Abstract
It is often of interest to understand how the structure of a genetic network differs between two conditions. In this paper, each condition-specific network is modeled using the precision matrix of a multivariate normal random vector, and a method is proposed to directly estimate the difference of the precision matrices. In contrast to other approaches, such as separate or joint estimation of the individual matrices, direct estimation does not require those matrices to be sparse, and thus can allow the individual networks to contain hub nodes. Under the assumption that the true differential network is sparse, the direct estimator is shown to be consistent in support recovery and estimation. It is also shown to outperform existing methods in simulations, and its properties are illustrated on gene expression data from late-stage ovarian cancer patients.
Keywords: Differential network, Graphical model, High dimensionality, Precision matrix
1 Introduction
A complete understanding of the molecular basis of disease will require characterization of the network of interdependencies between genetic components. There are many types of networks that may be considered, such as protein-protein interaction networks or metabolic networks (Emmert-Streib and Dehmer, 2011), but the focus in this paper is on transcriptional regulatory networks. In many cases, interest centers not on a particular network but rather on whether and how the network changes between disease states. Indeed, differential networking analysis has recently emerged as an important complement to differential expression analysis (de la Fuente, 2010; Ideker and Krogan, 2012). For example, Hudson et al. (2009) studied a mutant breed of cattle known to differ from wild-type cattle by a mutation in the myostatin gene. Myostatin was not differentially expressed between the two breeds, but Hudson et al. (2009) showed that a differential network analysis could correctly identify it as the gene containing the causal mutation. In another example, using an experimental technique called differential epistasis mapping, Bandyopadhyay et al. (2010) demonstrated large-scale changes in the genetic networks of yeast cells after perturbation by a DNA-damaging agent.
Transcriptional networks are frequently modeled as Gaussian graphical models (Markowetz and Spang, 2007). Gene expression levels are assumed to be jointly Gaussian, so that two expression levels are conditionally independent given the other genes if and only if the corresponding entry of the precision matrix, or the inverse covariance matrix, is zero. Representing gene expression levels as nodes and conditional dependency relationships as edges in a graph results in a Gaussian graphical model (Lauritzen, 1996). A differential network can be modeled as changes in this graph structure between two conditions.
However, there may be cases where the conditional dependency relationships between pairs of genes change in magnitude but not in structure. For example, two genes may be positively conditionally dependent in one group but negatively conditionally dependent in the other. The supports of the precision matrices of the two groups would be identical, and would not reflect these potentially biologically significant differences in magnitude. Instead, in this paper two genes are defined to be connected in the differential network if the magnitude of their conditional dependency relationship changes between two groups. More precisely, consider independent observations of expression levels of p genes from two groups of subjects: Xi = (Xi1, . . . , Xip )T for i = 1, . . . , nX from one group and Yi = (Yi1, . . . , Yip )T for i = 1, . . . , nY from the other, where Xi ~ N(μX, ΣX) and Yi ~ N(μY , ΣY ). The differential network is defined to be the difference between the two precision matrices, denoted . The entries of Δ0 can also be interpreted as the differences in the partial covariances of each pair of genes between the two groups. This type of model for a differential network has been adopted by others as well, for example in Li et al. (2007), Danaher et al. (2013), and an unpublished technical report by Städler and Mukherjee (arxiv:1308.2771).
2 Previous approaches
There are currently two main types of approaches to estimating Δ0. The most straightforward one is to separately estimate and and then to subtract the estimates. A naive estimate of a single precision matrix can be obtained by inverting the sample covariance matrix. However, in most experiments the number of gene expression probes exceeds the number of subjects. In this high-dimensional data setting, the sample covariance matrix is singular and alternative methods are needed to estimate the precision matrix. Theoretical and computational work has shown that estimation is possible under the key assumption that the precision matrix is sparse, meaning that each row and each column has relatively few nonzero entries (Friedman et al., 2008; Ravikumar et al., 2008; Yuan, 2010; Cai et al., 2011).
The second type of approach is to jointly estimate and , taking advantage of an assumption that they share common features. For example, Chiquet et al. (2011), Guo et al. (2011), and Danaher et al. (2013) penalized the joint log-likelihood of the Xi and Yi using penalties such as the group lasso (Yuan and Lin, 2006) and group bridge (Huang et al., 2009; Wang et al., 2009), which encourage the estimated precision matrices to have similar supports. Danaher et al. (2013) also introduced the fused graphical lasso, which uses a fused lasso penalty (Tibshirani et al., 2005) to encourage the entries of the estimated precision matrices to have similar magnitudes.
However, most of these approaches assume that both and are sparse, but real transcriptional networks often contain hub nodes (Barabási and Oltvai, 2004; Barabási et al., 2011), or genes that interact with many other genes. The rows and columns of and corresponding to hub nodes have many nonzero entries and violate the sparsity condition. The method of Danaher et al. (2013) is one exception that does not require individual sparsity. Its estimates and minimize
| (1) |
where and are sample covariance matrices of the Xi and Yi, and are the (j, k)th entries of and , and det(·) and tr(·) are the determinant and trace of a matrix, respectively. The first term of (1) is the joint likelihood of the Xi and Yi and the second and third terms comprise a fused lasso-type penalty. The parameters λ1 and λ2 control the sparsity of the individual precision matrix estimates and the similarities of their entries, respectively, and when λ1 is set to zero (1) does not require or to be sparse. A referee also pointed out that a recently introduced method (Mohan et al., 2012) were also designed for estimating networks containing hubs. However, theoretical performance guarantees for these methods have not been derived.
