Abstract
Purpose of review
Many pediatric lung diseases are characterized by infection. These infections are generally diagnosed, studied, and treated using standard culture methods to identify “traditional pathogens”. Based on these techniques, healthy lungs have generally been thought to be sterile. However, recent advances in culture-independent microbiological techniques challenge this paradigm by identifying diverse microbes in respiratory specimens (respiratory microbiomes) from both healthy people and those with diverse lung diseases. In addition, growing evidence suggests a link between gastrointestinal microbiomes and inflammatory diseases of various mucosal surfaces, including airways.
Recent findings
This article reviews the rapidly developing field of respiratory microbiome research, emphasizing recent progress made employing increasingly sophisticated technologies. While many of the relevant studies have focused on adults with cystic fibrosis (CF), recent research has included children and adults with other respiratory diseases, as well as healthy subjects. These studies suggest that even healthy children have airway microbiomes, and that both respiratory and gastrointestinal microbiomes often differ between healthy people and those with different types and severities of airway disease. The causal relationships between microbiomes, disease type and progression, and treatments such as antibiotics must now be defined.
Summary
The advent of culture-independent microbiological techniques has transformed how we think about the relationship between microbes and airway disease. More research is required to translate these findings to improved therapies and preventive strategies.
Keywords: Microbiome, airway, respiratory, gastrointestinal, lung disease
1) Introduction
Much of the earliest work in microbiology, largely performed in the 17th–18th centuries, included microscopic observations of the diverse microbes in human tissues and excretions [1]. Medical microbiology, a field that has largely existed for the past century and a half, built on those early studies by focusing on the identification, cultivation, and analysis of specific microbial “pathogens”. This “pathogen-oriented” approach, while enormously helpful and successful for many diseases, often ignored the presence and activities of the numerous other microbes that we now know to inhabit us. Many statements published throughout the 20th-century literature attest to the general sterility of healthy human airways, tacitly asserting that mechanisms must exist to filter or clear the copious microbes inhaled during normal respiration ([2] and references therein). Meanwhile, in the gastrointestinal (GI) tract, comparisons of the bacteria identified in human fecal samples using microscopic versus cultivation-based methods revealed striking disparities in the types and abundances of microbes present [3,4]. Thus, in these two organs—the lung and the GI tract—there were discrepancies between what was observed using culture (the absence of microbes in airways, the balance of bacteria in stools) and what was expected based on other considerations (constant inhalation of microbes, the microscopic analyses of stools).
In the mid-20th century, remarkably similar observations were made by environmental microbiologists comparing microscopic findings with routine culture of aquatic specimens. These studies identified major discrepancies in microbial content now collectively referred to as “the great plate count anomaly” [5]. Such discordances underscored the inadequacies of routine culture techniques (which, it is now generally accepted, easily identify only ~1% of known bacteria) [6] and led to the adaptation, refinement, and application of “culture-independent” microbiological techniques that do not rely on microbial growth, but rather detect their molecular signatures (most often their genetic material). It was only a matter of time before medical microbiologists would take notice of these advances. These newer techniques, often referred to as microbiome methods, have provided fascinating new findings that have forced us to revise our notions of how microbes inhabit our bodies in various states of health. Our models of how these microbiota promote health and cause disease are continuously updated as fresh data become available.
In this review, we will briefly describe these new techniques, and the current view they have afforded of both lung and GI microbiomes in pediatric respiratory health. While most of the research in this area has been performed in people with cystic fibrosis (CF) and asthma, the implications for other diseases, and for healthy mucosal surfaces, are also discussed. While this article will occasionally (and necessarily) include technical details, our hope is to briefly and simply illustrate how recent technological advances have “pulled back the curtain” on our internal microbiological world, revealing a new view of pediatric respiratory health and disease pathogenesis. Before reviewing these recent advances, we will first define some of the most common, and often most confusing, terms and techniques used in the microbiome field.
2) Definitions
The term microbiome, originally coined by microbiologist Joshua Lederberg, signifies the “community of commensal, symbiotic, and pathogenic microorganisms that…share our body space” [7]. “Microbiome” can be more succinctly defined as “the totality of microbes, their genes, and their interactions in a given environment” [8], therefore referring not only to “who’s there”, but also “what they can do”. By contrast, the term “microbiota” refers only to the actual microorganisms that are found in a particular location (“niche”) in the human body [9] (or “who’s there”). Therefore, because much of what we consider to be “microbiome” research often focuses on identifying the microbes in a niche, “microbiota” is probably a more accurate term for many studies. For simplicity, however, and in keeping with common practice, we will use the term “microbiome” to refer to most research in this review. Furthermore, while a microbiome technically includes all microbe types—viruses, bacteria, fungi, and others—most studies thus far focused solely on bacteria. For this reason, much of the following discussion focuses on bacteria, acknowledging that more work must be done to characterize the fungal and viral contributions to the human microbiome.
