Abstract
A number of lower vertebrates including urodele amphibians and teleost fish are remarkably adept at repairing and regenerating damaged tissues and organs. Freshwater planarians are even more amazing, capable of regenerating entire body plans from small amputated fragments. In contrast, mammalian regenerative capacity is quite limited but of intense interest, especially related to human health and disease. For those organisms capable of robust regeneration, a common theme is the use of stem cells to replace complex tissues. Key questions remain as to the origin of these cells, whether there are pools of such cells that migrate to injured regions or whether they are generated on site. Beyond their origin, how are the genetic pathways that enable differentiation into multiple cell types and tissues regulated? microRNAs (miRNAs) are small noncoding RNAs that have recently been shown to play important roles in controlling stem cell self-renewal, proliferation and differentiation. Some of these are thought to be required to maintain “stemness”. Here, we summarize recent work on the role of miRNAs in stem cells and their roles during regeneration.
Keywords: regeneration, microRNAs, stem cells, zebrafish
Small RNAs in Regeneration
miRNA biogenesis
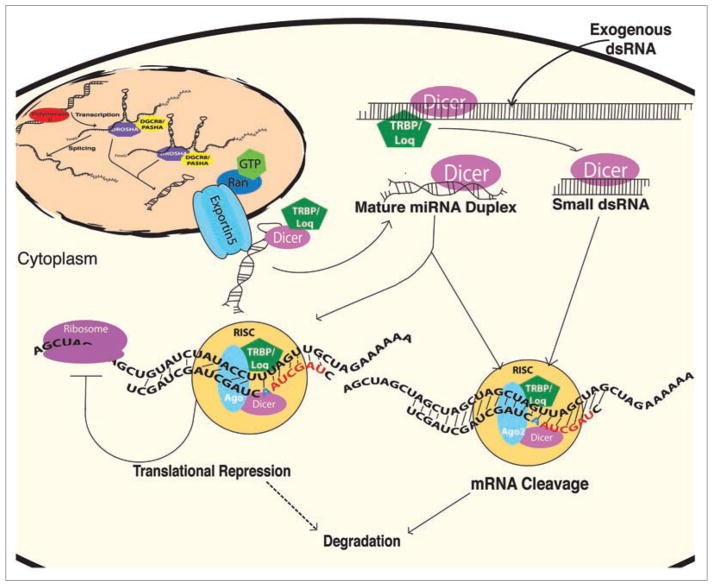
Recently, a new class of regulatory molecules, small noncoding RNAs referred to as microRNAs (miR-NAs), has been implicated in regeneration. miRNAs have been shown to regulate many cellular functions including cell specification, differentiation, proliferation and metabolism.1–11 A key to understanding their role is to determine how many are encoded in animal genomes and what genes they regulate. miRNA biogenesis begins with primary transcripts synthesized by RNA polymerase II that form extended hairpin structures containing one or more miRNAs. Some of these primary transcripts originate from independent transcripts whereas others are encoded within the introns of mRNA transcripts. Primary transcripts are recognized in the nucleus by an RNase III enzyme, Drosha, which cleaves hairpin precursors for subsequent export to the cytoplasm in a Ran-GTP dependent process.12,13 In the cytoplasm, hairpin precursors are recognized by another RNase III enzyme, Dicer, that produces RNA duplexes (~22 nucleotide) that are incorporated into RNA Induced Silencing Complexes (RISCs).14–16 Either coincident or subsequent to RISC assembly, miRNAs are guided to their target mRNAs where they most often pair with 3′UTR sequences.17,18 Partial complementarity leads to translational repression whereas perfect complementarity leads to cleavage of the target mRNA (Fig. 1). This process is highly conserved throughout development.
