Abstract
Baseline physiological function of the mammalian heart is under the constant threat of environmental or intrinsic pathological insults. Cardiomyocyte proteins are thus subject to unremitting pressure to function optimally and this depends upon them assuming and maintaining proper conformation. This review explores the multiple defenses a cell may employ for its proteins to assume and maintain correct protein folding and conformation. There are multiple quality control mechanisms to ensure that nascent polypeptides are properly folded and mature proteins maintain their functional conformation. When proteins do misfold, either in the face of normal or pathologic stimuli or because of intrinsic mutations or post-translational modifications, they must either be refolded correctly or recycled. In the absence of these corrective processes, they may become toxic to the cell. Herein, we explore some of the underlying mechanisms that lead to proteotoxicity. The continued presence and chronic accumulation of misfolded or unfolded proteins can be disastrous in cardiomyocytes as these misfolded proteins can lead to aggregation or the formation of soluble peptides that are proteotoxic. This in turn leads to compromised protein quality control and precipitating a downward spiral of the cell's ability to maintain protein homeostasis. Some underlying mechanisms are discussed and the therapeutic potential of interfering with proteotoxicity in the heart is explored.
Keywords: Autophagy, proteasome, protein
Introduction: Proteotoxicity
Protein homeostasis, or proteostasis, plays an essential role in maintaining overall cellular health. Perturbations to proteostasis must be adeptly controlled to avoid pathological consequences. In the absence of external or cell autonomous compensatory mechanisms, alterations to proteostasis the cell's overall health may rapidly deteriorate, leading to necrosis, apoptosis or caspase-independent cell death, all of which appear to involve regulated cellular and molecular programs.1,2
Many factors can affect protein folding and misfolding, but the cellular pathogenic aspects of protein misfolding and aggregation are collectively termed “proteotoxicity.” The term is relatively recent, with the first references in the cited literature occurring in a 1991 meeting report and was coined to “describe damage to proteins caused by chemical and physical agents.”3 The concept of proteotoxicity was firmly established over the next 10-20 years;4 broadly defined, it is any impairment of cellular function as a result of protein misfolding or aggregation. Its importance has been emphasized and remains a particular focus of study for various neurodegenerative diseases in which protein misfolding and the occurrence of proteinaceous aggregates is a recurring theme.5-8 Proteotoxicity is also a normal occurrence during the aging process (a universal risk factor) and indeed, many of the proteins that function in maintaining proteostasis4 have been implicated in cellular aging,9 offering an intriguing rationale for the age-dependent onset of many protein conformation dependent diseases, including some of the neurodegenerative pathologies.10
The general importance and prevalence of protein misfolding and its impact on human disease was illustrated by the first mechanistic understanding of an inherited disease, sickle cell anemia when, in 1957-1959, Ingram and Hunt showed that a glutamate to valine mutation in hemoglobin's β-globulin chain resulted in a conformational change in the protein and aberrant polymerization.11,12 However, it rapidly became apparent that there is no single resultant pathogenic mechanism that occurs as a result of protein folding.13 Rather, the resultant pathologies are protein-specific: 1) improper folding or maintenance of structure can lead to a protein's improper degradation, such as occurs in cystic fibrosis (cystic fibrosis transmembrane conductance regulator protein) and Gaucher disease (β-glucosidase),14,15; 2) a dominant negative mutation that leads to a hypo-functional peptide, which occurs in epidermolysis bullosa simplex (keratin),16; 3) a gain of toxicity function, such as occurs with certain variants of APOE4 in Alzheimer disease (AD),17; and 4) the accumulation of multiple, misfolded species resulting in formation of toxic polypeptides and aggregates, such as occurs with many of the neurodegenerative diseases.18,19
Thus, protein misfolding disorders are complex, multi-factorial and can be cell autonomous or non-cell autonomous with neighboring cells affected.20 In this review, we necessarily focus on a subset of processes that underlie an important part of proteostasis in the heart.
A Network of Cellular Defense
Cells possess innate mechanisms to sense protein unfolding/misfolding and restore normal conformations. The mechanisms that comprise cellular Protein Quality Control (PQC) are complex and multifaceted, consisting not only of constitutive and inducible chaperones (discussed below), but also engaging compartment-specific mechanisms. But even PQC represents only a small part of the overall process of proteostasis, which can be portrayed as an entire landscape, or all-encompassing network, that takes into account all aspects of the processes that underlie the cell's stable or unstable protein complements. Thus, transcriptional, translational and post-translational regulation all form a part of this network as do the dynamic processes of folding, unfolding, post-translational modifications, trafficking and finally, degradation. Rather than being static, proteostasis itself changes over time and is thought to break down as the organism ages.4 While the network can often handle processes that lead to damaged proteins by subjecting these peptides to various facets of the PQC such as degradation via the proteasome,21 eventually the cell can be overwhelmed, either as part of the normal aging process or, earlier in life, as a part of a disease process. Thus, an aberrantly misfolded protein that is competent to provoke pathogenesis may be tolerated for years or even decades, but as the overall competence of the proteostasis network declines as a result of age or some other extrinsic or intrinsic stimulus, the pathology begins to present clinically. Consistent with this view is the compromised ability of aged cells to maintain proteins in their proper conformation,22 as well as compromised PQC.23 Thus, an aging cell, particularly one that does not divide, may be faced with an ever-increasing burden of protein misfolding in the face of compromised systems for decreasing the pathogenic load.
That protein misfolding can lead to cardiovascular disease has been known for many years; transthyretin amyloidosis, caused by the by-product of transthyretin tetramer disassembly and the subsequent extracellular deposition of the resulting amyloid fibrils, has been well characterized24 and extensively reviewed.5,25,26 However, only recently have cardiomyocyte-based proteotoxic entities become a focus of mechanistic studies, which has led to the realization that proteotoxicity may represent a convergent pathway in heart disease and the development of heart failure.27-29 In this review, we will consider the particular sensitivity of the cardiomyocyte cell population to proteotoxic insult and introduce the relevant pathways that constitute aspects of the cardiomyocyte's PQC response. Some specific proteotoxic stimuli will be considered and the cardiomyocyte's ability to respond to these insults will be outlined, as well as the pathogenic sequelae that can ensue. Finally, we will consider whether and how our developing understanding of these processes might be translated into new therapeutic initiatives for decreasing cardiac morbidity and improving clinical outcomes.
Protective Pathways
Preventing the accumulation of misfolded proteins is the job of every cell in an organism. An estimated 30% of translated proteins never reach their final cellular destination,30 and these newly synthesized nascent proteins are recognized and degraded almost immediately after translation; degradation of some proteins occurs co-translationally. This requires constant monitoring of protein fidelity, and has led to evolutionarily conserved, efficient mechanisms of protein degradation to keep misfolded proteins from exerting cytotoxic effects. However, the risk of proteotoxic insult is not equal among all cell types. Not surprisingly, post-mitotic cells appear to be particularly susceptible to proteotoxic effects. When faced with a proteotoxic insult, mitotic cells can asymmetrically divide the misfolded protein load, thus protecting one daughter cell while sacrificing the other.31 Post-mitotic cells do not have this luxury; the relative inability of these cells to regenerate puts the entire organ at increased risk of proteotoxic threats, as the cells cannot readily divide to replace cells that are lost to injury. In this respect, the cardiomyocyte is similar to another post-mitotic cell in a vital organ: the neuron.32 These two cell types are the most susceptible to proteotoxic insults; neuronal cell death can result in neurological dysfunction and neurodegeneration,33 just as loss of cardiomyocytes due to cell death leads to cardiomyopathy and heart failure. Indeed, recent data have strengthened the idea that cardiomyocyte proliferation and regeneration of the adult heart is a rare occurrence34-36 as is the case for human neocortical neurons.32
Proteotoxic insults can lead to cell death and dysfunction, and give rise to severe phenotypes in tissues such as the brain and heart. The inability of a cell to divide its way out of trouble has led to the evolutionary development of sophisticated, multi-tiered and interconnected mechanisms of PQC; these have evolved to maintain protein fidelity in the face of intrinsic and extrinsic insults and prevent misfolded proteins from accumulating. Among these are the chaperones, which include the heat shock proteins (HSP), the ubiquitin proteasome system (UPS), the autophagy-lysosome pathway and the ER-associated degradation (ERAD) stress response (Figure 1). These four pathways comprise the cellular PQC response. Together, they normally ensure that the balance of protein synthesis to protein degradation is maintained,37 functioning independently and in concert with one another to recognize and eliminate proteotoxic threats.
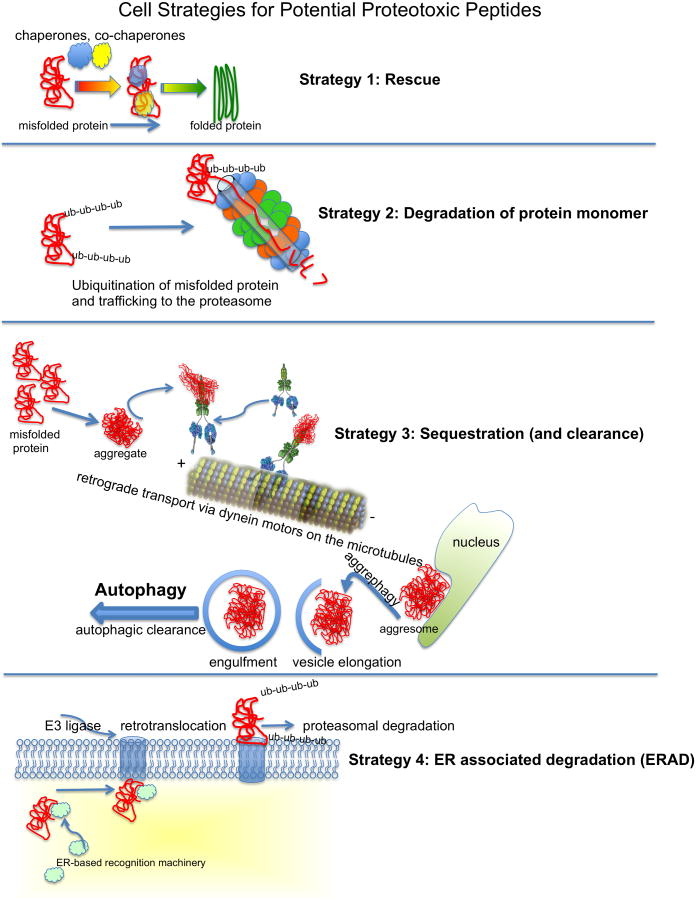
Figure 1.
Schematic diagram of selected protection strategies or proteotoxic pathways in the cardiomyocyte. Figure 1. Schematic diagram of selected protection strategies in the cardiomyocyte for dealing with misfolded or potentially proteotoxic peptides. Strategy 1) [Rescue], the cell's chaperones, which can be constitutively expressed or inducible, act alone or in concert to maintain or restore the peptide's functional conformation. Strategy 2) [Degradation], the misfolded protein is recognized by the Protein Quality Control machinery, ubiquitinated and trafficked to the proteasome for degradation and amino acid recycling. Strategy 3) [Sequestration], the misfolded protein forms larger aggregates, which are unable to be processed by the proteasome. These aggregates can be attached to dynein motors and transported on microtubules to a perinuclear location where the potentially proteotoxic peptides are sequestered. These aggregates can be cleared from the cytoplasm by autophagy (see text for details).89,109,110 Strategy 4) [ERAD] The endoplasmic reticulum associated degradation pathway is quite complex and a simplified schematic diagram simply outlines the process. Mutated or terminally misfolded proteins are recognzied by ER assocaited proteins that may have either enzymatic or chaperone activities. The protein is then sheparhaerded to the retrotranslocation machinery whoese exact identity(ies) remain obscure. Membrane associated E3 ligases and associated proteins are able to mediate ubiquitination of the protein, which may partially drive its retrotranslocation. There are multiple checkpoints and components underlying these processes as well (see text for details). The ubiquitinated proteins are then targeted for proteasomal degradation.
The Heat Shock Response
Chaperones represent a first line of defense in the post-translational processes that conserve protein tertiary and quaternary structure and conformation. Chaperones are constitutively expressed, but the expression of a small subset can also be upregulated in response to unfolded or misfolded protein accumulations; this response represents part of the heat shock response (HSR).38,39 The molecular identities, functions and genetics of HSPs were first rigorously explored using the power of fly genetics.40 As the field has grown and matured over the past 35 years, the importance of the chaperones and their essential role in the correct folding of multiple proteins has become well established.1 The chaperones are represented by multiple classes and have diverse natures.41 Their many functional roles are underscored by changes they may undergo as a result of post-translational modification,42 the potential for their trans-cellular activities43 and the role they play in a wide variety diseases including but not limited to Pompe disease, Huntington disease, cancer, pulmonary and cardiovascular disease.6,44-51 Not surprisingly, considering their role in these processes, targeting the chaperones has become a legitimate therapeutic avenue.49,52
The HSR is the most evolutionarily ancient of the PQC pathways, enlisting molecular chaperones (Figure 1, top panel), such as the HSPs, to manage two primary functions within the cell: transiently interact with nascent unfolded polypeptides to guide them towards their proper folded conformation and target terminally misfolded proteins for degradation. Many or even a majority of proteins cannot spontaneously fold to assume a functional conformation; during or immediately after translation on the polysomes, exposed hydrophobic stretches are prone to aggregate in the aqueous environment of the cytoplasm. HSPs bind to these hydrophobic stretches and prevent them from aggregating before the proper folded conformation can be attained.53 A primary driver of the HSR is heat shock factor-1 (HSF-1), a powerful transcription factor that is normally bound in an inactive complex in the cytosol containing other stress-responsive proteins such as Hsp90 and histone deacetylase (HDAC)6.54 Misfolded protein stress facilitates dissolution of this complex and HSF-1 is translocated into the nucleus and induces expression of other HSPs such as Hsp70 and Hsp40. In the heart, chaperones are critical for ensuring that the proteins of the contractile apparatus arrive at their proper folded conformation and location within the cell. For example, αB-crystallin (CryAB), a member of the small HSP family, ensures proper folding of desmin, an intermediate filament protein with critical roles in cardiomyocyte structure and function.55,56
The Ubiquitin-Proteasome System
The second line of defense in the PQC response is the ubiquitin-proteasome system (UPS) (Figure 1, panel 2). The UPS mediates the turnover of most cellular proteins, as well as degrading terminally misfolded proteins, via proteasomal proteolysis. The proteasome is a large, multi-subunit complex tasked with the degradation of monomeric proteins; the majority of misfolded protein species are eliminated from the cell by way of the proteasome.57 Proteins destined for degradation via the proteasome are poly-ubiquitinated by a cadre of enzymes (E1, E2 and E3 enzymes),57 which engage free ubiquitin and covalently conjugate ubiquitin via a K48 linkage to the target protein substrate.58 Multiple rounds of this process occur until a poly-ubiquitin chain is attached to the substrate, which is then recognized by transport proteins and trafficked to the proteasome. The proteasome contains a cavity, or bore, that is equipped with a variety of proteases, which have cleavage activities similar to trypsin, chymotrypsin and caspases, providing the ability to non-selectively degrade most cellular proteins. The fidelity of this process is essential to cellular homeostasis; malfunction of proteasomal proteolysis will lead to misfolded protein accumulation and cellular dysfunction.57 In fact, proteasomal inhibition as a therapy for selected cancers is being intensively studied, with the goal of inducing proteotoxicity-induced cell death in the metastasizing cell population.59 However, the efficacy towards cancer cells comes at the expense of the heart,60 possibly due to the same induced proteotoxicity.
