Abstract
The flavivirus nonstructural protein 3 (NS3) is a protease and helicase, and on the basis of its similarity to its homologue encoded by the hepatitis C virus (HCV), the flavivirus NS3 might be a promising drug target. Few flavivirus helicase inhibitors have been reported, in part, because few specific inhibitors have been identified when nucleic acid unwinding assays have been used to screen for helicase inhibitors. To explore the possibility that compounds inhibiting NS3-catalyzed ATP hydrolysis might function as antivirals even if they do not inhibit RNA unwinding in vitro, we designed a robust dengue virus (DENV) NS3 ATPase assay suitable for high-throughput screening. Members of two classes of inhibitory compounds were further tested in DENV helicase-catalyzed RNA unwinding assays, assays monitoring HCV helicase action, subgenomic DENV replicon assays, and cell viability assays and for their ability to inhibit West Nile virus (Kunjin subtype) replication in cells. The first class contained analogues of NIH molecular probe ML283, a benzothiazole oligomer derived from the dye primuline, and they also inhibited HCV helicase and DENV NS3-catalyzed RNA unwinding. The most intriguing ML283 analogue inhibited DENV NS3 with an IC50 value of 500 nM and was active against the DENV replicon. The second class contained specific DENV ATPase inhibitors that did not inhibit DENV RNA unwinding or reactions catalyzed by HCV helicase. Members of this class contained a 4-hydroxy-3-(5-methylfuran-2-carbonyl)-2H-pyrrol-5-one scaffold, and about 20 μM of the most potent pyrrolone inhibited both DENV replicons and West Nile virus replication in cells by 50%.
Keywords: nonstructural protein 3, ATPase, motor protein, direct acting antiviral, Dengue fever, West Nile virus, yellow fever virus, positive sense single-stranded RNA ((+)ssRNA) virus
Flaviviruses comprise a genus of positive-sense single-stranded RNA ((+)ssRNA) viruses. They include the important human pathogens yellow fever virus (YFV), Japanese encephalitis virus (JEV), Dengue virus (DENV), and West Nile virus (WNV). Many of these mosquito-borne viruses are endemic in tropical regions. Vaccines exist for only YFV and JEV. DENV is probably the most noteworthy human health threat because it infects an estimated 390 million people each year, causing “break-bone” fever, an extraordinarily painful disease with symptoms ranging from mild fever to a fatal hemorrhagic syndrome.1
The recent development of direct-acting antiviral (DAA) drugs to treat the hepatitis C virus (HCV), which is in the same Flaviviridae family as the flaviviruses, suggests that similar compounds might be useful to treat flavivirus infections or hemorrhagic fevers.2 DAA targets encoded by flaviviruses include the viral structural proteins (capsid protein C, membrane protein M, envelope protein E) and nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). DAAs have been reported that inhibit the flavivirus capsid protein,3 the NS5 polymerase,4 the NS2B-NS3 protease/helicase,5,6 the NS4B protein,7 the NS5 guanylyltransferase,8 and the NS5 polymerase.4
NS3 is one of the most thoroughly studied antiviral drug targets, and this work has led to approval of several new antiviral drugs to treat HCV.9 NS3 proteins are unlike any others found in nature because they combine an N-terminal protease needed for viral polyprotein processing with a C-terminal domain functioning as an ATP-fueled RNA helicase. The new HCV drugs inhibit the NS3 protease function, but compounds that inhibit the helicase10–13 and compounds that bind between the helicase and protease domains14 also function as antivirals in cell culture.
The viruses in Flaviviridae encode the only known proteins that are both helicases and proteases. The helicase portion of these NS3 proteins is composed of two motor domains that are similar in all superfamily 1 and 2 helicases and a third domain very different from other helicases. ATP binds between the motor domains, and RNA binds between the motor domains and domain 3.15 Detailed mechanistic studies have revealed how the NS3 helicase moves like an inchworm in a 3′ to 5′ direction along one strand of RNA while actively displacing the complementary strand in an ATP-fueled reaction.10,16 Similar structural15 and enzymatic analyses of the DENV helicase suggest that the hepacivirus and flavivirus proteins share a similar mechanism of action.