Figure 1.
miRNA Biogenesis. Long mono- and poly-cistronic miRNA primary transcripts are synthesized by RNA Polymerase II. These transcripts are processed in the nucleus by Drosha and exported in a Ran-GTP dependent process to the cytoplasm. Dicer recognizes cytoplasmic hairpin precursors, cleaves the loop regions, and mature miRNAs are incorporated into RISC complexes. RISC guides the miRNA to its target 3′UTRs. If the miRNA binds with perfect complementarity, mRNA cleavage occurs. If the miRNA binds with partial complementarity, translation repression occurs with possible subsequent degradation. siRNAs utilize the same cytoplasmic pathway to silence RNAs after delivery of exogenous double stranded RNAs (dsRNAs).
A number of studies have examined the expression patterns of miRNAs during development, differentiation and regeneration. A common theme is that more and more miRNAs are expressed as differentiation proceeds.19,20 A smaller number are expressed in stem cells and at early stages of development, including some that are thought to be required to maintain “stemness”.21–25 For animals that are adept at repairing or regenerating damaged tissues, the role of stem cells during regeneration can be simplistically boiled down to two nonexclusive mechanisms. First, a reserve of stem cells are somehow recruited to the site of damage and differentiate to restore complex tissues and organs. Second, local stem-cell like cells near the site of damage de-differentiate to restore and repair the damage. Both scenarios propose the existence of some kind of stem cell which responds to signals that drive regeneration. Here, we will focus on recent work illustrating the role that miRNAs play in stem cell maintenance and proliferation followed by a review of recent work on the role of miRNAs during regeneration.
miRNAs in Stem Cells Maintenance
One intriguing possibility for how or why miRNAs may play a key role in regeneration is related to the finding that only a subset of miRNAs are expressed in stem cells.8,26–28 Dicer deficient mice lack almost all pluripotent stem cells and die around embryonic day 7.5.29 Similarly, mouse Dicer hypomorphs die mid-gastrulation30 and in cultured cells, loss of Dicer inhibits embryonic stem cell proliferation.31 Deletion of a DGCR8, a protein involved in nuclear processing of miRNA primary transcripts also leads to slowly proliferating ES cells that accumulate in G1.24 This is consistent with the fact that an intact miRNA biogenesis pathway is required for development and raises the possibility that early steps in embryonic development require the action of specific miRNAs to properly regulate stem cells. In line with this, restoration of multiple members of the miR-290 family could rescue proliferation in DGCR8 deficient ES cells although they could not restore differentiation defects.32 These miRNAs are referred to as ESCC miRNAs (ES cell-specific Cell Cycle regulating miR-NAs). Interestingly, proliferation and self-renewal can be blocked in ES cells by expression of let-7 miRNAs indicating that distinct subsets of miRNAs regulate stem cell maintenance, proliferation and differentiation.22 Broadly, this is consistent with findings that overall miRNA complexity increases with development. More and more miRNAs are expressed as cells differentiate into tissues and organs.19,20,33,34 For let-7, this is additionally interesting because its expression is regulated by control of miRNA processing and maturation.35–39 Mature let-7 is readily detected in more differentiated cell types whereas the precursor accumulates in ES cells and de-differentiating tumor cells.40
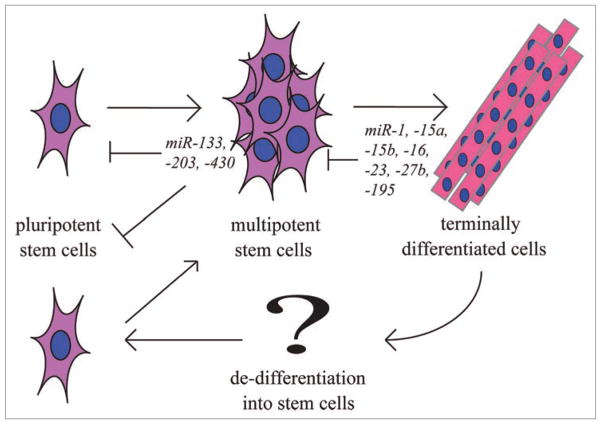
One possibility for the above results is that let-7 functions as an anti-stemness factor.22 Since regeneration relies on recruitment of stem cells or de-differentiation of cells into stem-cell like cells, it is logical to expect that let-7 or similar miRNAs will need to be turned off during regeneration whereas opposing sets of miRNAs will need to be turned on. Two examples are miR-196, whose expression is elevated during early salamander tail regeneration.41 In contrast, miR-203 must be turned off to initiate fin regeneration and allow Lef1 expression.23 For miR-203, the Fuchs and Candi groups showed that it represses stem cell activity in mice by downregulating p63, thereby repressing ‘stemness’.21,25 Further identification and understanding of the role that miRNAs play in ES self-renewal and proliferation is likely to shed light on similar involvement in regeneration and vice versa (Fig. 3).
