SUMMARY
Intracellular transport and distribution of organelles play important roles in diverse cellular functions, including cell polarization, intracellular signaling, cell survival and apoptosis. Here we report an optogenetic strategy to control the transport and distribution of organelles by light. This is achieved by optically recruiting molecular motors onto organelles through the heterodimerization of Arabidopsis thaliana cryptochrome 2 (CRY2) and its interacting partner CIB1. CRY2 and CIB1 dimerize within subseconds upon blue light exposure, which requires no exogenous ligands and low intensity of light. We demonstrate that mitochondria, peroxisomes, and lysosomes can be driven towards the cell periphery upon light-induced recruitment of kinesin, or towards the cell nucleus upon recruitment of dynein. Light-induced motor recruitment and organelle movements are repeatable, reversible and can be achieved at subcellular regions. This light-controlled organelle redistribution provides a new strategy for studying the causal roles of organelle transport and distribution in cellular functions in living cells.
INTRODUCTION
In mammalian cells, important proteins and organelles are actively transported to their appropriate locations by a two-way microtubule-based traffic system. In this system, the molecular motors dyneins and kinesins convert chemical energy into mechanical work to deliver cargoes. Dyneins walk towards the minus end of microtubules and thus carry cargos to the cell nucleus. In the opposite direction, kinesins walk towards the plus end of microtubules and take cargos to the cell periphery. Accumulating evidence indicate that distinct spatial distributions of organelles exist under defined conditions and play important roles in various cellular functions in cells. For example, it is observed that nutrients induce peripheral positioning of lysosomes while starvation causes perinuclear clustering of lysosomes (Korolchuk et al., 2011). Localization of the mitochondria to the vicinity of plasma membrane is proposed to be crucial for sustained Ca2+ influx across plasma membrane and T-cell activation (Quintana et al., 2006; Schwindling et al., 2010). On the other hand, the perinuclear clustering of mitochondria, observed during hypoxia, is proposed to be indispensable for high nuclear reactive oxygen species level under hypoxia and regulate hypoxia-induced gene expression (Al-Mehdi et al., 2012). At subcellular level, non-symmetric transport and distribution of organelles are crucial for the development and functioning of highly polarized cells (Mellman and Nelson, 2008). For example, in cultured hippocampal neurons, the preferential delivery of post-Golgi vesicles to an immature neurite biases the morphological polarization of the neurite into an axon (Bradke and Dotti, 1997). However, a direct link between organelle distributions and cellular functions is missing due to the lack of effective and controllable means to manipulate organelle transport in living cells.
A desirable method to control the organelle distribution in cells should have the following characteristics. First, it should be able to be conducted in living cells so that the functional consequences of organelle redistribution can be investigated in vivo. Second, it should be reversible so that the regulation can be stopped at a desired time point. Finally, it should permit temporal and spatial control in order to study intracellular activities in certain temporal patterns and/or in distinct subcellular regions. Recently, a rapamycin-induced FKBP-FRB heterodimerization system has been developed that enables inducible recruitment of molecular motors to specific organelles and drives organelle redistribution inside the cell (Kapitein et al., 2010a; Kapitein et al., 2010b; van Spronsen et al., 2013).This technique provides the unique capability of controlling organelle distribution in living cells. However, it requires a rapamycin cofactor which needs to penetrate the cell membrane and cannot be applied with subcellular precision. In addition, the rapamycin-induced binding is not reversible.
The rapidly emerging area of optogenetic actuators provides new opportunities to control motor activities and organelle redistribution. Several pairs of light-induced dimerization proteins have been developed, including light-oxygen-voltage (LOV) domain (Harper et al., 2003; Renicke et al., 2013; Strickland et al., 2012; Strickland et al., 2010; Wu et al., 2009), phytochrome B (Levskaya et al., 2009; Shimizu-Sato et al., 2002; Toettcher et al., 2013) and cryptochrome 2 (CRY2) (Bugaj et al., 2013; Kennedy et al., 2010). Among them, the CRY2 and the LOV systems do not need exogenous cofactors for dimerization. In a very recent study, the light-induced heterodimerization between the LOV2 domain from Avena sativa phototropin 1 and an engineered PDZ domain was utilized to optically control organelle redistribution (van Bergeijk et al., 2015). Here in this work, using similar strategy, we exploit the light-controlled binding of CRY2 to its interacting partner CIB1 to achieve optogenetic control of organelle redistribution in living cells. Previously, light-inducible CRY2-CIB1 binding has been utilized to control endogenous transcription (Konermann et al., 2013), phosphoinositide metabolism (Idevall-Hagren et al., 2012) and activation of signaling pathways such Raf/MEK/ERK (Zhang et al., 2014) and PI3K signaling pathway (Kakumoto and Nakata, 2013). The light-induced heterodimerization of CRY2-CIB1 requires low level of light and thus introduce minimal light toxicity in long-term studies (Zhang et al., 2014). By tethering CRY2 to specific organelles and CIB1 to molecular motors, we demonstrate light-induced transport of organelles to the perinuclear region by recruiting dyneins and light-induced transport of organelles to the cell plasma membrane by recruiting kinesins. The strategy is generally applicable to many types of organelles including mitochondria, peroxisomes and lysosomes. The manipulation of organelle distribution is reversible and spatial control of organelle distribution can be achieved in subcellular regions.
RESULTS
Design Scheme for Optogenetic Control of Molecular Motors and Organelle Distribution
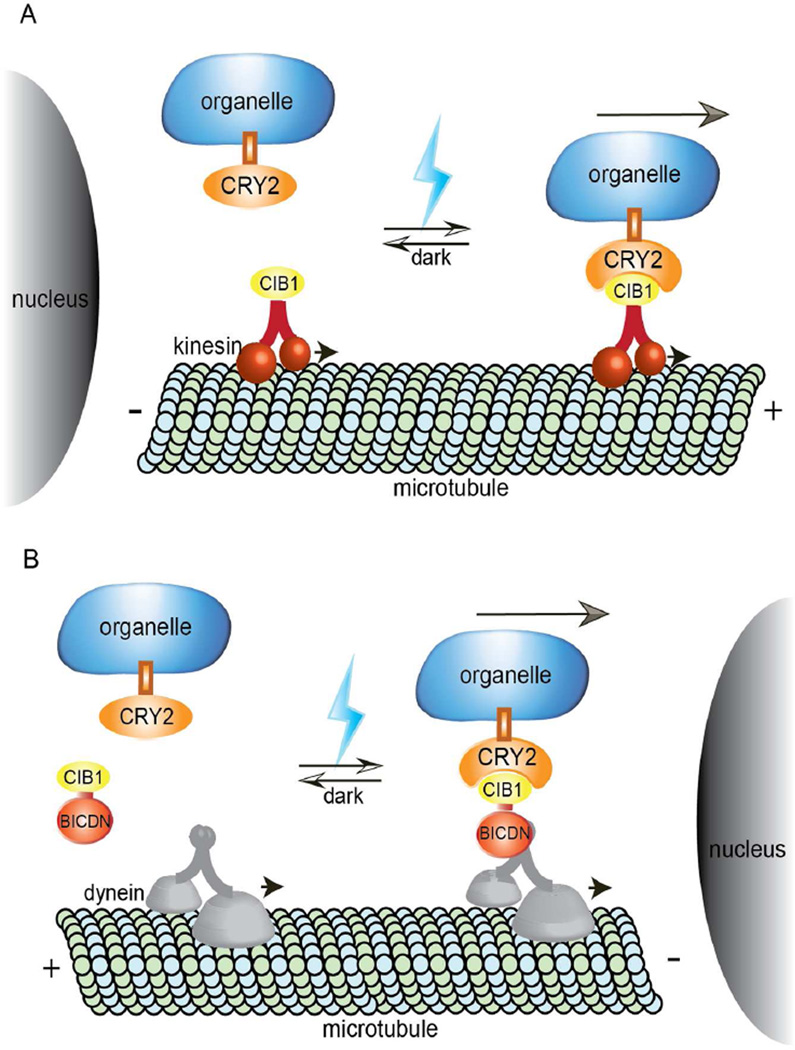
We have designed a generic method that uses light to control motor recruitment to organelle membrane and thus controls organelle distributions in living cells. Intracellular transport of organelles is mainly propelled by microtubule-based motor proteins including kinesins and dyneins. It was previously reported that utilizing FKBP-rapalog-FRB heterodimerization system, recruitment of motors can drive specific cargo movement and organelle redistribution (Kapitein et al., 2010b). Inspired by this study, we hypothesized that reversible and spatial control of inducible organelle movement can be achieved if we were able to use light to recruit motors to specific organelles. To this end, we chose the CRY2-CIB1 heterodimerization module that can bind within subseconds in the presence of blue light and dissociate in a few minutes after turning off the blue light. In this study, we employed the photolyase homology region of CRY2 (amino acids 1–498) and N-terminal region of CIB1 (amino acids 1–170) (Kennedy et al., 2010). In our design (Figure 1), CRY2 was localized to different organelle via cargo-specific membrane linkers, while to control motors, CIB1 was fused to either KIF5A, a truncated kinesin protein, or BICDN, the N-terminal of the dynein/dynactin interacting protein BICD. Truncated kinesin KIF5A without the tail domains is known to be unable to bind to cargos(Cai et al., 2007). When truncated KIF5A are recruited to organelles upon CRY2/CIB1 dimerization under blue light, the organelles should move towards the plus-ends of microtubules which are mostly localized at the cell periphery. On the other hand, the recruitment of BICDN to organelles will recruit dynein motors and induce transport of organelles towards the minus-ends of microtubules, which usually reside around the cell nucleus.