The Autophagy-Lysosome Pathway
Recent work suggests that the autophagy-lysosome pathway plays a major role in the cardiac stress response61 and here we consider it in some detail. Macroautophagy (hereafter referred to as autophagy) involves a budding membrane called a phagophore, which envelopes cargo destined for degradation into a maturing double-membrane vesicle, the autophagosome. The autophagosome travels along microtubules and fuses with lysosomes, forming a distinct structure, the autolysosome. There, the vesicle is acidified and lysosomal enzymes degrade the enclosed cargo. Membrane components to form these vesicles come from diverse cellular locations such as the plasma membrane, mitochondria, golgi and the ER with multiple proteins precisely controlling all aspects of the process.62 For a complete description of the autophagic process and its relevance in cardiovascular biology, we direct the reader to a recent comprehensive set of reviews on the topic.62-65
Under basal conditions, autophagy functions at low levels, helping the cell to remodel when necessary and generally functions to maintain a healthy homeostatic balance. However, autophagy can be quickly induced under conditions of cellular stress.66 For example, subjecting cardiomyocytes to starvation, via either calorie deprivation or fasting in mice or serum/glucose deprivation in vitro, results in a rapid upregulation of autophagic flux.67 This allows cellular components to be degraded and maintains the pool of free amino acids so that the nutrient and metabolic demands of the cell continue to be satisfied. Indeed, autophagy has been shown to be necessary in all stages of life; for example, autophagy is markedly upregulated in neonates during the latent period between birth and establishment of the maternal milk supply.68 Ablation of the autophagy gene atg5, which encodes an E3 ubiquitin ligase that plays an essential role in autophagosome formation, resulted in neonatal lethality within one day after birth, an effect that could be prevented by forced milk feeding.68 Autophagy is also reduced in aging, suggesting that a deficit in the breakdown of damaged proteins and organelles may contribute to the general aging process.69
Autophagy is essential for normal heart maintenance and function, and defects in any part of the autophagic process can lead to cardiomyopathy. Deletion of atg5 in the adult mouse led to increased pathological hypertrophic signaling, the development of hypertrophy and cardiac dysfunction as well as increased cell death.61 Induction of parallel PQC mechanisms, such as proteasomal activity and ER stress markers, was also observed. These pathways may be induced in order to compensate for loss of autophagy, although the response clearly could not fully mitigate the damage. These data are underscored by deficits in the autophagic pathways being shown to directly cause human cardiac disease. For example, a deficiency in the lysosomal membrane protein Lamp2 causes Danon disease, a severe cardiomyopathy.70 Lamp2 knockout mice show significant accumulations of autophagic vacuoles, implying a deficit in lysosomal degradation of autophagic cargo, a phenotype that leads to increased mortality and cardiac dysfunction.71
In addition to the role played in degrading organelles and pathogens, autophagy is the primary pathway for degrading and recycling long-lived proteins, and particularly those that form larger aggregates that cannot be processed through the proteasomal bore. In some cases, these aggregates can actively form by a process that was well characterized in the neurodegenerative diseases, in which small proto-aggregates or misfolded polypeptides are actively transported on dynein motors retrogradely via microtubules to a perinuclear location to form proteinaceous bodies known as aggresomes (Figure 1, panel 3).72 As autophagy is the only direct mechanism that can degrade these large, misfolded protein aggregates within cells, this pathway is particularly consequential in combating proteotoxic phenotypes.
Endoplasmic Reticulum Associated Degradation (ERAD) and Stress Response
The ER stress response, also referred to as the unfolded protein response, is another important PQC mechanism in the heart. This pathway plays a critical role in maintaining proteostasis, as it serves as a cellular monitor of misfolded protein load and, through induction of ER stress genes, tailor the cellular homeostatic balance in response to increased levels of misfolded protein in the ER. Upon sensing changes to the misfolded protein load in the ER/SR micro-environment, three transmembrane ER proteins (PERK, IRE1 and ATF6) are activated. Activation stimulates nuclear translocation of transcription factors ATF4, ATF6 and XBP1, which upregulate the proteins that comprise the ER stress response, including chaperones and other proteins involved in misfolded protein stress. When a protein misfolds and cannot be rescued, these ER stress response genes efficiently target aberrantly-folded proteins for degradation through the mechanism of ER-associated degradation (ERAD) (Figure 1, panel 4) in which multiple ER lumen or ER membrane associated proteins bind to the misfolded protein, the end result being export of the peptide into the cytoplasm where it is ubiquitinated and targeted to the proteasome or for chaperone-assisted refolding. This process is crucial for dealing with terminally misfolded proteins and preventing their accumulation. Unsurprisingly, ER stress is induced with cardiac injury including ischemia, pathological hypertrophy and heart failure, and helps to manage the cellular protein imbalance induced by these conditions in an attempt to mitigate cellular damage.73
Recent data have linked ER stress to autophagy activation,74,75 suggesting the two pathways might work in concert in a cardioprotective mechanism designed to restore homeostasis. However, the intersections, interplay and coordinating control mechanisms between these pathways remain largely obscure. These pathways are indispensable to ensure that the degradative capacity of the cardiomyocyte is met. However, it is important to note that more protein degradation is not protective in all cases. To the contrary, these pathways are only protective if they maintain the appropriate balance of protein synthesis and degradation such that proteostasis is achieved. Unconstrained or inappropriate catabolism can be equally damaging to the heart. For example, the mechanistic target of rapamycin (mTOR) is a negative regulator of autophagy, and the use of mTOR inhibitors can induce autophagy. However, ablation of mTOR leads to unrestrained autophagy and severe cardiac dysfunction.76 Similarly, the chemotherapeutic drug, doxorubicin, can be cardiotoxic.77 Intriguingly, recent data show that doxorubicin can augment proteasomal activity, suggesting that increased proteasomal degradation can also be deleterious to the heart.78,79
Choices: Selective Autophagy
The cell must have efficient mechanisms for selecting the correct, misfolded proteins for either degradation or sequestration. In this section, we focus on the process of autophagy and discuss mechanisms that make it more selective. Autophagy was first thought to be a relatively non-specific process, but we now know that it is highly regulated and cargo directed.80,81 In fact, the process can be quite selective, with specialized pathways existing for specific degradation of old or damaged organelles such as mitochondria, peroxisomes and ribosomes, although the specific players and processes of these pathways are just beginning to be understood.82 This process, known as selective autophagy, uses specific cargo proteins, such as p62 and Nbr1, which bind ubiquitinated proteins and bring them into contact with the protein LC3, which is associated with the autophagosomal membrane, thus targeting ubiquitinated proteins for autophagic degradation (Figure 2).82 For example, upon polyubiquitination of mitochondrial membrane proteins by the E3 ligase, parkin, p62 binding facilitates degradation of the mitochondria by mitophagy. As damaged mitochondria can quickly begin to produce cellular oxidative stress and other inducers of cell death, the fidelity of these processes is crucial to overall cell health. Indeed, similar autophagic mechanisms can also be induced to quickly recognize and eliminate cellular pathogens, thus playing a critical role in the host response to viral infection.83,84
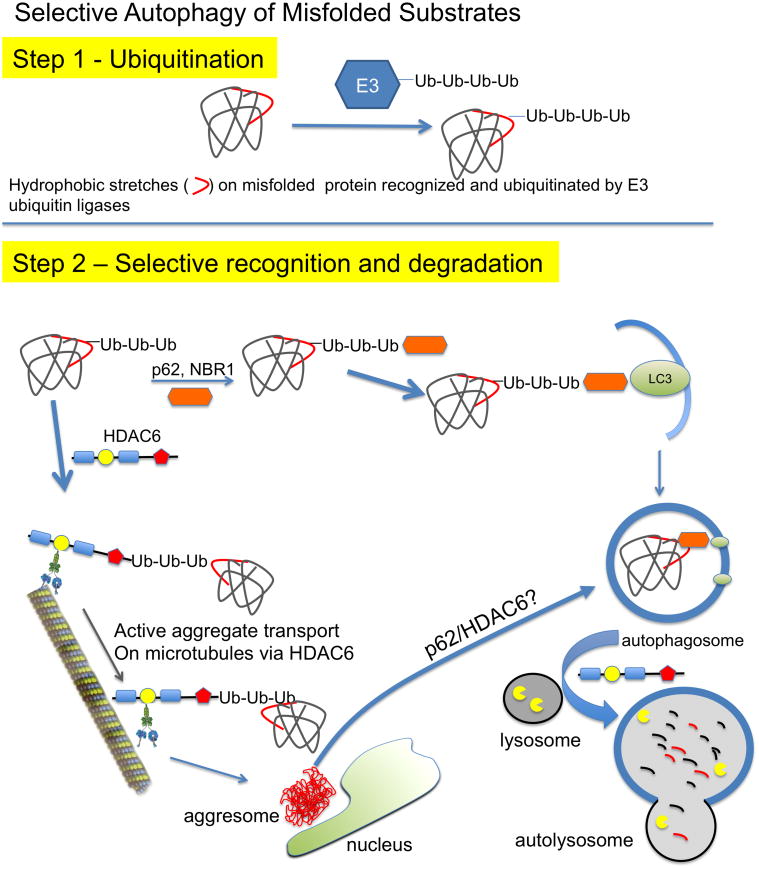
Figure 2.
Selective cargo recognition and degradation by autophagy. Step 1, Ubiquitin binding is necessary for most modes of selective autophagy. Polyubiquitin conjugation onto a substrate protein is accomplished via an E3 ubiquitin ligase. Many of these proteins are ubiquitinated via a K63 linkage, which signals lysosomal degradation rather than the K48 linkage that is typical for proteasomal targeting. Step 2, Ubiquitinated proteins can be recognized by cargo receptors, such as p62, NBR1 and/or HDAC6 for transport towards the lysosome or aggresome. p62 (
 ) binds ubiquitinated proteins for interaction with LC3 in a growing autophagosome, thus selectively targeting the protein for autophagic degradation after fusion with lysosomes. In addition, HDAC6 can bind ubiquitinated proteins through its ubiquitin binding domain (
) binds ubiquitinated proteins for interaction with LC3 in a growing autophagosome, thus selectively targeting the protein for autophagic degradation after fusion with lysosomes. In addition, HDAC6 can bind ubiquitinated proteins through its ubiquitin binding domain (
 ) and transport them in a retrograde fashion along microtubules by binding to the motor protein dynein through the dynein motor binding domain (
) and transport them in a retrograde fashion along microtubules by binding to the motor protein dynein through the dynein motor binding domain (
 ). These can be single misfolded species or soluble proto-aggregates. This event can happen many times, causing the proteins to coalesce and form aggresomes in perinuclear region of the cell. Aggresomal proteins can also be degraded by autophagy, and these cargo transport proteins may be involved in this process as well, particularly HDAC6 with known roles in mediating the autophagosome-lysosome fusion event.
). These can be single misfolded species or soluble proto-aggregates. This event can happen many times, causing the proteins to coalesce and form aggresomes in perinuclear region of the cell. Aggresomal proteins can also be degraded by autophagy, and these cargo transport proteins may be involved in this process as well, particularly HDAC6 with known roles in mediating the autophagosome-lysosome fusion event.
Cargo adapter proteins such as p62 may hold the key to selective autophagy: p62 colocalizes with ubiquitin in punctate structures, and is found in both membrane-bound autophagosomes and protein aggregates, and is degraded by autophagy.85 Interestingly, when p62 levels are reduced, LC3 puncta, indicative of autophagosomes and aggresomal structures are not formed, suggesting that a cargo adapter like p62 is necessary to bind misfolded ubiquitinated structures and transport them for autophagic and/or aggresomal sequestration (Figure 2).85
Zheng et al probed the role of p62 in cardiac proteinopathy and found increased p62 levels in two mouse models that exhibit proteotoxicity asa result of cardiomyocyte specific expression of either a mutated desmin or an associated protein, a mutated α-B crystallin.86 This compensatory induction is likely an attempt to bind ubiquitinated substrates for autophagic degradation, as the aggregates in CryABR120G contain ubiquitinated proteins.87 Indeed, knockdown of p62 not only reduced protein levels of the autophagic marker, LC3-II, but also reduced aggresome formation in a manner that was cytotoxic to the cardiomyocytes.86 These data suggest that p62 is a necessary factor in inducing aggregate formation and autophagic degradation of these aggregates. Indeed, upon Atg7-mediated autophagy induction in CryABR120G hearts, p62 levels increased, which correlated with reduced aggregates and improved cardiac function.88 We speculate this increase in p62 allows more ubiquitinated misfolded protein to be sequestered in autophagosomes by a selective autophagic mechanism, consistent with a hypothesis that aggregate formation process may, in some cases, be protective, which is discussed in detail in the following sections.