Figure 3.
Stem cell maturation. During normal development and during regeneration, stem cells play key roles in self-renewal, differentiation and specification. Shown are several miRNAs that have been implicated in the maintenance and differentiation of stem cells.
miRNAs in Planaria
Planarians are small flatworms often found in freshwater and marine environments that are capable of remarkable feats of regeneration. Numerous stem-cell like cells, called neoblasts, make up nearly 30% of the total cells in planarians and their regenerative capacity allows re-creation of whole animals, even when cut into numerous pieces.42–44 Using microarrays and in situ localization, a unique set of ~50–60 miRNAs were found to be highly expressed in neoblasts.45,46 Key protein components of miRNA effector complexes (RISC) are members of the Argonaute family. Interestingly, two genes encoding members of the Argonaute family are expressed in neoblasts and knockdown of mRNAs encoding one of these proteins (smedwi-2) blocked regeneration.43 It is not yet clear what small RNAs are affected by loss of smedwi-2, whether miRNAs or a closely related set of small RNAs referred to as piwi-associated RNAs (piRNAs),42 but the findings are consistent with a key role for small RNAs in controlling stem cell gene expression pathways. In this case, the loss of smedwi-2 did not alter neoblast maintenance but affected one or more steps of differentiation. By identifying the target genes controlled by these and other small RNAs, the hope is that key genetic pathways can be identified and dissected to potentially facilitate regeneration in higher organisms and/or to possibly allow reprogramming of adult stem cells.
miRNAs Regulate Fin Regeneration
Zebrafish are able to regenerate many damaged tissues and organs including fins, hearts, retinas and spinal cords.47 Zebrafish caudal fins are relatively simple, composed of bony rays consisting of two hemirays that create a protective shell around nerves, blood vessels and mesenchymal cells. Fin regeneration involves five steps: wound healing, mesenchymal disorganization or reorganization, blastema formation, outgrowth and termination (Fig. 2). The key is the formation of a blastema, a structure containing stem cell-like cells that are either recruited to the damaged area or originate from the de-differentiation of cells in the area. How fin amputation sets in motion the five steps above and results in the formation of blastema cells remains largely unknown.
Figure 2.
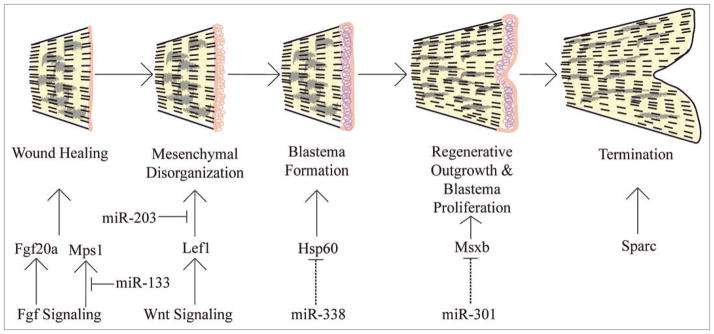
Stages of zebrafish fin regeneration. Five stages of zebrafish fin regeneration are illustrated. Initial wound healing responses apparently stimulate a number of signaling pathways that ultimately result in the formation of a blastema, a structure that contains stem cell-like cells which divide and differentiate to re-form complex tissues. miRNA expression analyses have shown that miR-203 and miR-133 regulate targets as part of the Fgf and Wnt cascades,23,48 other miRNAs are predicted to regulate downstream targets (dashed lines23).