The direct estimation method proposed in this paper does not require and to be sparse and does not require separate estimation of these precision matrices. Theoretical performance guarantees are provided for differential network recovery and estimation, and simulations show that when the separate networks include hub nodes, direct estimation is more accurate than fused graphical lasso or separate estimation.
3 Direct estimation of difference of two precision matrices
3.1 Constrained optimization approach
Let |·| denote element-wise norms and let ∥·∥ denote matrix norms. For a p × 1 vector a = (a1, . . . , ap)T, define |a|0 to be the number of nonzero elements of a, , and |a|∞ = maxj |aj|. for a p × p matrix A with entries ajk, define |A|0 be the number of nonzero entries of A, |A|1 = Σj,k |ajk|, |A|∞ = maxj,k |ajk|, ∥A∥1 = maxk Σj |ajk|, ∥A∞ = maxj Σk |ajk|, ∥A∥2 = sup|a|2≤1 |Aa|2, and .
Let , where , and let be defined similarly. Since the true Δ0 satisfies ΣX Δ0ΣY − (ΣX − ΣY) = 0, a sensible estimation procedure would solve for Δ. When min(nX, nY ) < p there are an infinite number of solutions, but accurate estimation is still possible when Δ0 is sparse. Motivated by the constrained minimization approach to precision matrix estimation of Cai et al. (2011), one estimator can be obtained by solving
and then symmetrizing the solution. This is equivalent to a linear program, as for any three p × p matrices A, B, and C, , where denotes the Kronecker product and vec(B) denotes the p2 × 1 vector obtained by stacking the columns of B. Therefore Δ0 could be estimated by solving and then symmetrizing
| (2) |
This approach directly estimates the difference matrix without even implicitly estimating the individual precision matrices. The key is that sparsity is assumed for Δ0 and not for or . Direct estimation thus allows the presence of hub nodes in the individual networks and can still achieve accurate support recovery and estimation in high dimensions, as will be discussed in Section 4. A similar direct estimation approach was also proposed by Cai and Liu (2011) for high-dimensional linear discriminant analysis. Linear discriminant analysis depends on the product of a precision matrix and the difference between two mean vectors, and Cai and Liu (2011) showed that direct estimation of this product is possible even in cases where the precision matrix or the mean difference are not individually estimable.
3.2 A modified problem
The linear program (2) has a p2 × p2 constraint matrix and can become computationally demanding for large p. A modified procedure can alleviate this burden by requiring the estimate to be symmetric. Denote the (j, k)th entry of a matrix Δ by δjk, and define β to be the p(p + 1)/2 × 1 vector with β = (δjk)1≤j≤k≤p. Estimating a symmetric Δ is thus equivalent to estimating β, which has only p(p + 1)/2 parameters. Define the p2 × p(p + 1)/2 matrix S with columns indexed by 1 ≤ j ≤ k ≤ p and rows indexed by l = 1, . . . , p and m = 1, . . . , p, so that each entry is labeled by Slm,jk. For j ≤ k, let Sjk,jk = Skj,jk = 1, and set all other entries of S equal to zero. For example, when p = 3,
When Δ is symmetric some calculation shows that . Furthermore, if , where is the (j, k)th entry of Δ0, by Lemma A1 β0 is the unique solution to . Therefore one reasonable approach to estimate a sparse β0 in high dimensions is to solve
However, the inequality constraints can be improved. Let E be the p × p matrix such that . These constraints treat the diagonals and off-diagonals of differently, with the diagonals constrained roughly half as much as the off-diagonals.
Therefore the remainder of this paper considers the estimate of Δ0 obtained by solving
| (3) |
where , , and for a p(p+1)/2 × 1 vector c, |c|O∞ denotes the sup-norm of the entries of c corresponding to the off-diagonal elements of its matrix form, and |c|D∞ is the sup-norm of the entries corresponding to the diagonal elements. The matrix form of will be denoted by . Compared to (2), (3) requires only a p(p + 1)/2 × p(p + 1)/2 constraint matrix, but requires a stronger theoretical condition to guarantee support recovery and estimation consistency, which is discussed in Section 4.
3.3 Implementation
The estimator (3) can be computed by slightly modifying code from the R package flare, recently developed by X. Li, T. Zhao, X. Yuan, and H. Liu to implement a variety of high dimensional linear regression and precision matrix estimation methods. Their method uses the alternating direction method of multipliers; for a thorough discussion see Boyd et al. (2011). To apply their algorithm, rewrite (3) as
where f(r) equals infinity if |r|O∞ > λn or |r|D∞ > λn/2 and zero otherwise. The augmented Lagrangian is then
where u is the Lagrange multiplier and ρ > 0 is a penalty parameter specified by the user. The alternating direction method of multipliers obtains the solution using the updates
for each iteration t. The flare package incorporates several strategies to speed convergence, such as using a closed-form expression for rt+1, using a hybrid coordinate descent and linearization procedure to obtain βt+1, and dynamically adjusting ρ at each iteration.