Microbiomes are generally very complex with respect to the identities and abundances of specific microbes. For this reason, comparing microbiomes from different sources can be difficult, and statistical descriptors have been developed for simplified characterization and comparison of microbiomes. For example, “richness” refers to the number of different species present in a microbiome; “evenness” is the degree to which those species are of equal abundance; and “dominance” is the extent to which one or more species is numerically superior [9]. These concepts and measures allow for relatively straightforward comparison of microbiomes from different people or anatomic locations, and to describe changes within one microbiome under stress or perturbation, such as with disease or medical treatments [10]. These situations are often accompanied by dysbiosis, or an imbalance in the microbes present in a particular niche that can frequently be found with changes in health [9].
3) Methods
As noted above, new molecular diagnostic techniques have revealed that traditional cultures only identify a small percentage of the microbes present in the airway [11,12]. Several methods have been used to more comprehensively define microbial community membership, each of which has advantages, disadvantages, and potential biases or errors, which will be briefly described here but have been reviewed in detail elsewhere [13]. Real-time quantitative polymerase chain reaction (qPCR) remains the most common technique for amplifying and quantifying abundance of specific DNA or RNA sequences. To elucidate spatial characteristics and abundance of organisms in clinical specimens, fluorescent in situ hybridization (FISH) can be performed. FISH uses fluorescently labeled probes for specific bacterial sequences, followed by fluorescent microscopy to identify their targets and localize them within tissues.
Most microbiome research has relied on techniques that identify and characterize a gene carried by all bacteria, the 16S ribosomal RNA (rRNA) gene. This gene contains some sequences that are well-conserved among bacteria (allowing for identification of a cell as a bacterium) and others that are highly variable (allowing for identification of specific bacterial types, or taxa, often at the phylum, genus or species level); it is this quality of mixed sequence conservation that makes this gene so useful for bacterial taxonomic identification [14]. Once the DNA is purified from a sample of interest, PCR is usually used to amplify a large portion of the rRNA gene using primers targeting conserved regions (i.e., to generate a pool of 16S rRNA “amplicons”—the amplified rRNA genes--from all detectable bacteria), and then various methods are used to probe or sequence the variable regions of those amplicons to identify and quantify specific taxa. Over the last several decades this work has revealed many previously unculturable bacteria [15].
In the earliest days of microbiome research, gel electrophoretic methods were most often used to define the microbial diversity of samples. These methods generally provide a qualitative “footprint” of microbiomes, allowing for comparison of microbial communities between different samples, but providing relatively limited quantitative or taxonomic information relative to newer sequencing techniques. Denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP) analyses exploit the differences in rRNA amplicon sequences to provide these types of “footprints”.
Recently, the gradually wider availability and dropping costs of next-generation sequencing have dramatically changed the field, generally supplanting other techniques. High-throughput sequencing of 16S rRNA gene amplicons, followed by computational comparison of the resulting sequence “reads” to taxonomic databases, has yielded high-resolution snapshots of microbiomes [13]. With phylogenetic microarrays, such as the 16S rRNA PhyloChip, microbiomes are characterized by hybridizing their 16S amplicons to arrays of taxon-specific probes. These information provided is similar to that available from sequencing, with some differences in specificity and quantitative capacity but that is limited to the probes available [16]. Sophisticated and powerful computational algorithms are required to analyze the sequence data, thus far limiting these techniques largely to research rather than clinical practice.
On the cutting edge are metagenomic techniques, which sequence all of the DNA (not just the 16S rRNA gene) in a specimen [13]. Various computational methods can then be used to identify the microbial sequences in the resulting data, allowing for the identification of not only bacteria, but also fungi, viruses (again, “who’s there”), as well as the specific genes or gene classes that they carry (“what they can do”). These developing technologies show promise as the next step towards comprehensively defining the biology of the lung.
