Abstract
The unprecedented scale of the Ebola outbreak in West Africa has, as of 29 April 2015, resulted in more than 10,884 deaths among 26,277 cases. Prior to the ongoing outbreak, Ebola virus disease (EVD) caused relatively small outbreaks (maximum outbreak size 425 in Gulu, Uganda) in isolated populations in central Africa. Here, we have compiled a comprehensive database of estimates of epidemiological parameters based on data from past outbreaks, including the incubation period distribution, case fatality rate, basic reproduction number (R0), effective reproduction number (Rt) and delay distributions. We have compared these to parameter estimates from the ongoing outbreak in West Africa. The ongoing outbreak, because of its size, provides a unique opportunity to better understand transmission patterns of EVD. We have not performed a meta-analysis of the data, but rather summarize the estimates by virus from comprehensive investigations of EVD and Marburg outbreaks over the past 40 years. These estimates can be used to parameterize transmission models to improve understanding of initial spread of EVD outbreaks and to inform surveillance and control guidelines.
Subject terms: Epidemiology, Ebola virus, Viral epidemiology, Viral infection
Background & Summary
Ebola virus disease (EVD), formerly known as Ebola hemorrhagic fever, is caused by a zoonotic virus first discovered in 1976 in remote villages of Democratic Republic of Congo (DRC, formerly Zaire) and Sudan1–4. The virus was again identified in the mid-1990s, when it re-emerged in Gabon and Kikwit, DRC5,6. Since then, there have been sporadic outbreaks in human and non-human-primate populations, primarily in remote areas in central Africa (Table 1)7–9. There are five Ebola viruses: Zaire ebolavirus, Sudan ebolavirus, Bundibugyo ebolavirus, Tai Forest ebolavirus and Reston ebolavirus. The most lethal is the Ebola Zaire virus. Ebola Reston is unique among the five Ebola viruses in that it is not known to cause disease in humans10 and Ebola Tai Forest has only been reported in 1 human case11. Together with Marburg virus, Ebola forms the Filoviridae family (filovirus)8.
Table 1. Human Outbreaks of Ebola Zaire, Ebola Sudan and Ebola Bundibugyo from 1976 to present.
| Outbreak | Year | Ebola virus | Number of confirmed, probable and suspected cases |
|---|---|---|---|
| Outbreaks with more than 1 case are included in the table. |
|||
| DRC=Democratic Republic of Congo; The outbreak in West Africa includes cases from the following countries: Guinea, Liberia, Mali, Nigeria, Senegal and Sierra Leone, Spain, United Kingdom, and United States. |
|||
| Yambuku, DRC2 | 1976 | Zaire | 318 |
| South Sudan1 | 1976 | Sudan | 284 |
| Nzara, Sudan24 | 1979 | Sudan | 34 |
| Gabon6 | 1994–1995 | Zaire | 49 |
| Kikwit, DRC5 | 1995 | Zaire | 315 |
| Maybout, Gabon* | 1996 | Zaire | 37 |
| Booue, Gabon6 | 1996–1997 | Zaire | 60 |
| South Africa74 | 1996 | Zaire | 2 |
| Gulu, Uganda45,52,53 | 2000–2001 | Sudan | 425 |
| Republic of Congo and Gabon75 | 2001–2002 | Zaire | 65 in Gabon, 59 in Congo |
| Republic of Congo25,72 | 2002–2003 | Zaire | 143 |
| Mbomo, Republic of Congo25 | 2003 | Zaire | 35 |
| Yambio, South Sudan† | 2004 | Sudan | 17 |
| Etoumbi, Republic of Congo25 | 2005 | Zaire | 12 |
| Kasai Occidental Province, DRC23 | 2007 | Zaire | 264 |
| Bundibugyo District, Uganda47 | 2007–2008 | Bundibugyo | 116 |
| Kasai Occidental Province, DRC‡ | 2008–2009 | Zaire | 32 |
| Orientale Province, DRC§ | 2012 | Bundibugyo | 77 |
| Kibaale District, Uganda|| | 2012 | Sudan | 24 |
| Luwero District, Uganda¶ | 2012–2013 | Sudan | 7 |
| Équateur province, DRC49 | 2014 | Zaire | 69 |
| West Africa# | 2014–2015 | Zaire | >26,000 |
*Reference: WHO 26 April 1996 Disease Outbreak News. 1996- Ebola haemorrhagic fever in Gabon- Update3: www.who.int/csr/don/1996_04_26b/en/.
†Reference: WHO 7 August 2004: WHO announces end of Ebola Outbreak in Southern Sudan: www.who.int/csr/don/2004_08_07/en/.
‡Reference: WHO Disease Outbreak News: www.who.int/csr/don/2009_02_17/en/.
||Reference: WHO Disease Outbreak News: www.who.int/csr/don/2012_09_03/en/.
¶Reference: WHO Disease Outbreak News: www.who.int/csr/don/2012_11_30_ebola/en/.
#Outbreak is ongoing in Guinea, Liberia and Sierra Leone. Case count reflects cases reported as of 29 April 2015.
The primary reservoir of the Ebola virus is believed to be fruit bats12,13. However, non-human primates, including chimpanzees, gorillas, and cynomolgus monkeys, and forest antelopes have been reported as possible vectors in transmission to humans14, and EVD has caused devastating mortality in non-human-primate populations15. Once infected, the symptoms of human EVD are non-specific and typically include fever, headache, joint or muscle pain, sore throat, vomiting, and/or diarrhea15–17. More severe cases involve hemorrhagic manifestations, shock and other neurological symptoms14,16–21.
While it has been difficult to trace the source of human outbreaks, it is believed that EVD outbreaks usually start from a zoonotic source with subsequent human-to-human transmission22,23. Transmission between humans occurs through exposure to infectious bodily fluids, typically from close contact with infectious individuals when caring for EVD patients (e.g., sharing of contaminated needles, family home care, insufficient protective measures among health care workers in health care settings6,24,25) or with fatal EVD patients in preparation for burial19,20. Control measures for EVD are well documented and include identification, isolation and care of suspected patients, strict infection prevention and control among those caring for patients and safe burials26,27.
At the start of an infectious disease outbreak, it is critical to understand the transmission dynamics of the pathogen and to determine those at highest risk for infection or severe outcomes in the population(s) affected28,29. This information is needed to develop interventions to reduce the spread of disease and to reduce morbidity and mortality in the affected populations. Real-time analysis of any ongoing outbreak by analyzing detailed information collected on the confirmed, probable and suspected cases and deaths provides an opportunity to determine the stages of disease and areas where control measures can be applied. For example, knowledge of the incubation period distribution of the pathogen will inform the duration of time required to follow up the contacts of cases to evaluate whether or not they become secondary cases. Additionally, information on the timing of symptom onset, isolation, hospitalization and outcome (either death or recovery) are important to understand EVD progression. Mathematical models which make use of available data early in an outbreak to estimate the outbreak’s potential impact are increasingly used by public health policy makers to inform decision making around emerging and re-emerging pathogens28–30.
The purpose of this review was to collect all published epidemiological parameter estimates (reprinted in detailed tables containing estimates, and corresponding confidence intervals) estimated from past EVD outbreaks. Our aim was not to perform a meta-analysis, but rather to compile and document the available parameter estimates based on data from EVD outbreaks over the past 40 years. In order to estimate any of the parameters referenced in our manuscript, we would need detailed case data of each of the cohorts studied in the original papers, which we do not have. We also reprint parameter estimates from past Marburg outbreaks and the ongoing outbreak in West Africa for comparison. This information is valuable for public health organizations that need to quickly evaluate the early behavior of a new outbreak and estimate the potential impact, in terms of morbidity, mortality and geographic spread. We highlight how the parameter estimates we have examined improve our understanding of EVD epidemiology. Our results help to put the ongoing EVD outbreak in West Africa into context and to assess the likely effects of ongoing and novel interventions.
Methods
Data collection
All searches using the following search terms (Ebola, Marburg, EHF, EVD, MHF, EBOV, Ebola Zaire, Ebola Sudan, Ebola Reston, Ebola Bundibugyo, outbreak, model, parameterization, incubation period, case fatality rate, case fatality rate (CFR), risk factors, basic reproduction number, R0, effective reproduction number, serial interval, delay distributions, generation time) were carried out on 1 August 2014, 15 September 2014 and again in February 2015 using the following databases: ScienceDirect, ResearchGate, Google, GoogleScholar, BioOne, Web of Science and PubMed. Our searches aimed to find primary reports describing and analyzing data collected from investigations of EVD and Marburg outbreaks since the virus was identified in 1976. The criteria for inclusion were: sample size of EVD cases described in the study ≥5, studies of human outbreaks, studies which evaluated potential risk factors had to report prevalence proportion ratios, odds ratios or relative risks. Reviews, commentaries, case reports on individual cases, and policy pieces were excluded. Additionally, literature evaluating non-human outbreaks or the potential for international (human) spread of EVD outside of an outbreak zone was excluded.