Figure 1. Schematic of Light-Controlled Motor Recruitment and Organelle Transport in Cells.
(A) CRY2 is anchored to organelles via specific organelle targeting transmembrane domain. CIB1 is linked to truncated kinesin KIF5A. Upon blue light exposure, CIB1-CRY2 binding recruits kinesin motors to organelles, which drives organelles towards the plus end of microtubule.
(B) CIB1 domain is linked to BICDN, a dynein/dynactin adaptor protein. When illuminated with blue light, CIB1 binds with CRY2 and thus recruits dynein to organelles. Consequently, organelles are propelled towards the minus end of microtubule.
Light Induces Redistribution of Cellular Mitochondria by Recruiting Molecular Motors
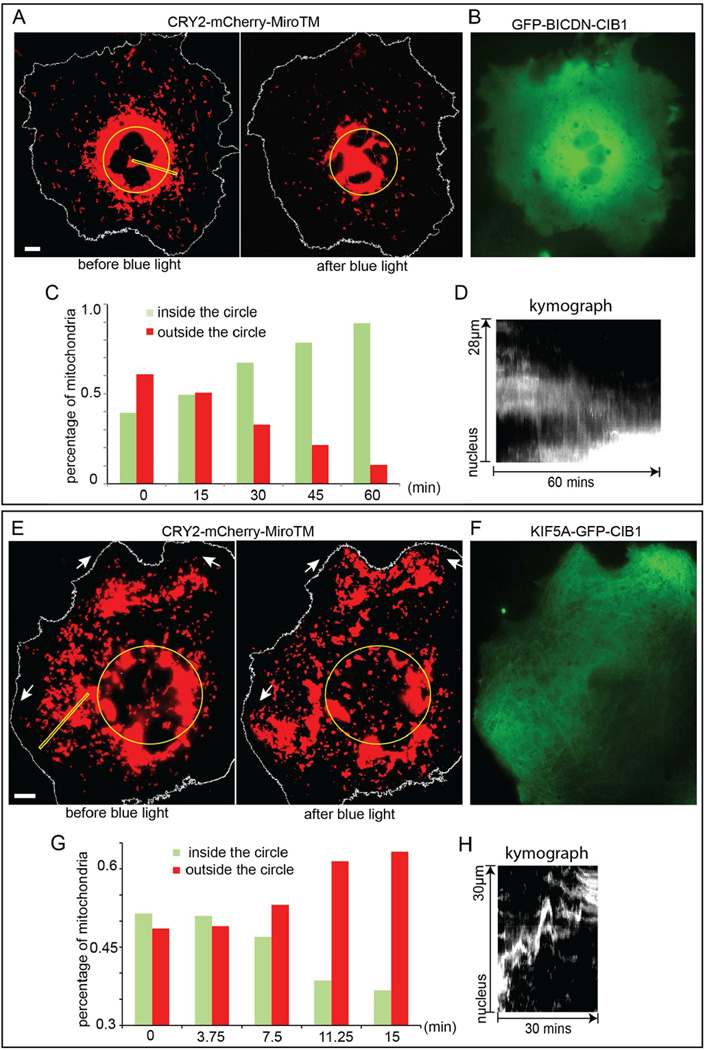
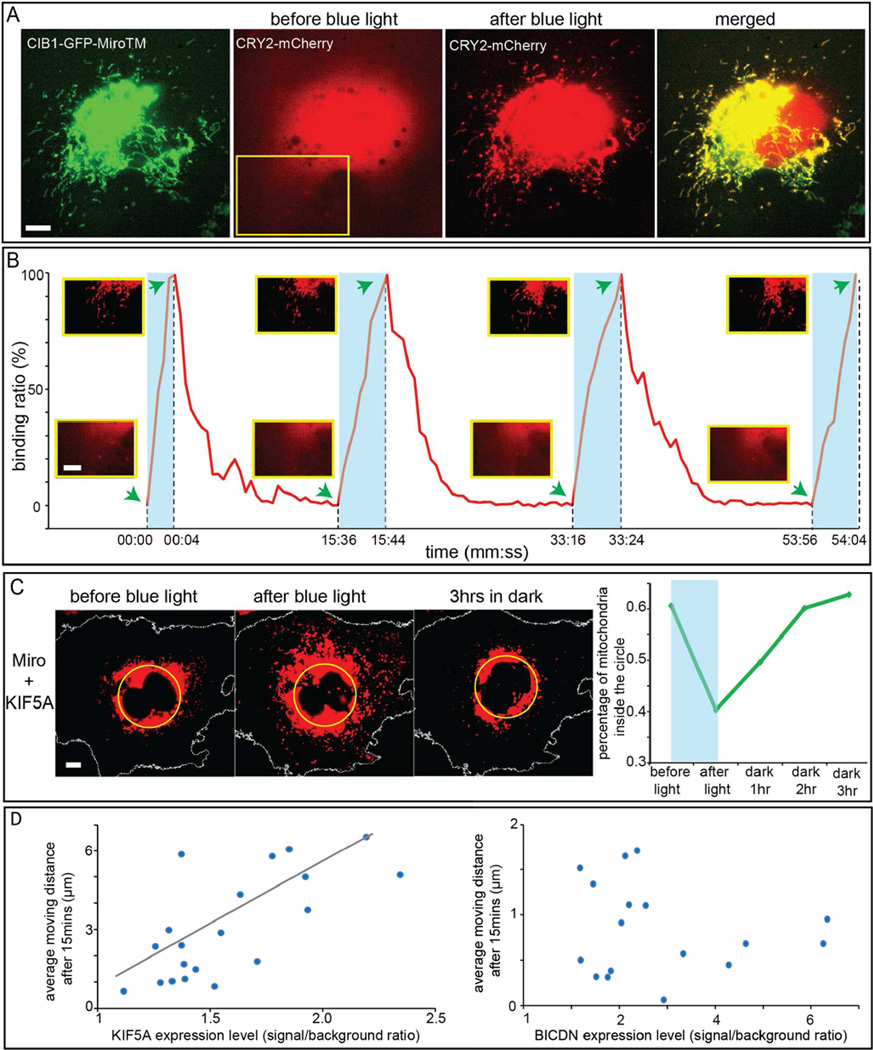
We first demonstrated that mitochondria could be moved toward the perinuclear region by light-induced recruitment of BICDN. In this report, intermittent 200-ms exposure of blue light at 9.7 W/cm2 per 10-second interval was used to stimulate the dimerization of CRY2-CIB1 unless otherwise noted. CRY2 was targeted to mitochondria by CRY2-mCherry-MiroTM, where MiroTM is the transmembrane domain of Miro1, a component of the motor/adaptor complex that anchors to the mitochondria outer membrane (Fransson et al., 2006). The specific mitochondria localization of CRY2-mCherry-MiroTM was confirmed by the colocalization between CRY2-mCherry-MiroTM and Mito-YFP, a known mitochondria targeting sequence from cytochrome C oxidase (Rizzuto et al., 1989) (Figure S1). As shown in Figure 2A, in the COS-7 cell co-transfected with CRY2-mCherry-MiroTM and GFP-BICDN-CIB1, some mitochondria were positioned close to the nucleus while others spread throughout the cell. The distribution of GFP-BICDN-CIB1 in the same cell was cytosolic and diffusive (Figure 2B). After intermittent blue light stimulation for 1 hour, mitochondria became highly clustered around cell nucleus (Figure 2A), driven by light-recruited dynein motors (Movie S1 shows the transport dynamics). The cytosolic distribution of GFP-BICDN-CIB1 in this cell did not change by blue light illumination (Figure S2A), likely due to the much higher expression of cytosolic GFP-BICDN-CIB1 than mitochondria-localized CRY2-mCherry-MiroTM. Indeed, the colocalization between GFP-BICDN-CIB1 and CRY2-mCherry-MiroTM was observed in cells with very low expression level of GFP-BICDN-CIB1 (Figures S2B–S2E). To quantify the global mitochondria movements, the percentage of mitochondria inside the yellow perinuclear circle was calculated at different time points using a Matlab program (Experimental Procedures and Figure S3). The perinuclear circle was set to enclose the whole nucleus. The diameter was adjusted to include approximately ~50% fluorescence intensity of the whole cell at the beginning of the experiment, which will give the best contrast if organelles move in or out of the circle. In the cell shown in Figure 2A, before blue light illumination, only 39% of mitochondria were localized inside the circle. Over the 60-minute intermittent illumination of blue light, the percentage of mitochondria inside the circle gradually increased to 89% (Figure 2C). The kymograph extracted from the time-lapse movie (along the yellow line in Figure 2A) demonstrated that mitochondria were actively transported to the proximity of cell nucleus (Figure 2D).