4) Lung microbiome
By comparison with traditional culture methods, culture-independent techniques have revealed surprisingly diverse bacterial microbiomes in the respiratory tracts of even healthy humans [17,18]. Thus far, most respiratory microbiome analyses have been performed on bronchoalveolar fluid, oropharyngeal swabs, or sputum. Because bronchoscopes and sputum must traverse the upper airway, and oropharyngeal swabs directly target an upper airway site, it is difficult to predict a priori how accurately these specimens reflect lower airway microbiology. Recently, studies that directly analyzed CF lung tissue have identified diverse microbiomes [19–21], and that upper and lower respiratory tracts have distinct but related microbiomes [14,22]. In adults with end-stage CF lung disease, oropharyngeal swabs were shown to reflect lung tissue microbiomes poorly, and sputum somewhat more accurately [19,23]. Similarly, lung tissue from a toddler with CF contained diverse and anatomically variable, populations of microorganisms, indicating a high degree of spatial heterogeneity in early CF lung disease, and little similarity between lung tissue and oropharyngeal microbiomes [24]. These findings provide a view of the respiratory tract as an anatomically heterogenous ecosystem stretching from the nares to the most distal airways, providing a diversity of niches for microbes. The microbiome compositions on these surfaces is likely determined by several factors, including rates of microbial immigration or delivery by inhalation; microbial elimination by cough, mucociliary clearance, or immunity; local physicochemical characteristics; the relative microbial growth rates; and interactions between different microbes [17]. As all of these characteristics may change over time, with different disease states and treatments, or among anatomic locations, so may the respiratory microbiota. These findings and considerations underscore the early state of lung microbiome research, and the need for more work to better understand the determinants and consequences of microbiome composition within healthy lungs. However, respiratory microbiome research owes its origins to studies of respiratory diseases, with CF accounting for the lion’s share of studies, followed by chronic obstructive lung disease (COPD), asthma, non-CF bronchiectasis, and other generally obstructive diseases.
5) CF
The role of microbes in CF lung disease has been a topic of intense research and therapeutic importance since the earliest descriptions of this disorder [25], and it is fitting that the earliest respiratory microbiome research focused on CF. The airway surfaces of people with CF have compromised mucociliary clearance and altered local antimicrobial activities, usually leading to chronic bacterial infection and inflammation [26–28].
Antibiotic treatment has improved quality of life and survival for people with CF, generally targeting a small group of bacterial species traditionally associated with CF pulmonary disease, including Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and Burkholderia cepacia complex [29]. One might therefore hypothesize that respiratory symptoms derive from an increased burden of these culturable pathogens [30]. However, neither the respiratory specimen densities of traditional CF pathogens, nor of all bacteria, have been found to reproducibly correlate with disease severity or symptomatic changes [31,32]. Many children with CF exacerbations requiring IV antibiotics do not have culturable pathogens, [32] and while treatment with antibiotics has been associated with clinical improvement [33], research has not identified a significant relationship between clinical response and the in vitro susceptibilities of cultured bacteria [34,35]. These observations further highlight the limited understanding of CF lung disease pathogenesis provided by the identification and therapeutic targeting of traditional pathogens, prompting a great deal of interest in microbiome-oriented approaches to CF lung disease.
Early studies used T-RFLP to examine sputum from adults with CF. These studies provided ample, reproducible evidence for diverse microbes, many not identified by clinical cultures. Since then, dozens of studies using progressively advanced methods have confirmed, clarified, and expanded those findings (reviewed in [36]), and ongoing research continues to investigate how microbes relate to CF lung disease progression, and the impact of antibiotics [37]. In short, there is ample evidence that CF airway microbiomes frequently include bacteria, such as obligate and facultative anaerobes, that are not readily cultured with standard techniques [11,38–40].