Recently, miRNAs have been shown to play a role in regulating fin regeneration in fish and tail regeneration in salamanders.23,41,48 We showed that an intact miRNA biogenesis pathway is required for caudal fin regeneration in zebrafish; inhibition of Dicer by injection of anti-sense morpholinos blocked regeneration.23 Given that miRNA expression patterns change, both up and down, during regeneration, it appears that overall gene regulation requires proper control of both positive and negative regulators of regeneration. Consistent with this, some of the miRNAs that get downregulated during regeneration, including miR-203 and miR-133, are predicted to target genes that are upregulated during regeneration (lef1, msxb, fgfr1, fgf20a and hsp60).23,48 Conversely, a subset of miRNAs whose expression levels increase are predicted to target genes that are downregulated during regeneration.23 One example is miR-200b which increases during regeneration. One of its predicted targets, bmp3, is correspondingly downregulated during regeneration.49 More broadly, genes that are downregulated during regeneration are likely involved in preserving the terminally differentiated state and are therefore normally tightly controlled, reflecting the balance between growth and possible out of control proliferation.
Another interesting aspect of recent discoveries involving miRNAs during fin regeneration is that the first two identified miRNAs regulate key players involved in Wnt and Fgf signaling.23,48 One of the earliest events upon damage is wound healing which results in early activation of Fgf signaling cascades, perhaps activated by hydrogen peroxide synthesis at the wound site.50 Downstream, or coincident with Fgf signaling, a number of signaling cascades are activated including wnts, hedgehogs and TGFβ.51 One possible role for miRNA regulation is to ensure that the gradients or readouts formed by these signaling cascades are properly initiated, maintained and finally, terminated. In many ways, this may be similar to such regulation during normal development, such as the role of miR-214 to control muscle differentiation.1
miRNAs Regulate Muscle Regeneration
miRNAs play key roles in muscle, controlling specification, proliferation and differentiation.52 miR-1 and miR-133 are expressed in cardiac and skeletal muscle where they are transcriptionally regulated by the well known myogenic factors MyoD, myogenin, Mef2 and SRF (serum response factor).53–57 Expression of miR-1 in mammals terminates the cell cycle in cardiac and skeletal muscle progenitors and promotes terminal differentiation.54,57,58 miR-133 has the opposite effect to maintain proliferating progenitor cells.53 miR-206 is expressed only in skeletal muscles and is induced by MyoD and myogenin to promote muscle differentiation.59,60 This is consistent with the notion that specific subsets of miRNAs are expressed that can act in opposition to either promote or repress renewal versus differentiation (Fig. 3).
Pax3 and Pax7 are known to be expressed in skeletal muscle stem cells and represent a self-renewing population of cells that are critical for muscle growth and to maintain satellite cells that are required for the regeneration of adult skeletal muscle.61 Pax3 is essential for maintenance of these cells and its migration to sites of myogenic differentiation and is regulated by miR-27b to rapidly initiate terminal muscle differentiation.62
miRNAs in Pancreas/β-cell Regeneration
Three major pancreatic transcription factors are necessary for early development, FoxA2, Hes1 and Pdx1.63–65 Conditional Dicer deletion in pancreatic progenitor cells adversely affects the development of β-, δ-, exocrine and duct cells, indicating that proper miRNA-mediated gene regulation is critical for normal pancreas development, glucose metabolism and insulin secretion.66 Consistent with this, miR-124a downregulates foxa2 expression which in turn regulates the targets of FoxA2 including pdx1, kr6.2, sur-1.64
Several groups have shown that NGN3 is a “master-controller” of pancreas development.63,67–70 Hairy/enhancer of split (hes1), a notch signaling effector, tightly controls the number of Neurogenin3 (NGN-3) producing cells65 and knockout of hes1 can lead to ectopic pancreas development. miRNA profiling of the regenerating mouse pancreas has indicated that miR-15a, -15b, -16 and -195 negatively regulate translation of ngn3 (Fig. 3).71 miR-23 downregulates hes1 during neuronal development and it may have a similar role in the pancreas.72 In the regenerating pancreas, NGN3 levels are low, despite the fact that a number of activating transcription factors upstream of NGN3 are expressed. One possibility is that miRNA control of ngn3 limits its expression. Interestingly, NGN3 is required to regenerate hormone-producing cells and it is possible that understanding how to alter NGN3 levels might help to treat diabetes through promoting regeneration of insulin-secreting cells.