Although they share key similarities, NS3 proteins from the hepacivirus and flavivirus genera are also remarkably different. First, NS3 proteases are most active only when combined with another viral peptide, but the HCV NS3 protease is activated by HCV NS4A,17,18 and flavivirus NS3 is activated by NS2B.19,20 Unlike the analogous NS4A protein in HCV, NS2B wraps like a belt around NS3 such that part of the cofactor contributes residues to the active site of the protease.21,22 Second, the NS3 helicase and protease domains are oriented differently relative to each other in the two genera. In all X-ray crystal structures reported to date, the HCV NS3 protein folds in a compact conformation where the protease domain does not contact the known RNA- or ATP-binding sites on the helicase.23 However, the DENV NS3 has been observed twice in more extended conformations. In the first observed conformation,19 the DENV protease partially blocks the ATP-binding site. In the second conformation, the protease is rotated by about 180° so that both ATP and RNA can access the helicase active site.24 These conformations appear to be biologically relevant because mutations that affect the flexibility of the helicase–protease linker also affect the ability of DENV to replicate in cells.24 Third, the HCV helicase is capable of separating both DNA25 and RNA26 duplexes in vitro, but the flavivirus NS3 clearly prefers RNA in most assays. Finally, unlike the HCV proteins, the flavivirus helicase ATP-binding site can also accommodate RNA so that the protein can cleave the terminal 5′ phosphate to prepare genomic RNA for capping.25
A few flavivirus NS3 helicase inhibitors have been reported previously.27 Recent examples include ivermectin, which was discovered to inhibit the helicase using molecular modeling,28 ST-610, which was discovered using assays monitoring DENV cytopathic effects,29 and suramin, which was discovered to inhibit DENV by screening compound libraries with a molecular beacon-based helicase assay.30 Ndjomou et al.12 also recently showed that some compounds resembling NIH molecular probe ML28331 are potent inhibitors of DENV NS3-catalyzed ATP hydrolysis. We have therefore designed a new DENV helicase assay, optimized the new assay for high-throughput screening, and used the assay to analyze a small library of additional ML283 analogues and other compounds. Here, we report that some of the more specific ML283 analogues exert an antiviral effect against DENV and that another class of helicase inhibitors, which share a pyrrolone scaffold, specifically inhibit the ability of DENV NS3 to cleave ATP and DENV and WNV replication in cells.
RESULTS
After comparing several different ATPase assay protocols with several different flavivirus helicases, we found that the most cost-effective (at ~2.5¢/assay) was a colorimetric inorganic phosphate assay using DENV helicase. The recombinant flavivirus helicase we found to be most stable in vitro was a truncated enzyme from dengue virus serotype 2 (TSV01 strain). In this NS3 protein (called NS3h), the protease domain is replaced by thioredoxin to enhance solubility. NS3 residues 171–618 are present, and thioredoxin can be removed with enterokinase so the protein can be examined using X-ray crystallography.15,32
The colorimetric assay included poly(U) to mimic the stimulatory effect of RNA binding to the helicase (Figure 1A). In this way, compounds that restrict ATP hydrolysis, ATP binding, or RNA binding could be discovered. Like all helicases, NS3h hydrolyzes ATP more rapidly in the presence of nucleic acids than in their absence. ATP hydrolysis fuels helicase movements on RNA. In the procedure for automated HTS, assays were assembled, compounds added, and reactions initiated with 3 μL of an MgCl2 solution and terminated after 30 min with 50 μL of the Biomol Green reagent (Enzo Life Sciences). Phosphate was then determined from absorbance at 620 nm (Figure 1B). The sensitivity of the Biomol reagent allowed the assays to be conducted at subsaturating ATP concentrations near the Km observed for the enzyme (i.e., 100 μM ATP). At this ATP concentration, compounds that either affect the turnover number of the enzyme (kcat) or the affinity of the enzyme for ATP (Km) could, in theory, be detected in an HTS.
Figure 1.