In addition to p62, HDAC6 may also be a central player in the cellular response to proteotoxic stress. HDAC6 regulates many aspects of PQC;54 for example, HDAC6 deacetylation of Hsp90 helps to maintain a complex between HDAC6, Hsp90 and HSF1, which serves to repress the HSR; this complex is dissociated in response to proteasomal inhibition.89,90 HDAC6 is central to aggresome formation and, in its absence, aggregate-prone proteins have a reduced ability to form aggresomes, which increases cell death.91 HDAC6 expression rescued neurodegenerative phenotypes in a Drosophila model of polyglutamine expansion in an autophagy-dependent manner. The rescue was abolished in autophagy-deficient flies, 92 consistent with a role for HDAC6 in autophagic degradation.93
Not coincidentally, some of the same proteins involved in selective autophagy appear to contribute to aggresome formation as well,93,94 suggesting that process and autophagic degradation of large aggregates may directly intersect (Figure 2). Recent data bear this out: small aggregates can be degraded by both basal (ie, that which occurs under normal cellular conditions) and induced autophagy by starvation, whereas aggresomal proteins are only degraded under conditions of induced autophagy and not by autophagy that occurs at the basal state.95
Cardiac Proteotoxicity
In light of the potential for varying etiologies and mechanisms, it is not surprising that a unifying mechanism of cardiac proteotoxicity is not yet apparent. At this point, it is clear that protein misfolding in the heart can be either extracellular, as is the case for transthyretin amyloidosis,24,96 or intracellular as occurs in the Desmin Related (Cardio)Myopathies (DRM).97 Diverse pathogenic insults can also lead to proteotoxicity including, but not limited to: chronic synthesis of a compromised protein,87 organ-specific pathogenic stimuli such as cardiac infarction, pressure overload-induced hypertrophy or the production of reactive oxygen species98-100 and generalized systemic insults such as diabetes101-103 or aging.4 These different primary etiologies can have multiple effects on either the production of misfolded proteins, their accumulation in critical cellular compartments or the cell's machinery for decreasing the misfolded protein/protein aggregate load.
Mechanistic Basis for Protein Aggregation Toxicity
Amyloidosis remains the classic manifestation of protein aggregate-based disease but, as many proteins have amyloidogenic potential, its diagnosis is dependent upon general histological criteria, including positive staining with Congo Red, which produces an “apple-green” characteristic appearance when viewed under birefringent microscopy.104 Early on, the discovery that neurodegenerative diseases such as AD were invariably accompanied by the accumulation of amyloid aggregates or plaques prompted the conclusion that the aggregates themselves were toxic but, as murine models of the disease were developed in the recent past, a more nuanced view has emerged.105
As noted above, amyloidosis in the cardiac system is well-characterized, with multiple proteins such as transthyretin, the immunoglobulin light and heavy chains, serum amyloid A, atrial natriuretic factor and a subset of the apolipoproteins confirmed as being amyloidogenic in the heart.106 However, the study of other cardiac diseases that are characterized by the intracellular deposition of proteinaceous aggregates has been instructive as well in determining the pathogenic sequelae associated with abnormal peptide aggregates in the cardiomyocyte. Approximately a decade ago, others and we began an intensive study of an aggregation-prone form of the small HSP-like CryAB and its expression in the heart. Originally identified as disease-causing through the genetic analysis of a French family,107 disease causation of the mutated protein, CryABR120G, was confirmed by its cardiomyocyte-directed expression in transgenic animals.99,108 As is the case in human patients, expression of CryABR120G in the mice led to the gradual development of restrictive, hypertrophic or dilated cardiomyopathy87,109 and was characterized by the formation of large, granulofilamentous protein aggregates in the myoplasm that were easily visible by both light and electron microscopy (Figure 3). Interestingly, the aggresomal and autophagic machinery and transport mechanisms have many common players, and we are beginning to understand a sophisticated mechanism that can recognize and degrade protein aggregates by autophagy, termed aggrephagy. Similar to other forms of selective autophagy, aggrephagy involves recognition of ubiquitinated proteins by cargo adaptors such as p62 and NBR1, which recruit other autophagic proteins such as Atg5-Atg12 and Alfy, a proposed component of the specific aggrephagic response.110,111
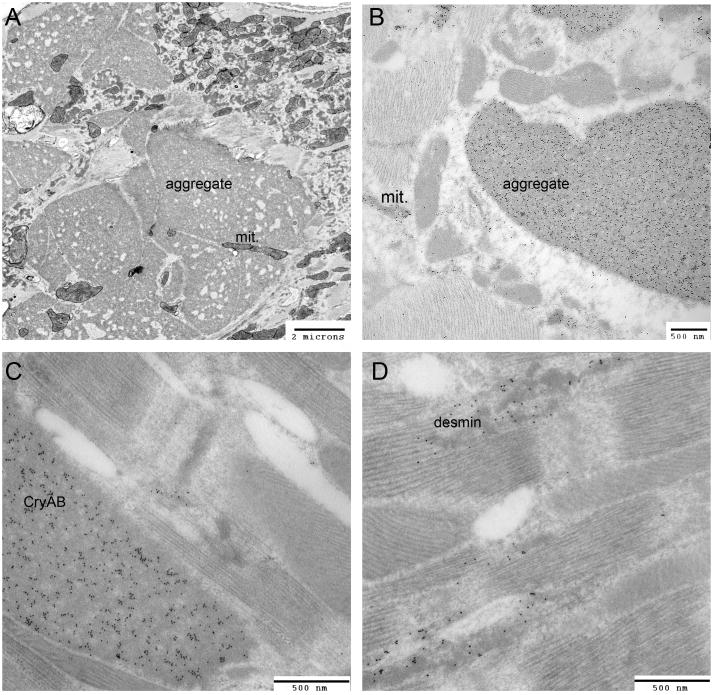
Figure 3.
Anatomy of aggregates. A, Shown is a typical, mature aggregate in a CryABR120G cardiomyocyte derived from a 4 month old, symptomatic heart. Note the fenestrated organization and the mitochondria trapped within the aggregate. B, Immunogold staining (dark grains) showing the strong presence of CryAB within both the large, well defined aggregates and the smaller, more amorphous masses, some of which are mitochondria (mit). C, D, High magnification, immunogold staining using cardiomyocytes derived from 6 week old CryABR120G hearts showing the internalization of CryAB, but also (D) the presence of desmin. mit; mitochondria. Photomicrographs courtesy of H. Osinska. Mice, fixation techniques and details of the transmission electron microscopy were carried out as described.219
Are aggresomes intrinsically toxic? Like many of the amyloid diseases, DRM is caused by a toxic gain-of-function protein, in the sense that it is dominant over the wild type protein and causes aggregates to form even in the presence of near normal amounts of wild type CryAB. For many years, these protein deposits in general, and amyloid plaques in particular, were thought to be the toxic species that caused the clinical disease as, in the different neurodegenerative diseases, they were invariably present in high concentrations upon post-mortem assessment of the affected tissues. But closer examination in animal models that appeared to contain benign amyloid deposits, or even deposits that were posited to have positive phenotypic effects remained a nagging contradiction to this hypothesis.112,113 A variety of experimental avenues from different neurodegenerative diseases including AD and the polyglutamine expansion-based diseases indicated that the visible aggregates were, in fact, not the toxic entity; rather, a soluble, oligomeric precursor that assumed a characteristic conformation might be the primary culprit (Figure 4).4,114,115 Development of an antibody capable of recognizing the conformer, which forms as a result of a folding process that many diverse proteins can undergo,115-118 revealed the presence of the “pre-amyloid oligomers” (PAO) in many protein aggregation-based diseases, including DRM.87 Although the toxic oligomer, as opposed to the insoluble plaques, is now widely thought of as being the causative agent in many neurodegenerative diseases, the concept is not universally accepted.119 That, and the lack of an agreed upon structure for the oligomer (which can apparently be formed by many diverse proteins through an as yet ill-defined process), continue to hamper progress towards a coherent therapeutic approach and a final determination as to whether the toxic oligomer truly is the pathogenic species.120
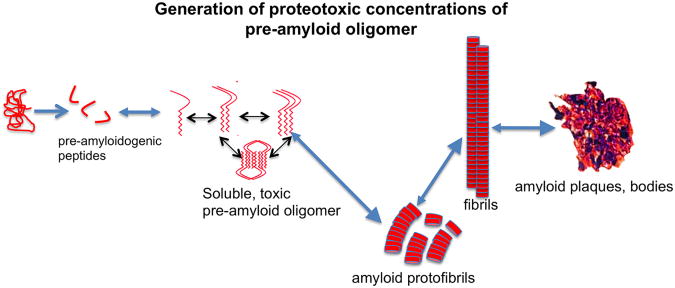
Figure 4.
Proteotoxic peptides. Shown is the formation of what are thought to be the toxic entities in a wide variety of proteotoxic diseases; small, soluble pre-amyloid oligomers. These are metastable entities in slow or rapid equilibriums with one another and, at least for some entities, the dimers and trimers are the most toxic form. Enhanced autophagy is correlated with reduction in toxic pre-amyloid levels but the mechanism of clearance remains obscure (see text for details).
Although often associated with the same misfolding phenomena, aggresome formation (Figure 1) is fundamentally different from PAO production (Figure 4), even though the same protein or proteins may or may not be involved. In a mouse model of AD, cellular toxicity is correlated with the presence of PAO, not amyloid fibrils.121 Cellular toxicity in Huntington disease, which is caused by the accumulation of mutant huntingtin protein aggregates within intranuclear inclusion bodies, is associated not with these large aggregates but with the presence of a soluble form of the mutant protein.122 Similarly, the large, neuronal intranuclear inclusions in spinobulbar muscular atrophy, which is caused by polyglutamine expansion (CAG repeat expansion) in exon 1 of the androgen receptor, consist of proteinaceous aggregates that do not correlate well with cellular toxicity. Instead, the morbidity in this degenerative disease of the lower motor neurons and skeletal muscle correlated with high levels of the soluble intermediates.123 In fact, cellular toxicity presented in the absence of visible aggregates when androgen receptor containing expanded CAG repeats were expressed in a neuroblastoma cell line.124 Similar data concerning the toxicity of the soluble oligomer were obtained in an AD mouse model in which an oligomer-capable amyloid β (Aβ) species was synthesized and the animals were showed neurodegenerative processes even the absence of visible fibrils.125 These data were also recapitulated in human and mouse models of Type 2 diabetes, which is characterized by a loss of β cell mass and extracellular accumulations of islet amyloid derived from the islet amyloid polypeptide, expressed by the β cells along with insulin. When islet amyloid polypeptide accumulates as oligomers, it is a potent cytotoxin but its accumulation within inclusion bodies in the form of “inert” fibrils appears to be largely benign.126 Cardiac data are consistent with the hypothesis that aggresomes are not necessarily cytotoxic as, in inducible PAO-genic models, rescue of the animal was accompanied by decreased PAO levels, although aggresome concentrations appeared to be unaffected.127,128
To determine the relevancy of proteotoxicity in cardiac muscle, an analysis of normal and diseased human hearts for the presence of PAO was undertaken in order to determine if PAO was present in diseased cardiomyocytes. While three non-diseased hearts showed no or minimal traces of PAO, samples from nine hypertrophic or dilated cardiomyopathic hearts showed high, but variable, levels of PAO internal in the cardiomyocytes.87 The presence of PAO in diseased hearts suffering from presumably different primary causes implies that PAO formation may be a general and potentially widespread phenomenon in a number of different heart failure presentations. However, this remains to be formally demonstrated in a rigorous, carefully controlled manner and sample collection becomes a key factor in quantitating the degree of pathology as characterized by PAO levels.
PAO Causality in Heart Failure
Are the soluble PAOs observed in diseased hearts sufficient to cause heart disease and failure? To address this question, ectopic expression of a PAO-genic polypeptide specifically in cardiomyocytes was carried out using transgenesis. Expression of a PAO-genic CAG repeat at very low concentrations in the cardiomyocytes of transgenic mice was invariably toxic, with all mice dying of heart failure by 7 months.129 Although aggresomes were also present, they appeared to be frequently engulfed by autophagic vesicles, consistent with the hypothesis that the aggresomes were in the process of being degraded. This experiment confirmed that synthesis of a PAO-genic polypeptide in cardiomyocytes was sufficient to cause death by heart failure and causality for the PAO was thus established.
Pre-Amyloid Structure and Toxicity
The nature of these soluble oligomers has been extensively studied but, as alluded to above, the processes leading to their generation and structure or structures130 remain elusive as they are detected by either polyclonal or monoclonal antibodies that recognize conformers that do not depend upon a specific linear amino acid sequence.115,130-133 This diversity has led to an unsettled literature as to the toxic mechanism or mechanisms by which their pathogenic effects are mediated.134 Although the oligomerization pathways (Figure 1, panel 4) are not straightforward due to the diversity of primary sequences in the many proteins with amyloidogenic potential, there do appear to be some commonalities. Many of the oligomers show, upon biophysical examination, ordered assemblies with a well-defined pattern of intermolecular contacts and at least some of these contacts involve regions that form a β-sheet core in the fibrillar state, should that conformation be achieved.135 Thus, while these diverse proteins form very different structures in their soluble state, their fibril states are distinguished by structural commonalities, thought to be due to the ability of the primary sequence to be stabilized by hydrogen bond interactions in the polypeptide, such that β-strands of the oligomer can stack in register and be arranged perpendicular to the long axis of the ultimate fibril to generate a cross-β structure.136-138
The Aβ oligomers characteristic of AD have been studied most intensively and their levels correlate well with disease severity. Subsequently, the monomers can reversibly undergo conformational transitions, forming dimers or trimers of low molecular weight, soluble s oligomers that, in turn, can form spherical oligomers consisting of 12-24 monomers. These oligomers are found external and internal to the cell, and can go on to form protofibrils (Figure 4) and, ultimately, the characteristic insoluble amyloid fibril.139
In an elegant series of experiments, Diociaiuti and colleagues attempted to dissect out the toxic contributions of the metastable intermediates that form during this aggregation process. Taking advantage of the very slow aggregation properties of an amyloidogenic protein, salmon calcitonin, they generated the different oligomeric forms and “froze” them in the various intermediate states via photo-induced cross linking in SDS PAGE gels.140 Using a combination of biochemical and imaging techniques, they were then able to purify the different forms and individually test their neurotoxicity in cultured primary hippocampal neurons. They found that the globular dimers, trimers and tetramers were the most toxic species, resulting in calcium influx and apoptosis while the monomers and other, larger aggregates appeared to be biologically inert. Importantly, in mixtures, the presence of very low concentrations of the toxic species were sufficient to render the entire mixture neurotoxic, as one would expect in vivo when all the various forms would be present.
