease the background while increasing the likelihood of detecting a false positive response (Fig. 5A). This also defies the logic of including a negative control to determine baseline or background responses. Additionally, changing gates between donors might make it difficult to compare results between different groups of donors within a specific experiment or in longitudinal studies. However, no consensus on a general strategy for applying uniform gates between donors exists, and a decision has to be made based on the study question and setup.
After the final report and conclusions were released to all participants, a webinar was hosted, and 46 of the participating laboratories joined to discuss the gating panel results. The use of uniform gates generated an extended discussion. Many laboratories change the lymphocyte, dump, and/or T-cell gates both between and within donors as each donor is unique, and the cell populations might look slightly different between donors. However, alteration of the penultimate gate, the cytokine-positive cells, led to further debate. Although all laboratories agreed that using uniform gates within a donor was necessary, many laboratories did not agree that uniform gates should be applied across donors. It was argued that as there can be donor-to-donor variability in the appearance of cell populations, it is important to adjust the cytokine gate between donors to include the appropriate cells. Laboratories that prefer to not change the cytokine gate across donors indicated that they do so to more easily compare results within an experiment.
Biexponential Scaling
Difficulty in fully visualizing all populations due to the populations being pushed against the axis, as shown in Figure 5B, may result in detecting fewer responses. However, how to properly adjust scaling to clearly visualize all populations needs to be clearly defined. There are several tutorials and papers that help to understand biexponential transformation of flow cytometry data (23,24); FlowJo Transformation Overview).
Conclusions and Next Steps
This gating proficiency panel is a landmark panel as many different laboratories and immune monitoring networks came together from diverse backgrounds to address a common challenge. The results shown here demonstrate a marked benefit to harmonizing gating strategies to decrease variability and to increase accuracy and comparability of results across laboratories in ICS assays. Although the conclusions and recommendations in this article are based on our observations during this gating panel, these results should be used by others, with expertise in the area of computational and mathematical approaches, to help us identify specific software and algorithms that may be used to more precisely identify and quantify rare antigen-specific cells of interest. Our current analysis tools are not adequate for the types of assays we are running or too complex for the average user.
New software is being developed that uses complex algorithms for automated gating (10,11,25,26). One such automated gating program was among the best performers in this panel. However, more work is needed to make these programs easy to use, amenable to differences in experimental parameters (such as antibody–fluorochromes) between laboratories, less computationally intense, and readily available to the entire flow cytometry community. FlowCAP and other groups are actively working on standardizing these efforts (27).
To further study the harmonization of ICS gating, a second gating panel is being rolled out in 2013 to confirm the recommendations from the first panel and to increase reproducibility of results across laboratories. Another aspect of analysis of ICS experiments that affects reproducibility across laboratories is the question of how to define a positive response. Criteria for determining a positive response can range from simply subtracting background to twofold, threefold, or even fourfold above background to more complex statistical analyses. Several groups are now addressing the need for standardized methods to determine cytokine positivity. These efforts in combination with other ongoing projects in the immune monitoring field will critically impact the role and positioning of immune monitoring in the field of basic research and translational medicine.
Supplementary Material
Acknowledgments
Grant sponsors: Cancer Research Institute
The authors thank the participants of the ICS Gating Proficiency Panel.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Cebon J, Knights A, Ebert L, Jackson H, Chen W. Evaluation of cellular immune responses in cancer vaccine recipients: Lessons from NY-ESO-1. Expert Rev Vaccines. 2010;9:617–629. doi: 10.1586/erv.10.58. [DOI] [PubMed] [Google Scholar]
- 2.Nagorsen D, Scheibenbogen C, Thiel E, Keilholz U. Immunological monitoring of cancer vaccine therapy. Expert Opin Biol Ther. 2004;4:1677–1684. doi: 10.1517/14712598.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 3.van der Burg SH, Kalos M, Gouttefangeas C, Janetzki S, Ottensmeier C, Welters MJ, Romero P, Britten CM, Hoos A. Harmonization of immune biomarker assays for clinical studies. Sci Transl Med. 2011;3:108ps44. doi: 10.1126/scitranslmed.3002785. [DOI] [PubMed] [Google Scholar]
- 4.Maecker HT, Rinfret A, D’Souza P, Darden J, Roig E, Landry C, Hayes P, Birungi J, Anzala O, Garcia M, et al. Standardization of cytokine flow cytometry assays. BMC Immunol. 2005;6:13. doi: 10.1186/1471-2172-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nomura L, Maino VC, Maecker HT. Standardization and optimization of multiparameter intracellular cytokine staining. Cytometry Part A. 2008;73A:984–991. doi: 10.1002/cyto.a.20602. [DOI] [PubMed] [Google Scholar]