Using these search terms, a total of 49 papers were determined eligible for inclusion. In addition, for context we included additional published information on EVD including the final outbreak sizes as reported by the World Health Organization (WHO) Disease Outbreak News following declaration that each outbreak was over.
From the relevant EVD and Marburg literature, we extracted the following details for all parameter estimates (as provided): point estimates, confidence intervals, ranges, sample size used to estimate the parameter (total numbers of cases encompassing confirmed, suspected, and retrospectively diagnosed cases, depending on the study), EVD virus, and inferential methods. We then compiled the parameter estimate database into tables. Table 1 and Data Citation 1 list the human outbreaks of Ebola Zaire, Ebola Sudan and Ebola Bundibugyo that have occurred in Africa from 1976 to present. We have not provided detailed information on the outbreaks as these have been previously described9. Table 2 (available online only) summarizes the literature we used in this review.
Table 2. List of studies used in the review and the estimated parameters.
|
Estimated Parameters
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study | Virus | Year ofoutbreak | Incubation PeriodDistribution | R0 | Rt | Serial Interval*Distribution | GenerationTime Distribution | Delay Distributions | CFR | RFI |
| CFR refers to case fatality rate. |
||||||||||
| RFI refers to risk factor for infection. |
||||||||||
| International Commission2 | EBOV Zaire | 1976 | x | x | x | |||||
| Camacho et al. 38 | EBOV Zaire | 1976 | x | x | x | x | ||||
| WHO International Study Team1 | EBOV Sudan | 1976 | x | x | ||||||
| Baron et al. 24 | EBOV Sudan | 1979 | x | x | x | |||||
| Dowell et al. 39 | EBOV Zaire | 1995 | x | x | x | x | ||||
| Bwaka et al. 37 | EBOV Zaire | 1995 | x | x | ||||||
| Chowell et al. 44 | EBOV Zaire and Sudan | 1995; 2000–2001 | x | x | x | x | ||||
| Lekone & Finkenstädt42 | EBOV Zaire | 1995 | x | x | ||||||
| Eichner et al. 40 | EBOV Zaire | 1995 | x | |||||||
| Ndambi et al. 43 | EBOV Zaire | 1995 | x | x | ||||||
| Khan et al. 5 | EBOV Zaire | 1995 | x | x | x | |||||
| Muyembe & Kipasa70 | EBOV Zaire | 1995 | x | x | ||||||
| Legrand et al. 59 | EBOV Zaire and Sudan | 1995; 2000–2001 | x | |||||||
| Ferrari et al. 58 | EBOV Zaire | 1995 | x | |||||||
| Sadek et al. 50 | EBOV Zaire | 1995 | x | x | x | |||||
| Rowe et al. 20 | EBOV Zaire | 1995 | x | |||||||
| Bertherat et al. 71 | EBOV Zaire | 1995 | x | |||||||
| Ndanguza et al. 35 | EBOV Zaire | 1995 | x | x | ||||||
| White & Pagano60 | EBOV Zaire | 1995 | x | x | ||||||
| Georges et al. 6 | EBOV Zaire | 1996 | x | x | ||||||
| Okware et al. 53 | EBOV Sudan | 2000–2001 | x | x | x | x | ||||
| Borchert et al. 51 | EBOV Sudan | 2000 | x | x | ||||||
| Lamunu et al. 52 | EBOV Sudan | 2000–2001 | x | x | ||||||
| Francesconi et al. 45 | EBOV Sudan | 2000–2001 | x | x | ||||||
| Nkoghe et al. 25 | EBOV Zaire | 2005 | x | x | ||||||
| MacNeil et al. 46 | EBOV Bundibugyo | 2007 | x | x | ||||||
| Wamala et al. 47 | EBOV Bundibugyo | 2007 | x | x | x | x | ||||
| Roddy et al. 19 | EBOV Bundibugyo | 2007 | x | x | ||||||
| Althaus36 | EBOV Zaire | 2014–2015 | x | x | ||||||
| Gomes et al. 62 | EBOV Zaire | 2014–2015 | x | |||||||
| Fisman et al. 30 | EBOV Zaire | 2014–2015 | x | x | ||||||
| Nishiura & Chowell65 | EBOV Zaire | 2014–2015 | x | |||||||
| WHO Ebola Response Team16 | EBOV Zaire | 2014–2015 | x | x | x | x | x | x | ||
| WHO Ebola Response Team17 | EBOV Zaire | 2014–2015 | x | x | x | x | x | |||
| Althaus et al. 61 | EBOV Zaire | 2014–2015 | x | x | x | x | ||||
| Fasina et al. 54 | EBOV Zaire | 2014–2015 | x | x | x | |||||
| Faye et al. 41 | EBOV Zaire | 2014–2015 | x | x | x | x | x | |||
| Lewnard et al. 64 | EBOV Zaire | 2014–2015 | x | |||||||
| Towers et al. 66 | EBOV Zaire | 2014–2015 | x | |||||||
| Maganga et al. 49 | EBOV Zaire | 2014 | x | x | x | x | ||||
| Webb et al. 68 | EBOV Zaire | 2014–2015 | x | |||||||
| White et al. 69 | EBOV Zaire | 2014–2015 | x | |||||||
| Yamin et al. 76 | EBOV Zaire | 2014–2015 | x | |||||||
| Rivers et al. 48 | EBOV Zaire | 2014–2015 | x | x | x | x | ||||
| Colebunders et al. 57 | Marburg | 1998 | x | x | ||||||
| Bausch et al. 56 | Marburg | 1998 | x | |||||||
| Ajelli & Merler55 | Marburg | 2005 | x | x | x | x |
*Some of the studies refer to this parameter estimate as generation time but actually estimate serial interval.
Our manuscript and tables include estimates, confidence intervals and ranges obtained from the referenced publications (Table 2 (available online only) and Data Citation 2).
Definition of key parameters recorded
The incubation period is the interval between exposure to a pathogen and initial occurrence of symptoms and signs28,29. The incubation period distribution is usually characterized using the mean or the median incubation period.
The CFR is the proportion of cases (infected symptomatic individuals) within a designated population who die as a result of their infection. For past EVD and Marburg outbreaks, we report on the CFR estimated after the outbreak was declared over (estimated at least 42 days after the last case experienced symptom onset) by taking the number of deaths among cases divided by the total number of cases recorded during the outbreak. However, during outbreaks, the CFR is often estimated before all cases have been identified and before some cases have either recovered or died.
Risk factors for infection include demographic factors, medical conditions and behavioral exposures or practices that are associated with an individual’s risk of becoming infected with Ebola.
The basic reproduction number (R0) is used to measure the transmission potential of a disease. It is the average number of secondary infections produced by an infected case in a susceptible population31. If R0 >1, then once established the outbreak will continue, whereas if R0<1, then the outbreak will die out.
The effective reproduction number (Rt) is similar to R0 but relates to a particular calendar time t (after the start of the outbreak). Like R0, if Rt>1, then the outbreak will continue, whereas if Rt<1, then the outbreak will die out. Rt can be reduced through the use of successful control measures (e.g., by limiting contacts between susceptible and infectious individuals). Rt can also be reduced due to the depletion of susceptible individuals whether through extensive transmission or through the immunization of susceptible individuals32.
The serial interval is the interval between symptom onset in an index case and symptom onset in a secondary case infected by that index case33.
The generation time is the interval between infection of an index case and infection of a secondary case infected by that index case. The serial interval is more frequently estimated than the generation time and is often assumed to be the same duration as the generation time34.
Delay distributions
Symptom onset to hospitalization (also referred to as onset to clinical assessment): The interval between symptom onset and hospitalization.
Hospital admission to day of first blood sample: The interval between admission to hospital or medical facility for treatment of EVD and when a biological sample is collected for diagnosis.
Symptom onset to recovery/discharge: The interval between symptom onset and recovery or hospital discharge.
Symptom onset to death: The interval between symptom onset and death.
Duration of admission (survivors)—hospitalization to discharge: The interval between admission to a hospital or medical facility for treatment of EVD and discharge from the facility.
Duration of admission (fatal cases)—hospitalization to death: The interval between admission to a hospital or medical facility for treatment of EVD and death.
Data Records
The data from this analysis are summarized in two types of data format. Four data tables detail the methods and parameter estimates from each study included in our review. Our data tables:
Table S1: Human Outbreaks of Ebola Zaire, Ebola Sudan and Ebola Bundibugyo from 1976 presents compiled data on the year and location of the each human outbreak, the Ebola Virus causing the outbreak and number of cases reported (Data Citation 1).
Table S2: Parameter Estimates by Outbreak presents a comprehensive list of parameter estimates, including incubation period distribution, reproduction number, serial interval distribution, generation time distribution, delay distributions, and CFR by Ebola virus and study (Data Citation 2).
Table S3: Parameter estimates for the ongoing EVD outbreak in West Africa presents published estimates of delay distributions and CFR for the ongoing outbreak in West Africa (Data Citation 3).