Figure 2. Light-Induced Re-Distribution of Mitochondria in COS-7 Cells by Recruiting Molecular Motors.
(A–D) The COS-7 cell was transfected with CRY2-mCherry-MiroTM and GFP-BICDN-CIB1. (A) Mitochondria were visualized in the red channel by CRY2-mCherry-MiroTM. Before blue light illumination, some mitochondria were positioned close to the nucleus while others spread throughout the cell. After blue light illumination, most mitochondria moved towards cell nucleus as indicated by the yellow circle. (N=20) (B) The green channel showed GFP-BICDN-CIB1 diffusing in the cytosol. (C) The percentage of mitochondria inside the yellow circle in (A) increased from 39% to 89% after intermittent light exposure for 60 minutes. (D) The kymograph extracted from the area indicated by yellow rectangle in (A) showed the clear trend of mitochondria moving toward the nucleus.
(E–H) The COS-7 cell was transfected with CRY2-mCherry-MiroTM and KIF5A-GFP-CIB1. (E) After blue light illumination, many mitochondria moved away from the perinuclear region (yellow circle) and some clustered at cell edge as indicated by the arrows. (N=73) (F) KIF5A-GFP-CIB1 in the same cell distributed along microtubules. (G) The percentage of mitochondria at the perinuclear region (inside the yellow circle) decreased from 52% to 36% after intermittent light exposure for 30 minutes. (H) The kymograph extracted from the area indicated by yellow rectangle in (E) showed that over time, mitochondria were actively moved towards the cell periphery.
Likewise, we showed that mitochondria could be moved outward to the cell periphery by light-induced recruitment of KIF5A (Figures 2E–2H; Movie S2). The COS-7 cell was co-transfected with CRY2-mCherry-MiroTM and KIF5A-GFP-CIB1. Unlike GFP-BICDN-CIB1 that was diffusive, KIF5A-GFP-CIB1 retained kinesins’ microtubule binding affinity and its distribution showed microtubule tracks (Figure 2F). Upon blue light illumination, mitochondria spread out towards cell periphery and some clustered at cell edge as indicated by the white arrows (Figure 2E). KIF5A-GFP-CIB1 distribution in this cell did not change noticeably after blue light exposure (Figure S2F). The percentage of mitochondria inside the yellow circle gradually decreased from 52% to 36% after blue light illumination for 15 minutes (Figure 2G). The kymograph along the yellow rectangular region indicated that mitochondria were actively transported towards the cell periphery during light illumination (Figure 2H).
As negative controls, without blue light stimulation, COS-7 cell co-transfected with either CRY2-mCherry-MiroTM & GFP-BICDN-CIB1 or KIF5A-GFP-CIB1 & CRY2-mCherry-MiroTM did not have significant change in mitochondria spatial distribution after 1 hour (Figures S4A and S4B). Furthermore, in co-transfected cells that lacked CIB1(i.e. CRY2-mCherry-MiroTM & GFP-BICDN, CRY2-mCherry-MiroTM & KIF5A-GFP) or in cells single-transfected with CRY2-mCherry-MiroTM, mitochondria distribution did not change after blue light stimulation for 1 hour (Figures S4C–S4E).
Multiple Types of Organelles Can Be Relocated by Light-Induced Recruitment of Motors
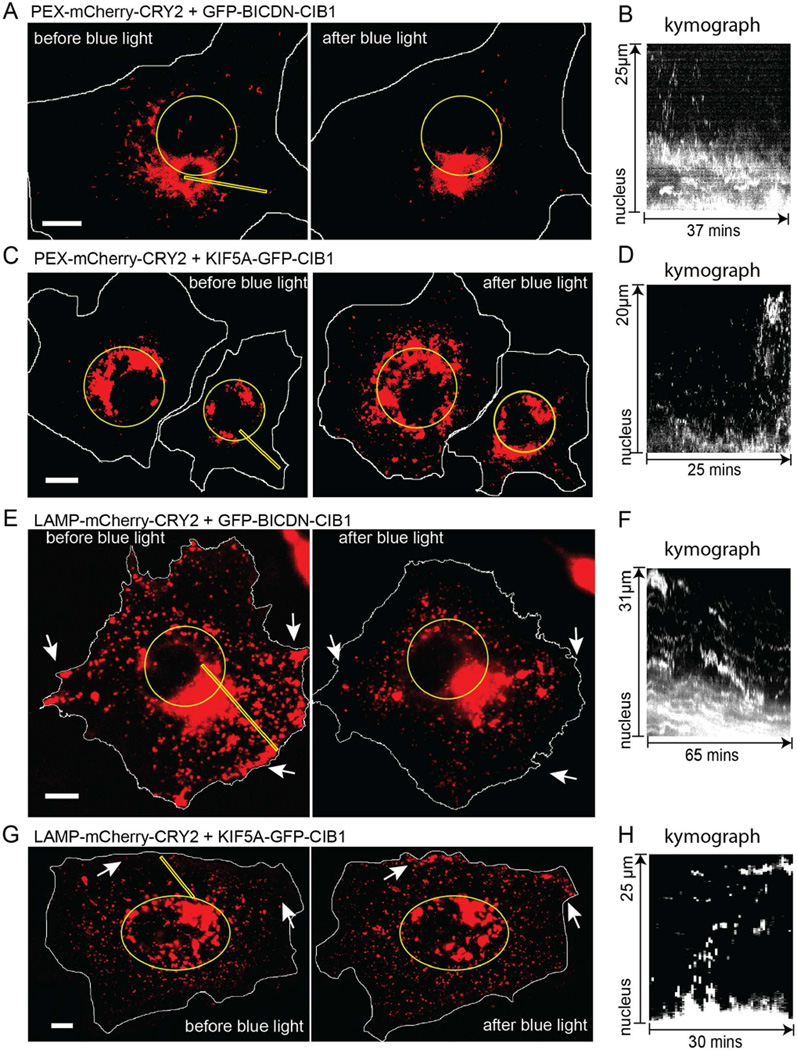
In addition to the ability to induce redistribution of mitochondria, our method can also initiate directional transport of other organelles by tethering CRY2 to their outer membranes. In this regard, we attached CRY2 to the membrane of peroxisomes by linking to the N-terminal of PEX (amino acids 1–42), which is the peroxisome membrane targeting domain of a peroxisomal membrane protein (Kammerer et al., 1998). In the cell transfected with PEX-mCherry-CRY2 and GFP-BICDN-CIB1 as shown in Figure 3A, some peroxisomes were located close to the nuclear region and some were distributed throughout the cell. After intermittent blue light exposure for 37 minutes, almost all peroxisomes were relocated very close to nucleus. The kymograph in Figure 3B showed the clear trend of peroxisomes moving toward the nucleus. In the opposite microtubule direction, we demonstrated that, in the cell transfected with PEX-mCherry-CRY2 and KIF5A-GFP-CIB1 as shown in Figure 3C, blue light stimulation drove the peroxisomes moving toward the cell periphery, similar to the behavior of mitochondria as described in the previous section. After 25-minute blue light stimulation, the distribution of peroxisomes was much more spread out in the cell as compared before blue light stimulation. The kymograph shown in Figure 3D (generated from the yellow line in Figure 3C) clearly showed the trend that peroxisomes cargoes were moving outward. The distribution of GFP-BICDN-CIB1 and KIF5A-GFP-CIB1 did not change after light-induced peroxisome redistribution (Figures S5A and S5B).