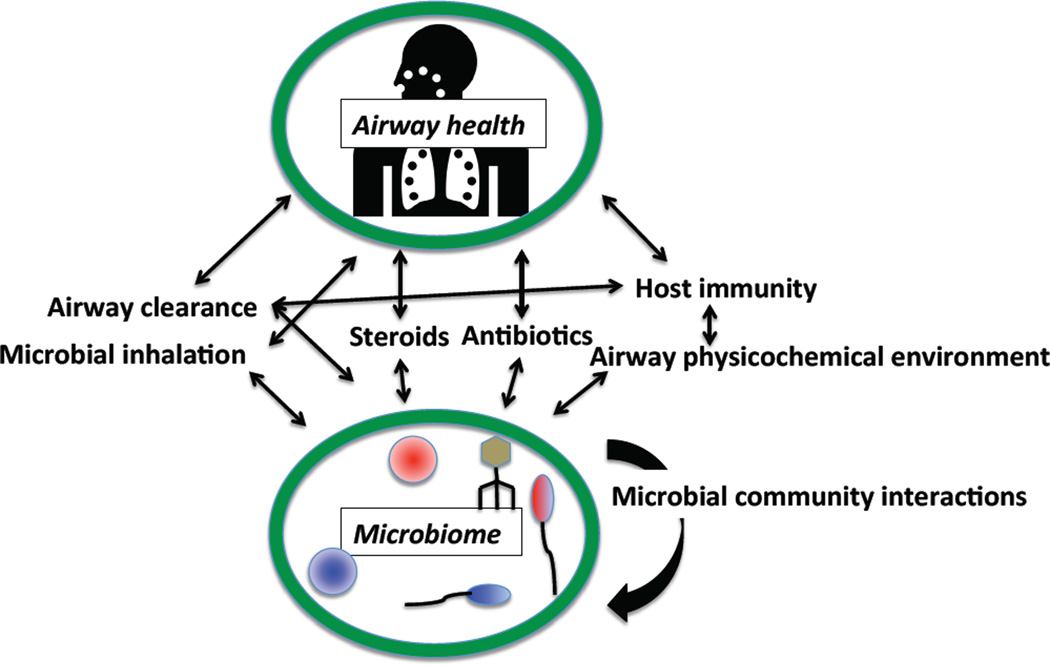
Relatively few studies of CF respiratory microbiomes have focused on children. Existing evidence suggests that pediatric airway microbiomes initially increase in diversity, peak in early adulthood, and then decline [16]. This decline in diversity has been associated with more severe lung disease [40], both of which are in turn associated with increased antibiotic treatment burden [40–43]. While studies have attempted to define a reliable microbiome “marker” heralding the onset of CF respiratory exacerbations, no universal or reproducible biomarker has been identified [39]. Therefore, the relationships between disease severity, respiratory microbiomes, and antibiotics must be clarified by longitudinal, modeling and/or interventional studies (Figure 1).
Figure 1.
A schematic illustrating how many features of pediatric lung disease interact, confounding researchers’ attempts to understand how microbiomes relate to disease symptoms and severity. For example, treatments such as steroids and antibiotics given to children with more severe disease can alter airway immunity, which may alter the observed microbiome. Alternatively, different microbiomes may lead to worse disease, leading to treatment with those medications. Unravelling these causal relationships is a notoriously difficult but critical step in identifying effective treatments targeting the microbiome.
6) Non-CF pediatric lung diseases
In addition to CF, other chronic respiratory diseases—including asthma, non-CF bronchiectasis, and primary ciliary dyskinesia—have also been characterized using microbiome methods. This collective research has revealed remarkable parallels between the observations from CF and other airway diseases. The limited pediatric research to date has emphasized the importance of studying the earliest stages of disease development to understand pathogenesis. One recent study [44] compared sputum and bronchoalveolar lavage microbiota among children with three different chronic airway diseases: CF, non-CF bronchiectasis, and protracted bacterial bronchitis. The “core” microbiota (the most common and abundant bacteria) for each group were remarkably similar among all children, but were different in adults with the same diagnoses. As with other studies [18,45–47], specimens from “controls” without airway disease also had identifiable microbiota, but the focus on children here afforded a new finding: Overlapping core respiratory microbiota in health and early disease. These findings suggest a model in which all children have diverse airway microbiota that can change over time with disease.
While the lung diseases discussed above have customarily been associated with infection, the role of microbes in asthma is less well understood. While asthma exacerbations are strongly associated with viral respiratory infections [17], until recently very few studies examined the relationship between asthma and other microbes in the respiratory tract. It was recently shown that airway specimens from patients with asthma have distinct bacterial microbiota from those of healthy people, suggesting an association between respiratory microorganisms, airway inflammation, disease control, and even susceptibility to exacerbations [17,48,49]. One study comparing the respiratory specimen microbiota of poorly controlled asthmatics to controls found increased bacterial burden and bacterial diversity among asthmatic subjects, showing a correlation between diverse bacterial species and severity of bronchial hyper-responsiveness [49]. Another study identified a relationship between patients’ baseline microbiota compositions and their clinical responses to systemic corticosteroids [50]. However, as with CF, much more work will be needed to define the causal relationships between asthma disease severity, treatments (such as steroids), and airway microbiota. Nevertheless, the similarities that are emerging from respiratory microbiome research in different diseases are instructive. For example, airway microbiota do generally change yet remain remarkably complex during acute exacerbations of all of the above, diverse chronic inflammatory respiratory conditions. These observations contrast with what would be predicted according to pathogen-oriented models, wherein one bacterium invades or overtakes the local microbiota; instead, exacerbations may be caused by dysbiosis, or disruptions in microbiome structures or behaviors.