Table S4: Risk factors for EVD and Marburg infection presents published results from outbreak (Data Citation 4).
Using these four tables, we then summarized the parameter database in six tables and two figures presented in this article. The parameters estimated for Ebola Zaire, Ebola Sudan and Ebola Bundibugyo outbreaks, including the incubation period distribution, serial interval distribution, R0, delay distributions and CFR, are shown in Tables 3 (available online only), 4 (available online only), 5, respectively. Parameter estimates for the ongoing outbreak in West Africa are summarized in Table 6 (available online only) and for Marburg outbreaks are presented in a single table (Table 7). Risk factors for Ebola and Marburg infection are summarized in Table 8 (available online only). Estimates of the incubation period distribution and CFR are presented in Figs 1 and 2, respectively.
Table 3. Parameter Estimates for Ebola Zaire excluding the ongoing outbreak in West Africa.
| Parameter | Estimate |
|---|---|
| Med=median; s.d.=standard deviation. |
|
| Incubation Period Distribution (range of central estimates, (range)) * | 5.3–12.7 (1–21) days |
| Serial interval Distribution (range of mean estimates) | 10–16.1 days |
| Khan et al. (mean) † 5 | 14 days‡ |
| Muyembe & Kipasa (approximation) † 70 | 10 days ‡ |
| Dowell et al. (median, (range)) † 39 | Med=17 days (9–25) |
| White & Pagano (mean, (IQR)) 60 | 5.82 days (5.43–7.60) |
| Maganga et al. (median, (range), mean, s.d.) 49 | Med=16 days (3–27), 16.1 days, 4.4 |
| R 0 (range of estimates) | 1.36–4.71 |
| Chowell et al. (estimate, s.d.) 44 | 1.83, 0.06 |
| Ferrari et al. (estimate, (95% CI) 58 | 3.65 (3.05, 4.33) |
| Legrand et al. (estimate, (95% CI)) 59 | 2.7 (1.9, 2.8) |
| Lekone & Finkenstädt (estimate, s.d.) 42 | 1.36, 0.13 |
| Ndanguza et al. (estimate, (95% CI)) 35 | 2.22 (1.90, 2.73) |
| White & Pagano (estimate, (IQR)) 60 | 1.93 (1.74–2.78) |
| Camacho et al. (estimate, (95% CI)) 38 | 4.71 (3.92, 5.66) |
| R t (range of estimates) | 0.84–1.29 |
| Maganga et al. (estimate, (95% CI) 49) | 1.29 (−4.72, 7.29)0.84 (−0.38, 2.06) |
|
Delay Distributions (days)
|
|
| Infectious period |
|
| Chowell et al. (mean, s.d.) 44 | 5.61,0.19 |
| Symptom onset to… | |
| … hospitalization (range of mean and median estimates) | 4–5; Med=3–4 |
| Khan et al. (mean, median, (range), n) 5 | 5, Med=4 (0–19), n=219 |
| Rowe et al. (mean, s.d., median, (range)) 20 | 4, 3.3, Med=3 (0–14) |
| Camacho et al. (median, (95% CI)) 38 | Med=3.00 (2.81, 3.20) |
| … death (range of mean estimates) | 6–10.1 |
| Camacho et al. (median, (95% CI)) 38 | Med=7.49 (7.30, 7.69) |
| Bwaka et al. (mean, (range), n) 37 | 10.1 (3–21), n=86 |
| Dowell et al. (median) 39 | Med=10 |
| Khan et al. (mean, median, (range), n) 5 | 9.6, Med=9 (0–34), n=224 |
| Nkoghe et al. (mean, (range), n) 25 | 6.2 (3–13), n=12 |
| Med=6 (<15 years old) | |
| Med=9 (15–29 years old) | |
| Sadek et al. (median, n) 50 | Med=10 (30–44 years old) |
| Med=8 (45–59 years old) | |
| Med=9.5 (>59 years old), overall n=226 | |
| Georges et al. (range) 6 | 12–18 |
| Maganga et al. (median, (range), mean, s.d.) 49 | 11 (1–30), 11.3, 6.8 |
| … recovery | 10 |
| Camacho et al. (median, (95% CI)) 38 | 10.00 (9.80, 10.19) |
| Hospitalization to… | |
| … discharge | 17 |
| Khan et al. (mean, median, (range), n) 5 | 17, Med=14 (0–56), n=34 |
| … death | 4.6 |
| Khan et al. (mean, median, (range), n) 5 | 4.6, Med=4 (0–20), n=185 |
| Death to burial | |
| Camacho et al. (median, (95% CI)) 38 | Med=0.99 (0.8, 1.18) |
| Overall Case Fatality Rate (range of estimates)α | 0.69–0.88 |
| International Commission (estimate, n, year of outbreak) 2 | 0.88, n=318, 1976 |
| Muyembe & Kipasa (estimate, n, year of outbreak) 70 | 0.74, n=136, 1995 |
| Ndambi et al. (estimate, n, year of outbreak) 43 | 0.78, n=23, 1995 |
| Khan et al. (estimate, n, year of outbreak) 5 | 0.81, n=315, 1995 |
| Overall: 0.807, n=310, 1995 | |
| 0.778 (<15 years old) | |
| Sadek et al. (estimate, n, year of outbreak) 50 | 0.69 (15–29 years old) |
| 0.796 (30–44 years old) | |
| 0.89 (45–59 years old) | |
| 0.957 (>59 years old) | |
| Chowell et al. (estimate, n, year of outbreak) 44 | 0.81, n=315, 1995 |
| Nkoghe et al. (estimate, n, year of outbreak) 25 | 0.83, n=12, 2005 |
| Camacho et al. (estimate, n, year of outbreak) 38 | 0.88, n=262, 1976 |
| Maganga et al. (estimate, n, year of outbreak) 49 | 0.74, n=69, 2014 |
*See Fig. 1 for individual estimates; extreme outlier estimate from Ndanguza et al. 35 not included.
†generation time estimated as serial interval.
‡No methodology provided.
α*See Fig. 2 for CFR estimates.
Table 4. Parameter Estimates for Ebola Sudan .
| Parameter | Estimate |
|---|---|
| Med=Median; s.d.=standard deviation. |
|
| Incubation Period Distribution (range of central estimates, (range)) * | 3.35–14 (2–21) days |
| R 0 (range of estimates) | 1.34–2.7 |
| Chowell et al. (estimate, s.d.)44 | 1.34, 0.03 |
| Legrand et al. (estimate, (95% CI)) 59 | 2.7 (2.5, 4.1) |
| Ferrari et al. (estimate, (95% CI)) 58 | 1.79 (1.52, 2.30) |
|
Delay Distributions (days)
|
|
| Infectious period… |
|
| Chowell et al. (mean, s.d.) 44 | 3.50,0.67 |
| Symptom onset to… |
|
| … hospitalization | 2 |
| Borchert et al. (mean, (range), n) 51 | 2 (0–8), n=26 |
| … death | 9 |
| Baron et al. (median, (range), n) 24 | Med=9 (5–15), n=22 |
| … discharge | 12 |
| Okware et al. (mean, (range), median) 53 | 12, (2–35), 13 |
| Hospitalization to… |
|
| … discharge (range of mean estimates) | 8–10 |
| Borchert et al. (mean, (range), n) 51 | 8.0 (2–11) n=8 |
| Okware et al. (mean, (range)) 53 | 10 (1–29) |
| … death | 6.1 |
| Borchert et al. (mean, (range), n) 51 | 6.1 (2–13) n=18 |
| Case Fatality Rate † (range of estimates) | 0.53–0.69 |
| WHO International Study Team (estimate, n, year of outbreak) 1 | 0.53, n=284, 1976 |
| Baron et al. (estimate, n, year of outbreak) 24 | 0.65, n=34, 1979 |
| Okware et al. (estimate, n, year of outbreak) 53 | 0.53, n=425, 2000–2001 |
| Lamunu et al. (estimate, n, year of outbreak) 52 | 0.53, n=425, 2000–2001 |
| Chowell et al. (estimate, n, year of outbreak) 44 | 0.53, n=425, 2000–2001 |
| Borchert et al. (estimate, n, year of outbreak) 51 | 0.69, n=26, 2000 |
*See Fig. 1 for individual estimates.
†See Fig. 2 for CFR estimates.
Table 5. Parameter Estimates for Ebola Bundibugyo .
| Parameter Estimate | Estimate |
|---|---|
| Med=median; (NP)=Not Provided. |
|
| Incubation Period Distribution (days) (mean, (95% CI), median, (range)) * | 6.3 (5.2, 7.3), Med=7 (2–20) |
|
Delay Distributions (days)
|
|
| Symptom onset to… |
|
| … hospitalization | 3.5 |
| Roddy et al. (median, (range), n) 19 | Med=3.5 (0–8), n=26 |
| … death (range of medians) | 9–10 |
| Roddy et al. (median, (range), n) 19 | Med=9 (3–20), n=11 |
| Wamala et al. (median, (range), n) 47 | Med=10 (3–21), n=(NP) |
| …recovery | 10 |
| Wamala et al. (median, (range), n) 47 | Med=10 (2–26), n=(NP) |
| Case Fatality Rate † (range of estimates) | 0.34–0.42 |
| MacNeil et al. (estimate, n, year of outbreak) 46 | 0.40, n=43‡, 2007 |
| Wamala et al. (estimate, n, year of outbreak) 47 | 0.34, n=116, 2007 |
| Roddy et al. (estimate, n, year of outbreak) 19 | 0.42, n=26 (hospitalized confirmed), 2007–08 |
*See Fig. 1 for individual estimates.