For at least for some oligomers, including Aβ, their toxicity is partially mediated via their action on specific receptors.141 It is not clear whether this due to direct binding or to some other action of the oligomer on one or more signaling pathways, but binding to multiple receptors has been observed.134 The propensity of oligomers to be “sticky” and exhibit detergent-like qualities has been remarked upon by a number of groups and the oligomer's ability to interact with different membrane systems has been proposed to be intrinsic to their toxicity.138,142-145
In the heart, membrane integrity is obviously critical for the maintenance of cell viability. Disruption of either the plasma membrane or the other vital membrane-enclosed organelles such as mitochondria or T-tubules would clearly affect cardiomyocyte function. Data from model membrane systems demonstrate the ability of oligomers to form cation-specific channels that destroy calcium homeostasis with concomitant cytosolic elevations of the ion.146 Consistent with this hypothesis, the mutant form of CryAB, CryABR120G, preferentially associates with mitochondrial membrane proteins (Figure 3, panel B) and the early ablation of mitochondrial membrane integrity occurred almost immediately upon expression of the CryABR120G in cardiomyocytes, with cytotoxicity quickly following despite abundant mitochondrial biogenesis. 72 On the basis of all these observations, we can conclude that PAO interactions at the membrane can clearly impair ion and redox homeostasis in the affected cells but the specific interactions that result in cytotoxicity will differ depending upon the membrane system and, to some extent, the oligomeric form assumed by the amyloidogenic polypeptide.
In cardiomyocytes, inducible expression of PAO-genic CryABR120G was used to determine if the cardiomyocyte had the capacity to clear PAO after it accumulated.128 Pharmacologic intervention via doxycycline treatment effectively terminated expression of the mutant CryAB in symptomatic mice (11-12 weeks of CryABR120G expression) and PAO was subsequently measured after 1 month. Decreases of approximately 40% were observed, implying that either the PAO is intrinsically unstable during this time period or the cardiomyocyte is able to clear the toxic entity.
As the above discussion makes clear, soluble oligomers can interact with membrane systems and it was therefore reasonable to test whether autophagy could play a role in PAO clearance as the process involves interaction with and engulfment by membrane vesicles.62,69 Upregulating autophagy by either exercise or Atg7 overexpression in cells did, in fact, lead to decreased PAO accumulation.28,127,147 Similarly, loss of function for autophagy led to increased PAO accumulation28,147 although a direct demonstration of toxic PAO trafficking through autophagosomes was lacking.
Are Aggresomes also Toxic?
The toxic role that aggresomes play may be more nuanced, as they represent the end result of a process by which misfolded proteins and protein aggregates are sequestered from the general cytoplasm (Figure 1, panel 3). If the misfolded protein is indeed toxic, then within limits, aggresome formation might be thought of as a protective mechanism for isolating a potentially toxic entity, as noted above for the islet amyloid polypeptide.126 Indeed, inclusion body formation in the α-synuclein accumulation diseases such as Parkinson disease does appear to be cytoprotective.148,149 Aggresomes can, in many cases, successfully protect the cell against proteotoxicity, likely by sequestering the toxic species until the aggresome or inclusion body can be cleared by autophagy.89,109 Sequestration of toxic oligomers by the chaperone, HspB1, resulted in removal of the Aβ oligomers from the soluble fraction into large, nontoxic aggregates.150 However, one could envision these “garbage dumps” becoming cytotoxic if PAO was not sufficiently sequestered in the aggregate interior or the aggregate becomes excessively large. This is particularly relevant in cardiomyocytes as function depends upon coordinated, compliant contraction over the entire cell. Generation of ever-increasing amounts of large proteinaceous aggregates could also compromise the mechanical characteristics of the cytoplasm. Indeed, in a cell-based study in which cardiomyocytes rapidly expressed high levels of CryABR120G and large aggregates formed, determination of compliance in three dimensions by two-photon microtomy151 and magnetic bead microrheology at the single cardiomyocyte level showed significant changes in the cytoskeleton's mechanical characteristics and deficits in passive cytoskeletal stiffness.152 Additionally, although some commonalities in protein content across different aggresomes occur,87 there appear to distinct morphologies as well, with some larger aggregates showing a fenestrated appearance and other, smaller aggregates showing a less defined morphology (Figure 3, panels A, B). In the CryABR120G model of proteotoxicity, the aggresomes contain high concentrations of CryAB. The intermediate filament protein, desmin, is also present (Figure 3, panel D), a result consistent with data obtained from diverse systems in which intermediate filament proteins, including vimentin and keratin,153 form a cage around the aggresome, presumably functioning to sequester off the potentially proteotoxic aggregated protein species.
Restoration of Cardiac PQC Reduces Proteotoxicity
The soluble and insoluble inclusions present in proteotoxic diseases will invariably invoke the cellular PQC response. However, the dysfunction of these processes is as much a part of the proteotoxic phenotype as the oligomers and aggregates themselves. To understand how failure of the proteolytic pathways can cause proteotoxic cardiac disease, it is useful to examine known proteotoxic disease models and understand the specific defects in the cardiac PQC response, and the subsequent, functional consequences.
Generally speaking, there are two somewhat predictable responses to PQC pathways as a result of increased proteotoxicity: compensatory induction or inhibition. In the CryABR120G model, both of these effects are observed when examining proteasomal function. Proteasomal activity is targeted to small peptides and smaller misfolded proteins that have not yet formed oligomeric quaternary structures, as the small proteasomal bore dimensions (∼ 13Å) disallow entry and degradation of larger structures (Figure 1, panel 2).154 Using a GFP-tagged proteasomal degron sequence (GFPdgn)155 as a reporter of proteasomal degradation, Chen et al showed increased levels of GFPdgn in the hearts of two DRM models as well as increased levels of ubiquitinated proteins, both of which point to decreased degradation of proteasomal substrates.154,156 In addition, they noted dysregulation of the expression of individual proteasomal subunits, suggesting abnormalities in the proteasome itself. Despite this, the specific enzymatic activities of the proteasome were increased, suggesting a functional proteasome that was simply unable to degrade the misfolded protein load. To test this concept, they treated cardiomyocytes infected with CryABR120G with Congo Red, a compound that can prevent aggregate formation by binding misfolded proteins and preventing increased intermolecular interaction. After this treatment, they found reduced aggregate formation and a concomitant increase in proteasomal degradation, shown by decreased levels of GFPu (a GFPdgn analog). This study demonstrated a deficit in proteasomal degradation in cardiac proteotoxicity that appeared to be the direct result of misfolded protein accumulation.156 This situation, coined “proteasome functional insufficiency,”157 can activate the pathological calcineurin-NFAT pathway,158 a deleterious pathway in the heart. 159 Of note, GFPu accumulation was also observed in the brain of an AD mouse model,160 providing further evidence of the similarities in PQC dysfunction, particularly proteasome functional insufficiency, in different proteinopathies.
These data imply that proteasomal degradation is critical in preventing proteotoxicity and that increased misfolded protein load in the sarcoplasm can negatively affect the proteasome from efficiently degrading proteins. Increasing proteasomal function may be beneficial to keep misfolded protein accumulation in check and prevent pathogenesis. To test this hypothesis, Wang and colleagues used transgenesis to induce overexpression of proteasomal components. As expected, overexpression of PA28α, a component of the 11S proteasome, increased proteasomal proteolytic function as measured by reduced GFPdgn levels. When crossed with CryABR120G mice, they observed reduced protein aggregation and hypertrophy, which led to an extension of lifespan.161 Further, when they subjected PA28α mice to ischemia/reperfusion, they noted reductions in infarct size. Consistent with these data, they noted that activation of protein kinase G could increase proteasomal activity and degradation of a proteasomal substrate, and reduce pathologic aggregates in the CryABR120G mouse model.162 Recently, Gupta et al demonstrated that increased levels of Ubc9, an E2-like ligase in the SUMOylation cascade, also enhanced proteasomal function and degradation, leading to reduced levels of both soluble and insoluble CryABR120G protein aggregates. Convincingly, this protection was abolished upon proteasomal inhibition, confirming that increased proteasomal degradation was causative in the protection.163 These data show an important role for the proteasome in reducing misfolded protein accumulation and imply that inducing proteasomal function might be a legitimate therapeutic avenue in cardiac proteinopathies.
Autophagy and Cardiac Disease
Autophagic degradation is required for normal cardiac development and function; however, whether autophagy is protective or detrimental during the development and progression of cardiac disease is still a matter of some debate. As with most cardiac PQC mechanisms, autophagic function can be altered under disease conditions, which can lead to an induction or inhibition of autophagic degradation. Some of the first genetic studies of autophagy in the heart focused on beclin1, a gene involved in autophagosome formation. Reduction of beclin1 by heterozygous deletion reduced levels of autophagy and led to a reduction in infarct size in mice after reperfusion injury. Reduced cell death was also noted, so it is not completely clear that the reduction in autophagy was necessarily the driving force behind reduced ischemic injury.67 Similarly, beclin1 deletion improved hypertrophic phenotypes in a model of pressure overload induced hypertrophy. In contrast, other studies have shown beneficial effects resulting from up-regulating autophagy. Increased autophagy was found to improve cell viability in HL-1 cells subjected to ischemia-reperfusion in culture.66,164 In addition, evidence shows that inducing autophagy may be partly responsible for the cardioprotective effects of ischemic preconditioning (IPC). IPC caused increased levels of the autophagic proteins LC3-II and Beclin1, and electron microscopy showed elevated levels of autophagosomes containing mitochondria.165 To determine the importance of autophagy in cardioprotection after IPC, Huang et al inhibited autophagy using a dominant-negative Atg5, atg5K130R. They found that the cardioprotection afforded by IPC was almost completely abolished when autophagy was disrupted.166 Indeed, mTOR inhibition and autophagy stimulation were observed in both chronic and acute IPC.167 In addition, recent data in a large animal model showed beneficial effects of pharmacologic autophagy induction after reperfusion injury.168
While some groups have reported that autophagy is activated during a myocardial infarction and upregulating autophagy can be cardioprotective in this context by limiting pathological remodeling and cellular injury,169 others have published conflicting data and this issue has not yet been completely resolved.170 Indeed, the apparent discrepancies between different groups' data may be both apparent and real, with the net effect of autophagy being dependent upon the exact environmental insult, its duration, the particular mouse strain, when the autophagic stimulus is induced and the methodology used to induce it. Similar conflicting data have been obtained concerning the role of autophagy in mediating pressure overload induced hypertrophy61,171 and the controversies have recently been reviewed.170 Carefully controlled studies are needed to resolve these apparent conflicts and the use of pure mouse strains and standardized protocols across the different labs will be needed to bring much needed clarity to the questions being asked. While the effects of autophagy may not be universally protective, a consensus is beginning to be reached that autophagy is a protective mechanism in many cardiac pathophysiologies, particularly in those phenotypes with a proteotoxic component. There are multiple steps in autophagy and the process may be compromised at a single or multiple sites. If only markers of the early autophagic processes are measured, an investigator might be misled and before any firm conclusions about increased or decreased autophagy are reached, it is important to measure the overall activity of the entire process.64
The effects of autophagy on cardiac proteotoxicity have been comprehensively studied in the CryABR120G model. As misfolded proteins begin to accumulate and aggregates coalesce into aggresomes (Figure 1, panel 3), one might expect compensatory induction of autophagy as the cardiomyocyte adapts to the insult. Indeed, in proteotoxicity induced by pressure-overload and CryABR120G expression, compensatory induction of autophagy was observed.172 However, as the disease progressed, the autophagic system became impaired, leading to decreased autophagic flux and an inability to clear newly-formed and pre-existing aggregates.28,88 These data suggest that autophagic activity is necessary for aggregate clearance and impairment is a significant aspect of the proteotoxic phenotype.
The hypothesis that increased autophagy might be beneficial in the setting of cardiac proteotoxic disease was tested using the DRM model. Taking advantage of the known pathways that underlie macroautophagy, Pattison et al. were able to increase autophagic flux in isolated cardiomyocytes by expressing high levels of Atg7, a critical, ubiquitin-like modifier-activating enzyme (E1) that functions in the initial stages of vesicle elongation, a necessary step for vesicular engulfment of the aggregates or damaged organelles destined for degradation and recycling.173,174 When Atg7 was expressed either in cultured cardiomyocytes28 or subsequently in transgenic mouse hearts during the proteotoxic insult of CryABR120G expression,88 increased autophagic flux resulted in decreased aggregate accumulation, decreased levels of toxic PAO and, in the whole animals, improved cardiac function and decreased early mortality. Thus, increasing compromised levels of autophagy to basal or even greater than basal activities can be cardioprotective during proteotoxic disease. These data unambiguously demonstrate a beneficial effect of autophagy stimulation on cardiac proteotoxicity in the DRM model and are considered below in terms of potential therapies.