The direct estimation approach can be tuned using an approximate Akaike information criterion. For the loss functions
| (4) |
where makes explicit the dependence of the estimator on the tuning parameter, λn is chosen to minimize
| (5) |
where L(λn) represents either L∞ or LF and k is the effective degrees of freedom, which can be approximated by , or the number of nonzero elements in the upper triangle of . The loss functions (4) focus on the supremum and Frobenius norms in light of Theorems 2 and 3 in Section 4, but other norms could be used as well.
4 Theoretical properties
Let and be the (j, k)th entries of ΣX and ΣY , respectively. Define and . Good performance of direct estimation requires the following conditions.
Condition 1
The true difference matrix Δ0 has s < p nonzero entries in its upper triangle, and |Δ0|1 ≤ M, where M does not depend on p.
Condition 2
With s defined as in Condition 1, the constants and must satisfy , where .
Condition 1 requires the difference matrix to have essentially constant sparsity, which is reasonable because genetic networks are not expected to differ much between two conditions. Condition 2 requires that the true covariances between the covariates are not too high, and can hold even when and are not sparse. Actually it is sufficient to require only that the magnitude of the largest off-diagonal entry of be less than , but Condition 2 is more interpretable.
Condition 2 is closely related to the mutual incoherence property introduced by Donoho and Huo (2001), but is more complicated in the current setting because it involves a linear function of the Kronecker product of two covariance matrices. Solving (2) instead of (3) would require only , with s̃ equal to the total number of nonzero entries of Δ0. If in addition for all j, max(μX, μY) ≤ (2s̃)−1 would be required, which is similar to imposing the usual mutual incoherence condition on ΣX and ΣY . Condition 2 is more restrictive, but (3) is easy to compute and still gives good finite-sample results.
Under these conditions, a thresholded version of the direct estimator can successfully recover the support of Δ0. Let the (j, k)th entries of Δ0 and be and , respectively. For a threshold τn > 0 define the estimator
Let the (j, k)th entry of be , and define the function
Then if and are vectors of the signs of the entries of the estimated and true difference matrices, respectively, the following theorem holds.
Theorem 1
Suppose Conditions 1 and 2 hold, and let and μ be defined as in Condition 2. If min(nX, nY ) > log p,
and , then with probability at least 1 − 8p−τ, where C is defined in Lemma A2 in the Appendix.
Theorem 1 states that with high probability, can recover not only the support of Δ0 but also the signs of its nonzero entries, as long as those entries are sufficiently large. In other words, in the context of genetic networks, can correctly identify genes whose conditional dependencies change in magnitude between two conditions, as well as the directions of those changes, as long as min(nX, nY ) is large relative to log p. In practice the threshold τn can be treated as a tuning parameter. In simulations and data analysis τn was set to 0 · 0001.
A thresholding step is natural in practice because small entries of are most likely noisy estimates of zero. This step could be avoided by imposing an irrepresentability condition on , similar to those assumed in the proofs of the selection consistencies of the lasso (Meinshausen and Bühlmann, 2006; Zhao and Yu, 2006) and the graphical lasso (Ravikumar et al., 2008). However, these types of conditions are stronger than the mutual incoherence-type property assumed in Condition 2, as discussed in Lounici (2008). The thresholded estimators are pursued in this paper because of their milder theoretical requirements.
In addition to identifying the entries of and that change, can correctly quantify these changes, in the sense of being consistent for Δ0 in the Frobenius norm.
Theorem 2
Suppose Conditions 1 and 2 hold and define and μ as in Condition 2. If min(nX, nY ) > log p and
then
with probability at least 1 − 8p−τ, where C is defined in Lemma A2.
The proofs of Theorems 1 and 2 rely on the following bound on the element-wise norm of the estimation error.
Theorem 3
Suppose Conditions 1 and 2 hold, and define and μ as in Condition 2. If min(nX, nY ) > log p and
then
with probability at least 1 − 8p−τ, where C is defined in Lemma A2 in the Appendix.
Similar theoretical properties have been derived for separate and joint approaches to estimating differential networks (Cai et al., 2011; Guo et al., 2011). However, these require sparsity conditions on each Σ−1, such as ∥Σ−1∥∞ ≤ M′ < ∞, which can be violated if the individual networks contain hub nodes. In contrast, Theorems 1–3 can still hold in the presence of hubs.
5 Simulations
5.1 Settings
Simulations were conducted to compare direct estimation (3), fused graphical lasso (1), and separate estimation using the procedure of Cai et al. (2011). Data were generated with p = 40, 60, 90, and 120 and X1, . . . , XnX and Y1, . . . , YnY were generated from N(0, ΣX) and N(0, ΣY ), respectively, with nX = nY = 100.