†See Fig. 2 for CFR estimates.
‡Confirmed cases only.
Table 6. Parameter estimates for the ongoing outbreak in West Africa.
| Parameter | All countries * | Guinea | Liberia | Nigeria | Sierra Leone | References |
|---|---|---|---|---|---|---|
| Rt refers to effective reproduction number. |
||||||
| μ=mean; Med=median; s.d.=standard deviation. |
||||||
| WHO ERT is the World Health Organization Ebola Response Team16,17. |
||||||
|
Incubation Period Distribution (days)
|
||||||
| Multi-day exposures, observed (mean) | 11.4, n=155 | 10.9, n=20 | 11.7, n=79 | — | 10.8, n=48 | 16 |
| Single-day exposures, observed, (median (IQR)) | 9 (5–13) | 17 | ||||
| Fitted single-day exposures (mean, (95% CI)) | 10.3 (9.9, 10.7) | 17 | ||||
| Fitted values (mean) | 12 | 10 | 48 | |||
|
Serial interval Distribution (days)
|
||||||
| WHO ERT 2014 (gamma fit mean, s.d., n) | 15.3, 9.3, n=92) | 19.0, 11.2, n=40) | 13.1, 7.8, n=26) | — | 11.6, 6.3, n=25) | 16 |
| WHO ERT 2015 (gamma fit mean, (95% CI), n) | 14.2 (13.1, 15.3), n=305 | 17 | ||||
| Fisman et al. 30 | 15 days (derived) | 30 | ||||
| Faye et al. 2014 (mean, (range), s.d., (95% CI for s.d.)) | 14.2 (13.1–15.5), 7.1 (6.2, 8.2) | 41 | ||||
|
R0 (estimate, (95% CI))
|
||||||
| With estimated serial interval 15.3 d | — | 1.71 (1.44, 2.01) | 1.83 (1.72, 1.94) | 2.02 (1.79, 2.26) | 16 | |
| With fixed incubation period 5.3 d | — | 1.51 (1.50, 1.52) | 1.59 (1.57, 1.60) | 9.0 (5.2, 15.6) † | 2.53 (2.41, 2.67) | 36,61 |
| Overall, data driven | 1.8 (1.5, 2.0) | — | — | — | — | 62 |
| SEIR model | 2.1 (1.9, 2.4) | — | — | — | — | 62 |
| Community using Legrand et al. 59 model | 0.8 (0.3, 0.9) | — | — | — | — | 62 |
| Hospital using Legrand et al. 59 model | 0.4 (0.2, 1.4) | — | — | — | — | 62 |
| Funeral using Legrand et al. 59 model | 0.6 (0.2, 1.0) | — | — | — | — | 62 |
| Assumed fixed generation time of 15d | 1.78 | 2.46 | 1.72 | — | 8.33 | 30 |
| Estimated mean serial interval 15.3d | — | 1.71 (1.44, 2.01) | 1.83 (1.72, 1.94) | 1.2 (0.67, 1.96) | 2.02 (1.79, 2.26) | 16 |
| Overall (Conakry) | 1.7 (1.2, 2.3) | 41 | ||||
| Community (Conakry) | 1.0 (0.6, 1.5) | 41 | ||||
| Hospital (Conakry) | 0.4 (0.2, 0.7) | 41 | ||||
| Funeral (Conakry) | 0.3 (0.1, 0.6) | 41 | ||||
| Montserrado only | 2.49 (2.38, 2.60) | 64 | ||||
| Webb et al. 2015 | 1.54 | 1.26 | 68 | |||
| Rivers et al. 48 (overall) | 2.22 | 1.78 | 48 | |||
| Rivers et al. 48 (community) | 1,35 | 1.11 | 48 | |||
| Rivers et al. 48 (hospitals) | 0.35 | 0.24 | 48 | |||
| Rivers et al. 48 (funerals) | 0.53 | 0.43 | 48 | |||
| Khan et al. 63 (raw data) | 1.76 | 1.49 | 63 | |||
| Khan et al. 63 (corrected for underreporting) | 1.9 | 1.37 | 63 | |||
|
Rt (estimate, (95% CI))
|
||||||
| Estimated mean serial interval 15.3 d | — | 1.81 (1.60, 2.03) | 1.51 (1.41, 1.60) | <1† | 1.38 (1.27, 1.51) | 16,36 |
| Estimated mean generation time 12 d‡ | — | 1 | 1.7 | — | 1.4 | 65 |
| Sierra Leone (56 days of data) | 1.1 (0.95, 1.24) | 69 | ||||
| Montserrado only | 1.73 (1.66, 1.83) | 76 | ||||
| Survivors Montserrado only | 0.66 (0.10, 1.69) | 76 | ||||
| Non-survivors Montserrado only | 2.36 (1.72, 2.80) | 76 | ||||
|
Observed Delay Distributions (days)
|
||||||
| Symptom onset to…
|
||||||
| … hospitalization
|
||||||
| WHO ERT 2014 (mean, s.d., n) | 5.0, 4.7, n=1135 | 5.3, 4.3, n=484 | 4.9, 5.1, n=245 | 4.1, 1.4, n=11 | 4.6, 5.1, n=395 | 16 |
| WHO ERT 2015 (mean, (95% CI), n) | 5.0 (4.9, 5.1), n=5616 | 17 | ||||
| Faye et al. 2014 (mean, s.d., n) | 5.0, 3.9, n=152 | |||||
| Rivers et al. 48 (mean) | 3.24 | 4.12 | 48 | |||
| ... hospital discharge
|
||||||
| WHO ERT 2014 (mean, s.d., n) | 16.4, 6.5, n=267 | 16.3, 6.1, n=152 | 15.4, 8.2, n=41 | — | 17.2, 6.2, n=70 | 16 |
| WHO ERT 2015 (mean, (95% CI)) | 15.1 (14.6, 15.6) | 17 | ||||
| … death
|
||||||
| WHO ERT 2014 (mean, s.d., n) | 7.5, 6.8, n=594 | 6.4, 5.3, n=248 | 7.9, 8, n=212 | — | 8.6, 6.9, n=128 | 16 |
| WHO ERT 2015 (mean, (95% CI)) | 8.2 (7.9, 8.4) | 17 | ||||
| Faye et al. 2014 (mean, s.d., n) | 8.9, 4.0, n=82) | 41 | ||||
| Althaus et al. 61 (mean, range) | 7.41 (4–17), n=17 | 61 | ||||
| … WHO notification | 6.1, 8.5, n=2185 | 7.5, 10.4, n=743 | 6, 8.7, n=797 | — | 4.5, 5, n=634 | 16 |
|
WHO notification to…
|
||||||
| … hospital discharge (mean, s.d., n) | 11.8, 7.2, n=312 | 11.1, 5.8, n=164 | 11, 8, n=41 | — | 12.7, 8.4, n=102 | 16 |
| … death (mean, s.d., n) | -3.0, 13.8, n=584 | -4.4, 14.4, n=300 | -1.8, 13.6, n=221 | — | -1.6, 9.2, n=58 | 16 |
|
Hospitalization to…
|
||||||
| … discharge
|
||||||
| WHO ERT 2014 (mean, s.d., n) | 11.8, 6.1, n=290 | 11, 5.4, n=159 | 12.8, 8.1, n=40 | — | 12.4, 5.8, n=86 | 16 |
| WHO ERT 2015 (mean, (95% CI)) | 11.2 (10.8, 11.7) | 17 | ||||
| Rivers et al. 48 (recovery) (mean) | 15.88 | 15.88 | 48 | |||
| … death
|
||||||
| WHO ERT 2014 (mean, s.d., n) | 4.2, 6.4, n=121 | 2.5, 3.4, n=36 | 4.5, 6, n=63 | — | 4.4, 6, n=17 | 16 |
| WHO ERT 2015 (mean, (95% CI)) | 4.3 (4.1, 4.5) | 17 | ||||
| Rivers et al. 48 (mean) | 10.07 | 6.26 | 48 | |||
|
Infectious period
|
||||||
| Rivers et al. 48 (mean) | 15.00 | 18.00 | 48 | |||
|
Infection to death
|
||||||
| Rivers et al. 48 (mean) | 13.31 | 10.38 | 48 | |||
|
Case Fatality Rate (estimate, (95% CI), n)
|
||||||
| WHO ERT 2014 (based on current status) | 37.7 (36.1, 39.2), n=3747 | 57.5 (53.7, 61.1), n=677 | 34.7 (32.4, 37.1), n=1616 | 40.0 (19.8–64.3), n=15 | 31.6 (29.3, 34.1), n=1439 | 16 |
| Althaus 36 (based on reported deaths) | 74 (72, 75), n=607 | 71 (69, 74), n=1082 | 48 (47, 50), n=910 | 36 | ||
|
All cases (based on definitive outcome)
|
||||||
| WHO ERT 2014 | 70.8 (68.6, 72.8), n=1737 | 70.7 (66.7, 74.3), n=542 | 72.3 (68.9, 75.4), n=739 | 45.5 (21.3, 72), n=11 | 69.0 (64.5, 73.1), n=445 | 16 |
| WHO ERT 2015 | 70.4 (69.2, 71.6), n=5616 | 17 | ||||
| Althaus et al. 61 | 39 (14, 71), n=20 | 61 | ||||
| Fasina et al. 54 | 40 (22, 61), n=20 | 54 | ||||
|
All hospitalized cases
|
||||||
| WHO ERT 2014 (based on definitive outcome) | 63.5 (60.5, 66.3), n=1100 | 63.4 (58.6, 67.9), n=434 | 66.6 (61.3, 71.6), n=338 | 100 (2.5, 100), n=1 | 61 (55.4, 66.4), n=318 | 16 |
| WHO ERT 2015 (based on definitive outcome) | 60.7 (59.2, 62.3), n=3839 | 17 | ||||
| Rivers et al. 48 | 0.500 | 0.750 | 48 | |||
|
Subset of Cases
|
||||||
| Faye et al. 2014 (Conakry) | 54 (49, 63), n=152 | 41 |
*here all countries refer to Guinea, Liberia and Sierra Leone.