- 6.Britten CM, Gouttefangeas C, Welters MJ, Pawelec G, Koch S, Ottensmeier C, Mander A, Walter S, Paschen A, Muller-Berghaus J, et al. The CIMT-monitoring panel: A two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol Immunother. 2008;57:289–302. doi: 10.1007/s00262-007-0378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaimes MC, Maecker HT, Yan M, Maino VC, Hanley MB, Greer A, Darden JM, D’Souza MP. Quality assurance of intracellular cytokine staining assays: Analysis of multiple rounds of proficiency testing. J Immunol Methods. 2011;363:143–157. doi: 10.1016/j.jim.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welters MJ, Gouttefangeas C, Ramwadhdoebe TH, Letsch A, Ottensmeier CH, Britten CM, van der Burg SH. Harmonization of the intracellular cytokine staining assay. Cancer Immunol Immunother. 2012;61:967–978. doi: 10.1007/s00262-012-1282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aghaeepour N, Jalali A, O’Neill K, Chattopadhyay PK, Roederer M, Hoos HH, Brinkman RR. RchyOptimyx: Cellular hierarchy optimization for flow cytometry. Cytometry Part A. 2012;81A:1022–1030. doi: 10.1002/cyto.a.22209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan C, Feng F, Ottinger J, Foster D, West M, Kepler TB. Statistical mixture modeling for cell subtype identification in flow cytometry. Cytometry Part A. 2008;73A:693–701. doi: 10.1002/cyto.a.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson J, Kidd P, Mandy F, Livnat D, Kagan J. Three-color supplement to the NIAID DAIDS guideline for flow cytometric immunophenotyping. Cytometry. 1996;26:227–230. doi: 10.1002/(SICI)1097-0320(19960915)26:3<227::AID-CYTO8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 13.Schenker EL, Hultin LE, Bauer KD, Ferbas J, Margolick JB, Giorgi JV. Evaluation of a dual-color flow cytometry immunophenotyping panel in a multicenter quality assurance program. Cytometry. 1993;14:307–317. doi: 10.1002/cyto.990140311. [DOI] [PubMed] [Google Scholar]
- 14.Schnizlein-Bick CT, Mandy FF, O’Gorman MR, Paxton H, Nicholson JK, Hultin LE, Gelman RS, Wilkening CL, Livnat D. Use of CD45 gating in three and four-color flow cytometric immunophenotyping: Guideline from the National Institute of Allergy and Infectious Diseases, Division of AIDS. Cytometry. 2002;50:46–52. doi: 10.1002/cyto.10073. [DOI] [PubMed] [Google Scholar]
- 15.Britten CM, Janetzki S, Butterfield LH, Ferrari G, Gouttefangeas C, Huber C, Kalos M, Levitsky HI, Maecker HT, Melief CJ, et al. T cell assays and MIATA: The essential minimum for maximum impact. Immunity. 2012;37:1–2. doi: 10.1016/j.immuni.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Lee JA, Spidlen J, Boyce K, Cai J, Crosbie N, Dalphin M, Furlong J, Gasparetto M, Goldberg M, Goralczyk EM, et al. MIFlowCyt: The minimum information about a flow cytometry experiment. Cytometry Part A. 2008;73A:926–930. doi: 10.1002/cyto.a.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janetzki S, Price L, Britten CM, van der Burg SH, Caterini J, Currier JR, Ferrari G, Gouttefangeas C, Hayes P, Kaempgen E, et al. Performance of serum-supplemented and serum-free media in IFNγ ELISPOT assays for human T cells. Cancer Immunol Immunother. 2010;59:609–618. doi: 10.1007/s00262-009-0788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suni MA, Ghanekar SA, Houck DW, Maecker HT, Wormsley SB, Picker LJ, Moss RB, Maino VC. CD4(+)CD8(dim) T lymphocytes exhibit enhanced cytokine expression, proliferation and cytotoxic activity in response to HCMV and HIV-1 antigens. Eur J Immunol. 2001;31:2512–2520. doi: 10.1002/1521-4141(200108)31:8<2512::aid-immu2512>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 19.Itoh Y, Germain RN. Single cell analysis reveals regulated hierarchical T cell antigen receptor signaling thresholds and intraclonal heterogeneity for individual cytokine responses of CD4+ T cells. J Exp Med. 1997;186:757–766. doi: 10.1084/jem.186.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novo D, Wood J. Flow cytometry histograms: Transformations, resolution, and display. Cytometry Part A. 2008;73A:685–692. doi: 10.1002/cyto.a.20592. [DOI] [PubMed] [Google Scholar]
- 21.Bitmansour AD, Douek DC, Maino VC, Picker LJ. Direct ex vivo analysis of human CD4(+) memory T cell activation requirements at the single clonotype level. J Immunol. 2002;169:1207–1218. doi: 10.4049/jimmunol.169.3.1207. [DOI] [PubMed] [Google Scholar]
- 22.Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, Persaud N, Trigona W, Fu TM, Sinclair E, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 23.Herzenberg LA, Tung J, Moore WA, Parks DR. Interpreting flow cytometry data: A guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 24.Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry Part A. 2006;69A:541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 25.Chan C, Lin L, Frelinger J, Herbert V, Gagnon D, Landry C, Sekaly RP, Enzor J, Staats J, Weinhold KJ, et al. Optimization of a highly standardized carboxyfluorescein succinimidyl ester flow cytometry panel and gating strategy design using discriminative information measure evaluation. Cytometry Part A. 2010;77A:1126–1136. doi: 10.1002/cyto.a.20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zare H, Bashashati A, Kridel R, Aghaeepour N, Haffari G, Connors JM, Gascoyne RD, Gupta A, Brinkman RR, Weng AP. Automated analysis of multidimensional flow cytometry data improves diagnostic accuracy between mantle cell lymphoma and small lymphocytic lymphoma. Am J Clin Pathol. 2012;137:75–85. doi: 10.1309/AJCPMMLQ67YOMGEW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.FlowCAP Flow Cytometry. Critical assessment of population identification methods. [Accessed date: 06-Jun-2013]; Available from: http://flowcap.flowsite.org//
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.