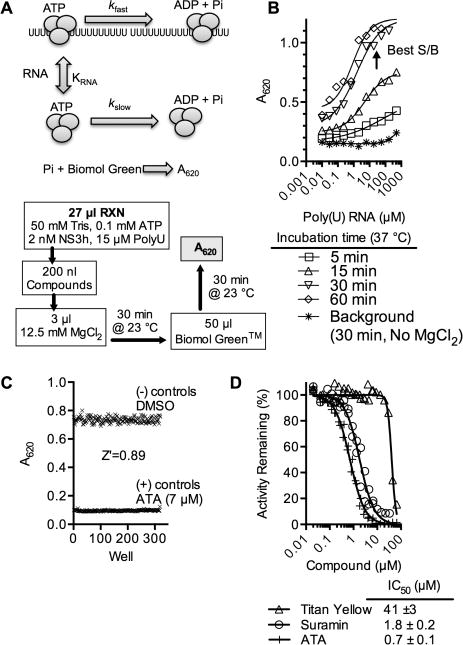
DENV helicase high-throughput ATPase assay. (A) Assay setup. DENV helicase (gray) cleaves ATP at a slow basal rate (kslow) and more rapidly in the presence of RNA (kfast). The Biomol Green reagent reacts with inorganic phosphate to produce a product that absorbs light at 620 nm. In automated screening, compounds are added to a reaction mix, and ATP hydrolysis is initiated with Mg2+. In addition to key reagents noted, all assays were performed at pH 7.5 and contained 5 μg/mL BSA, 0.01% (v/v) Tween 20, 5 mM DTT, and 0.3% DMSO (final concentration). (B) Effect of poly(U) RNA on inorganic phosphate released from ATP in the presence of 2 nM DENV NS3h after indicated times. Conditions with the best signal compared to control assays where reactions were not initiated with MgCl2 were chosen for HTS. (C) To assess assay quality, a Z′ factor33 was calculated by performing 192 positive control reactions (7 μM aurintricarboxylic acid (ATA)) and 192 negative control reactions (DMSO only) arranged in a checkerboard fashion in a 384-well plate. (D) Concentration–response assays using nonspecific helicase inhibitors.11, 34
Under these conditions, pilot screens comparing DMSO and the nonspecific helicase inhibitor aurintricarboxylic acid (ATA)11,34 led to excellent Z′ factors (Figure 1C).33 Concentration–response assays with ATA and other nonspecific helicase inhibitors such as suramin11 and titan yellow35 yielded half-maximal inhibitory concentrations similar to those reported with other assays with the related NS3h protein from HCV (Figure 1D). The assay was also insensitive to DMSO concentrations up to 10% (v/v) (data not shown).
The assay was used to screen a library of 253 compounds that were reported to inhibit other helicases. Most of the compounds in this focused helicase inhibitor library were either previously reported to inhibit the homologous HCV helicase or the SV40 Tag helicase, or they were nucleic acid binding compounds reported to inhibit one or more helicases. When tested at 7 μM, only 41 of the compounds inhibited DENV NS3h-catalyzed ATP hydrolysis >50% in two separate assays (Figure 2A).
Figure 2.
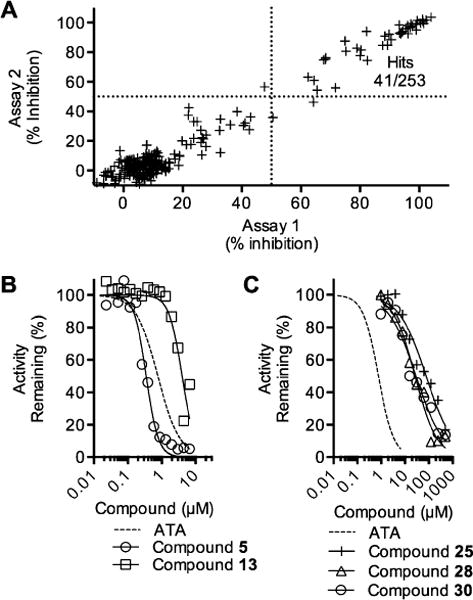
Screen of a focused helicase inhibitor library (A) Each compound was tested twice, and the result from the first assay (x-axis) was compared to the result from the second (y-axis) to assess assay reproducibility. (B) Concentration–response assays with selected inhibitors in the benzothiazole class. (C) Concentration–response assays with representatives of the pyrrolone class of compounds. In panels B and C results are compared with data obtained with the potent nonspecific helicase inhibitor ATA (dotted lines).
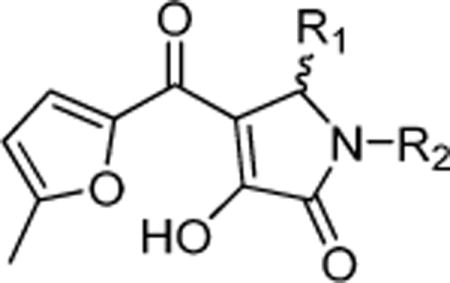
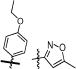
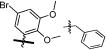
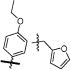
Two sets of hit compounds were selected for further analysis. The first set (Tables 1 and 2) were analogues of NIH molecular probe ML283 (compound 13), which was developed to target the HCV helicase.31 ML283 and most of its analogues were synthesized from a benzothiazole dimer purified from the yellow dye primuline.36 The second set contained pyrrolones (Table 3, see the Supporting Information for synthesis). The most potent compound in the benzothiazole class (compound 5, Table 1) inhibited DENV NS3h-catalyzed ATP hydrolysis with a half-maximal inhibitory concentration (IC50) that was about 100 times lower than the IC50 observed with the most potent compound in the pyrrolone class (compound 26, Table 3).