†R0 of the index case arriving by air travel from Liberia; Rt estimated to be below 1 within 15 days (95% CI 11–21 days) after arrival of the index case from Liberia36
‡Authors refer to generation time, but have estimated the serial interval and assumed serial interval is the same as generation time.
Table 7. Parameter Estimates for Marburg.
| Parameter | Estimate |
|---|---|
| Med=median; s.d.=standard deviation; (NP)=Not Provided. |
|
| Generation Time Distribution (days) (mean, 95% CI, s.d., n) * 55 | 9 (8.2, 10), 5.4, n=374 |
| R 0 (estimate, 95% CI) | 1.59 (1.53, 1.66) |
|
Delay Distributions (days)
|
|
| Symptom onset to death (range of medians) | 7–8 |
| Ajelli & Merler (median, range, n) 55 | Med=7 (5–9), n=329 |
| Colebunders et al. (median, range, n) 57 | Med=8 (2–16), n=40 |
| Case Fatality Rate † (range of estimates) | 0.78–0.88 |
| Ajelli & Merler (estimate, 95% CI, n, year of outbreak) 55 | 0.88 (0.84–0.91), n=374, 2005 |
| Bausch et al. (estimate, n, year of outbreak) 56 | 0.83, n=(NP), 1998 |
| Colebunders et al. (estimate, n, year of outbreak) 57 | 0.78, n=77, 1998 |
*Authors refer to generation time, but have estimated the serial interval and assumed serial interval is the same as generation time.
†See Fig. 2 for CFR estimates.
Table 8. Risk factors for human-to-human transmission for infection with EVD.
| Risk Factors for infection | OR, PPR, or RR | 95% CI | Virus | References |
|---|---|---|---|---|
| PPR=Prevalence Proportion Ratio; RR=Risk Ratio; OR=Odds Ratio. | ||||
|
Physical Contact
|
||||
| Sharing bed with a patient during their late stage of illness | RR=2.2 | 1.2, 4.2 | Zaire | 39 |
| Slept in the same hut on the same mat | PPR=2.78 | 1.15, 6.70 | Sudan | 45 |
| Touch cadaver | RR=2.1 | 1.1, 4.2 | Zaire | 39 |
| Touch body of deceased person | PPR=1.95 | 0.91, 4.17 | Sudan | 45 |
| Contact with known suspected case | OR=2.7 | 1.35, 5.24 | Bundibugyo | 47 |
| Contact with confirmed case | RR=3.21 | 1.53, 6.75 | Zaire | 71 |
| Intimate contact: nursing care | OR=5.1 | 1.31, 15.48 | Sudan | 24 |
| Receiving injections | OR=7.4 | 1.6, 33.2 | Marburg | 56 |
| Caring for patients at early stage of illness | PPR=6.0 | 1.32, 27.10 | Sudan | 45 |
| Cared for the patient until the patient’s death at hospital | PPR=8.57 | 1.95, 37.66 | Sudan | 45 |
| Cared for the patient until the patient’s death at home | PPR=13.33 | 3.20, 55.59 | Sudan | 45 |
| Contact with body fluids | PPR=5.3 | 2.14, 13.14 | Sudan | 45 |
| Direct physical contact with a sick person | PPR=3.53 | 0.52, 24.11 | Sudan | 45 |
|
Non-Physical Contact
|
||||
| Age (>18 years old) | RR=3.6 | 1.3, 10.1 | Zaire | 39 |
| Sharing a meal with a patient during their late stage of illness | RR=2.2 | 1.2, 4.0 | Zaire | 39 |
| Conversation with a patient during their late stage of illness | RR=3.9 | 1.2, 12.2 | Zaire | 39 |
| Shared meals with a sick person | PPR=1.94 | 0.89, 4.22 | Sudan | 45 |
| Washed clothes of a sick person | PPR=1.68 | 0.78, 3.60 | Sudan | 45 |
| Slept in the same hut | PPR=2.16 | 0.90, 5.19 | Sudan | 45 |
| Profession- Miner | OR=13.9 | 3.1, 62.1 | Marburg | 56 |
| Travelled before illness | OR=2.1 | 1.0, 4.5 | Bundibugyo | 47 |
| Hospitalized or visited hospital | OR=2.6 | 1.4, 4.9 | Bundibugyo | 47 |
| Consulted traditional healer | OR=0.2 | 0.01, 1.5 | Bundibugyo | 47 |
|
Funeral-associated contact
|
||||
| Ritual hand washing during funeral | PPR=2.25 | 1.08, 4.72 | Sudan | 45 |
| Communal meal during funeral | PPR=2.84 | 1.35, 5.98 | Sudan | 45 |
| Funeral rites participation | OR=4.22 | 2.2, 8.2 | Bundibugyo | 47 |
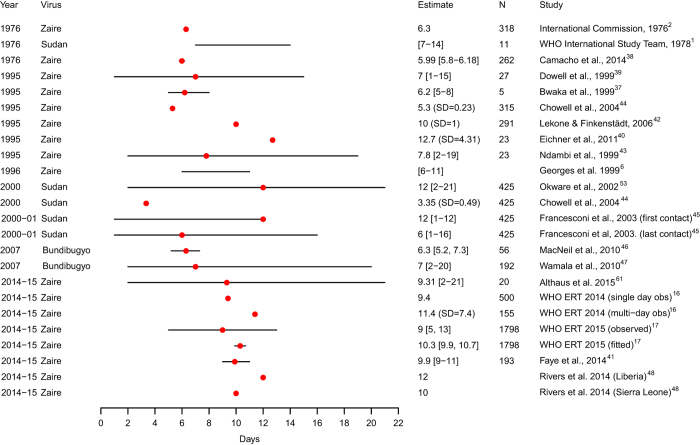
Figure 1. Estimates of the incubation period distribution by virus, year of outbreak and study.
The dots represent the mean or median estimate and the lines illustrate the range, for all studies, with the exception of MacNeil et al. 46 and WHO Ebola Response Team (ERT) 201517 (fitted) where the line represents the 95% CI for the estimate. Chowell et al. 44, Eichner et al. 40, WHO ERT 2014 (multi-day observed)16 and Lekone and Finkenstädt42 provide standard deviation (s.d.).
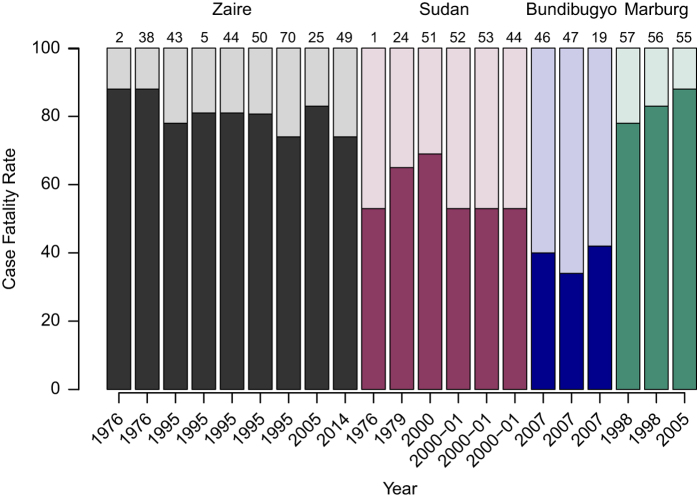
Figure 2. Overall case fatality rate (CFR) for Ebola by virus and Marburg virus; we include only those outbreaks that have been declared over.