7) GI microbiota and lung diseases
Increasing evidence implicates the GI microbiome in key developmental, metabolic and immunologic activities that, in turn, impact the development of respiratory disease. The gut microbiome increases dramatically in diversity in the first three years of life, stabilizing thereafter [51]. Culture-independent methods have demonstrated that the mode of delivery at birth, diet, and antibiotic use affect the pattern of infant gut colonization [52–55].
An growing body of literature suggests that the GI microbiome impacts the developing immune system and later health outcomes. For example, the GI microbiome appears to play a key role in the development and severity of a range of allergic diseases, including asthma, suggesting a mechanistic connection between GI microbiomes and health of the airway and other mucosal surfaces [56,57]. Clinical studies [56] have shown that early infant exposures known to alter infant gut microbiomes were associated with increased risks of asthma and eczema. However, other exposures, including to dog–associated household dust, were associated with distinct GI microbiota, and decreased risk of atopic disease [58]. These results provide an emerging view of a complex relationship between early exposures, GI microbiomes, and atopic diseases such as asthma.
Another example of a relationship between the GI system and respiratory disease comes from studies of children with CF. Early nutritional status in cystic fibrosis has been correlated with later severity of lung disease and overall survival [59]. Recent work has identified a marked fecal dysbiosis in the pediatric CF gut microbiota characterized by greater abundances of Escherichia coli noted when compared to controls, irrespective of antibiotic exposure, and correlating with markers for GI dysfunction [60]. A longitudinal study found that bacteria in the GI microbiomes of infants with CF tend to appear later in their oropharyngeal microbiomes, suggesting the GI tract may serve as a reservoir for respiratory infection [55]. Further linking GI and respiratory health, clinical trials of probiotics in CF patients that have demonstrated improvements in gastrointestinal inflammation, function, and decreased rates of pulmonary exacerbations [61–63]. Therefore, the GI microbiome offers an enticing target for modifying respiratory microbiology and health.
8) Conclusions and the future of the field
It is now clear that, even in healthy children, diverse communities of microbes inhabit tissue surfaces exposed to the environment, including the airways and GI tract. With regards to the sites relevant to lung disease, the bulk of human microbiome research in the past decade has largely focused on tabulating “who’s there”—the types of microbes in these sites and how they vary between similar and different people, within a single person over time, and with perturbations such as diseases and medical treatments. The challenge we now face is to define how these microbiomes relate to health and disease: Specifically, what roles do our microbes play in the natural development and health of our mucosal surfaces? To what extent do the medications we give in response to disease (e.g., antibiotics, steroids) account for the changes we observe? As in the past century, answering these questions will involve surprising findings, revisions of prevailing models, and the aid of ever-advancing technologies.
The next decade will likely see a great deal of transformative research, and hopefully new insights, in these critical areas of causality (Figure 1). The impact of treatments that promote “healthy” microbiota, such as probiotics, prebiotics, and even inter-person microbial transfers, on disease pathogenesis are a topic of intense interest. We may thus identify novel, rational ways to shape the microbiota of our young patients in ways that will promote or restore their respiratory health. One fascinating side benefit likely to result from this quest for better therapies is a more complete understanding of the microbes that rapidly colonize our bodies, and how to distinguish between beneficial and detrimental microbe-host relationships.
Bullet points.
Thanks to advances in sequencing technologies, it is now known that the healthy and diseased lung contain diverse microbes that are often not identified by traditional culture methods.
The relationship between lung microbes and disease is a topic of intense research.
These same technologies have also provided evidence that the microbiota in the gastrointestinal tract can impact the development and severity of lung diseases such as asthma.
Continued research on the microbiomes of the lung and GI tract in health and disease will clarify the microbial contributions to the pathogenesis of diverse diseases, and may lead to improved treatments.
Acknowledgements
The authors are grateful to D. Cornfield for helpful advice.
Financial support and sponsorship:
Support: This work was supported by a grant from the NIH: K02HL105543
Footnotes
Conflicts of interest:
None.