Figure 3. The Distributions of Peroxisomes and Lysosomes in Cells Can Be Modulated by Light-Induced Recruitment of Motors.
(A) In the cell co-transfected with GFP-BICDN-CIB1 and PEX-mCherry-CRY2, blue light illumination caused peroxisomes to become highly clustered around the cell nucleus indicated by the circle.
(B) Kymographs generated along the yellow rectangular region in (A) shows the trajectories of peroxisomes moving toward the nucleus.
(C) In the cell transfected with KIF5A-GFP-CIB1 and PEX-mCherry-CRY2, blue light illumination caused peroxisomes to move away from the cell nucleus. The circles marked the cell nuclei.
(D) Kymographs generated along the yellow rectangular region in (C) shows peroxisomes moving away from the nucleus.
(E) In the cell transfected with GFP-BICDN-CIB1 and LAMP-mCherry-CRY2, blue light illumination induced lysosomes clustering around the nucleus marked by the yellow circle.
(F) Kymographs generated along the yellow rectangular region in (E) shows lysosomes moving toward the nucleus.
(G) In the cell transfected with KIF5A-GFP-CIB1 and LAMP-mCherry-CRY2, lysosomes moved towards cell periphery with blue light exposure. The cell nucleus was indicated by the eclipse.
(H) Kymographs generated along the yellow rectangular region in (G) shows lysosomes moving toward the cell periphery.
Scale bars, 10µm. See also Figure S5.
We also demonstrated the optogenetic control of cargo distribution for lysosomes. To activate the light-induced movement of lysosomes, mCherry-CRY2 was fused to the N-terminus of LAMP, an integral membrane protein specific to lysosomes(Chen et al., 1988). When blue light was turned on, the recruitment of BICDN to lysosomes resulted in translocation of lysosomes to perinuclear region in COS-7 cells co-transfected with GFP-BICDN-CIB1 and LAMP-mCherry-CRY2 (Figure 3E). In particular, the amount of lysosomes near cell edge was drastically reduced as indicated by the white arrows, corroborated by the kymograph in Figure 3F. On the other hand, the light-induced recruitment of KIF5A-GFP-CIB1 to lysosomes resulted in lysosome translocation to the cell periphery (Figure 3G and 3H). The distribution of GFP-BICDN-CIB1 and KIF5A-GFP-CIB1 did not change obviously after light-induced lysosome redistribution (Figures S5C and S5D).
We noticed that the instantaneous moving speeds of translocation were similar for mitochondria, lysosomes and peroxisomes. However, the redistribution of peroxisomes, on average, occurred much faster than mitochondria when induced by blue light exposure. We also noticed that most light-induced mitochondria movements were in a zigzag and stop-and-go patterns (see Movies S1 and S2), while peroxisome moved more smoothly in large distances and in straight lines (see Movies S3 and S4). One possibility is that peroxisomes are smaller in size than mitochondria and thus maneuver more easily through dense cellular environment. It is also possible that some mitochondria are tethered to cellular structures, for example cytoskeleton (Boldogh and Pon, 2006), and not able to be moved even when molecular motors are recruited to them. The successful demonstration of light-induced organelle movement and redistribution of mitochondria, peroxisomes, and lysosomes shows that our method is generally applicable to induce polarized transport and differential distribution patterns of different organelles in cells.
Light-Recruited Molecular Motors Move Along Microtubules
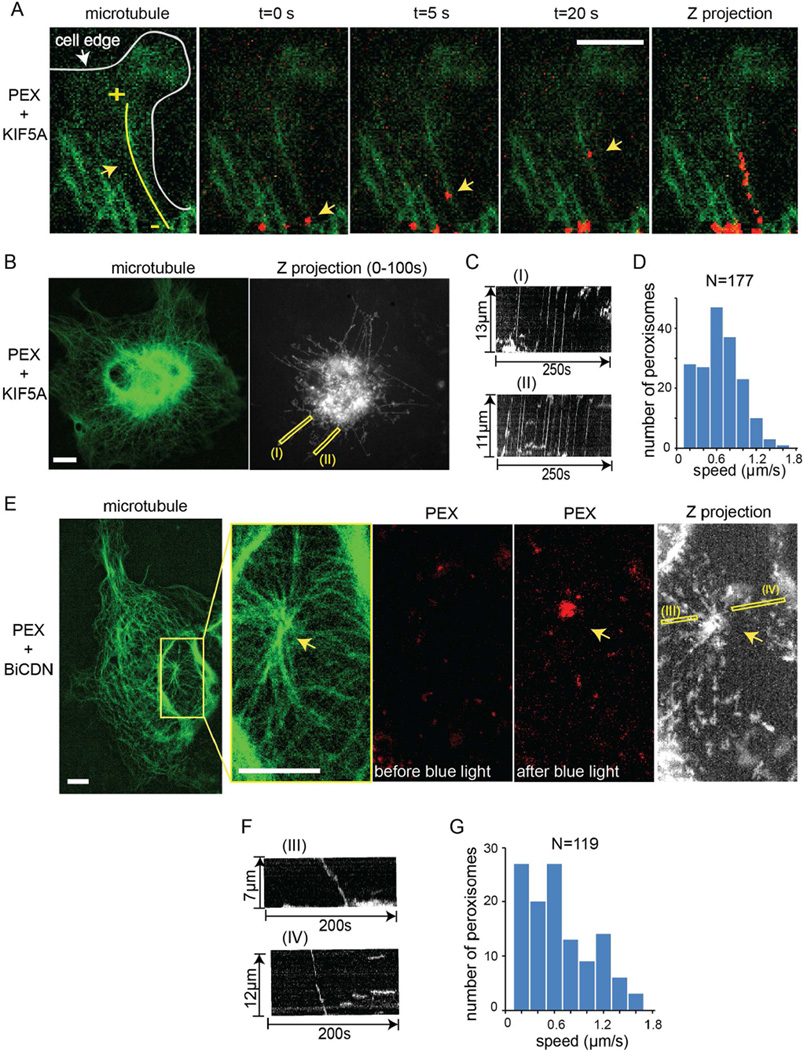
Though light-induced recruitment of molecular motors drives organelle redistribution, it is interesting to know if the artificially recruited motors retain their native properties. Therefore, we investigated whether motors recruited via CRY2-CIB1 dimerization system move along microtubules and also measured the speeds of cargo movement propelled by these artificially recruited motors. For this purpose, we chose peroxisomes as cargoes because peroxisomes are smaller and fewer in numbers than mitochondria or lysosomes, which make it easier to track their individual cargo movements. First, we showed that light-induced recruitment of kinesins resulted in peroxisome transporting along microtubules. Cells were triple-transfected with PEX-mCherry-CRY2, KIF5A-CIB1 and TAU-YFP, among which TAU-YFP was used to label microtubules (Samsonov et al., 2004). In this section, KIF5A-CIB1 was not labeled with any fluorescence tag. Figure 4A showed that a peroxisome moved along a microtubule after blue light stimulation and headed towards the microtubule plus end that pointed to the cell edge. The overlap could be seen more clearly in the maximum intensity Z projection of the time lapse movie.
Figure 4. Characterization of Organelle Transport by Light-Induced Recruitment of Molecular Motors.
(A) The peroxisome indicated by the yellow arrow moved along the microtubule and towards the cell periphery by light-induced recruitment of kinesins. The cell was triple-transfected with KIF5A-CIB1, TAU-YFP and PEX-mCherry-CRY2. Microtubules were labeled by TAU-YFP. Z projection of peroxisomes was obtained by projecting the maximum intensity of PEX-mCherry-CRY2 during 20 frames acquisition at 1frame/second and was merged with GFP image, which overlaps with the underlying microtubule.
(B) Z projection of light-induced peroxisome trajectories spreads radially from the nucleus to cell periphery, resembling the distribution of microtubule network. The cell was triple-transfected with KIF5A-CIB1, TAU-YFP and PEX-mCherry-CRY2. Z projection of peroxisomes was obtained by recording the maximum intensity of PEX-mCherry-CRY2 during 100 frames acquisition at 1frame/second.