For each p, the support of was first generated according to a network with p(p − 1)/10 edges and a power law degree distribution with an expected power parameter of 2, which should mimic real-world networks (Newman, 2003). This still gives a relatively sparse network, since only 20% of all possible edges are present, but the power law structure creates hub nodes, which make certain rows and columns nonsparse.
The value of each nonzero entry of was next generated from a uniform distribution with support [−0 · 5, −0 · 2] ∪ [ [0 · 2, 0 · 5]. To ensure positive-definiteness, each row was divided by two when p = 40, three when p = 60, four when p = 90, and five when p = 120. The diagonals were then set equal to one and the matrix was symmetrized by averaging it with its transpose. The differential network Δ0 was generated such that the largest 20%, by magnitude, of the connections of the top two hub nodes of changed sign between and . In other words, Δ0 was a sparse matrix, with zero entries everywhere except for 20% of the entries in two rows and columns.
Each method was tuned using an approximate Akaike information criterion. Direct estimation was tuned using (5) and one of the loss functions in (4). For a fair comparison fused graphical lasso was tuned in the same way, after searching across all combinations of three values of λ1 and 10 values of λ2. Small values of λ1 were used because the true precision matrices were nonsparse. Separate estimation was tuned by searching across 10 different values of the tuning parameter to minimize , where was the sample covariance matrix of the X and was the estimated precision matrix. The same was done for the Yi, with AICY defined similarly. Results were averaged over 250 replications.
5.2 Results
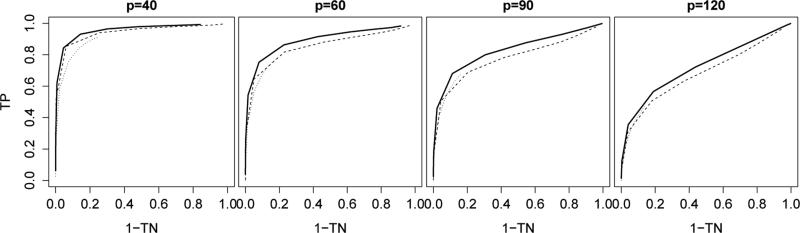
Figure 1 illustrates the receiver operating characteristic curves of the three estimation methods. Let be the (j, k)th entry of a given estimator and let be the (j, k)th entry of the true Δ0. The true positive and negative rates of were defined as
respectively. Different points on the curves correspond to different tuning parameter values. The curves for the fused graphical lasso estimator (1) were plotted by varying λ2. The λ1 parameter, which controls the sparsity of the individual precision matrix estimates, was fixed at a small value because the individual matrices were not sparse. For a fair comparison with the thresholded direct estimator, the fused graphical lasso was thresholded at 0 · 0001. The separate estimator performed poorly and its curves were not plotted. Figure 1 shows that direct estimation compared favorably to fused graphical lasso.
Figure 1.
Receiver operating characteristic curves for support recovery of ; solid line: thresholded direct estimator; dashed line: thresholded fused graphical lasso estimator with λ1 = 0; dotted line: fused graphical lasso estimator with λ1 = 0 · 1
The true discovery and nondiscovery rates of the three estimators were studied as well, and were defined as
respectively. These rates were taken to be zero when their denominators were equal to zero. In the analysis of genomic data minimizing the number of false discoveries is a major concern. The direct and fused graphical lasso estimators were thresholded at 0 · 0001 and the separate estimator was thresholded at 0 · 0002. In all settings, the true nondiscovery rates of the direct and fused graphical lasso estimators were close to 100%; separate estimation frequently did not identify any zero entries in the differential network. The true discovery rates are reported in Table 1, which also compares the effects of tuning using different loss functions (4). For direct estimation, tuning using L∞ gave the best true discovery rates for smaller p while LF was preferrable for larger p. For fused graphical lasso, L∞ was always the better choice. Using either L∞ or LF , direct estimation performed well compared to fused graphical lasso and separate estimation, especially for larger p.
Table 1.
Average true discovery rates over 250 simulations. Standard errors are in parentheses
| p | L∞ | LF | L∞ | LF | |
|---|---|---|---|---|---|
| 40 | 77(16) | 29(16) | 83(12) | 27(17) | 2(0) |
| 60 | 76(19) | 66(21) | 74(19) | 65(30) | 1(0) |
| 90 | 66(25) | 80(31) | 55(26) | 12(28) | 1(0) |
| 120 | 48(39) | 61(45) | 33(25) | 1(7) | 1(0) |
The Frobenius norm estimation accuracies of the unthresholded estimators tuned using different loss functions are reported in Table 2. The results of tuning using L∞ or LF were comparable. Direct estimation was much more accurate than separate estimation and slightly more accurate than fused graphical lasso. It is possible for direct estimation to simultaneously give markedly better support recovery but similar estimation compared to fused graphical lasso because estimation error depends on the magnitudes of the estimated entries, while support recovery depends only on whether they are nonzero. For example, suppose had the same support as the true Δ0, but each nonzero entry had magnitude 0 · 01. Then under the p = 120 simulation setting, . The estimation error of this is even higher than to those in Table 2, but it exactly recovers the true support.