Table 1.
Ability of Selected ML283 Analogues To Inhibit DENV Helicase-Catalyzed ATP Hydrolysis and RNA Unwinding
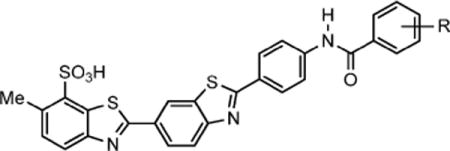
| ||||||
|---|---|---|---|---|---|---|
| compd (CID)a | R | DENV ATPase IC50b (μM) | DENV RNA- helicase IC50c (μM) | HCV ATPase IC50d (μM) | HCV DNA- helicase IC50e (μM) | HCV RNA-helicase IC50f (μM) |
| 1 (49849280) | H | 5.3 ± 1.2 | ndg | 69 ± 6 | 11 ± 1.5 | 8.5 ± 2 |
| 2 (49849300) | 4-NH2 | 2.8 ± 0.6 | 7.6 ± 1.6 | 58 ± 10 | 10 ± 2.4 | 7.0 ± 1* |
| 3 (49849294) | 4-F | 3.0 ± 0.4 | 6.6 ± 0.9 | 32 ± 7 | 5.2 ± 0.6 | 4.2 ± 2* |
| 4 (49849282) | 4-OCH3 | 3.0 ± 0.5 | nd | 115 ± 14 | 10 ± 2.6 | nd |
| 5 (49849290) | 4-CO2CH3 | 0.4 ± 0.1 | 7.9 ± 2.5 | >200 | 9.7 ± 4.6 | nd |
| 6 (49849302) | 4-Cl | 1.4 ± 0.2 | 5.6 ± 1.2 | 53 ± 4 | 3.4 ± 0.3 | 3.3 ± 1 |
| 7 (49849286) | 4-CH3 | 1.1 ± 0.2 | 4.3 ± 1.9 | 61 ± 1 | 3.3 ± 0.3 | 3.9 ± 1 |
| 8 (49849276) | 4-CF3 | 1.2 ± 0.2 | 5.2 ± 0.6 | 111 ± 60 | 1.8 ± 0.4 | 2.8 ± 2 |
| 9 (49849299) | 4-t-Bu | 4.0 ± 0.5 | 10 ± 3.4 | >200 | 8.2 ± 1 | 8.8 ± 3* |
| 10 (49849284) | 4-N(CH3)2 | 3.4 ± 0.3 | nd | >200 | 11 ± 6.7 | 8.6 ± 2 |
| 11 (50930740) | 4-Br | 1.1 ± 0.1 | 5.0 ± 1.2 | 21 ± 6 | 5.2 ± 4 | 3.8 ± 0.4* |
| 12 (46839370) | 4-NHFmoc | 1.3 ± 0.3 | 3.5 ± 1.7 | 39 ± 31 | 5.4 ± 1 | 2.8 ± 1 |
| 13 (50930730) | 3-Cl | 4.0 ± 0.8 | 5.1 ± 0.9 | 24 ± 5 | 2.6 ± 1 | 5.9 ± 2 |
| 14 (50930737) | 3,4-di-Cl | 2.7 ± 0.2 | 3.6 ± 0.9 | 53 ± 3 | 3.7 ± 1 | 3.9 ± 2* |
| 15 (50930748) | 2-CF3 | 5.4 ± 0.8 | nd | 71 ± 23 | 14 ± 1 | 22.8 ± 5 |
| 16 (50930755) | 3-CF3 | 9.7 ± 3.1 | nd | >200 | 20 ± 12 | 22.4 ± 8 |
| 17 (50930745) | 2-F,6-CF3 | 5.3 ± 6.8 | nd | 93 ± 16 | 17 ± 6 | 32.5 ± 7 |
| 18 (50930751) | 2-F,3-CF3 | 4.3 ± 0.3 | nd | 42 ± 14 | 9.2 ± 3 | 11.9 ± 4* |
| 19 (50930743) | 3-F,4-CF3 | 3.5 ± 1.1 | nd | 55 ± 42 | 17 ± 7 | 5.4 ± 2 |
| 20 (50930749) | 3,5-di-CF3 | 63 ± 20 | 7.7 ± 1.8 | >200 | 22 ± 4 | 14.2 ± 2* |
| 21 (50930741) | 2-F,5-CF3 | 3.8 ± 0.2 | 5.4 ± 1.3 | 16 ± 2 | 6.4 ± 2 | 7.5 ± 2 |
| 22 (50930733) | 3-F,6-CF3 | 7.2 ± 0.8 | nd | 65 ± 10 | 19 ± 15 | nd |
| 23 (50930732) | 3-F,5-CF3 | 11 ± 5.7 | nd | >200 | 28 ± 7 | 9.8 ± 3 |
PubChem40 compound identification number (CID).