(C) Kymographs of the indicated yellow lines (I) and (II) in (B) show that many peroxisomes moved towards cell periphery on the same microtubules and at a relatively constant speed.
(D) Histogram of peroxisomes moving speeds for 177 peroxisomes in 4 cells shows an average speed of 0.55 µm/s.
(E) Trajectories of light-induced peroxisome retrograde movement resemble the distribution of microtubules around the center where many microtubules merged. The cell was triple-transfected with BICDN-CIB1, TAU-YFP and PEX-mCherry-CRY2.
(F) Kymographs of the indicated lines (III) and (IV) show that peroxisomes moved along microtubules towards cell nucleus.
(G) Histogram of peroxisomes speeds was obtained by calculating the speeds for 119 peroxisomes in 4 cells shows an average speed of 0.57 µm/s.
Scale bars, 5 µm (A), 10µm (B, E). See also Movie S3 and S4.
Next, we examined the movement speed of individual peroxisome by blue light-recruitment of KIF5A (Figure 4B). In cells co-transfected with PEX-mCherry-CRY2, KIF5A -CIB1 and TAU-YFP, we recorded time-lapse movies at 1frame/second for peroxisome movement over a period of 400 seconds. Z projection of the movie yielded peroxisome trajectories that demonstrated an outward radial distribution overlapping with some microtubules. Interestingly, we often observed many peroxisomes moving on the same microtubules (Movie S3). Two kymographs (I) and (II) in Figure 4C were generated along the two microtubules shown as yellow rectangles in Figure 4B. There were respectively 8 and 16 peroxisomes transporting along these two microtubules in 250 seconds (Figure 4C). However, peroxisomes seemed to move on selected subsets of microtubules and only a small fraction of cellular microtubules were used. Throughout the 400-second movie acquisition, there were only around 50 out of hundreds of microtubules that had peroxisomes moving on. Most peroxisomes moved along these trajectories continuously and at a relatively constant speed. We measured the moving speeds of 177 peroxisomes and the distribution of transport velocity showed a mean value at 0.55 µm/s with a spread between 0 to 2.0 µm/s (Figure 4D).
Next, we also confirmed that light-induced dynein recruitment drove cargos moving along microtubules. In Figure 4E, the cell triple-transfected with BICDN-CIB1, TAU-YFP and PEX-mCherry-CRY2 showed a region where many microtubule ends merged as the arrow indicated, likely a microtubule organizing center. Blue light illumination triggered the retrograde transport of peroxisomes which resulted in the accumulation of peroxisomes at the microtubule nucleation site (Movie S4). The trajectories of peroxisomes in the Z projection image clearly overlap with the microtubule patterns surrounding the microtubule nucleation site. Kymographs were generated from the two trajectories indicated as (III) and (IV) and showed that peroxisomes moved along the microtubules towards the cell center (Figure 4F). Transport speeds of 119 peroxisomes in 4 cells were measured and presented as the histogram of speeds in Figure 4G, showing a mean value at 0.57 µm/s and a spread from 0 to 2.0 µm/s (Figure 4G). The average speed for peroxisome transport driven by light-induced molecular motors recruitment in this study is in line with speed measurements reported in previous in vivo peroxisome transport study (Kural et al., 2005).
Light-Induced Organelles Redistribution Is Reversible
Reversibility is a highly desirable property to regulate motor activity and organelle transport. Here we showed that the binding of molecular motors and light-induced movement of mitochondria was reversible upon turning off the blue light. First, we demontrated that the binding between CRY2 and CIB1 on mitochondria was reversible using cells co-transfected with CIB1-GFP-MiroTM and CRY2-mCherry. Here to demonstrate the CRY2-CIB1 binding reversibility, we use organelle-localized CIB1 and cytosolic CRY2 instead of organelle-localized CRY2 and cytosolic CIB1. This is because CRY2 usually has much lower expression level than CIB1. Binding of CRY2-mCherry to organelle-localized CIB1 will visually deplete cytosolic CRY2 and thus the colocalization can be clearly observed. In this experiment, during blue light illumination, the cell was stimulated with 200-ms exposure of blue light at 9.7 W/cm2 per 1-second interval. As shown in Figure 5A, the initially diffusive and cytosolic CRY2-mCherry were quickly recruited to mitochondria after blue light illumination. The CIB1-CRY2 binding is reversible after the blue light is turned off and the process is highly repeatable. Figure 5B showed 4 consecutive cycles of the CIB1-CRY2 association/dissociation in the yellow rectangle area indicated in the cell (see Movie S5). The CYR2-CIB1 binding was usually completed within 10 seconds while the dissociation took around 10 minutes. Repeated process did not change the binding/unbinding dynamics.
Figure 5. Light-Induced Organelle Redistribution Is Reversible and Is Dependent on Motor Expression Level.
(A) CRY2 and CIB1 binding on mitochondria membrane is reversible and repeatible. The cell was transfected with CIB1-GFP-Miro1TM and CYR2-mCherry. Before blue light, CIB1-GFP-Miro1TM was targeted to mitochondria while CYR2PHR-mCherry was cytosolic. Upon blue light exposure, CRY2-mCherry bound to CIB1-GFP-Miro1TM and localized to mitocondria.
(B) The same transfected cell shown in (A) was repeatedly subjected to four cycles of blue light and dark periods. For each cycle, CRY2-mCherry was recruited to mitochondria within seconds of blue light illumination, while it returned to its cytosolic distribution after incubating in dark for 10–15 minutes. Within each period of blue light illumination, the cell was stimulated with 200-ms exposure of blue light at 9.7 W/cm2 per 1-second interval.
(C) In the cell transfected with CRY2-mCherry-MiroTM and KIF5A-GFP-CIB1, mitochondria were driven towards the cell periphery by intermittent blue light exposure (one 200-ms pulse per 10 second) for 30 minutes. Then the cell was incubated without blue light stimulation and mitochondria were observed to gradually move back towards the cell nucleus. Quantification of the percentage of the mitochondria inside the yellow circle shows the percentage decreased from 61% to 40% after blue light stimulation and increased back to 63% after 3-hour dark incubation.
(D) The average moving distances of mitochondria after 15-minute intermittent blue light illumination were calculated and plotted along with motor expression level. In 20 cells double-transfected with CRY2-mCherry-MiroTM and KIF5A-GFP-CIB1, the higher the KIF5A expression level was, the further the mitochondria moved towards cell periphery at the end of 15-minute period (left graph). However, in 17 cells transfected with CRY2P-mCherry-MiroTM and GFP-BICDN-CIB1, the expression level of BICDN had no obvious influence on how far mitochondria moved towards cell nucleus during 15-minute period (right graph).
Scale bars, 10µm. See also Movie S5.
We also demonstrated that the light-induced mitochondria redistribution was reversible (Figure 5C). In the cell transfected with KIF5A-GFP-CIB1 and CRY2-mCherry-MiroTM, mitochondria were observed to move away from nucleus with light-induced recruitment of kinesins. In this cell, the percentage of mitochondria inside the yellow circle was 61% initially and dropped to 40% after blue light illumination. However, during the following 3 hours’ dark incubation, more and more mitochondria moved back towards cell nucleus and the percentage increased from 40% to 63%. After 10 minutes of dark incubation, CRY2 and CIB1 would dissociate and thus kinesins would be no longer bound to mitochondria. Afterward, other motors such as dyneins or myosins, may bind to mitochondria and drive them back towards the cell nucleus. We noted that not all light-induced organell redistribution were reversible within the 3-hour time frame that we were measuring, suggesting complex intracellular processes.
We found that the expression level of KIF5A affected the overall rate of light-induced organelle redistribution, while the expression level of BICDN did not. We compared light-induced mitochondria redistribution in cells with different motor expression levels but with similar MiroTM (cargo) expression levels. Before and after blue light illumination for 15 minutes, the average distance of mitochondria movement was calculated using a custom-written Matlab progam (see Experimental Procedures for details). Figure 5D shows the movement of 20 cells at different KIF5A levels. It is clear that the higher the KIF5A expression level, the further the mitochondria moved towards the cell pheriphery. Therefore, more KIF5A motors recruited to mitochondria will drive mitochondria further in the same time frame. On the other hand, the expression level of BICDN did not appear to affect mitochondria moving distance. It is likely that, in cells transfected with GFP-BICDN-CIB1, the translocation of mitochondria is limited by the total amount of endogenous dyneins.