Table 2.
Average estimation errors in Frobenius norm over 250 simulations. Standard errors are in parentheses
| p | L∞ | LF | L∞ | LF | |
|---|---|---|---|---|---|
| 40 | 1·67(0·13) | 1·46(0·22) | 1·72(0·06) | 1·50(0·15) | 12·96(0·78) |
| 60 | 1·68(0·07) | 1·62(0·11) | 1·68(0·05) | 1·69(0·06) | 28·45(2·01) |
| 90 | 1·68(0·04) | 1·71(0·03) | 1·68(0·03) | 1·72(0·03) | 57·30(2·75) |
| 120 | 1·55(0·02) | 1·55(0·01) | 1·54(0·02) | 1·55(0·00) | 102·84(4·06) |
The good performance of direct estimation came at the price of some computational convenience. The memory required by the large constraint matrix behaves like O(p4), though the simulations generally needed no more than one gigabyte of memory when p = 120. In an unpublished technical report, Hong and Luo (arxiv:1208.3922) proved the global linear convergence of the alternating direction method of multipliers applied to problems like (3). However, each iteration of the proposed algorithm requires roughly O(sp4) computations, where s is the number of nonzero entries in the upper triangle of Δ0, as defined in Condition 1. The simulations required on average 51, 853, 6231, and 51589 seconds when p = 40, 60, 90, and 120, respectively. On the other hand, these memory and time requirements are still reasonable in practice. Recently, H. Pang, H. Liu, and R. Vanderbei have developed an even faster algorithm for constrained -minimization problems such as (3), available in the R package fastclime, which should reduce the computational burden of direct estimation.
6 Gene expression study of ovarian cancer
The proposed approach was applied to gene expression data collected from patients with stage III or IV ovarian cancer. Using these data, Tothill et al. (2008) identified six molecular subtypes of ovarian cancer, which they labeled C1 through C6. They found that the C1 subtype, characterized by differential expression of genes associated with stromal and immune cell types, was associated with much shorter survival times.
The proposed direct estimation procedure was applied to investigate whether this poor prognosis sub-type was also associated with differential wiring of genetic networks. The subjects were divided into a C1 group, with 78 patients, and a C2–C6 group, with 113 patients. Several pathways from the KEGG pathway database (Ogata et al., 1999; Kanehisa et al., 2012) were studied to determine if any differences in the conditional dependency relationships of the gene expression levels existed between the subtypes. All probesets corresponding to the same gene symbol were first averaged to get gene-level expression measurements.
Direct estimation and fused graphical lasso were tuned using (5) with the loss function L∞ because in simulations this gave the best results for fused graphical lasso and good results for direct estimation. Separate estimation was tuned as described in Section 3.3. The direct and fused graphical lasso estimators were thresholded at 0 · 0001 to recover the differential network. The separate estimator was not simply thresholded at 0 · 0002 because simulations showed that this method gave poor true discovery rates. Instead, two genes were defined as being linked in the differential network if they were connected in one group but not the other, or if they were connected in both groups but their conditional dependency relationship changed sign. The procedure of Cai et al. (2011) thresholded at 0 · 0001 was used to recover the individual networks.
Two illustrative examples are reported in Fig. 2. Only genes included in at least one edge, or which saw a change in partial variance between the two subtypes, were included in the figures. In the results of separate estimation, only genes in the differential network estimated by direct estimation were labeled. To interpret the results, the most highly connected genes in the differential networks were considered to be important.
Figure 2.
Estimates of the differential networks between ovarian cancer subtypes. The direct and fused graphical lasso estimators were thresholded and the separate estimator was further sparsified; see text. Black edges show increase in conditional dependency from ovarian cancer subtype C1 to subtypes C2–C6, gray edges show decrease. (a)–(c): KEGG 04350, TGF-β pathway, (d)–(f): KEGG 04210, Apoptosis pathway. (a)–(e) tuned using L∞ with (5), (f) tuned with λ1 = 0 and λ2 to give the same number of edges as (d); see text. Separate estimator not shown for apoptosis pathway.
Figures 2(a)–2(c) illustrate estimates of the differential network of the TGF-β signaling pathway, which at 82 genes is larger than the sample size of the C1 group. Direct estimation suggested the presence of two hub genes, COMP and THBS2, which have both been found to be related to resistance to platinum-based chemotherapy in epithelial ovarian cancer (Marchini et al., 2013). Fused graphical lasso gave the same number of edges in the differential network as direct estimation and only suggested the importance of COMP. It was hard to draw meaningful conclusions from the results of separate estimation because the denseness of the estimated network made it difficult to identify a small number of important genes.