Half-maximal inhibitory concentration observed in assays monitoring DENV NS3h-catalyzed ATP hydrolysis.
Half-maximal inhibitory concentration observed in assays monitoring DENV NS3h RNA unwinding.
Half-maximal inhibitory concentration observed in assays monitoring HCV NS3h-catalyzed ATP hydrolysis.
Half-maximal inhibitory concentration in assays monitoring HCV NS3h DNA unwinding. Data from Li et al.36
Half-maximal inhibitory concentration in assays monitoring HCV NS3h RNA unwinding. Values marked with asterisks (*) were published previously in Ndjomou et al.12
nd, not determined.
Table 2.
ML283 Analogue with Optimal Antiviral Activity in Cells
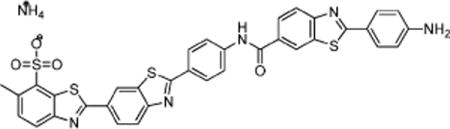
| ||||
|---|---|---|---|---|
| compd (CID) | DENV ATPase IC50a (μM) | DENV helicase IC50a (μM) | replicon EC50b (μM) | TIc |
| 24 (49849289) | 0.5 ± 0.1 | 1.5 ± 0.2 | 7.1±3 | 17 |
As defined in Table 1.
Half-maximal inhibitory concentration observed in assays monitoring BHK cell DENV replicon content.
Half-maximal inhibitory concentration observed in BHK cell viability assays (CC50)/replicon EC50.
Table 3.
DENV Helicase Inhibitors Active against DENV Replicons but Not the HCV Helicase
Representatives of each class of inhibitory compounds were next tested in helicase assays in which the ability of the enzyme to separate duplex RNA was monitored (Figure 3A). All of the compounds tested from the benzothiazole class also inhibited the ability of DENV NS3h to separate an RNA duplex in a concentration-dependent manner, but none of the pyrrolones inhibited RNA unwinding (Figure 3C; Tables 1–3). Most of the IC50 values obtained in the unwinding assays were similar to the IC50 values obtained in ATPase assays with a few exceptions. About 20 times more of compound 5 was needed to inhibit RNA unwinding to the same degree as was needed to inhibit ATP hydrolysis (Table 1), and about 8 times less of compound 20 was needed to inhibit RNA unwinding than ATP hydrolysis.
Figure 3.
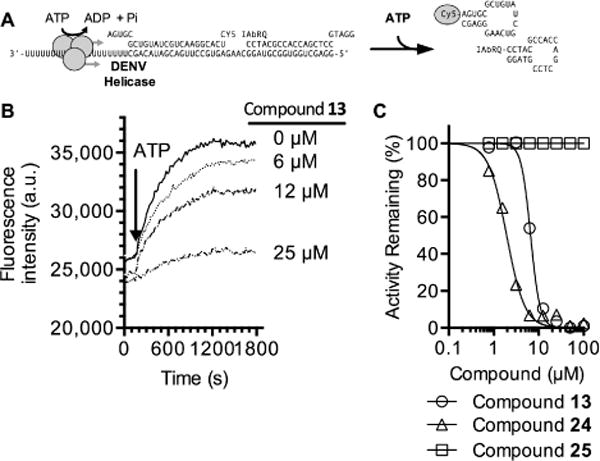
Ability of hit compounds to inhibit DENV helicase-catalyzed RNA strand separation. (A) In the assay, the helicase loads on the 3′ single-stranded tail of the substrate, and translocates 3′ to 5′ upon addition of ATP to separate a Cy5-labeled annealed RNA strand from an annealed DNA strand labeled with the fluorescence quencher Iowa black RQ (IAbRQ, Integrated DNA Technologies).12,37 (B) Concentration-dependent inhibition of DENV helicase-catalyzed RNA unwinding by ML283 (compound 13). Fluorescence traces with only three ML283 concentrations are shown for clarity. (C) Concentration–response plots obtained for select inhibitors. Activity was normalized by first calculating initial rates of fluorescence increase and then normalizing these rates to those observed in negative control reactions containing DMSO alone. IC50 values obtained by fitting data with GraphPad PRISM are shown in Table 1.