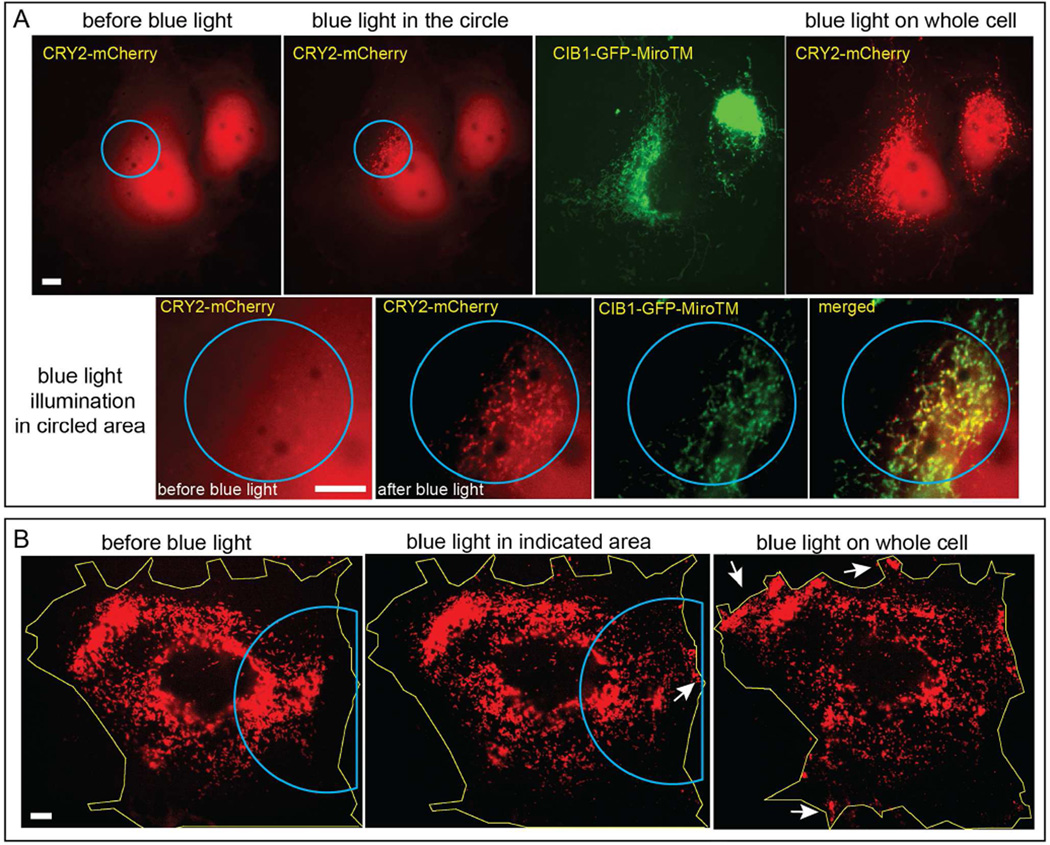
Organelle Redistribution Can Be Spatially Controlled at Subcellular Regions
The optogenetic control affords high spatial resolution and we demonstrated that light-induced mitochondria movement could be spatially controlled at subcellular regions. First, we showed that CRY2/CIB1 heterodimerization can be controlled spatially in the cell co-transfected with CRY2-mCherry and CIB1-GFP-MiroTM (Figure 6A). When blue light was restricted to the circled area, CRY2-mCherry was observed to colocalize with mitochondria only in the circled subcellular area, while CRY2-mCherry remained diffusive and cytosolic in the rest of the cell. After the subcellular recruitment demonstration, the entire cell was subsequently illuminated with blue light and CRY2-mCherry was observed to colocalize with mitochondria throughout the cell, indicating that the previous subcellular recruitment of CIB1-GFP-MiroTM was indeed due to the restricted light illumination.
Figure 6. Subcellular Spatial Control of Light-Induced Mitochondria Movement.
(A) The COS-7 cell was transfected with CRY2-mCherry and CIB1-GFP-MiroTM. Blue light illumination was restricted within a subcellular region of the cell (indicated by the blue circle). Upon blue light activation, CRY2-mCherry was recruited to mitochondria only in the targeted region, whereas it remained cytosolic in the rest of the cell. The local recruitment can be more clearly seen in the zoomed-in images shown in the lower panel. Later, the whole cell was illuminated with blue light and CRY2-mCherry got colocalized with mitochondria in the whole cell (the rightmost image).
(B) In the cell co-expressing CRY2-mCherry-MiroTM and KIF5A-GFP-CIB1, only the marked region was illuminated with intermittent blue light for 1 minute at 5-second interval. Only in this region, mitochondria moved outwards to the cell periphery as indicated by the white arrow while in the remaining part of the cell, mitochondria distribution remained mostly unchanged (the middle image). Later, the whole cell was illuminated with intermittent blue light for 15 minutes and mitochondria moved out towards cell periphery throughout the cell as indicated by the white arrows.
Scale bars, 10µm.
Next, we demonstrated that the distribution of organelles could be spatially controlled by confining the illumination area to subcellular regions. As shown in Figure 6B, the cell was co-transfected with CRY2-mCherry-Miro1TM and KIF5A-GFP-CIB1. Before any blue light illumination, there were few mitochondria in the proximity of cell membrane. Blue light was confined to the marked area and illuminated for 1 minute. Mitochondria in the illuminated area were observed to move out towards cell membrane, while in other areas, mitochondria remained mostly stationary. To confirm that mitochondria in other areas could also be translocated by light, we removed the spatial restriction. After the blue light activated the whole cell for 15 minutes, the outward movement of mitochondria was seen throughout the cell.
DISCUSSION
In this report, we have developed a method to optically control directional transport and redistribution of organelles in living cells by utilizing light-induced CRY2-CIB1 interaction. Light-induced recruitment of kinesins initiates the transport of organelles to microtubule plus-ends (towards the cell edge), while light-induced recruitment of dynein adaptors propels organelles towards microtubule minus-ends (towards the cell nucleus). Light-induced redistributions of multiple types of organelles have been demonstrated, including mitochondria, peroxisome and lysosome. The use of light as a control switch provides unique advantages. First, light-induced CRY2-CIB1 binding is reversible, which is highly desirable for reversible motor recruitment and organelle redistribution. The reversibility also ensures complete termination of light-inducible motor recruitment in the absence of blue light, thus providing an ending point of regulation. Second, light affords highly localized spatial control. We have demonstrated that the organelles in the light illuminated subcellular area became motile while organelles in non-illuminated areas remained unchanged (Figure 6). The spatial control of motor activities and organelle movement would be particularly important in investigating the cargo transport in polarized cells, which are structurally and functionally asymmetric.
In this study, we used CRY2-CIB1 optogenetic system that requires low intensity of blue light and would not induce significant phototoxicity for long-term studies. As we have shown previously, CRY2-CIB1 can dimerize at light intensity as low as 25 µW/cm2 and 36-hour continuous illumination of light at 200 µW/cm2 induced minimal phototoxicity to PC12 cells (Zhang et al., 2014). We have carried out preliminary test of the suitability of our method for long-term investigation of organelle redistribution. When COS-7 cells transfected with either CRY2-mCherry-MiroTM/BICDN-GFP-CIB1 or CRY2-mCherry-MiroTM/KIF5A-GFP-CIB1 were placed under 150 µW/cm2 blue light for 24 hours, the death rates for cells with sustained molecular motor recruitments were similar with those for dark controls (Figure S6). The suitability for long-term experiments would be particularly useful in studying the role of organelle redistributions in cell events that proceed over a relatively long span of time, such as development of cell polarity and elongation of axons which usually take days to complete.
It is worth noting that CRY2 undergoes homo-oligomerization in addition to CRY2-CIB1 heterodimerization upon blue light exposure (Bugaj et al., 2013; Kim et al., 2014; Lee et al., 2014), which could potentially complicate the results obtained from CRY2-CIB1 interaction. However, in this work, the CRY2 homo-oligomerization would not interfere with our results. No matter whether organelle-localized CRY2 undergoes homo-oligomerization or not, it does not prevent the binding between CRY2 and CIB1. Therefore, our strategy based on CRY2-CIB1 binding to drive organelles in cells is robust and reproducible.