Mitophagy
Mechanistically, autophagy appears to be protective in part through degradation of damaged organelles. The process of mitochondrial turnover by autophagy, known as mitophagy, is particularly important in the heart, as cardiomyocyte function absolutely requires steady production and supply of ATP. When damaged, mitochondria can become tagged for degradation through a variety of pathways.175,176 The best understood signaling cascade for mitophagy involves PINK1/Parkin, wherein PINK1, a serine/threonine kinase, phosphorylates proteins on the mitochondrial outer membrane and recruits parkin, an E3 ubiquitin ligase, to ubiquitinate these proteins through K63 linkages, which are thought to signal lysosomal rather than proteasomal degradation.176 Adaptor proteins such as p62 bind and signal autophagosome recruitment and engulfment of the mitochondrion by binding to LC3.176 Upon ischemic insult or IPC, parkin translocates to mitochondria, which in turn signals the autophagic adaptor p62 to the mitochondria, suggesting that ischemic insult is inducing mitophagy.177 That similar signaling cascades are activated in both ischemic injury and IPC imply this pathway is cardioprotective. Indeed, the recruitment of p62 to mitochondria after IPC is abolished in parkin ablated mice, suggesting the PINK1/parkin route is essential for cardioprotective mitophagy post-ischemia.177
Pathway Coordination
The above data confirm that the UPS and autophagic pathways are instrumental in trying to manage proteotoxic phenotypes in the cardiomyocyte. From a therapeutic perspective, factors that can modulate both pathways would be of particular interest. Carboxy terminus of Hsc70-interacting protein (CHIP) is one such protein and functions as a molecular co-chaperone or E3 ubiquitin ligase.104,178 Unsurprisingly, CHIP is very highly expressed in tissues that require an intensive PQC response, such as the heart and brain:104 CHIP plays a major role in the cardiac stress response.179 Recently, in addition to these functions with the UPS, Schisler et al showed that CHIP is required for activation of AMP kinase (AMPK) in response to pressure overload. They identified CHIP as a constituent of the AMPK complex and found decreased AMPK phosphorylation in the absence of CHIP, leading to an exacerbated hypertrophic phenotype180 AMPK is a known regulator of autophagy, as activated AMPK inhibits mTOR, allowing autophagy to proceed.181 Proteins such as CHIP, which mediate crosstalk among multiple protective pathways, may prove especially useful for treating proteotoxic diseases of the heart.
While modulation of the UPS and autophagic degradation have clear benefits towards modulating cardiac proteotoxicity, other strategies, such as manipulation of the oxido-reductive or ER stress pathways, also show promise. In the CryABR120G model, abnormalities in the HSR have been observed, including induction of chaperone proteins182 and sequestration of HSPs inside aggregates.87 In transgenic mice expressing human CryABR120G in the cardiomyocytes, increased levels of Hsp25, glucose-6-phosphate dehydrogenase (G6PD), catalase and glutathione reductase led to the hypothesis that reductive stress via overcompensation of the oxidative stress response was pathogenic. Indeed, reducing the levels of G6PD by crossing human CryABR120G mice with G6PD mutant mice, which have reduced G6PD activity, reduced hypertrophy and aggregate formation, although the exact mechanism by which this occurs was not determined.182
Therapeutic Modulation of PQC to Curtail Proteotoxicity
Taken together, the above data demonstrate that there are a number of protective mechanisms that could potentially be targeted for therapeutic intervention in cardiac proteotoxicity. The specifics of potential therapeutic targets differ for the different proteotoxic diseases but may broadly be divided into two approaches: enhancing the intrinsic clearance mechanisms capable of degrading and recycling the proteinaceous aggregates or preventing their formation. Herein, we explore a few promising candidates, illustrating their actions on autophagy and underscoring the therapeutic potential of interfering with these proteotoxic pathways.
It is becoming clear that different clearance mechanisms are affected in a number of protein misfolding diseases. For example, the UPS is compromised in both the neurodegenerative diseases183 as well as in DRM.154 Conceptually, it is rather straightforward to appreciate that large aggregates of ubiquitinated proteins may negatively impact on a subset of the cell's clearance pathways as they are trafficked to the proteasome but are too large to pass through the constraints of the proteasomal bore (Figure 1, panel 2).184 Indeed, impairment of the UPS by protein aggregation in the heart has been well documented, both here and elsewhere.57 The proteasome is already a target in treatment of different blood cancers, with inhibitors of the 20S proteasome such as bortezomib or marizomib validated in clinical trials.185 However, for cardiac disorders, inhibition of the proteasomal activity is clearly detrimental in the context of proteotoxicity.57,186 Therefore, efforts to augment proteasomal activity are underway to determine if cardiac function can be maintained in the face of an acute or chronic proteotoxic insult.
The modulation of another major clearance pathway, autophagy, is also being explored as a potential therapeutic target.187 As outlined in detail above, in the face of a proteotoxic insult, autophagy can serve as the primary clearance mechanism for proteinaceous aggregates that are too large for proteasomal degradation. Continuing the analogy to the proteotoxic neurodegenerative diseases, accumulating evidence suggests that the autophagic pathways are significantly compromised in cardiac proteotoxic environments.188 Unambiguous data show autophagy is reduced in Parkinson disease, with decreased autophagic flux resulting in α-synuclein accumulation, mitochondrial dysfunction and neuronal cell death.189 By analogy, compromised autophagy in the context of proteotoxic heart disease might very well serve as an increased pathogenic insult and lead to increased morbidity and hasten heart failure. Decreased autophagic flux has been noted in many disease processes and restoration of normal or even enhanced autophagy appears to be beneficial in a wide spectrum of proteotoxic diseases ranging from the neurodegenerative diseases to Type 2 diabetes.126,190-193
Because of the diverse roles that autophagy can play in both normal and disease metabolism, it should be emphasized that these data cannot be broadly applied to cardiac disease in general or even throughout the natural history of the same disease. But, if upregulation of autophagy is beneficial in certain proteotoxic contexts, how might it be upregulated as part of a directed therapeutic approach? There are many small molecule effectors of autophagy,194-197 with some FDA-approved drugs having potent effects on autophagy in the cardiovascular system.198 However, non-pharmacologic approaches may also be possible. The beneficial effects of exercise are far-reaching and well documented across human physiology and there are extensive data concerning the impact of physical activity on certain neurodegenerative diseases such as AD. In different AD mouse models, environmental enrichment199,200 or voluntary exercise201 was beneficial, resulting in either the delayed onset of overt symptoms or progression of the disease. The physical component of environmental enrichment had particular impact, with voluntary exercise delaying the onset of neurological deficits in a mouse model of Huntington disease202 and long-term exercise in a mouse model of AD, decreasing Aβ and amyloid deposition levels.200 A recent computational analysis involving genome wide expression data from Parkinson and Huntington disease brain tissue was entirely consistent with the importance of autophagy in these disease processes.203
On the basis of these data and the parallels between the neurodegenerative disorders and proteotoxicity-induced cardiomyopathy, we tested the effects of long-term voluntary exercise in the CryABR120G DRM model. Voluntary exercise was quantitated with the use of running wheels with which both duration and distance could be continuously monitored in an isolated setting, free from any overt stress over a period of 6 months.127 The results were striking: mice that were not placed in cages where they could voluntarily exercise were all dead by approximately 7 months while the mice that were exercising showed 100% survival at that time point. Although those mice did eventually succumb to heart disease, lifespan increased by approximately 30% in later trials.88 It is well established that exercise results in increased autophagy,204-208 and, indeed, exercise does result in numerous autophagic markers being upregulated (Figure 5). In contrast, when autophagy is induced by Atg7 expression in cardiomyocytes, a general upregulation of genes associated with autophagy does not occur.88 This raised the intriguing possibility that, although both interventions (voluntary exercise and Atg7 overexpression), led to the same outcome in terms of increased autophagic flux, the two interventions might be synergistic. This appeared to be the case and when the Atg7 mice were allowed to voluntarily exercise, survival was prolonged as compared to either the Atg7 overexpressors or the exercise-only DRM animals.88
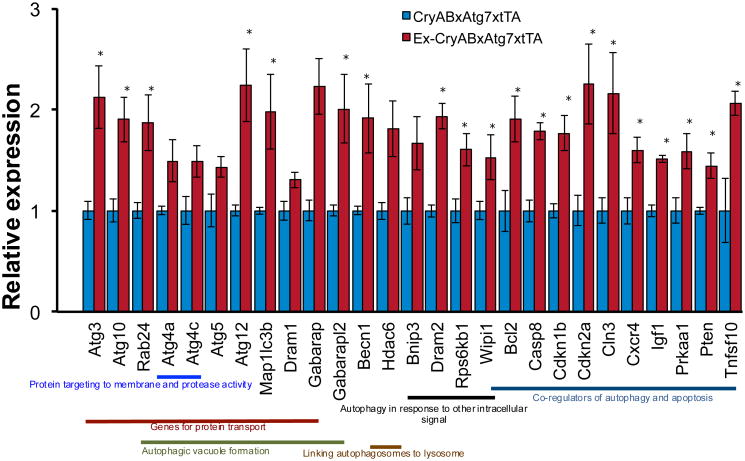
Figure 5.
Autophagy PCR array in voluntary exercised CryABR120GxAtg7xtTA hearts. Graph representing direct group wise comparison of fold change in mRNA levels in male voluntary exercise CryABR120GxAtg7xtTA and control non-exercised CryABR120GxAtg7xtTA hearts at 4.5 months (fold change versus non-exercised CryABR120GxAtg7xtTA control, n = 3 per group). All values are reported as mean ± S.E.M. P<0.05 and P<0.01 by Student's t test. Mice and experimental treatments were carried out as described.previously.88
Pharmacological Therapy
As outlined previously, HDAC inhibition can also be cardioprotective, due in part to anti-hypertrophic effects.209 Several laboratories have now shown a positive effect on cardiac autophagy when using the FDA-approved pan-HDAC inhibitor suberoylanilide hydroxamic acid (SAHA). Hill and colleagues administered SAHA to rabbits before and/or during the course of ischemia/reperfusion injury. SAHA treated rabbits showed a reduction in infarct size, which led to an improvement in cardiac fractional shortening.168 They noted LC3-II, a reliable autophagic marker, was significantly increased in the infarct border zone of SAHA treated mice compared to controls, which led to a reduction in cell death in this region of the myocardium. Indeed, studies in cardiomyocytes corroborate that SAHA could increase autophagic flux under normal and ischemic conditions.168
SAHA had similar effects when applied to a pure model of proteotoxicity. Chronic SAHA administration to CryABR120G mice showed attenuation in sarcoplasmic aggregates and a concomitant improvement in cardiac function compared to animals treated with vehicle.210 Similarly, it was found that SAHA increased autophagic flux in CryABR120G expressing cardiomyocytes, suggesting that increased autophagy plays an important role in instigating the observed cardioprotective effects. However, further study is needed to ascertain which HDACs are directly involved in the autophagic response.210
HDAC6 knockout mice displayed improved systolic function after chronic stimulation with angiotensin II,211 suggesting that HDAC6 function may be deleterious in the heart under some stress conditions, but the protective mechanisms are unclear. To determine the role of HDAC6 in a proteotoxic disease model, we assessed the role of HDAC6 function in DRM. We found that, despite increases in HDAC6 protein and catalytic activity, deacetylation of its microtubule substrate was decreased,210 underscoring that it is essential to measure the downstream signaling effects of the protein's activity rather than merely quantitating protein levels. As per its canonical function, HDAC6 mediated aggresome formation in cardiomyocytes. Chronic treatment with SAHA improved cardiac function and reduced cellular aggregates while also increasing acetylation of the HDAC6 substrate α-tubulin. Interestingly, in this case, inhibiting HDAC6-mediated tubulin deacetylation led to increased autophagic flux. While seemingly at odds with HDAC6's reported function, reduction in tubulin deacetylation may be necessary for autophagic activity.212 Indeed, increased levels of tubulin acetylation were found in CryABR120G mice after 5 months of voluntary exercise, despite no change in HDAC6 catalytic activity.210 While more data are needed to assess the precise roles of HDAC6 in cardiac physiology and pathobiology, it remains an intriguing therapeutic target.
Final Thoughts: a Next Step
Currently, there exist no available therapeutics that will effectively dissolve pre-existing protein inclusions and reverse a proteotoxic disease state. The existence of protein inclusions continues to pose a constant and progressive threat. This reality is a particularly distressing one: many proteotoxic phenotypes do not present in patients until significant tissue damage has occurred. Thus, new therapeutics that can fully reverse proteotoxic phenotypes are needed.
Disease progression appears to proceed through successive accumulation of newly misfolded protein. As outlined above, this likely occurs due to reduced protein clearance through the PQC mechanisms, which leads to aggregate buildup, cellular dysfunction and ultimately cell death. However, data suggest that blunting the expression of the aggregate-prone protein can halt the disease progression.111,128 In fact, reversal of disease and functional improvement upon clearance suggests that the continued production of the aggregate-prone protein serves as a feed-forward loop that may be necessary for disease progression and an ever-worsening prognosis. Looking at this situation optimistically, therapeutic opportunities could be directed at several points along the pathogenic sequence: removing aggregated species, targeting the soluble PAO or reducing causative protein expression.
A striking feature of protein aggregate based disorders is the remarkable similarity between the aggregates despite myriad differences in cell type and affected protein. 213,214 Therapeutically, this may be a fortuitous feature: if the cell biology governing aggregate formation is conserved, then disaggregation mechanisms that work for one proteotoxic model might translate to another. Hypothesis-driven science has provided several promising strategies for combating aggregate formation, but none have yet reached fruition. Our current understanding of the protein aggregation process is still limited, underscoring the broad need for discovery-driven science that can identify new factors, and potentially intersecting and novel pathways that could effectively reduce or reverse protein aggregation.