References
- 1.Institute of Medicine (U.S.) The human microbiome, diet, and health: workshop summary. National Academies Press; 2013. [PubMed] [Google Scholar]
- 2.Laurenzi GA, Berman L, First M, Kass EH. A QUANTITATIVE STUDY OF THE DEPOSITION AND CLEARANCE OF BACTERIA IN THE MURINE LUNG. J. Clin. Invest. 1964;43:759–768. doi: 10.1172/JCI104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J-C, Million M, Hugon P, Armougom F, Raoult D. Human Gut Microbiota: Repertoire and Variations [Internet] Front. Cell. Infect. Microbiol. 2012;2 doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore WE, Holdeman LV. Special problems associated with the isolation and identification of intestinal bacteria in fecal flora studies. Am. J. Clin. Nutr. 1974;27:1450–1455. doi: 10.1093/ajcn/27.12.1450. [DOI] [PubMed] [Google Scholar]
- 5.Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 6.Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of “unculturable” bacteria. FEMS Microbiol. Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 7.NIH HMP Working Group. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am. J. Respir. Crit. Care Med. 2013;187:1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rogers GB, Shaw D, Marsh RL, Carroll MP, Serisier DJ, Bruce KD. Respiratory microbiota: addressing clinical questions, informing clinical practice. Thorax. 2015;70:74–81. doi: 10.1136/thoraxjnl-2014-205826. ** This review on respiratory microbiota provides a particularly informative glossary of key microbiome definitions, concepts, and newer, sophisticated microbial identification techniques.
- 10.Beck JM. ABCs of the lung microbiome. Ann. Am. Thorac. Soc. 2014;11(Suppl 1):S3–S6. doi: 10.1513/AnnalsATS.201306-188MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tunney MM, Klem ER, Fodor AA, Gilpin DF, Moriarty TF, McGrath SJ, Muhlebach MS, Boucher RC, Cardwell C, Doering G, et al. Use of culture and molecular analysis to determine the effect of antibiotic treatment on microbial community diversity and abundance during exacerbation in patients with cystic fibrosis. Thorax. 2011;66:579–584. doi: 10.1136/thx.2010.137281. [DOI] [PubMed] [Google Scholar]
- 12.Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, Ranganathan SC, Waters VJ, Ratjen FA. Antibiotic Management of Lung Infections in Cystic Fibrosis: Part I. The Microbiome, MRSA, Gram-Negative Bacteria, and Multiple Infections. Ann. Am. Thorac. Soc. 2014 doi: 10.1513/AnnalsATS.201402-050AS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox MJ, Cookson WOCM, Moffatt MF. Sequencing the human microbiome in health and disease. Hum. Mol. Genet. 2013;22:R88–R94. doi: 10.1093/hmg/ddt398. * This review highlights the applications of 3 key sequencing techniques for microbiome research.
- 14.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev. Respir. Med. 2013;7:245–57. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, Curtis JL, Erb-Downward J, Lynch SV, Sethi S, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67:456–463. doi: 10.1136/thoraxjnl-2011-201183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly Ra, Karaoz U, Andersen GL, Brown R, Fujimura KE, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PloS One. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384:691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blainey PC, Milla CE, Cornfield DN, Quake SR. Quantitative analysis of the human airway microbial ecology reveals a pervasive signature for cystic fibrosis. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004458. 153ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012;6:471–474. doi: 10.1038/ismej.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudkjøbing VB, Thomsen TR, Alhede M, Kragh KN, Nielsen PH, Johansen UR, Givskov M, Høiby N, Bjarnsholt T. The microorganisms in chronically infected end-stage and non-end-stage cystic fibrosis patients. FEMS Immunol. Med. Microbiol. 2012;65:236–244. doi: 10.1111/j.1574-695X.2011.00925.x. [DOI] [PubMed] [Google Scholar]
- 22.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, Bushman FD. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PloS One. 2012;7:e42786. doi: 10.1371/journal.pone.0042786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, Fligner CL, Singh PK. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown PS, Pope CE, Marsh RL, Qin X, McNamara S, Gibson R, Burns JL, Deutsch G, Hoffman LR. Directly sampling the lung of a young child with cystic fibrosis reveals diverse microbiota. Ann. Am. Thorac. Soc. 2014;11:1049–1055. doi: 10.1513/AnnalsATS.201311-383OC. ** This study obtained lung tissue from a pediatric CF patient and used bacterial ribosomal RNA gene pyrosequencing and computation phylogenetic analysis to define its microbiota. The study found diverse and anatomically heterogenous bacterial populations in different portions of the lung, suggesting the presence of spatial heterogeneity within an individual host.