Figures 2(d)–2(f) give the estimates for the apoptosis pathway, which at 87 genes was also larger than sample size of the C1 group. The separate estimator again resulted in a dense network and is not included in Fig. 2. Direct estimation pointed to BIRC3 and TNFSF10 as being important genes. Indeed, TNFSF10 encodes the TRAIL protein, which has been studied a great deal because of its potential as an anticancer drug (Yagita et al., 2004; Bellail et al., 2009), and in particular as a therapy for ovarian cancer (Petrucci et al., 2012; Kipps et al., 2013). BIRC3 can inhibit TRAIL-induced apoptosis (Johnstone et al., 2008) and has also been considered for use as a therapeutic target in cancer (Vucic and Fairbrother, 2007). Figure 2(e) shows the fused graphical lasso estimator tuned in the same way as the direct estimator, and suggests only BIRC3 as being important. For a fairer comparison with direct estimation, Fig. 2(f) depicts the fused graphical lasso estimator after fixing λ1 = 0 and adjusting λ2 to achieve the same level of sparsity as Fig. 2(d). The result is similar to Fig. 2(d), though it suggests that BIRC3 and PRKAR2B, a protein kinase, are important, rather than BIRC3 and TRAIL.
7 Discussion
Instead of modeling a differential network as the difference of two precision matrices, as proposed above, another possibility is to use the difference between two directed acyclic graphs. These graphs are natural models for single transcriptional regulatory networks, with nodes representing gene expression levels and edges indicating how the nodes are causally related to each other. Biological changes to a network can be thought of as interventions on some of its nodes, which results in changes to the graphical structure; see the unpublished technical report by Hauser and Bühlmann (arxiv:1303.3216) and references therein. Frequently, however, only observational data are available for gene expression, so it is difficult to estimate the underlying causal structures (Kalisch and Bühlmann, 2007; Maathuis et al., 2009). It would be interesting to develop a direct estimation method for differential networks with interventional data.
For method (3) to have good properties in high dimensions, Δ0 must be sparse. While reasonable, this assumption will be violated if the biological differences between two groups manifest as global changes that affect a large number of gene-gene dependencies. If some proportion of these global changes are of sufficient magnitude, the method should still be able to detect their presence, though it may not recover all of the changes or accurately estimate their magnitudes. The most challenging case for the proposed method occurs when the network changes are numerous but small. A new statistic could be defined to quantify the degree of global change between two precision matrices, but so far there is little consensus as to what statistic might be most biologically meaningful.
Finally, while the focus has been on directly estimating the difference between two precision matrices, there are situations where interest may center on how a transcriptional regulatory network differs between K conditions, where K > 2. The proposed method could of course be used to estimate all pairwise differential networks, but this could be time-consuming. Another possibility would be to estimate the difference between each precision matrix and some common precision matrix, which could be taken to be the inverse of the pooled covariance matrix of all K groups. In other words, if were the sample covariance matrix of the kth group, let be some weighted average of the and consider solving
If the difference matrix were sparse, where and is the precision matrix of the kth group, would be a direct estimate of Δk. The differential network between the jth and kth group could then be estimated as .
Acknowledgments
This research is supported by National Institute of Health grants and a National Science Foundation grant. The authors thank Xingguo Li for his expertise regarding the computational complexity of the proposed algorithm.
APPENDIX: PROOFS OF THEOREMS
Lemma 1
The matrix is invertible, where ΣY and ΣX are p × p covariance matrices and S is defined in Section 3.2.
Proof
Since ΣX and ΣY are positive-definite, there exists a full-rank matrix Σ1/2 such that . Furthermore, from its construction S has full column rank, so rank(S) = p(p+ 1)/2. Therefore
Since is p(p + 1)/2 × p(p + 1)/2, it is full rank and therefore invertible.
The next lemma comes from the proofs of Theorems 1(a) and 4(a) in Cai et al. (2011).
Lemma 2
Let Xi = (Xi1, . . . , Xip)T for i = 1, . . . , n be independent and identically distributed random vectors with E(Xi) = (μ1, . . . , μjT , and let X̄ = n−1Xi and . If there exists some 0 < η < 1/4 such that log p/n ≤ η and for all |t| ≤ η and j = 1, . . . , p, then
with probability at least 1 − 4p−τ, where C = 2η −2(2 + τ + η−1 e2K2)2 and τ > 0.
Lemma 3
Let . Label the entries of STΣS as . Then
Proof
Label the entries of Σ as σl′m′,lm(l′ = 1, . . . , p; m′ = 1, . . . , p; l = 1, . . . , p; m = 1, . . . , p) and the entries of S as Slm,jk(l = 1, . . . , p; m = 1, . . . , p; 1 ≤ j ≤ k ≤ p), as in Section 3.3. By the definition of the Kronecker product, , and , so the lemma follows from the definition of the entries of S.
Proof of Theorem 3
Let the entries of Δ0 be denoted , and define the p(p + 1)/2 × 1 vector . Define Σ as in Lemma A3, , b = vec(ΣX − ΣY ), and . The bound on is obtained following Lounici (2008).
Denote the ath component of ST ΣSh by (STΣSh)a, the (a, b)th entry of STΣS by , and the bth component of h by hb. Also let . Then , which implies
| (6) |
The diagonal terms can be relabeled as , where j may equal k, and from Lemma A3 must satisfy , with defined in Condition 2. The off-diagonal terms , a ≠ b can be relabeled as with j′ ≠ j or k′ ≠ k, and from Lemma A3 must satisfy , with μX and μY defined in Condition 2. Using these facts, and Condition 2, (6) becomes
| (7) |
The method of Cai et al. (2010b) is used to bound |h|1. Let T0 be the set of indices corresponding to the support of β0, and for any p × 1 vector a = (a1, . . . , ap)T let aT0 be the vector with components aT0j = 0 for j ∉/ T0 and aT0j = aj for j ∈ T0.