In this report, in order to control cargo movements, CIB1 was fused to either truncated kinesin or a dynein adaptor protein BICDN. The truncated kinesin is known to be constitutively active, which means they may occupy microtubule binding sites excessively and thus possibly prevent endogenous molecular motors from attaching to microtubules (Cai et al., 2007). By comparison, overexpression of BICDN won't induce competition between transfected proteins and endogenous molecular motors. However, in our experiments, we did not observe any obvious toxicity induced from the overexpression of either plasmid after 2 days.
The work we present here agrees very well with a very recent report that controlling organelle redistribution in cells is feasible through optogenetics (van Bergeijk et al., 2015). The optogenetic systems used in the two studies are different: van Bergeijk et. al. utilized the dimerization of LOV2 and engineered PDZ while our method is based on the binding of CRY2/CIB1. In a benchmarking study that directly compared the LOV and the CRY2/CIB1 systems, CRY2 system showed a lower background activation and less toxicity in some systems (Pathak et al., 2014). Our study also shows that CRY2/CIB1 system is suitable for long-term experiments.
In conclusion, we provide an optogenetic strategy for controlling motor activities and organelle distribution in living cells with reversibility, spatiotemporal control and minimal toxicity. This method will be useful in studying organelle transport and the biological importance of organelle distribution.
SIGNIFICANCE
Increasing evidence indicate that organelle transport and distribution plays important roles in various cellular activities. However, establishing a direct link between organelle distribution and cellular functions is hindered by the lack of means to manipulate molecular motors in the complex intracellular environment. Here we provide an optogenetic strategy to control the transport and distribution of organelles by light, in which blue light induces recruitment of molecular motors onto organelles based on the heterodimerization of Arabidopsis thaliana cryptochrome 2 and CIB1. This optical approach has outstanding advantages, such as minimal side effects, reversibility and precise spatiotemporal control. In this article, we demonstrated that various organelles, including mitochondria, peroxisomes and lysosomes, can be driven towards the cell periphery or towards the cell nucleus upon recruitment of specific molecular motors. We confirmed that light-recruited molecular motors move along microtubules. It was also shown that the light-inducible motor recruitment and organelle movements are repeatable and reversible. We also demonstrated that organelle redistribution can be spatially controlled at subcellular regions. In summary, the optogenetic strategy in this report offers precise and reversible light control of molecular motors and organelle transport in vivo, thus providing a valuable tool to unveil the role of distributions of various organelles in many cellular functions.
EXPERIMENTAL PROCEDURES
Plasmid Construction
All the plasmids used in this study were cloned in the mammalian expression vector pEGFPN1 or pmCherryC1. In this paper, light-induced dimerization was based on a truncated CRY2 consisting of the photolyase homology region alone and an N-terminal region of CIB1. Details of all DNA plasmids used in this work are summarized in Table S1A.
GFP-CIB1, CIB1-GFP and mCherry-CRY2 were firstly made by ligation and used as templates for construction of other plasmids that contain these genes. CRY2-mCherry-Miro1TM was made by inserting the transmembrane domain of Miro1 into CRY2-mCherry using InFusion cloning kit (Clontech, Mountain View, CA, USA). LAMP-mCherry-CRY2 and PEX-mCherry-CRY2 were constructed by inserting LAMP or PEX into mCherry-CRY2 using ligation and InFusion respectively. KIF5A segment was inserted into GFP-CIB1 to make KIF5A-GFP-CIB1 by ligation. GFP-BICDN-CIB1 was made by inserting BICDN into GFP-CIB1 by InFusion. CIB1-GFP-MiroTM was constructed by inserting MiroTM into CIB1-GFP-Caax by 2-step overlapping extension PCR (Bryksin and Matsumura, 2010). The details for plasmid construction in this paper are summarized in Table S1B.
Cell Culture and Transfection
COS-7 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. All the cell cultures were maintained in a standard humidified incubator at 37°C with 5% CO2. 1–2 days before transfection, CO7 cells were plated on PLL-coated coverslip and allowed to reach 80% confluency. Cells were transfected with desired DNA plasmids using Lipofectamine 2000 according to manufacturer’s protocol. The transfected cells were allowed to recover and to express the desired proteins. Fluorescence imaging of the transfected cells was carried out one day after transfection.
Live Cell Imaging
Live cell imaging was performed on an epi-fluorescence microscope (Leica DMI6000B microscope) equipped with an on-stage CO2 incubation chamber (Tokai Hit GM-8000) and a motorized stage (Prior, Cambridge). Cells plated on coverslips were maintained in standard medium at 37°C with 5% CO2 during imaging. An adaptive focus control was used to actively keep the image in focus for long-term imaging. A light-emitting diode (LED) light source (Lumencor Sola, Beaverton, OR) was used for fluorescence light source. For blue light stimulation, pulsed blue light (200-ms pulse duration per 10-second interval at 9.7 W/cm2) was used for initiating CRY2-CIB1 binding and GFP imaging unless noted otherwise. For mCherry imaging, pulsed green light (200-ms pulse duration) was used at intervals of 10 seconds or 1 second depending on frame rate of the movie acquisition. The green light used for imaging mCherry does not stimulate CRY2-CIB1 binding, confirming previous studies (Kennedy et al., 2010). Fluorescence signal from GFP was detected using the commercial GFP filter cube (Leica, excitation filter 472/30, dichroic mirror 495, emission filter 520/35); fluorescence signal from mCherry was detected using the commercial Texas Red filter cube (Leica, excitation filter 560/40, dichroic mirror 595, emission filter 645/75). All the images were acquired with an oil-immersion 100× objective (Leica, HCX PL APL, n.a. 1.4) and imaged with a sensitive CMOS camera (PCO.EDGE 5.5, PCO, Kelheim, Germany).
Quantitative Determination of Mitochondria Distribution from Cell Nucleus
Mitochondria were labelled by CRY2-mCherry-MiroTM. Using ImageJ, double-transfected cells were extracted from the original images and pasted to a black background in order to eliminate signal interference from neighboring cells. As shown in Figure S3, a ring that just covered the nucleus was picked and defined as the nucleus in measurement. Using a custom written Matlab program, CRY2-mCherry-MiroTM fluorescence signal was segmented into nucleocentric rings of equal width from the nucleus toward the periphery of the cells (Figure S3). The total intensity within each ring indicated the abundance of mitochondria located in that ring. The percentage of mitochondria within a certain ring pl was calculated as the ratio of the total intensity within that ring versus the total intensity of the whole cell, where the total intensity of the whole cell did not include the intensity inside the nuclear ring. The average distance of all the mitochondria in the cell with respect to the nucleus was calculated as the weighted distance . l was calculated as the difference between the radius of the nucleocentric ring and that of the nuclear ring.
Determination of KIF5A-GFP-CIB1 and GFP-BICDN-CIB1 Expression Level
Expression levels for GFP-BICDN-CIB1 and KIF5A-GFP-CIB1 were proportional to the GFP fluorescence intensity. Imaging conditions were kept consistent for GFP signal capture. After manually defining the whole cell area and selecting a background area, ImageJ “Measurement” function was used to measure the average intensity of the GFP fluorescence signal in a transfected cell Icell and that of the background Ibackground. The expression level was calculated as the ratio of GFP intensity inside the cell over the background intensity of the image (Icell/Ibackground).
Analysis of CRY2-CIB1 Binding Kinetics
For kinetic analysis of CRY2-CIB1 dimerization on organelle membrane, an individual mitochondrion was manually selected. The average mCherry fluorescence intensity of the selected mitochondrion was measured. The mCherry signal increases as the blue light recruits mCherry-CRY2 to the surface of mitochondria until the recruitment saturates at Imax. Then, the blue light illumination was stopped and the mCherry intensity will decrease as the CRY2-CIB1 dissociate in the absence of blue light until the dissociation is complete at Imin. The time-dependent binding percentage of CRY2-CIB1 was calculated as the ratio of the difference between mCherry intensity at time t I(t) and the minimum intensity to the difference between maximum mCherry intensity and minimum intensity: .
Cell Cultures and LED Array Setup for Long-Term Light Stimulation
COS-7 cells were plated in 6-well plate one day before transfection. 6 hours after transfection, cells in each well were split into three 12-well plates, one well per plate. In this way, we got three sets of COS-7 cells with the same cell passage number and under identical transfection conditions. Before next step, the cells were allowed to recover in dark for 12 hours.