- 25.Di Sant’ Agnese PEA, Andersen DH. Celiac syndrome; chemotherapy in infections of the respiratory tract associated with cystic fibrosis of the pancreas; observations with penicillin and drugs of the sulfonamide group, with special reference to penicillin aerosol. Am. J. Dis. Child. 1911. 1946;72:17–61. [PubMed] [Google Scholar]
- 26.Hoegger MJ, Fischer AJ, McMenimen JD, Ostedgaard LS, Tucker AJ, Awadalla MA, Moninger TO, Michalski AS, Hoffman EA, Zabner J, et al. Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science. 2014;345:818–822. doi: 10.1126/science.1255825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ, Hanfland RA, Wohlford-Lenane C, Dohrn CL, Bartlett JA, et al. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3000928. 29ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abou Alaiwa MH, Reznikov LR, Gansemer ND, Sheets KA, Horswill AR, Stoltz DA, Zabner J, Welsh MJ. pH modulates the activity and synergism of the airway surface liquid antimicrobials β-defensin-3 and LL-37. Proc. Natl. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1422091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LiPuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferkol T, Rosenfeld M, Milla CE. Cystic fibrosis pulmonary exacerbations. J. Pediatr. 2006;148:259–264. doi: 10.1016/j.jpeds.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Stressmann FA, Rogers GB, Marsh P, Lilley AK, Daniels TWV, Carroll MP, Hoffman LR, Jones G, Allen CE, Patel N, et al. Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation? J. Cyst. Fibros. Off. J. Eur. Cyst. Fibros. Soc. 2011;10:357–365. doi: 10.1016/j.jcf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Zemanick ET, Wagner BD, Harris JK, Wagener JS, Accurso FJ, Sagel SD. Pulmonary exacerbations in cystic fibrosis with negative bacterial cultures. Pediatr. Pulmonol. 2010;45:569–577. doi: 10.1002/ppul.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regelmann WE, Elliott GR, Warwick WJ, Clawson CC. Reduction of sputum Pseudomonas aeruginosa density by antibiotics improves lung function in cystic fibrosis more than do bronchodilators and chest physiotherapy alone. Am. Rev. Respir. Dis. 1990;141:914–921. doi: 10.1164/ajrccm/141.4_Pt_1.914. [DOI] [PubMed] [Google Scholar]
- 34.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest. 2003;123:1495–1502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
- 35.Hurley MN, Ariff AHA, Bertenshaw C, Bhatt J, Smyth AR. Results of antibiotic susceptibility testing do not influence clinical outcome in children with cystic fibrosis. J. Cyst. Fibros. Off. J. Eur. Cyst. Fibros. Soc. 2012;11:288–292. doi: 10.1016/j.jcf.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conrad D, Haynes M, Salamon P, Rainey PB, Youle M, Rohwer F. Cystic Fibrosis Therapy: A Community Ecology Perspective. Am. J. Respir. Cell Mol. Biol. 2013;48:150–156. doi: 10.1165/rcmb.2012-0059PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armougom F, Bittar F, Stremler N, Rolain J-M, Robert C, Dubus J-C, Sarles J, Raoult D, La Scola B. Microbial diversity in the sputum of a cystic fibrosis patient studied with 16S rDNA pyrosequencing. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2009;28:1151–1154. doi: 10.1007/s10096-009-0749-x. [DOI] [PubMed] [Google Scholar]
- 39. Carmody La, Zhao J, Schloss PD, Petrosino JF, Murray S, Young VB, Li JZ, LiPuma JJ. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann. Am. Thorac. Soc. 2013;10:179–187. doi: 10.1513/AnnalsATS.201211-107OC. ** This study attempted to characterize changes in airway bacterial communities around the time of CF pulmonary exacerbation, and overall found no significant differences in bacterial community diversity or bacterial density between baseline and exacerbations samples.