First it must be shown that β0 is in the feasible set with high probability. Since Xi and Yi are both Gaussian, they satisfy the conditions of Lemma A2 and thus and are both less than C{log p/ min(nX, nY)}1/2 with probability at least 1 − 8p−τ. Then
where ∥S∥1 = 2 by the definition of S and |β0|1 ≤ M by Condition 1. Next, from the proof of Lemma A3, each entry of Σ can be written as , so
since min(nX, nY ) > log p. Then β0 is feasible with probability at least 1 − 8p−τ if .
Now |h|1 can be bounded. By the definition of (3), . This implies that . Using the triangle inequality, , or in other words, . Therefore . To bound |hT0|2, observe following Cai et al. (2009) that for any s-sparse vector c,
This implies that
Together with , this implies that
so (7) becomes
Bounding |STΣSh|∞ uses the proof that β0 is feasible, because
when and are both less than C{log p/ min(nX, nY)}1/2.
Proof of Theorem 1
Let be the (j, k)th entry of . Then
Suppose . Then with probability going to 1, by Theorem 3, so . Next suppose . Then with probability going to 1, so . Finally for with probability going to 1, so .
Proof of Theorem 2
Solutions to this type of -constrained optimization problem have . Cai et al. (2010a) used this property of h, along with their Lemma 3, to show that , where T* is the set of indices corresponding to the s/4-largest components of . Then , and combining this with Theorem 3 completes the proof.
Contributor Information
Sihai Dave Zhao, Department of Biostatistics and Epidemiology, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania 19104, USA.
T. Tony Cai, Department of Statistics, The Wharton School, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA.
Hongzhe Li, Department of Biostatistics and Epidemiology, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania 19104, USA.
References
- Bandyopadhyay S, Mehta M, Kuo D, Sung M-K, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, Fiedler D, Dutkowski J, Guénolé A, van Attikum H, Shokat KM, Kolodner RD, Huh W-K, Aebersold R, Keogh M-C, Krogan NJ, Ideker T. Rewiring of genetic networks in response to DNA damage. Sci. Signal. 2010;330(6009):1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellail AC, Qi L, Mulligan P, Chhabra V, Hao C. TRAIL agonists on clinical trials for cancer therapy: the promises and the challenges. Rev. Recent Clin. Trials. 2009;4(1):34–41. doi: 10.2174/157488709787047530. [DOI] [PubMed] [Google Scholar]
- Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Foundat. Trends Mach. Learn. 2011;3(1):1–122. [Google Scholar]
- Cai T, Liu W. A Direct Estimation Approach to Sparse Linear Discriminant Analysis. J. Am. Statist. Assoc. 106(496):1566–1577. Dec. 2011. ISSN 0162-1459. doi: 10.1198/jasa.2011.tm11199. URL http://www.tandfonline.com/doi/abs/10.1198/jasa.2011.tm11199. [Google Scholar]
- Cai T, Xu G, Zhang J. On recovery of sparse signals via l 1 minimization. IEEE Trans. Inf. Theory. 2009;55(7):3388–3397. [Google Scholar]
- Cai T, Wang L, Xu G. Shifting inequality and recovery of sparse signals. IEEE Trans. Signal Process. 2010a;58(3):1300–1308. [Google Scholar]
- Cai T, Wang L, Xu G. Stable recovery of sparse signals and an oracle inequality. IEEE Trans. Inf. Theory. 2010b;56(7):3516–3522. [Google Scholar]
- Cai T, Liu W, Luo X. A constrained ℓ1 minimization approach to sparse precision matrix estimation. Am. Statist. Assoc. 2011;106(494):594–607. [Google Scholar]
- Chiquet J, Grandvalet Y, Ambroise C. Inferring multiple graphical structures. Statist. Comput. 2011;21(4):537–553. [Google Scholar]
- Danaher P, Wang P, Witten DM. The joint graphical lasso for inverse covariance estimation across multiple classes. J. Roy. Statist. Soc. Ser. B. 2013 doi: 10.1111/rssb.12033. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente A. From differential expression to differential networking – identification of dysfunctional regulatory networks in diseases. Trends Genet. 2010;26(7):326–333. doi: 10.1016/j.tig.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Donoho D, Huo X. Uncertainty principles and ideal atomic decomposition. IEEE Trans. Inf. Theory. 2001;47(7):2845–2862. [Google Scholar]
- Emmert-Streib F, Dehmer M. Networks for systems biology: conceptual connection of data and function. IET Syst. Biol. 2011;5(3):185–207. doi: 10.1049/iet-syb.2010.0025. [DOI] [PubMed] [Google Scholar]