For long-term blue light illumination setup, a 2-by-2 blue LED array was built by assembling 4 blue LEDs (B4304H96, Linrose Electronics) on a breadboard. The LED device was controlled by a LabVIEW program with a data acquisition board (National Instrument-DAQ, PCI-6035E). The LEDs were supplied with user-defined DC voltages that were controlled through the LabVIEW program. The breadboard was kept in an aluminum box. A light diffuser film was placed on top of the LED array to make the light intensity homogeneous. The average light intensity illuminated to cell cultures was measured by a power meter (Newark, 1931-C). In this study, the light intensity in LED array was set at 150 µW/cm2.
Data Acquisition and Analysis of Cell Death Rate
Using the COS-7 cell culturing method mentioned above, two 12-well pates were prepared each of which contained a set of two co-transfection groups. The two pairs of plasmids for transfection were CRY2-mCherry-MiroTM&BICDN-GFP-CIB1, CRY2-mCherry-MiroTM&KIF5A-GFP-CIB1. For cell death measurement, one plate was placed on the blue LED array for 24-hour continuous blue light illumination before imaging. The other plate was incubated in dark for 24 hours and imaged as the 24-hour dark control.
For cell death measurement, cells were incubated in DMEM containing 1µg/ml propidium iodide (PI) for 20 minutes. After washing with PBS twice, cells were changed to DMEM for imaging. Using the Leica on-stage automatic setup, an area close to the center of the well was imaged and 36 frames of images were acquired for both GFP and mCherry channels. For the mCherry images, CRY2-mCherry-MiroTM signal was so weak compared with PI signal that it can be filtered off by adjusting the image contrast. For almost all transfected cells, cells were co-transfected other than single-transfected. Thus, the GFP signal from BICDN-GFP-CIB1 or KIF5A-GFP-CIB1 was used to find co-transfected cells. GFP images and mCherry images were merged using ImageJ. Cells with PI stains inside nucleus were considered as dead cells. Otherwise, they were considered as living cells. Dead cells and living cells were counted using “CellCounter” function in ImageJ. Only transfected cells were counted. Cell death ratio was calculated as the number of dead transfected cells over the total number of transfected cells.
Supplementary Material
Highlights.
We developed an optogenetic tool to control organelle distributions in cells
Motor proteins can be recruited to organelle membrane by dimerization of CRY2-CIB1
Organelles can be driven towards cell nucleus or towards cell periphery by light
Light-induced organelle transport can be spatially controlled at subcellular region
ACKNOWLEDGEMENTS
We thank Dr. Chandra Tucker (University of Colorado Denver) for providing CIB1-GFP-Caax and CRY2-mCherry; we thank Dr. Xinnan Wang (Stanford University) for providing Miro1 plasmid; we thank Dr. Casper Hoogenraad (Utrecht University) for providing plasmids encoding KIF5A, BICDN and PEX. We also thank Zhuoluo Feng (Stanford University) for his help in constructing the controllable blue LED array. This work was supported by the US National Institute of Health (DP2-NS082125) and a Packard fellowship in Science and Engineering.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
L.D., D.C., B.C. conceived and designed the experiments. L.D. made the plasmids. L.D., D.C., K.Z., Q.O., S.G. performed the experiments. L.D. analyzed the data. D.C. wrote the Matlab program. L.D., D.C., B.C. wrote the paper.
REFERENCES
- Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF, Gillespie MN. Perinuclear Mitochondrial Clustering Creates an Oxidant-Rich Nuclear Domain Required for Hypoxia-Induced Transcription. Sci Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Bba-Mol Cell Res. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. Neuronal polarity: Vectorial cytoplasmic flow precedes axon formation. Neuron. 1997;19:1175–1186. doi: 10.1016/s0896-6273(00)80410-9. [DOI] [PubMed] [Google Scholar]
- Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- Cai DW, Hoppe AD, Swanson JA, Verhey KJ. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J Cell Biol. 2007;176:51–63. doi: 10.1083/jcb.200605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JW, Cha Y, Yuksel KU, Gracy RW, August JT. Isolation and Sequencing of a Cdna Clone Encoding Lysosomal Membrane Glycoprotein Mouse Lamp-1 - Sequence Similarity to Proteins Bearing Onco-Differentiation Antigens. J Biol Chem. 1988;263:8754–8758. [PubMed] [Google Scholar]
- Fransson A, Ruusala A, Aspenstom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Bioph Res Co. 2006;344:500–510. doi: 10.1016/j.bbrc.2006.03.163. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. P Natl Acad Sci USA. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumoto T, Nakata T. Optogenetic Control of PIP3: PIP3 Is Sufficient to Induce the Actin-Based Active Part of Growth Cones and Is Regulated via Endocytosis. PLoS One. 2013;8:e70861. doi: 10.1371/journal.pone.0070861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer S, Holzinger A, Welsch U, Roscher AA. Cloning and characterization of the gene encoding the human peroxisomal assembly protein Pex3p. Febs Lett. 1998;429:53–60. doi: 10.1016/s0014-5793(98)00557-2. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, Kuijpers M, Wulf PS, van Spronsen M, MacKintosh FC, Hoogenraad CC. Mixed Microtubules Steer Dynein-Driven Cargo Transport into Dendrites. Curr Biol. 2010a;20:290–299. doi: 10.1016/j.cub.2009.12.052. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, van der Zwan WA, Wulf PS, Keijzer N, Hoogenraad CC. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys J. 2010b;99:2143–2152. doi: 10.1016/j.bpj.2010.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–U948. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Kim JM, Lee M, Kim CY, Chang KY, Heo WD. Spatiotemporal Control of Fibroblast Growth Factor Receptor Signals by Blue Light. Chem Biol. 2014;21:903–912. doi: 10.1016/j.chembiol.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Kinesin and dynein move a peroxisome in vivo: A tug-of-war or coordinated movement? Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- Lee S, Park H, Kyung T, Kim NY, Kim S, Kim J, Heo WD. Reversible protein inactivation by optogenetic trapping in cells. Nat Methods. 2014;11:633–636. doi: 10.1038/nmeth.2940. [DOI] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Bio. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak GP, Strickland D, Vrana JD, Tucker CL. Benchmarking of optical dimerizer systems. ACS Synth Biol. 2014;3:832–838. doi: 10.1021/sb500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana A, Schwarz EC, Schwindling C, Lipp P, Kaestner L, Hoth M. Sustained activity of calcium release-activated calcium channels requires translocation of mitochondria to the plasma membrane. J Biol Chem. 2006;281:40302–40309. doi: 10.1074/jbc.M607896200. [DOI] [PubMed] [Google Scholar]
- Renicke C, Schuster D, Usherenko S, Essen LO, Taxis C. A LOV2 Domain-Based Optogenetic Tool to Control Protein Degradation and Cellular Function. Chem Biol. 2013;20:619–626. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi GM, Mengel T, Walsh F, Kadenbach B, Dimauro S, Schon EA. A Gene Specifying Subunit-Viii of Human Cytochrome-C Oxidase Is Localized to Chromosome-11 and Is Expressed in Both Muscle and Non-Muscle Tissues. J Biol Chem. 1989;264:10595–10600. [PubMed] [Google Scholar]
- Samsonov A, Yu JZ, Rasenick M, Popov SV. Tau interaction with microtubules in vivo. J Cell Sci. 2004;117:6129–6141. doi: 10.1242/jcs.01531. [DOI] [PubMed] [Google Scholar]
- Schwindling C, Quintana A, Krause E, Hoth M. Mitochondria Positioning Controls Local Calcium Influx in T Cells. J Immunol. 2010;184:184–190. doi: 10.4049/jimmunol.0902872. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–U392. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D, Yao XL, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7:623–U618. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA. Using Optogenetics to Interrogate the Dynamic Control of Signal Transmission by the Ras/Erk Module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergeijk P, Adrian M, Hoogenraad CC, Kapitein LC. Optogenetic control of organelle transport and positioning. Nature. 2015;518:111–114. doi: 10.1038/nature14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M, Mikhaylova M, Lipka J, Schlager MA, van den Heuve DJ, Kuijpers M, Wulf PS, Keijzer N, Demmers J, Kapitein LC, et al. TRAK/Milton Motor-Adaptor Proteins Steer Mitochondrial Trafficking to Axons and Dendrites. Neuron. 2013;77:485–502. doi: 10.1016/j.neuron.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–U111. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Duan L, Ong Q, Lin Z, Varman PM, Sung K, Cui B. Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLoS One. 2014;9:e92917. doi: 10.1371/journal.pone.0092917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.