Outbreaks with fewer than 10 fatal patients were excluded from this figure. For each outbreak, the bars represent the rate of fatal (dark color) and recovered (light color) patients. For country specific information see Table 1. The source of each estimate is denoted by the reference number at the top of each bar.
Technical Validation
Incubation period distribution
The incubation period distribution of EVD has been estimated for past EVD outbreaks (Fig. 1 and Tables 3 (available online only), 4 (available online only), 5; minimum sample size n=5, maximum sample size n=1,798). The mean (or median) incubation period (Fig. 1), for the different Ebola viruses ranged from 3.35 to 12.7 days (range 1–21 days), excluding an extreme outlier35. Central estimates for the incubation period distribution were between 5.3–12.7 days (range 1–21 days) for Ebola Zaire 5,6,16,17,36–43, 3.35–12 days (range 1–16 days) for Ebola Sudan 1,44,45, and 6.3–7 days (range 2–20 days) for Ebola Bundibugyo 46,47.
The mean incubation period for the ongoing Ebola outbreak in West Africa has been estimated to be between 9–12 days (Table 6 (available online only))16,17,41,48. The range of incubation periods observed in past EVD outbreaks supports the policy of contact tracing for 21 days following contact with an EVD patient. An outbreak is officially declared over after no new cases are identified 42 days (2 times the 21-day maximum incubation period) after the last EVD case is found.
Case fatality rate (CFR)
In Fig. 2 and Tables 3 (available online only), 4 (available online only), 5, we reprint the estimated CFR for each Ebola outbreak (by virus) and for Marburg virus. The Ebola Zaire virus is the most lethal with an overall estimated CFR ranging from 69 to 88%2,5,25,38,43,49,50 (Table 3 (available online only)). The CFR of outbreaks due to Ebola Sudan virus ranged from 53 to 69%1,24,51–53 (Table 4 (available online only)), and the CFR of outbreaks due to Ebola Bundibugyo ranged from 34 to 42%19,46,47 (Table 5). For the ongoing outbreak in West Africa due to Ebola Zaire, the estimated CFR, as measured among confirmed and probable cases with definitive outcome (recovered or fatal), is approximately 70%, and varies little among the three most affected countries (Guinea, Liberia and Sierra Leone; Table 6 (available online only) and Data Citation 2)38. The CFR among EVD cases reported by Nigeria (n=20) was 40%54). A second, unrelated EVD outbreak occurred in Équateur province, DRC between July and October 2014 resulting in 69 confirmed and probable cases with a CFR of 74%49. The CFR for Marburg is approximately 80%55–57).
R0 and Rt
Looking at past outbreaks, estimates of R0 for Ebola Zaire ranged from 1.4-4.735,42,44,58–60) (Table 3 (available online only) and Data Citation 2), and for Ebola Sudan ranged from 1.3-2.744,58–60 (Table 4 (available online only)). R0 has not been estimated for Ebola Bundibugyo.
In the ongoing outbreak in West Africa, estimates of R0 and Rt have been estimated for all countries combined, as well as separately for Guinea, Liberia, Nigeria and Sierra Leone16,30,36,41,48,54,61–69. All estimates of Rt are provided in Table 6 (available online only) and Data Citation 2 and Data Citation 3. Gomes et al. 62 reported several all-country R0 estimates (means ranging 1.8–2.1), depending on model choice and assumptions. Fisman et al. 30 reported all-country and country-specific R0 estimates depending on assumptions including action taken to mitigate infection. For the most part, R0 estimates for Guinea, Liberia, and Sierra Leone ranged from 1.2 to 2.5 with the striking exception of the Fisman et al. 30 R0 estimate of 8.3 for Sierra Leone. Nishiura and Chowell65 estimated Rt fluctuating around 1 for Guinea, 1.7 for Liberia and 1.4 for Sierra Leone. The WHO Ebola Response Team16 estimated Rt for Guinea (ranging from 1.6 to 2.0), for Liberia (ranging from 1.4 to 1.6) and for Sierra Leone (ranging from 1.3 to 1.5).
Several groups have also estimated R0 for specific geographic areas within the region (full details in Table 6 (available online only) and Data Citation 2). For example, Faye et al. estimated R0 for cases occurring in Conakry, Guinea at the start of the outbreak (n=193)41 as 1.7 (95% CI 1.2, 2.3); whereas Lewnard et al. 64 estimated R0 for EVD cases in Montserrado, Liberia as of October 2014 (R0=2.5 (2.4, 2.6)).
Serial interval distribution
The serial interval, defined as the time interval between symptom onset in an index case and symptom onset in a secondary case infected by that index case, has been infrequently estimated due to the paucity of data on epidemiologically linked pairs of index and secondary cases. For Ebola Zaire (Table 3 (available online only)), the mean serial interval was estimated to be 10–16.1 days5,49,60,70. In the ongoing outbreak in West Africa, the mean serial interval has been estimated to be approximately 14–15 days16,17,30,41 (Table 6 (available online only)).
Generation time distribution
Closely related to the serial interval, the generation time is defined as the time interval between infection of an index case and infection of a secondary case infected by that index case. As such, the generation time distribution nearly always needs to be inferred indirectly from serial interval observations and knowledge of the incubation period distribution. We found one such estimate of the mean generation time for Marburg of 9 days (95% CI 8.2, 10.0)55.
Delay distributions
For Ebola Zaire, including the ongoing outbreak, the mean time from symptom onset to hospitalization (Table 3 (available online only) and Table 6 (available online only)), ranged from 3.2 to 5.3 days5,16,17,20,38,41,48, whereas the mean time from symptom onset to death ranged from 6 to 10.1 days5,17,25,37–39,41,49,61. For Ebola Sudan (Table 4 (available online only)), the mean time from symptom onset to hospitalization was 2 days (range 0–8)51 and the median time from symptom onset to death was 9 days (range 5–15)24, respectively. The mean time from hospitalization to discharge for Ebola Sudan ranged from 8 to 10 days51,53 whereas the mean time from hospitalization to death was 6.1 days (range 2–13)51. For Ebola Bundibugyo (Table 5), the median time from symptom onset to hospitalization was 3.5 days (range 0–8)19 and the median time from symptom onset to death was 9–10 days (range 3–21 days)19,47.
Risk for developing EVD
Risk factors for human-to-human transmission of EVD or Marburg were evaluated from comparison of the exposures, behaviors and practices in cases compared to controls (including unaffected controls, defined to be suspected cases but negative serologic test results) and were described using a prevalence proportion ratio, an odds ratio or a relative risk (and the corresponding confidence interval). Significant risk factors associated with developing EVD are reported in Table 8 (available online only) and Data Citation 4 and include direct physical contact (sharing a bed, touching a cadaver or funeral preparations for an EVD patient, nursing care and contact with bodily fluids) and non-physical contact (sharing a meal, contact with a hospital where EVD patients were treated)24,39,45,47,56,71.
Usage Notes
The data presented in this review summarize estimates of the epidemiological parameters of EVD and Marburg. These results can facilitate parameterization and sensitivity analysis of transmission models examining surveillance, control and treatment strategies. The results can also inform epidemiological studies investigating human-to-human transmission during Ebola and Marburg outbreaks, deepening our understanding of the transmission process.
The number of parameters estimated for each outbreak has generally increased with time (Table 2 (available online only)). While the incubation period distribution was consistently assessed, R0 has increasingly been estimated, most notably with the ongoing outbreak in West Africa (Table 6 (available online only) and Data Citation 2). Fig. 1 shows the central estimates and ranges for the different studies that estimated the incubation period distribution for EVD outbreaks. While there are small differences in the central estimates of the incubation period distribution of the four Ebola viruses, the ranges around the mean or median are consistent, with a maximum of ≤21 days. Current EVD guidance states that EVD has an incubation period of 2–21 days, which is the basis for the recommended duration of contact tracing of 21 days26,27. This is supported by the findings in our review.
Figure 2 shows CFR for different Ebola Zaire, Ebola Sudan, Ebola Bundibugyo and Marburg outbreaks. While the CFR for Ebola and Marburg are high (compared to other infectious diseases), outbreaks caused by Ebola Zaire and Ebola Sudan have experienced the highest CFR amongst these three Ebola viruses causing outbreaks in humans1,2,5,19,24,25,38,43,46,47,50–53,72–76. The estimated CFR for the ongoing outbreak in West Africa, due to Ebola Zaire, is approximately 70%16,17,36, which falls within the range of CFRs for past outbreaks due to this virus2,5,25,38,43,50.
Figure 2 also illustrates that recent CFR estimates for Ebola Zaire remain comparable to those observed in the 1970s. While there are ongoing efforts to develop medical treatments for EVD, treatment remains mainly supportive. The massive scale of the ongoing outbreak has highlighted the urgent need to develop new treatments and to fast track the use of experimental medical interventions77.