- Friedman JH, Hastie TJ, Tibshirani RJ. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Levina E, Michailidis G, Zhu J. Joint estimation of multiple graphical models. Biometrika. 2011;98(1):1–15. doi: 10.1093/biomet/asq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ma S, Xie H, Zhang C-H. A group bridge approach for variable selection. Biometrika. 2009;96(2):339–355. doi: 10.1093/biomet/asp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NJ, Reverter A, Dalrymple BP. A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation. PLoS Comput. Biol. 2009;5(5):e1000382. doi: 10.1371/journal.pcbi.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Krogan N. Differential network biology. Mol. Syst. Biol. 2012;8(1):565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer. 2008;8(10):782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- Kalisch M, Bühlmann P. Estimating high-dimensional directed acyclic graphs with the pc-algorithm. J. Mach. Learn. Res. 2007;8:613–636. [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(D1):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipps E, Tan DSP, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat. Rev. Cancer. 2013:273–282. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen SL. Graphical Models. Oxford University Press; Oxford: 1996. [Google Scholar]
- Li K-C, Palotie A, Yuan S, Bronnikov D, Chen D, Wei X, Choi O-W, Saarela J, Peltonen L. Finding disease candidate genes by liquid association. Genome Biol. 2007;8(10):R205. doi: 10.1186/gb-2007-8-10-r205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounici K. Sup-norm convergence rate and sign concentration property of Lasso and Dantzig estimators. Electron. J. Statist. 2008;2:90–102. [Google Scholar]
- Maathuis MH, Kalisch M, Bühlmann P. Estimating high-dimensional intervention effects from observational data. Ann. Statist. 2009;37(6A):3133–3164. [Google Scholar]
- Marchini S, Fruscio R, Clivio L, Beltrame L, Porcu L, Nerini IF, Cavalieri D, Chiorino G, Cattoretti G, Mangioni C, Milani R, Torri V, Romualdi C, Zambelli A, Romano M, Signorelli M, di Giandomenico S, DIncalci M. Resistance to platinum-based chemotherapy is associated with epithelial to mesenchymal transition in epithelial ovarian cancer. Eur. J. Cancer. 2013;49:520–530. doi: 10.1016/j.ejca.2012.06.026. [DOI] [PubMed] [Google Scholar]
- Markowetz F, Spang R. Inferring cellular networks–a review. BMC Bioinformatics. 2007;8(Suppl 6):S5. doi: 10.1186/1471-2105-8-S6-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinshausen N, Bühlmann P. High-dimensional graphs and variable selection with the lasso. Ann. Statist. 2006;34(3):1436–1462. [Google Scholar]
- Mohan K, Chung M, Han S, Witten D, Lee S-I, Fazel M. Structured learning of Gaussian graphical models. Adv. Neural Inf. Process. Syst. 2012:629–637. [PMC free article] [PubMed] [Google Scholar]
- Newman ME. The structure and function of complex networks. SIAM Rev. 2003;45(2):167–256. [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucci E, Pasquini L, Bernabei M, Saulle E, Biffoni M, Accarpio F, Sibio S, Di Giorgio A, Di Do-nato V, Casorelli A, Benedetti-Panici P, Testa U. A small molecule SMAC mimic LBW242 potentiates TRAIL-and anticancer drug-mediated cell death of ovarian cancer cells. PloS One. 2012;7(4):e35073. doi: 10.1371/journal.pone.0035073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar PK, Raskutti G, Wainwright MJ, Yu B. Model Selection in Gaussian Graphical Models: High-Dimensional Consistency of [lscript]1-regularized MLE. Adv. Neural Inf. Process. Syst. 2008;21:1329–1336. [Google Scholar]
- Tibshirani RJ, Saunders M, Rosset S, Zhu J, Knight K. Sparsity and smoothness via the fused lasso. J. Roy. Statist. Soc. Ser. B. 2005;67(1):91–108. [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew Y-E, Haviv I, Australian D, Ovarian Cancer Study Group. deFazio Gertig, A., Bowtell DDL. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008;14:5198–208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- Vucic D, Fairbrother WJ. The inhibitor of apoptosis proteins as therapeutic targets in cancer. Clin. Cancer Res. 2007;13(20):5995–6000. doi: 10.1158/1078-0432.CCR-07-0729. [DOI] [PubMed] [Google Scholar]
- Wang S, Nan B, Zhu N, Zhu J. Hierarchically penalized Cox regression with grouped variables. Biometrika. 2009;96(2):307–322. [Google Scholar]
- Yagita H, Takeda K, Hayakawa Y, Smyth MJ, Okumura K. TRAIL and its receptors as targets for cancer therapy. Cancer Sci. 2004;95(10):777–783. doi: 10.1111/j.1349-7006.2004.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M. Sparse inverse covariance matrix estimation via linear programming. J. Mach. Learn. Res. 2010;11:2261–2286. [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J. Roy. Statist. Soc. Ser. B. 2006;68(1):49–67. [Google Scholar]
- Zhao P, Yu B. On model selection consistency of lasso. J. Mach. Learn. Res. 2006;7:2541–2563. [Google Scholar]