Recent studies have shown a requirement for notch signaling in neuronal stem cell maintenance in embryonic and adult mice.73 Hes1;Hes3;Hes5 triple-mutant mice lack neural stem cell populations in most of the central nervous system except the telencephalon.74 All other neuronal progenitor pools prematurely differentiated into neurons. In order to more completely deplete Notch signaling in the adult mouse brain, the Kageyama group knock-downed Rbpj in mice. The Notch intracellular domain (NICD) forms a complex with Rbpj to induce the expression of transcriptional repressors like Hes1.73 Without repression of neuronal genes, cells prematurely differentiate. Thus, proper Notch signaling is crucial to regulate and maintain stem cell populations. miRNAs are likely to play a role in properly regulating Notch signaling, consistent with the finding that a number of miRNAs exhibit altered expression patterns upon inhibition of Notch.19
Initiation and Termination of Regeneration
The parallels to regeneration and cancer or tumor formation are many. As an example, during fin regeneration, genes that maintain the differentiated state must be turned off while genes that promote proliferation and outgrowth must be turned on. At the end of regeneration, the regulatory switches need to be reversed. It is remarkable that zebrafish fin regeneration terminates at precisely the right size, despite the fact that fish grow throughout their lifetimes. This raises the question as to what mechanisms are in place to properly terminate regeneration. We have shown that continued repression of miR-203 during fin regeneration leads to fin overgrowth.23 It remains unknown what signals regulate the transcriptional activity of miR-203 but it is clear that specific miRNAs are needed for both initiation and termination. For unregulated growth in cancer, the similarities are striking. Overexpression of miR-34 induces apoptosis and inhibits cell growth and tumor invasion in the pancreas via targeting of bcl-2 (B-cell CLL/lymphoma-2) and Notch1/2.75–77 In pancreatic cancer, the action of miR-34 must be downregulated to escape such growth controls.
Conclusion
The processes and mechanisms that allow certain organisms to undergo regeneration of complex tissues are only just beginning to be understood. The key lies in understanding those mechanisms (Fig. 3) and especially the role that stem cells play in these processes. As more and more is learned about regeneration, key regulatory steps that control stem cells and regeneration will be uncovered opening the door to application in humans to repair injured or damaged tissues.