- 40.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao J, Murray S, Lipuma JJ. Modeling the impact of antibiotic exposure on human microbiota. Sci. Rep. 2014;4:4345. doi: 10.1038/srep04345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PloS One. 2012;7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stressmann FA, Rogers GB, van der Gast CJ, Marsh P, Vermeer LS, Carroll MP, Hoffman L, Daniels TWV, Patel N, Forbes B, et al. Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax. 2012;67:867–873. doi: 10.1136/thoraxjnl-2011-200932. [DOI] [PubMed] [Google Scholar]
- 44. Van der Gast CJ, Cuthbertson L, Rogers GB, Pope C, Marsh RL, Redding GJ, Bruce KD, Chang AB, Hoffman LR. Three clinically distinct chronic pediatric airway infections share a common core microbiota. Ann. Am. Thorac. Soc. 2014;11:1039–1048. doi: 10.1513/AnnalsATS.201312-456OC. ** This work compared the core microbiota of children and adults with cystic fibrosis, non-CF bronchiectasis, and protracted bacterial bronchitis. The authors found that each clinically distinct airway infection shared a common early core microbiota, but that disease-specific characteristics ultimately select for divergent microbiota by adulthood.
- 45.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maughan H, Cunningham KS, Wang PW, Zhang Y, Cypel M, Chaparro C, Tullis DE, Waddell TK, Keshavjee S, Liu M, et al. Pulmonary bacterial communities in surgically resected noncystic fibrosis bronchiectasis lungs are similar to those in cystic fibrosis. Pulm. Med. 2012;2012:746358. doi: 10.1155/2012/746358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 6:e16384. doi: 10.1371/journal.pone.0016384. [date unknown] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PloS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011;127:372.e1–3–381.e1–3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, Good JT, Gelfand EW, Martin RJ, Leung DYM. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am. J. Respir. Crit. Care Med. 2013;188:1193–1201. doi: 10.1164/rccm.201304-0775OC. ** These authors examined the relationships between the airway microbiome and corticosteroid response in asthma, and concluded that in a subset of patients who were considered to have corticosteroid-resistant asthma, more gram-negative bacteria, such as Haemophilus parainfluenzae, were identified.
- 51.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biesbroek G, Bosch AATM, Wang X, Keijser BJF, Veenhoven RH, Sanders EAM, Bogaert D. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am. J. Respir. Crit. Care Med. 2014;190:298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 54.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012;56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, Moore JH, Guill MF, Morrison HG, Sogin ML, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio. 2012;3 doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bisgaard H, Bønnelykke K, Stokholm J. Immune-mediated diseases and microbial exposure in early life. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2014;44:475–481. doi: 10.1111/cea.12291. [DOI] [PubMed] [Google Scholar]
- 57.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, Boushey HA, Zoratti E, Ownby D, Lukacs NW, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. U.S.A. 2014;111:805–810. doi: 10.1073/pnas.1310750111. ** This study uses murine models to show that exposure to dog-associated household dust results in protection against airway allergen challenge and creates a distinct GI microbiome composition. These findings support the emerging view of a complex relationship between environmental exposures, GI microbiomes, and atopic diseases such as asthma.
- 59. Martinez FD. The human microbiome. Early life determinant of health outcomes. Ann. Am. Thorac. Soc. 2014;11(Suppl 1):S7–S12. doi: 10.1513/AnnalsATS.201306-186MG. *This work examines the literature on the role of early environmental microbe exposure in creating a "microbial-mucosal unit" that impacts future health outcomes.
- 60.Hoffman LR, Pope CE, Hayden HS, Heltshe S, Levy R, McNamara S, Jacobs MA, Rohmer L, Radey M, Ramsey BW, et al. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014;58:396–399. doi: 10.1093/cid/cit715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fallahi G, Motamed F, Yousefi A, Shafieyoun A, Najafi M, Khodadad A, Farhmand F, Ahmadvand A, Rezaei N. The effect of probiotics on fecal calprotectin in patients with cystic fibrosis. Turk. J. Pediatr. 2013;55:475–478. [PubMed] [Google Scholar]
- 62.Weiss B, Bujanover Y, Yahav Y, Vilozni D, Fireman E, Efrati O. Probiotic supplementation affects pulmonary exacerbations in patients with cystic fibrosis: a pilot study. Pediatr Pulmonol. 45:536–540. doi: 10.1002/ppul.21138. [DOI] [PubMed] [Google Scholar]
- 63.Bruzzese E, Raia V, Gaudiello G, Polito G, Buccigrossi V, Formicola V, Guarino A. Intestinal inflammation is a frequent feature of cystic fibrosis and is reduced by probiotic administration. Aliment. Pharmacol. Ther. 2004;20:813–819. doi: 10.1111/j.1365-2036.2004.02174.x. [DOI] [PubMed] [Google Scholar]