High-throughput screening (HTS) is an advantageous and relatively unbiased method to find novel effectors for a given molecular and cellular process. Screens can take many forms but usually consist of an appropriate cellular or organismal model being probed with either a large pharmacologic-based compound library or via genetic tools, with a defined cellular phenotype or response (readout) being measured. The quality of the data in such a screen is reliant on the robustness of the model used, the intrinsic noise of the system and the strength of the signal constituting the readout. In the case of proteotoxic phenotypes, the readout may be relatively straightforward: augmented or decreased protein aggregates. For the proteotoxic diseases, laboratories are beginning to publish some nascent HTS aimed at identifying new pathways or finding new drugs to reduce aggregation and/or cellular toxicity.215,216
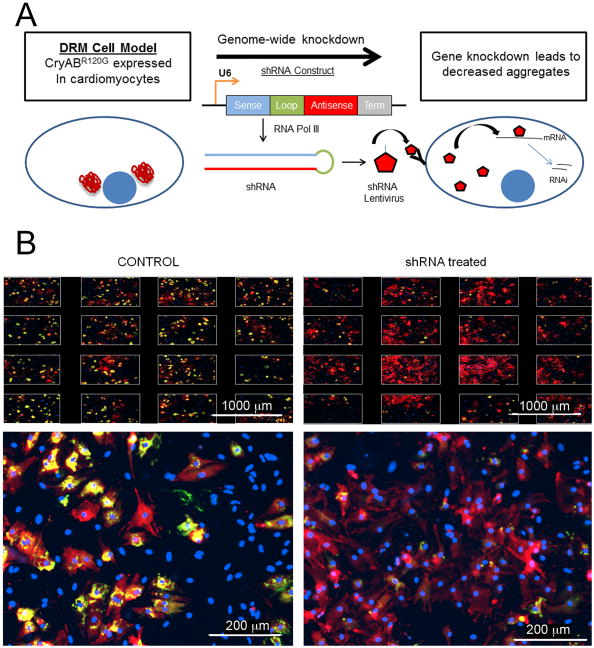
With the goal of finding novel pathways that impact on cardiac proteotoxicity, we have developed a cellular model of protein aggregation in primary mouse cardiomyocytes. Using a fluorescent, aggregate-prone protein (CryABR120G), we were able to produce a cellular model with a robust cardiac protein aggregation phenotype and a very high signal-to-noise ratio, such that phenotypic changes could be easily quantified.128 To find novel effectors of cardiac protein aggregation, we systematically knocked down each gene in the mouse genome using a library of short hairpin RNAs (shRNA) expressed via lentiviral transduction,217 and the phenotypic changes were measured using high-content imaging (Figure 6). Preliminary data from the screen have yielded several promising new inhibitors of cardiac protein aggregation that, once independently validated, will help to uncover novel pathways involved in protein aggregation in the heart.
Figure 6.
High-throughput assay to uncover novel effectors of cardiac protein aggregation. A, Screening Principle. A cell model of cardiac proteotoxicity was developed in primary cardiomyocytes. These cells were subjected to systematic, genome-wide knockdown by infection with lentiviruses expressing shRNAs capable of being processed such that selective degradation of their cognate mRNAs took place. The goal of the screen is to find genes that, when knocked down, lead to a reduction of aggregates. B, An example of a candidate gene or “hit” in the screen. The top panels show the images taken from one well, in which 16 images are acquired and aggregate content quantified. The lower panel depicts one image from a well. In the well treated with siRNA, a drastic reduction in aggregate content is observed compared to controls. This figure demonstrates the very high signal to noise ratio observed upon expression of CryABR120G, which allows one to robustly quantitate changes in aggregate content. Scale bars: top panel, 1000 μm; bottom panel, 200 μm.
These technology platforms are especially valuable due to the potential to uncover information about a disease state that is unattainable by more conventional methods. For example, as patient derived cells become more widely used in medicine, they will provide unprecedented potential for probing modulators in the pathobiology of a specific patient. This was recently achieved in ALS patients, in which patient fibroblasts were transformed via induced pluripotent stem cells into motor neurons that could then be used in a high-content drug screen to find appropriate compounds to reduce aggregate content.218 We are hopeful that ongoing and future screens may enable unbiased, discovery science that will not only find new therapeutic targets, but will generate testable hypotheses that will uncover the underlying mechanisms of proteotoxic disease in the heart.
Acknowledgments
Sources of Funding: This work was supported by NIH grants P01HL69779, P01HL059408, and R01HL105924 (J.R.) and The Transatlantic Network of Excellence Program grant from Le Fondation Leducq (to J.R.). P.M.M. is a postdoctoral fellow of Le Fondation Leducq.
Nonstandard Abbreviations and Acronyms
- HSP
heat shock protein
- AD
Alzheimer disease
- PQC
protein quality control
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- CryAB
alpha B crystallin
- mTOR
mammalian target of rapamycin
- DRM
desmin relation (cardio)myopathy
- PAO
pre-amyloid oligomer
- CHIP
carboxy terminus of Hsc70-interacting protein
- SAHA
suberoylanilide hydroxamic acid
- HTS
high throughput screen
- HSR
heat shock response
- UPS
unfolded protein response
- AMPK
5′ adenosine monophosphate-activated protein kinase
- HDAC
histone deacetylase
- HSF
heat shock factor
- IPC
ischemic preconditioning
- G6PD
glucose-6-phosphate dehydrogenase
Footnotes
Disclosures: None
References
- 1.Galluzzi L, Bravo-San Pedro JM, Vitale I, et al. Essential versus accessory aspects of cell death: Recommendations of the nccd 2015. Cell Death Differ. 2015;22:58–73. doi: 10.1038/cdd.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, Galluzzi L, Vandenabeele P, et al. Classification of cell death: Recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hightower LE. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- 4.Douglas PM, Cyr DM. Interplay between protein homeostasis networks in protein aggregation and proteotoxicity. Biopolymers. 2010;93:229–236. doi: 10.1002/bip.21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie NT, Lee AL, Fay HG, Gray AA, Kikis EA. Novel polyglutamine model uncouples proteotoxicity from aging. PLoS One. 2014;9:e96835. doi: 10.1371/journal.pone.0096835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox D, Carver JA, Ecroyd H. Preventing alpha-synuclein aggregation: The role of the small heat-shock molecular chaperone proteins. Biochim Biophys Acta. 2014;1842:1830–1843. doi: 10.1016/j.bbadis.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 7.Mulligan VK, Chakrabartty A. Protein misfolding in the late-onset neurodegenerative diseases: Common themes and the unique case of amyotrophic lateral sclerosis. Proteins. 2013;81:1285–1303. doi: 10.1002/prot.24285. [DOI] [PubMed] [Google Scholar]
- 8.Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 10.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 11.Hunt JA, Ingram VM. Human haemoglobin e: The chemical effect of gene mutation. Nature. 1959;184:870–872. doi: 10.1038/184870a0. [DOI] [PubMed] [Google Scholar]
- 12.Ingram VM. Gene mutations in human haemoglobin: The chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 13.Valastyan JS, Lindquist S. Mechanisms of protein-folding diseases at a glance. Dis Model Mech. 2014;7:9–14. doi: 10.1242/dmm.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu BH, Strickland E, Thomas PJ. Cystic fibrosis: A disease of altered protein folding. J Bioenerg Biomembr. 1997;29:483–490. doi: 10.1023/a:1022439108101. [DOI] [PubMed] [Google Scholar]
- 15.Demuro A, Mina E, Kayed R, Milton SC, Parker I, Glabe CG. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J Biol Chem. 2005;280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 16.Russell D, Andrews PD, James J, Lane EB. Mechanical stress induces profound remodelling of keratin filaments and cell junctions in epidermolysis bullosa simplex keratinocytes. Journal of cell science. 2004;117:5233–5243. doi: 10.1242/jcs.01407. [DOI] [PubMed] [Google Scholar]
- 17.Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies apoe epsilon4 effectors in alzheimer's disease. Nature. 2013;500:45–50. doi: 10.1038/nature12415. [DOI] [PubMed] [Google Scholar]
- 18.Dhungel N, Eleuteri S, Li LB, et al. Parkinson's disease genes vps35 and eif4g1 interact genetically and converge on alpha-synuclein. Neuron. 2015;85:76–87. doi: 10.1016/j.neuron.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaronen M, Goldsteins G, Koistinaho J. Er stress and unfolded protein response in amyotrophic lateral sclerosis-a controversial role of protein disulphide isomerase. Front Cell Neurosci. 2014;8:402. doi: 10.3389/fncel.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussbaum-Krammer CI, Morimoto RI. Caenorhabditis elegans as a model system for studying non-cell-autonomous mechanisms in protein-misfolding diseases. Dis Model Mech. 2014;7:31–39. doi: 10.1242/dmm.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Wang X. The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity. Biochim Biophys Acta. 2015;1852:188–194. doi: 10.1016/j.bbadis.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 24.Del Monte F, Agnetti G. Protein post-translational modifications and misfolding: New concepts in heart failure. Proteomics Clin Appl. 2014;8:534–542. doi: 10.1002/prca.201400037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sher T, Gertz MA. Recent advances in the diagnosis and management of cardiac amyloidosis. Future Cardiol. 2014;10:131–146. doi: 10.2217/fca.13.85. [DOI] [PubMed] [Google Scholar]
- 26.Yusuf SW, Solhpour A, Banchs J, et al. Cardiac amyloidosis. Expert Rev Cardiovasc Ther. 2014;12:265–277. doi: 10.1586/14779072.2014.876363. [DOI] [PubMed] [Google Scholar]
- 27.Kabakov AE, Budagova KR, Latchman DS, Kampinga HH. Stressful preconditioning and hsp70 overexpression attenuate proteotoxicity of cellular atp depletion. Am J Physiol Cell Physiol. 2002;283:C521–534. doi: 10.1152/ajpcell.00503.2001. [DOI] [PubMed] [Google Scholar]
- 28.Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in cryabr120g cardiomyocytes. Circ Res. 2011;109:151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 31.Rujano MA, Bosveld F, Salomons FA, et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhardwaj RD, Curtis MA, Spalding KL, et al. Neocortical neurogenesis in humans is restricted to development. Proc Natl Acad Sci U S A. 2006;103:12564–12568. doi: 10.1073/pnas.0605177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolozin B. Physiological protein aggregation run amuck: Stress granules and the genesis of neurodegenerative disease. Discov Med. 2014;17:47–52. [PMC free article] [PubMed] [Google Scholar]
- 34.Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Berlo JH, Kanisicak O, Maillet M, et al. C-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senyo SE, Steinhauser ML, Pizzimenti CL, et al. Mammalian heart renewal by preexisting cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--alzheimer's disease of the heart? N Engl J Med. 2013;368:455–464. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- 38.Taipale M, Jarosz DF, Lindquist S. Hsp90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 39.Taipale M, Tucker G, Peng J, et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loomis WF, Wheeler S. Heat shock response of dictyostelium. Dev Biol. 1980;79:399–408. doi: 10.1016/0012-1606(80)90125-6. [DOI] [PubMed] [Google Scholar]
- 41.Bustamante CJ, Kaiser CM, Maillard RA, Goldman DH, Wilson CA. Mechanisms of cellular proteostasis: Insights from single-molecule approaches. Annu Rev Biophys. 2014;43:119–140. doi: 10.1146/annurev-biophys-051013-022811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macario AJ, Conway de Macario E. Sick chaperones, cellular stress, and disease. N Engl J Med. 2005;353:1489–1501. doi: 10.1056/NEJMra050111. [DOI] [PubMed] [Google Scholar]
- 43.van Oosten-Hawle P, Morimoto RI. Organismal proteostasis: Role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev. 2014;28:1533–1543. doi: 10.1101/gad.241125.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth DM, Hutt DM, Tong J, et al. Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol. 2014;12:e1001998. doi: 10.1371/journal.pbio.1001998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azad AA, Zoubeidi A, Gleave ME, Chi KN. Targeting heat shock proteins in metastatic castration-resistant prostate cancer. Nat Rev Urol. 2015;12:26–36. doi: 10.1038/nrurol.2014.320. [DOI] [PubMed] [Google Scholar]
- 46.Bakthisaran R, Tangirala R, Rao CM. Small heat shock proteins: Role in cellular functions and pathology. Biochim Biophys Acta. 2014;1854:291–319. doi: 10.1016/j.bbapap.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 47.Graner MW, Lillehei KO, Katsanis E. Endoplasmic reticulum chaperones and their roles in the immunogenicity of cancer vaccines. Front Oncol. 2014;4:379. doi: 10.3389/fonc.2014.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang L, Liu X, Cheng B, Huang K. How our bodies fight amyloidosis: Effects of physiological factors on pathogenic aggregation of amyloidogenic proteins. Arch Biochem Biophys. 2015 doi: 10.1016/j.abb.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Kishnani PS, Beckemeyer AA. New therapeutic approaches for pompe disease: Enzyme replacement therapy and beyond. Pediatr Endocrinol Rev. 2014;12(1):114–124. [PubMed] [Google Scholar]
- 50.Margulis J, Finkbeiner S. Proteostasis in striatal cells and selective neurodegeneration in huntington's disease. Front Cell Neurosci. 2014;8:218. doi: 10.3389/fncel.2014.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taldone T, Patel HJ, Bolaender A, Patel MR, Chiosis G. Protein chaperones: A composition of matter review (2008 - 2013) Expert Opin Ther Pat. 2014;24:501–518. doi: 10.1517/13543776.2014.887681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calamini B, Morimoto RI. Protein homeostasis as a therapeutic target for diseases of protein conformation. Curr Top Med Chem. 2012;12:2623–2640. doi: 10.2174/1568026611212220014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patterson C, Hohfeld J. Protein degradation vol 2: The ubiquitin-proteasome system. Wiley; 2006. Molecular chaperones and the ubiquitin-proteasome system. [Google Scholar]
- 54.Truee O, Matthias P. Interplay between histone deacetylases and autophagy - from cancer therapy to neurodegeneration. Immunol Cell Biol. 2012;90:78–84. doi: 10.1038/icb.2011.103. [DOI] [PubMed] [Google Scholar]
- 55.Christians ES, Mustafi SB, Benjamin IJ. Chaperones and cardiac misfolding protein diseases. Curr Protein Pept Sci. 2014;15:189–204. doi: 10.2174/1389203715666140331111518. [DOI] [PubMed] [Google Scholar]
- 56.McLendon PM, Robbins J. Desmin-related cardiomyopathy: An unfolding story. Am J Physiol Heart Circ Physiol. 2011;301:H1220–1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Robbins J. Proteasomal and lysosomal protein degradation and heart disease. J Mol Cell Cardiol. 2014;71:16–24. doi: 10.1016/j.yjmcc.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams J. The proteasome: Structure, function, and role in the cell. Cancer Treat Rev. 2003;29(1):3–9. doi: 10.1016/s0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- 59.McConkey DJ, White M, Yan W. Chapter four - hdac inhibitor modulation of proteotoxicity as a therapeutic approach in cancer. In: Steven G, editor. Adv cancer res. Academic Press; 2012. pp. 131–163. [DOI] [PubMed] [Google Scholar]