Abbreviations
- miRNAs
microRNAs
- RISC
RNA induced silencing complex
- NGN3
neurogenin3
- ES cells
embryonic stem cells
References
- 1.Flynt A, Li N, Thatcher E, Solnica-Krezel L, Patton J. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–63. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flynt A, Thatcher E, Burkewitz K, Li N, Liu Y, Patton J. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J Cell Biol. 2009;185:115–27. doi: 10.1083/jcb.200807026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–81. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 5.Bartel D. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–8. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–8. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 9.Slack FJ, Weidhaas JB. MicroRNAs as a potential magic bullet in cancer. Future Oncol. 2006;2:73–82. doi: 10.2217/14796694.2.1.73. [DOI] [PubMed] [Google Scholar]
- 10.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 11.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miR-NA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 12.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein E, Denli Ahmet M, Hannon Gregory J. The rest is silence. RNA. 2001;7:1509–21. [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 16.Zamore P, Tuschl T, Sharp P, Bartel D. RNAi: double-stranded RNA directs the ATP-dependent dleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 17.Gu S, Rossi JJ. Uncoupling of RNAi from active translation in mammalian cells. RNA. 2005;11:38–44. doi: 10.1261/rna.7158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 19.Thatcher E, Flynt A, Li N, Patton J. MiRNA expression analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways. Dev Dyn. 2007;236:2172–80. doi: 10.1002/dvdy.21211. [DOI] [PubMed] [Google Scholar]
- 20.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA Expression in Zebrafish Embryonic Development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 21.Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–95. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 22.Melton C, Judson RL, Blelloch R. Opposing microR-NA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010:1–8. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thatcher E, Paydar I, Anderson KK, Patton J. Regulation of zebrafish fin regeneration by microR-NAs. Proc Natl Acad Sci USA. 2008;105:18384–9. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 27.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006;34:9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 30.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–5. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 31.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microR-NAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs Regulate Brain Morphogenesis in Zebrafish. Science. 2005;308:833–8. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 34.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–22. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 35.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–20. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 39.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson JM, Parker JS, Hammond SM. Microarray Analysis of miRNA Gene Expression. Methods Enzymol. 2007;427:107–22. doi: 10.1016/S0076-6879(07)27006-5. [DOI] [PubMed] [Google Scholar]
- 41.Sehm T, Sachse C, Frenzel C, Echeverri K. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev Biol. 2009;334:468–80. doi: 10.1016/j.ydbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–86. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–30. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 44.Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell. 2006;11:159–69. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Friedlander MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, et al. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci USA. 2009;106:11546–51. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Estevez C, Arseni V, Thambyrajah RS, Felix DA, Aboobaker AA. Diverse miRNA spatial expression patterns suggest important roles in homeostasis and regeneration in planarians. Int J Dev Biol. 2009;53:493–505. doi: 10.1387/ijdb.082825cg. [DOI] [PubMed] [Google Scholar]
- 47.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- 48.Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–33. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebrafish. ScientificWorldJournal. 2006;6:38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–9. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iovine MK. Conserved mechanisms regulate outgrowth in zebrafish fins. Nat Chem Biol. 2007;3:613–8. doi: 10.1038/nchembio.2007.36. [DOI] [PubMed] [Google Scholar]
- 52.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–9. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA. 2005;102:18986–91. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–6. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–54. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 59.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–73. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 62.Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA. 2009;106:13383–7. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heremans Y, Van De Casteele M, in’t Veld P, Gradwohl G, Serup P, Madsen O, et al. Recapitulation of embryonic neuroendocrine differentiation in adult human pancreatic duct cells expressing neurogenin 3. J Cell Biol. 2002;159:303–12. doi: 10.1083/jcb.200203074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282:19575–88. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 65.Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, et al. Ectopic pancreas formation in Hes1-knockout mice reveals plasticity of endodermal progenitors of the gut, bile duct and pancreas. J Clin Invest. 2006;116:1484–93. doi: 10.1172/JCI27704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–45. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 67.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97:1607–11. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu G, Brown JR, Melton DA. Direct lineage tracing reveals the ontogeny of pancreatic cell fates during mouse embryogenesis. Mech Dev. 2003;120:35–43. doi: 10.1016/s0925-4773(02)00330-1. [DOI] [PubMed] [Google Scholar]
- 69.Hara M, Dizon RF, Glick BS, Lee CS, Kaestner KH, Piston DW, et al. Imaging pancreatic beta-cells in the intact pancreas. Am J Physiol Endocrinol Metab. 2006;290:1041–7. doi: 10.1152/ajpendo.00365.2005. [DOI] [PubMed] [Google Scholar]
- 70.Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–47. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol. 2007;311:603–12. doi: 10.1016/j.ydbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 72.Kimura H, Kawasaki H, Taira K. Mouse microRNA-23b regulates expression of Hes1 gene in P19 cells. Nucleic Acids Symp Ser (Oxf) 2004:213–4. doi: 10.1093/nass/48.1.213. [DOI] [PubMed] [Google Scholar]
- 73.Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 30:3489–98. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–50. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 75.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 76.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]