The transmission potential, as measured by R0, is fairly consistent among the three Ebola viruses, ranging from approximately >1 to 4 (also mentioned in ref. 78). Previously, EVD typically affected villages in remote areas of central Africa35,38,42,44,58,59, and while devastating in these areas, the populations that are at risk are generally limited in number. The ongoing EVD outbreak had circulated for at least three months prior to discovery22,79 which allowed spread of the virus to go unchecked while it infected people in an area of Guinea that shares borders with Sierra Leone and Liberia. Recent experience in Nigeria, has shown that an Ebola virus with R0>1, even in a population of over 20 million, can be controlled with vigorous application of control methods49,54.
Differences in estimates of R0 and Rt are likely, at least in part, to be the result of the quality of data available and the inferential method. The focus on R0 estimation together with serial interval estimates may reflect a shift from data collection purely for surveillance to recognition of the epidemiological value of such data.
The specific factors that result in an EVD outbreak have been under investigation since the emergence of this virus and include examination of human and susceptible non-human populations living in close proximity with each other in remote areas of central Africa. Recent investigations into the first cases of the ongoing outbreak found that the outbreak may have begun in Meliandou, Guinea in a village where the inhabitants frequently came in contact with fruit bats in a hollowed out tree80. Although the current focus is on limiting human-to-human transmission and treating the infected, the challenging underlying factors that led to this large outbreak in West Africa will require long-term investments to improve both health care and surveillance for infectious diseases.
Our dataset is the most complete collection of published epidemiological parameter estimates from EVD outbreaks available at the time of writing and provides an evidence-based foundation for both retrospective analyses and responses to future outbreaks.
Additional Information
How to cite this article: Van Kerkhove, M. D. et al. A review of epidemiological parameters from Ebola outbreaks to inform early public health decision-making. Sci. Data 2:150019 doi: 10.1038/sdata.2015.19 (2015).
Supplementary Material
Acknowledgments
The authors would like to acknowledge the Medical Research Council, the Bill and Melinda Gates Foundation, the Wellcome Trust, the Health Protection Research Units of the National Institute for Health Research, and the European Union Seventh Framework Programme [FP7/2007–2013] under agreement Grant Agreement nu278433-PREDEMICS for funding.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. figshare. http://dx.doi.org/10.6084/m9.figshare.1381874 [DOI] [PMC free article] [PubMed]
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1381876 [DOI] [PMC free article] [PubMed]
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1381877 [DOI] [PMC free article] [PubMed]
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1381875 [DOI] [PMC free article] [PubMed]
References
- WHO International Study Team. Ebola haemorrhagic fever in Sudan, 1976. B World Health Organ 56, 247–270 (1978). [PMC free article] [PubMed] [Google Scholar]
- International Commission. Ebola haemorrhagic fever in Zaire, 1976. B World Health Organ 56, 271–293 (1978). [PMC free article] [PubMed] [Google Scholar]
- Bres P. The epidemic of Ebola haemorrhagic fever in Sudan and Zaire, 1976: introductory note. B World Health Organ 56, 245 (1978). [PMC free article] [PubMed] [Google Scholar]
- Breman J. et al. in Proceedings of the international colloquium on Ebola virus infections and other haemorrhagic fevers, ed. Pattyn S. R. 85–97 (Elsevier/North Holland Biomedical Press, 1978). [Google Scholar]
- Khan A. S. et al. The reemergence of Ebola hemorrhagic fever, Democratic Republic of the Congo, 1995. Commission de Lutte contre les Epidemies a Kikwit. J. Infect. Dis. 179, S76–S86 (1999). [DOI] [PubMed] [Google Scholar]
- Georges A. J. et al. Ebola hemorrhagic fever outbreaks in Gabon, 1994-1997: epidemiologic and health control issues. J. Infect. Dis. 179, S65–S75 (1999). [DOI] [PubMed] [Google Scholar]
- CDC. Outbreaks Chronology: Ebola Virus Disease. http://www.cdc.gov/vhf/ebola/outbreaks/history/chronology.html (2014).
- Kuhn J. H. Filoviruses A compendium of 40 years of epidemiological, clinical, and laboratory studies. Arch. Virol. Suppl. 20, 13–360 (2008). [PubMed] [Google Scholar]
- Mylne A. et al. A comprehensive database of the geographic spread of past human Ebola outbreaks. Sci.Data 1, 140042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M. et al. Epidemiology of Ebola (subtype Reston) virus in the Philippines, 1996. J. Infect. Dis. 179, S115–S119 (1999). [DOI] [PubMed] [Google Scholar]
- Le Guenno B. et al. Isolation and partial characterisation of a new strain of Ebola virus. The Lancet 345, 1271–1274 (1995). [DOI] [PubMed] [Google Scholar]
- Leroy E. M. et al. Fruit bats as reservoirs of Ebola virus. Nature 438, 575–576 (2005). [DOI] [PubMed] [Google Scholar]
- Pourrut X. et al. Spatial and temporal patterns of Zaire ebolavirus antibody prevalence in the possible reservoir bat species. J. Infect. Dis. 196, S176–S183 (2007). [DOI] [PubMed] [Google Scholar]
- Pourrut X. et al. The natural history of Ebola virus in Africa. Microbes and infection 7, 1005–1014 (2005). [DOI] [PubMed] [Google Scholar]
- Bermejo M. et al. Ebola outbreak killed 5000 gorillas. Science 314, 1564 (2006). [DOI] [PubMed] [Google Scholar]
- WHO Ebola Response Team. Ebola virus disease in West Africa–the first 9 months of the epidemic and forward projections. N. Engl. J. Med. 371, 1481–1495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Ebola Response Team. West African Ebola epidemic after one year–slowing but not yet under control. N. Engl. J. Med 372, 584–587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Sanchez A., Geisbert T. W. in Fields Virology, eds. Knipe D. M. & Howley P. M. (Lippincott, Williams and Wilkins, 2013). [Google Scholar]
- Roddy P. et al. Clinical manifestations and case management of Ebola haemorrhagic fever caused by a newly identified virus strain, Bundibugyo, Uganda, 2007-2008. PLoS One 7, e52986 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe A. K. et al. Clinical, virologic, and immunologic follow-up of convalescent Ebola hemorrhagic fever patients and their household contacts, Kikwit, Democratic Republic of the Congo. Commission de Lutte contre les Epidemies a Kikwit. J. Infect. Dis. 179, S28–S35 (1999). [DOI] [PubMed] [Google Scholar]
- Schieffelin J. S. et al. Clinical illness and outcomes in patients with Ebola in Sierra Leone. N. Engl. J. Med. 371, 2092–2100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baize S. et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 371, 1418–1425 (2014). [DOI] [PubMed] [Google Scholar]
- Leroy E. M. et al. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic. Dis. 9, 723–728 (2009). [DOI] [PubMed] [Google Scholar]
- Baron R. C., McCormick J. B. & Zubeir O. A. Ebola virus disease in southern Sudan: hospital dissemination and intrafamilial spread. B World Health Organ. 61, 997–1003 (1983). [PMC free article] [PubMed] [Google Scholar]
- Nkoghe D., Kone M. L., Yada A. & Leroy E. A limited outbreak of Ebola haemorrhagic fever in Etoumbi, Republic of Congo, 2005. Trans. R. Soc. Trop. Med. Hyg. 105, 466–472 (2011). [DOI] [PubMed] [Google Scholar]
- WHO Regional Office for Africa. Contact tracing during an outbreak of Ebola virus disease http://www.who.int/csr/resources/publications/ebola/contact-tracing/en/ (2014).
- World Health Organization. Fact Sheet N 103. http://www.who.int/mediacentre/factsheets/fs103/en/ (2014).