- 60.Nowis D, Maczewski M, Mackiewicz U, et al. Cardiotoxicity of the anticancer therapeutic agent bortezomib. Am J Pathol. 2010;176:2658–2668. doi: 10.2353/ajpath.2010.090690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 62.Gatica D, Chiong M, Lavandero S, Klionsky DJ. Molecular mechanisms of autophagy in the cardiovascular system. Circ Res. 2015;116:456–467. doi: 10.1161/CIRCRESAHA.114.303788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Meyer GR, Grootaert MO, Michiels CF, Kurdi A, Schrijvers DM, Martinet W. Autophagy in vascular disease. Circ Res. 2015;116:468–479. doi: 10.1161/CIRCRESAHA.116.303804. [DOI] [PubMed] [Google Scholar]
- 64.Gottlieb RA, Andres AM, Sin J, Taylor DP. Untangling autophagy measurements: All fluxed up. Circ Res. 2015;116:504–514. doi: 10.1161/CIRCRESAHA.116.303787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orogo AM, Gustafsson AB. Therapeutic targeting of autophagy: Potential and concerns in treating cardiovascular disease. Circ Res. 2015;116:489–503. doi: 10.1161/CIRCRESAHA.116.303791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gustafsson ÅB, Gottlieb RA. Autophagy in ischemic heart disease. Circulation Research. 2009;104:150–158. doi: 10.1161/CIRCRESAHA.108.187427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 68.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 69.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion 2008. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishino I, Fu J, Tanji K, et al. Primary lamp-2 deficiency causes x-linked vacuolar cardiomyopathy and myopathy (danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in lamp-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 72.Maloyan A, Sanbe A, Osinska H, et al. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-b-crystallin desmin-related cardiomyopathy. Circulation. 2005;112:3451–3461. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doroudgar S, Glembotski CC. New concepts of endoplasmic reticulum function in the heart: Programmed to conserve. Journal of Molecular and Cellular Cardiology. 2013;55:85–91. doi: 10.1016/j.yjmcc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 76.Zhang D, Contu R, Latronico MVG, et al. Mtorc1 regulates cardiac function and myocyte survival through 4e-bp1 inhibition in mice. The Journal of Clinical Investigation. 2010;120:2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chlebowski RT. Adriamycin (doxorubicin) cardiotoxicity: A review. Western Journal of Medicine. 1979;131:364–368. [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J, Zheng H, Tang M, Ryu YC, Wang X. A therapeutic dose of doxorubicin activates ubiquitin-proteasome system-mediated proteolysis by acting on both the ubiquitination apparatus and proteasome. American Journal of Physiology - Heart and Circulatory Physiology. 2008;295:H2541–H2550. doi: 10.1152/ajpheart.01052.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ranek M, Wang X. Activation of the ubiquitin-proteasome system in doxorubicin cardiomyopathy. Current Hypertension Reports. 2009;11:389–395. doi: 10.1007/s11906-009-0068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: Transcriptional, post-transcriptional, and post-translational regulation of autophagy. J Cell Biol. 2015 doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kraft C, Peter M, Hofmann K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 83.Orvedahl A, Sumpter R, Jr, Xiao G, et al. Image-based genome-wide sirna screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shoji-Kawata S, Levine B. Autophagy, antiviral immunity, and viral countermeasures. Biochim Biophys Acta. 2009;1793:1478–1484. doi: 10.1016/j.bbamcr.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bjorkoy G, Lamark T, Brech A, et al. P62/sqstm1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng Q, Su H, Tian Z, Wang X. Proteasome malfunction activates macroautophagy in the heart. Am J Cardiovasc Dis. 2011;1:214–226. [PMC free article] [PubMed] [Google Scholar]
- 87.Sanbe A, Osinska H, Saffitz JE, et al. Desmin-related cardiomyopathy in transgenic mice: A cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhuiyan MS, Pattison JS, Osinska H, et al. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boyault C, Zhang Y, Fritah S, et al. Hdac6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kovacs JJ, Murphy PJ, Gaillard S, et al. Hdac6 regulates hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 91.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase hdac6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 92.Pandey UB, Nie Z, Batlevi Y, et al. Hdac6 rescues neurodegeneration and provides an essential link between autophagy and the ups. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 93.Lee JY, Koga H, Kawaguchi Y, et al. Hdac6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Iwata A, Riley BE, Johnston JA, Kopito RR. Hdac6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 95.Wong E, Bejarano E, Rakshit M, et al. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat Commun. 2012;3:1240. doi: 10.1038/ncomms2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dharmarajan K, Maurer MS. Transthyretin cardiac amyloidoses in older north americans. J Am Geriatr Soc. 2012;60:765–774. doi: 10.1111/j.1532-5415.2011.03868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Spaendonck-Zwarts KY, van Hessem L, Jongbloed JD, et al. Desmin-related myopathy. Clin Genet. 2011;80:354–366. doi: 10.1111/j.1399-0004.2010.01512.x. [DOI] [PubMed] [Google Scholar]
- 98.Rothermel BA, Hill JA. The heart of autophagy: Deconstructing cardiac proteotoxicity. Autophagy. 2008;4:932–935. doi: 10.4161/auto.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rajasekaran NS, Connell P, Christians ES, et al. Human αb-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brewer AC, Mustafi SB, Murray TV, Rajasekaran NS, Benjamin IJ. Reductive stress linked to small hsps, g6pd, and nrf2 pathways in heart disease. Antioxid Redox Signal. 2013;18:1114–1127. doi: 10.1089/ars.2012.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pedersen JS. The nature of amyloid-like glucagon fibrils. J Diabetes Sci Technol. 2010;4:1357–1367. doi: 10.1177/193229681000400609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zemva J, Schubert M. Central insulin and insulin-like growth factor-1 signaling: Implications for diabetes associated dementia. Curr Diabetes Rev. 2011;7:356–366. doi: 10.2174/157339911797415594. [DOI] [PubMed] [Google Scholar]
- 103.Cohen E, Dillin A. The insulin paradox: Aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9:759–767. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pettersson T, Konttinen YT. Amyloidosis-recent developments. Semin Arthritis Rheum. 2010;39:356–368. doi: 10.1016/j.semarthrit.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 105.Franco R, Cedazo-Minguez A. Successful therapies for alzheimer's disease: Why so many in animal models and none in humans? Front Pharmacol. 2014;5:146. doi: 10.3389/fphar.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kholova I, Niessen HW. Amyloid in the cardiovascular system: A review. J Clin Pathol. 2005;58:125–133. doi: 10.1136/jcp.2004.017293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vicart P, Caron A, Guicheney P, et al. A missense mutation in the αb-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 108.Wang X, Osinska H, Dorn GW, 2nd, et al. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 109.Tanaka M, Kim YM, Lee G, Junn E, Iwatsubo T, Mouradian MM. Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. J Biol Chem. 2004;279:4625–4631. doi: 10.1074/jbc.M310994200. [DOI] [PubMed] [Google Scholar]
- 110.Filimonenko M, Isakson P, Finley KD, et al. The selective macroautophagic degradation of aggregated proteins requires the pi3p-binding protein alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: A role for aggrephagy in neurodegeneration. Neurobiol Dis. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 114.Berger Z, Roder H, Hanna A, et al. Accumulation of pathological tau species and memory loss in a conditional model of tauopathy. J Neurosci. 2007;27:3650–3662. doi: 10.1523/JNEUROSCI.0587-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kayed R, Head E, Thompson JL, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 116.Isas JM, Luibl V, Johnson LV, et al. Soluble and mature amyloid fibrils in drusen deposits. Invest Ophthalmol Vis Sci. 2010;51:1304–1310. doi: 10.1167/iovs.09-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Head E, Pop V, Sarsoza F, et al. Amyloid-beta peptide and oligomers in the brain and cerebrospinal fluid of aged canines. J Alzheimers Dis. 2010;20:637–646. doi: 10.3233/JAD-2010-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bacher M, Dodel R, Aljabari B, et al. Cni-1493 inhibits abeta production, plaque formation, and cognitive deterioration in an animal model of alzheimer's disease. J Exp Med. 2008;205:1593–1599. doi: 10.1084/jem.20060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Teplow DB. On the subject of rigor in the study of amyloid beta-protein assembly. Alzheimers Res Ther. 2013;5:39. doi: 10.1186/alzrt203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Benilova I, Karran E, De Strooper B. The toxic abeta oligomer and alzheimer's disease: An emperor in need of clothes. Nat Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 121.Shankar GM, Li S, Mehta TH, et al. Amyloid-beta protein dimers isolated directly from alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kitamura A, Kubota H, Pack CG, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 123.Takahashi T, Katada S, Onodera O. Polyglutamine diseases: Where does toxicity come from? What is toxicity? Where are we going? J Mol Cell Biol. 2010;2:180–191. doi: 10.1093/jmcb/mjq005. [DOI] [PubMed] [Google Scholar]
- 124.Avila DM, Allman DR, Gallo JM, McPhaul MJ. Androgen receptors containing expanded polyglutamine tracts exhibit progressive toxicity when stably expressed in the neuroblastoma cell line, sh-sy 5y. Exp Biol Med (Maywood) 2003;228:982–990. doi: 10.1177/153537020322800815. [DOI] [PubMed] [Google Scholar]
- 125.Tomiyama T, Matsuyama S, Iso H, et al. A mouse model of amyloid beta oligomers: Their contribution to synaptic alteration, abnormal tau phosphorylation, glial activation, and neuronal loss in vivo. J Neurosci. 2010;30:4845–4856. doi: 10.1523/JNEUROSCI.5825-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rivera JF, Costes S, Gurlo T, Glabe CG, Butler PC. Autophagy defends pancreatic beta cells from human islet amyloid polypeptide-induced toxicity. J Clin Invest. 2014;124:3489–3500. doi: 10.1172/JCI71981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Maloyan A, Gulick J, Glabe CG, Kayed R, Robbins J. Exercise reverses preamyloid oligomer and prolongs survival in alphaB-crystallin-based desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2007;104:5995–6000. doi: 10.1073/pnas.0609202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sanbe A, Osinska H, Villa C, et al. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102:13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pattison JS, Robbins J. Protein misfolding and cardiac disease: Establishing cause and effect. Autophagy. 2008;4:821–823. doi: 10.4161/auto.6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kayed R, Canto I, Breydo L, et al. Conformation dependent monoclonal antibodies distinguish different replicating strains or conformers of prefibrillar abeta oligomers. Mol Neurodegener. 2010;5:57. doi: 10.1186/1750-1326-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Glabe CG, Kayed R. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology. 2006;66:S74–78. doi: 10.1212/01.wnl.0000192103.24796.42. [DOI] [PubMed] [Google Scholar]
- 132.Kayed R, Glabe CG. Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 2006;413:326–344. doi: 10.1016/S0076-6879(06)13017-7. [DOI] [PubMed] [Google Scholar]
- 133.Kayed R, Head E, Sarsoza F, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kayed R, Lasagna-Reeves CA. Molecular mechanisms of amyloid oligomers toxicity. J Alzheimers Dis. 2013;33(1):S67–78. doi: 10.3233/JAD-2012-129001. [DOI] [PubMed] [Google Scholar]
- 135.Gallea JI, Celej MS. Structural insights into amyloid oligomers of the parkinson disease-related protein alpha-synuclein. J Biol Chem. 2014;289:26733–26742. doi: 10.1074/jbc.M114.566695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stefani M. Protein misfolding and aggregation: New examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta. 2004;1739:5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 137.Sunde M, Blake C. The structure of amyloid fibrils by electron microscopy and x-ray diffraction. Adv Protein Chem. 1997;50:123–159. doi: 10.1016/s0065-3233(08)60320-4. [DOI] [PubMed] [Google Scholar]
- 138.Cecchi C, Stefani M. The amyloid-cell membrane system. The interplay between the biophysical features of oligomers/fibrils and cell membrane defines amyloid toxicity. Biophys Chem. 2013;182:30–43. doi: 10.1016/j.bpc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 139.Glabe CG. Structural classification of toxic amyloid oligomers. J Biol Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Diociaiuti M, Macchia G, Paradisi S, et al. Native metastable prefibrillar oligomers are the most neurotoxic species among amyloid aggregates. Biochim Biophys Acta. 2014;1842:1622–1629. doi: 10.1016/j.bbadis.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 141.Yaar M, Zhai S, Fine RE, et al. Amyloid beta binds trimers as well as monomers of the 75-kda neurotrophin receptor and activates receptor signaling. J Biol Chem. 2002;277:7720–7725. doi: 10.1074/jbc.M110929200. [DOI] [PubMed] [Google Scholar]
- 142.Jang H, Connelly L, Arce FT, et al. Mechanisms for the insertion of toxic, fibril-like beta-amyloid oligomers into the membrane. J Chem Theory Comput. 2013;9:822–833. doi: 10.1021/ct300916f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Marshall KE, Marchante R, Xue WF, Serpell LC. The relationship between amyloid structure and cytotoxicity. Prion. 2014;8 doi: 10.4161/pri.28860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sebollela A, Mustata GM, Luo K, et al. Elucidating molecular mass and shape of a neurotoxic abeta oligomer. ACS Chem Neurosci. 2014;5:1238–1245. doi: 10.1021/cn500156r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun Y, Xi W, Wei G. Atomic-level study of the effects of o4 molecules on the structural properties of protofibrillar ass trimer: Ss-sheet stabilization, salt bridge protection, and binding mechanism. J Phys Chem B. 2015 doi: 10.1021/jp508122t. [DOI] [PubMed] [Google Scholar]