- Anderson R. M. & May R. M.. Infectious Diseases of Humans (Oxford University Press, 1992). [Google Scholar]
- Keeling M. J. & Rohani P. Modeling Infectious Diseases in Humans and Animals. (Princeton University Press, 2008). [Google Scholar]
- Fisman D., Khoo E. & Tuite A. Early epidemic dynamics of the West African 2014 Ebola outbreak: estimates derived with a simple two-parameter model. PLoS Curr. Outbreaks 6, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesterbeek J. A. A brief history of R0 and a recipe for its calculation. Acta Biotheor. 50, 189–204 (2002). [DOI] [PubMed] [Google Scholar]
- Nishiura H. & Chowell G. in Mathematical and Statistical Estimation Approaches in Epidemiology 103–121 (Springer, 2009). [Google Scholar]
- Fine P. E. The interval between successive cases of an infectious disease. Am. J. Epidemiol. 158, 1039–1047 (2003). [DOI] [PubMed] [Google Scholar]
- Vink M. A., Bootsma M. C. & Wallinga J. Serial intervals of respiratory infectious diseases: a systematic review and analysis. Am. J. Epidemiol. 180, 865–875 (2014). [DOI] [PubMed] [Google Scholar]
- Ndanguza D., Tchuenche J. & Haario H. Statistical data analysis of the 1995 Ebola outbreak in the Democratic Republic of Congo. Afrika Matematika 24, 55–68 (2013). [Google Scholar]
- Althaus C. L. Estimating the reproduction number of Ebola Virus (EBOV) during the 2014 outbreak in West Africa. PLoS Curr. Outbreaks 6, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bwaka M. A. et al. Ebola hemorrhagic fever in Kikwit, Democratic Republic of the Congo: clinical observations in 103 patients. J. Infect. Dis. 179, S1–S7 (1999). [DOI] [PubMed] [Google Scholar]
- Camacho A. et al. Potential for large outbreaks of Ebola virus disease. Epidemics 9, 70–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S. F. et al. Transmission of Ebola hemorrhagic fever: a study of risk factors in family members, Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 179, S87–S91 (1999). [DOI] [PubMed] [Google Scholar]
- Eichner M., Dowell S. F. & Firese N. Incubation period of ebola hemorrhagic virus subtype Zaire. Osong Public Health Res. Perspect 2, 3–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye O. et al. Chains of transmission and control of Ebola virus disease in Conakry, Guinea, in 2014: an observational study. Lancet Infect. Dis. 15, 320–326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekone P. E. & Finkenstädt B. F. Statistical inference in a stochastic epidemic SEIR model with control intervention: Ebola as a case study. Biometrics 62, 1170–1177 (2006). [DOI] [PubMed] [Google Scholar]
- Ndambi R. et al. Epidemiologic and clinical aspects of the Ebola virus epidemic in Mosango, Democratic Republic of the Congo, 1995. J. Infect. Dis. 179, S8–10 (1999). [DOI] [PubMed] [Google Scholar]
- Chowell G., Hengartner N. W., Castillo-Chavez C., Fenimore P. W. & Hyman J. M. The basic reproductive number of Ebola and the effects of public health measures: the cases of Congo and Uganda. J. Theor. Biol. 229, 119–126 (2004). [DOI] [PubMed] [Google Scholar]
- Francesconi P. et al. Ebola hemorrhagic fever transmission and risk factors of contacts, Uganda. Emerg. Infect. Dis. 9, 1430–1437 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil A. et al. Proportion of deaths and clinical features in Bundibugyo Ebola virus infection, Uganda. Emerg. Infect. Dis. 16, 1969–1972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamala J. F. et al. Ebola hemorrhagic fever associated with novel virus strain, Uganda, 2007-2008. Emerg. Infect. Dis. 16, 1087–1092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers C. M., Lofgren E. T., Marathe M., Eubank S. & Lewis B. L. Modeling the impact of interventions on an epidemic of Ebola in Sierra Leone and Liberia. PLoS Curr. Outbreaks 6, 1–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maganga G. D. et al. Ebola virus disease in the Democratic Republic of Congo. N Engl. J. Med. 371, 2083–2091 (2014). [DOI] [PubMed] [Google Scholar]
- Sadek R. F., Khan A. S., Stevens G., Peters C. J. & Ksiazek T. G. Ebola hemorrhagic fever, Democratic Republic of the Congo, 1995: determinants of survival. J. Infect. Dis. 179, S24–S27 (1999). [DOI] [PubMed] [Google Scholar]
- Borchert M. et al. Ebola haemorrhagic fever outbreak in Masindi District, Uganda: outbreak description and lessons learned. BMC Infect. Dis. 11, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamunu M. et al. Containing a haemorrhagic fever epidemic: the Ebola experience in Uganda (October 2000-January 2001). Int. J. Infect. Dis. 8, 27–37 (2004). [DOI] [PubMed] [Google Scholar]
- Okware S. I. et al. An outbreak of Ebola in Uganda. Trop. Med. Int. Health 7, 1068–1075 (2002). [DOI] [PubMed] [Google Scholar]
- Fasina F. O. et al. Transmission dynamics and control of Ebola virus disease outbreak in Nigeria, July to September 2014. Euro Surveill 19, 20920 (2014). [DOI] [PubMed] [Google Scholar]
- Ajelli M. & Merler S. Transmission potential and design of adequate control measures for Marburg hemorrhagic fever. PLoS One 7, e50948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch D. G. et al. Risk factors for Marburg hemorrhagic fever, Democratic Republic of the Congo. Emerg. Infect. Dis. 9, 1531–1537 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebunders R. et al. Marburg hemorrhagic fever in Durba and Watsa, Democratic Republic of the Congo: clinical documentation, features of illness, and treatment. J. Infect. Dis. 196, S148–S153 (2007). [DOI] [PubMed] [Google Scholar]
- Ferrari M. J., Bjornstad O. N. & Dobson A. P. Estimation and inference of R0 of an infectious pathogen by a removal method. Math Biosci. 198, 14–26 (2005). [DOI] [PubMed] [Google Scholar]
- Legrand J., Grais R. F., Boelle P. Y., Valleron A. J. & Flahault A. Understanding the dynamics of Ebola epidemics. Epidemiol. Infect. 135, 610–621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L. F. & Pagano M. A likelihood-based method for real-time estimation of the serial interval and reproductive number of an epidemic. Stat. Med. 27, 2999–3016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althaus C. L., Low N., Musa E. O., Shuaib F. & Gsteiger S. Ebola Virus Disease Outbreak in Nigeria: Transmission Dynamics and Rapid Control. Report No. 2167-9843. (PeerJ PrePrints, 2015). [DOI] [PubMed] [Google Scholar]
- Gomes M. F. et al. Assessing the international spreading risk associated with the 2014 west African ebola outbreak. PLoS Curr. Outbreaks 6, 1–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Naveed M., Dur E. A. M. & Imran M. Estimating the basic reproductive ratio for the Ebola outbreak in Liberia and Sierra Leone. Infect. Dis. Poverty 4, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewnard J. A. et al. Dynamics and control of Ebola virus transmission in Montserrado, Liberia: a mathematical modelling analysis. Lancet Infect. Dis. 14, 1189–1195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H. & Chowell G. Early transmission dynamics of Ebola virus disease (EVD), West Africa, March to August 2014. Euro Surveill 19, 1–6 (2014). [DOI] [PubMed] [Google Scholar]
- Towers S., Patterson-Lomba O. & Castillo-Chavez C. Temporal variations in the effective reproduction number of the 2014 west Africa ebola outbreak. PLoS Curr. Outbreaks 6, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E. & Pond S. Phylodynamic analysis of Ebola virus in the 2014 Sierra Leone epidemic. PLoS Curr. Outbreaks 10, 1–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb G. et al. A model of the 2014 ebola epidemic in west Africa with contact tracing. PLoS Curr. Outbreaks 7, 1–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. A. et al. Projected treatment capacity needs in Sierra Leone. PLoS Curr. Outbreaks 7, 1–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyembe T. & Kipasa M. Ebola haemorrhagic fever in Kikwit, Zaire. The Lancet 345, 1448 (1995). [DOI] [PubMed] [Google Scholar]
- Bertherat E., Renaut A., Nabias R., Dubreuil G. & Georges-Courbot M. C. Leptospirosis and Ebola virus infection in five gold-panning villages in northeastern Gabon. Am. J. Trop. Med. Hyg. 60, 610–615 (1999). [DOI] [PubMed] [Google Scholar]
- Formenty P. et al. Outbreak of Ebola hemorrhagic fever in the Republic of the Congo, 2003: a new strategy? Med. Trop. (Mars) 63, 291–295 (2003). [PubMed] [Google Scholar]
- Lefebvre A. et al. Case fatality rates of Ebola virus diseases: a meta-analysis of World Health Organization data. Med. Mal. Infect 44, 412–416 (2014). [DOI] [PubMed] [Google Scholar]
- World Health Organization. Ebola hemorrhagic fever–South Africa. Weekly Epidemiological Record 71, 359 (1996). [Google Scholar]
- World Health Organization. Outbreak(s) of Ebola haemorrhagic fever, Congo and Gabon, October 2001–July 2002. Weekly Epidemiological Record 78, 223–225 (2003). [PubMed] [Google Scholar]
- Yamin D. et al. Effect of Ebola progression on transmission and control in Liberia. Ann. Intern. Med. 162, 11–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Consultation on potential Ebola therapies and vaccines http://www.who.int/mediacentre/events/meetings/2014/ebola-interventions/en/ (2014).
- House T. Epidemiological dynamics of Ebola outbreaks. eLife 3, e03908 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch D. G. & Schwarz L. Outbreak of ebola virus disease in Guinea: where ecology meets economy. PLoS Negl.Trop. Dis. 8, e3056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí Saéz A. et al. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Molecular Medicine 7, 17–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. figshare. http://dx.doi.org/10.6084/m9.figshare.1381874 [DOI] [PMC free article] [PubMed]
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1381876 [DOI] [PMC free article] [PubMed]
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1381877 [DOI] [PMC free article] [PubMed]
- Van Kerkhove M., Bento A. I., Mills H. L., Ferguson N. M., Donnelly C. A. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1381875 [DOI] [PMC free article] [PubMed]