- 146.Kawahara M, Kuroda Y, Arispe N, Rojas E. Alzheimer's beta-amyloid, human islet amylin, and prion protein fragment evoke intracellular free calcium elevations by a common mechanism in a hypothalamic gnrh neuronal cell line. J Biol Chem. 2000;275:14077–14083. doi: 10.1074/jbc.275.19.14077. [DOI] [PubMed] [Google Scholar]
- 147.Pattison JS, Robbins J. Autophagy and proteotoxicity in cardiomyocytes. Autophagy. 2011;7:1259–1260. doi: 10.4161/auto.7.10.16882. [DOI] [PubMed] [Google Scholar]
- 148.Swinnen E, Buttner S, Outeiro TF, et al. Aggresome formation and segregation of inclusions influence toxicity of alpha-synuclein and synphilin-1 in yeast. Biochem Soc Trans. 2011;39:1476–1481. doi: 10.1042/BST0391476. [DOI] [PubMed] [Google Scholar]
- 149.Wakabayashi K, Tanji K, Odagiri S, Miki Y, Mori F, Takahashi H. The lewy body in parkinson's disease and related neurodegenerative disorders. Mol Neurobiol. 2013;47:495–508. doi: 10.1007/s12035-012-8280-y. [DOI] [PubMed] [Google Scholar]
- 150.Ojha J, Masilamoni G, Dunlap D, Udoff RA, Cashikar AG. Sequestration of toxic oligomers by hspb1 as a cytoprotective mechanism. Mol Cell Biol. 2011;31:3146–3157. doi: 10.1128/MCB.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang H, Macgillivray C, Kwon HS, et al. Three-dimensional cardiac architecture determined by two-photon microtomy. J Biomed Opt. 2009;14:044029. doi: 10.1117/1.3200939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Maloyan A, Osinska H, Lammerding J, et al. Biochemical and mechanical dysfunction in a mouse model of desmin-related myopathy. Circ Res. 2009;104:1021–1028. doi: 10.1161/CIRCRESAHA.108.193516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chávez Zobel AT, Loranger A, Marceau N, Thériault JR, Lambert H, Landry J. Distinct chaperone mechanisms can delay the formation of aggresomes by the myopathy-causing r120g αb-crystallin mutant. Human Molecular Genetics. 2003;12:1609–1620. doi: 10.1093/hmg/ddg173. [DOI] [PubMed] [Google Scholar]
- 154.Liu J, Chen Q, Huang W, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20:362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 155.Kumarapeli AR, Horak KM, Glasford JW, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J. 2005;19:2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 156.Chen Q, Liu JB, Horak KM, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 157.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol. 2011;301:H2207–2219. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tang M, Li J, Huang W, et al. Proteasome functional insufficiency activates the calcineurin-nfat pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88:424–433. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Olson EN, Molkentin JD. Prevention of cardiac hypertrophy by calcineurin inhibition: Hope or hype? Circ Res. 1999;84:623–632. doi: 10.1161/01.res.84.6.623. [DOI] [PubMed] [Google Scholar]
- 160.Liu Y, Hettinger CL, Zhang D, Rezvani K, Wang X, Wang H. The proteasome function reporter gfpu accumulates in young brains of the appswe/ps1de9 alzheimer's disease mouse model. Cell Mol Neurobiol. 2014;34:315–322. doi: 10.1007/s10571-013-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128:365–376. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gupta MK, Gulick J, Liu R, Wang X, Molkentin JD, Robbins J. Sumo e2 enzyme ubc9 is required for efficient protein quality control in cardiomyocytes. Circ Res. 2014;115:721–729. doi: 10.1161/CIRCRESAHA.115.304760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 165.Gurusamy N, Lekli I, Gorbunov NV, Gherghiceanu M, Popescu LM, Das DK. Cardioprotection by adaptation to ischaemia augments autophagy in association with bag-1 protein. J Cell Mol Med. 2009;13:373–387. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang C, Yitzhaki S, Perry CN, et al. Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res. 2010;3:365–373. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rohailla S, Clarizia N, Sourour M, et al. Acute, delayed and chronic remote ischemic conditioning is associated with downregulation of mtor and enhanced autophagy signaling. PLoS One. 2014;9:e111291. doi: 10.1371/journal.pone.0111291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xie M, Kong Y, Tan W, et al. Hdac inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.113.002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kanamori H, Takemura G, Goto K, et al. The role of autophagy emerging in postinfarction cardiac remodelling. Cardiovasc Res. 2011;91:330–339. doi: 10.1093/cvr/cvr073. [DOI] [PubMed] [Google Scholar]
- 170.Jimenez RE, Kubli DA, Gustafsson AB. Autophagy and mitophagy in the myocardium: Therapeutic potential and concerns. Br J Pharmacol. 2014;171:1907–1916. doi: 10.1111/bph.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cao DJ, Wang ZV, Battiprolu PK, et al. Histone deacetylase (hdac) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tannous P, Zhu H, Nemchenko A, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Noda NN, Satoo K, Fujioka Y, et al. Structural basis of atg8 activation by a homodimeric e1, atg7. Mol Cell. 2011;44:462–475. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 175.Strappazzon F, Nazio F, Corrado M, et al. Ambra1 is able to induce mitophagy via lc3 binding, regardless of parkin and p62/sqstm1. Cell Death Differ. 2015;22:419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Moyzis AG, Sadoshima J, Gustafsson AB. Mending a broken heart: The role of mitophagy in cardioprotection. Am J Physiol Heart Circ Physiol. 2015;308:H183–H192. doi: 10.1152/ajpheart.00708.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by parkin and p62/sqstm1. PLoS ONE. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. Chip-mediated stress recovery by sequential ubiquitination of substrates and hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ronnebaum SM, Patterson C, Schisler JC. Minireview: Hey u(ps): Metabolic and proteolytic homeostasis linked via ampk and the ubiquitin proteasome system. Mol Endocrinol. 2014;28:1602–1615. doi: 10.1210/me.2014-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schisler JC, Rubel CE, Zhang C, Lockyer P, Cyr DM, Patterson C. Chip protects against cardiac pressure overload through regulation of ampk. The Journal of Clinical Investigation. 2013;123:3588–3599. doi: 10.1172/JCI69080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Meley D, Bauvy C, Houben-Weerts JH, et al. Amp-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 182.Rajasekaran NS, Connell P, Christians ES, et al. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sulistio YA, Heese K. The ubiquitin-proteasome system and molecular chaperone deregulation in alzheimer's disease. Mol Neurobiol. 2015 doi: 10.1007/s12035-014-9063-4. [DOI] [PubMed] [Google Scholar]
- 184.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 185.Nishihori T, Baz R, Shain K, et al. An open label phase i/ii study of cyclophosphamide, bortezomib, pegylated liposomal doxorubicin and dexamethasone in newly diagnosed myeloma. Eur J Haematol. 2015 doi: 10.1111/ejh.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 187.Ryter SW, Choi AM. Autophagy in lung disease pathogenesis and therapeutics. Redox Biol. 2015;4C:215–225. doi: 10.1016/j.redox.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lynch-Day MA, Mao K, Wang K, Zhao M, Klionsky DJ. The role of autophagy in parkinson's disease. Cold Spring Harb Perspect Med. 2012;2:a009357. doi: 10.1101/cshperspect.a009357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang Y, James M, Middleton FA, Davis RL. Transcriptional analysis of multiple brain regions in parkinson's disease supports the involvement of specific protein processing, energy metabolism, and signaling pathways, and suggests novel disease mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2005;137B:5–16. doi: 10.1002/ajmg.b.30195. [DOI] [PubMed] [Google Scholar]
- 190.Sarkar S, Floto RA, Berger Z, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sarkar S, Perlstein EO, Imarisio S, et al. Small molecules enhance autophagy and reduce toxicity in huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Williams A, Sarkar S, Cuddon P, et al. Novel targets for huntington's disease in an mtor-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Aguado C, Sarkar S, Korolchuk VI, et al. Laforin, the most common protein mutated in lafora disease, regulates autophagy. Hum Mol Genet. 2010;19:2867–2876. doi: 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cheng Y, Ren X, Hait WN, Yang JM. Therapeutic targeting of autophagy in disease: Biology and pharmacology. Pharmacol Rev. 2013;65:1162–1197. doi: 10.1124/pr.112.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Martelli AM, Chiarini F, Evangelisti C, et al. Two hits are better than one: Targeting both phosphatidylinositol 3-kinase and mammalian target of rapamycin as a therapeutic strategy for acute leukemia treatment. Oncotarget. 2012;3:371–394. doi: 10.18632/oncotarget.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sarkar S. Chemical screening platforms for autophagy drug discovery to identify therapeutic candidates for huntington's disease and other neurodegenerative disorders. Drug Discov Today Technol. 2013;10:e137–144. doi: 10.1016/j.ddtec.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 197.Zhou M, Wang R. Small-molecule regulators of autophagy and their potential therapeutic applications. ChemMedChem. 2013;8:694–707. doi: 10.1002/cmdc.201200560. [DOI] [PubMed] [Google Scholar]
- 198.Salabei JK, Conklin DJ. Cardiovascular autophagy: Crossroads of pathology, pharmacology and toxicology. Cardiovasc Toxicol. 2013;13:220–229. doi: 10.1007/s12012-013-9200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jankowsky JL, Melnikova T, Fadale DJ, et al. Environmental enrichment mitigates cognitive deficits in a mouse model of alzheimer's disease. J Neurosci. 2005;25:5217–5224. doi: 10.1523/JNEUROSCI.5080-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lazarov O, Robinson J, Tang YP, et al. Environmental enrichment reduces abeta levels and amyloid deposition in transgenic mice. Cell. 2005;120:701–713. doi: 10.1016/j.cell.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 201.Adlard PA, Perreau VM, Pop V, Cotman CW. Voluntary exercise decreases amyloid load in a transgenic model of alzheimer's disease. J Neurosci. 2005;25:4217–4221. doi: 10.1523/JNEUROSCI.0496-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.van Dellen A, Blakemore C, Deacon R, York D, Hannan AJ. Delaying the onset of huntington's in mice. Nature. 2000;404:721–722. doi: 10.1038/35008142. [DOI] [PubMed] [Google Scholar]
- 203.Capurro A, Bodea LG, Schaefer P, Luthi-Carter R, Perreau VM. Computational deconvolution of genome wide expression data from parkinson's and huntington's disease brain tissues using population-specific expression analysis. Front Neurosci. 2014;8:441. doi: 10.3389/fnins.2014.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Grumati P, Coletto L, Schiavinato A, et al. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen vi-deficient muscles. Autophagy. 2011;7:1415–1423. doi: 10.4161/auto.7.12.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hulmi JJ, Oliveira BM, Silvennoinen M, et al. Exercise restores decreased physical activity levels and increases markers of autophagy and oxidative capacity in myostatin/activin-blocked mdx mice. Am J Physiol Endocrinol Metab. 2013;305:E171–182. doi: 10.1152/ajpendo.00065.2013. [DOI] [PubMed] [Google Scholar]
- 206.Jamart C, Naslain D, Gilson H, Francaux M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am J Physiol Endocrinol Metab. 2013;305:E964–974. doi: 10.1152/ajpendo.00270.2013. [DOI] [PubMed] [Google Scholar]
- 207.Mattson MP. Energy intake and exercise as determinants of brain health and vulnerability to injury and disease. Cell Metab. 2012;16:706–722. doi: 10.1016/j.cmet.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mattson MP. Interventions that improve body and brain bioenergetics for parkinson's disease risk reduction and therapy. J Parkinsons Dis. 2014;4:1–13. doi: 10.3233/JPD-130335. [DOI] [PubMed] [Google Scholar]
- 209.Antos CL, McKinsey TA, Dreitz M, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278:28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 210.McLendon PM, Ferguson BS, Osinska H, et al. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc Natl Acad Sci U S A. 2014;111:E5178–5186. doi: 10.1073/pnas.1415589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Demos-Davies KM, Ferguson BS, Cavasin MA, et al. Hdac6 contributes to pathological responses of heart and skeletal muscle to chronic angiotensin-ii signaling. Am J Physiol Heart Circ Physiol. 2014;307:H252–258. doi: 10.1152/ajpheart.00149.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Geeraert C, Ratier A, Pfisterer SG, et al. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J Biol Chem. 2010;285:24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bucciantini M, Giannoni E, Chiti F, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 214.Dobson CM. Protein folding and disease: A view from the first horizon symposium. Nat Rev Drug Discov. 2003;2:154–160. doi: 10.1038/nrd1013. [DOI] [PubMed] [Google Scholar]
- 215.Fuentealba RA, Marasa J, Diamond MI, Piwnica-Worms D, Weihl CC. An aggregation sensing reporter identifies leflunomide and teriflunomide as polyglutamine aggregate inhibitors. Hum Mol Genet. 2012;21:664–680. doi: 10.1093/hmg/ddr500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nollen EA, Garcia SM, van Haaften G, et al. Genome-wide rna interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Moffat J, Grueneberg DA, Yang X, et al. A lentiviral rnai library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 218.Burkhardt MF, Martinez FJ, Wright S, et al. A cellular model for sporadic als using patient-derived induced pluripotent stem cells. Mol Cell Neurosci. 2013;56:355–364. doi: 10.1016/j.mcn.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang X, Osinska H, Klevitsky R, et al. Expression of r120g-alpha-B-crystallin causes aberrant desmin and alpha B-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]