The specificity of each DENV helicase inhibitor was then evaluated by performing the same ATPase assay in the presence of HCV helicase instead of DENV helicase. The ability of each DENV helicase inhibitor to inhibit HCV helicase-catalyzed DNA and RNA unwinding was also evaluated using assays similar to those used for DENV, except that a different buffer system was used with HCV helicase because HCV helicase has poor unwinding activity at pH 7.5.38,39
None of the pyrrolone compounds inhibited any of the reactions catalyzed by HCV helicase, and more of each benzothiazole was needed to inhibit HCV helicase-catalyzed ATP hydrolysis than DENV-catalyzed ATP hydrolysis to the same extent (Table 1). When IC50 values obtained in DENV ATPase assays were compared with IC50 values obtained in various HCV helicase assays, the best correlations were between IC50 values obtained in HCV DNA unwinding assays (Table 1, column 6) and IC50 values obtained in DENV ATPase assays (Table 1, column 3). When these numbers were compared, most of the ML283 analogues displayed similar inhibitory potentials for HCV and DENV helicases with the noteworthy exception that compounds 5 and 24 were both more active against DENV than the HCV helicase (Tables 1 and 2).
For flavivirus helicase inhibitors to be used as molecular probes in cells, they must be relatively nontoxic and inhibit viral replication. Compounds in the focused helicase inhibitor library were therefore also tested in cell-based assays to determine the effects on cell viability and flavivirus replication (Figure 4).
Figure 4.
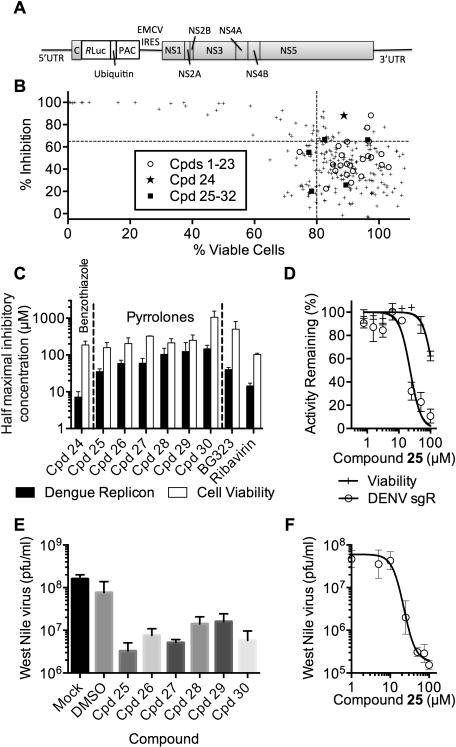
Effects of helicase inhibitors on cellular flavivirus replication. (A) Subgenomic DENV replicon used in this study. (B) Percent of DENV replicon remaining in cells treated with 20 μM of each compound in the focused helicase inhibitor library plotted against the effect of 20 μM of each compound on cell viability. Dotted lines mark arbitrary cutoffs. (C) Effects of compounds in Tables 2 and 3 on DENV replicon content and cell viability (av ± SD, n = 3). (D) Effect of various concentrations of 25 on cell viability (+) and subgenomic replicon (sgR) content (○). (E) BHK cells were infected with West Nile virus in the presence of 50 μM of indicated compounds and titered 72 h later (av ± SD, n = 3). (F) Effect of various concentrations of compound 25 on West Nile virus titers (av ± SD, n = 3).
The first set of assays used a subgenomic DENV-2 replicon designed by Whitby et al.41 In the DENV-2 replicon, DENV-2 nucleotides 180–2342 are replaced by fusing the first 28 amino acids of the capsid protein with the Renilla luciferase, ubiquitin, and puromycin N-acetyltransferase (Purr) genes. An encephalomyocarditis virus internal ribosome entry site is also present between the reporter and the NS1 region (Figure 4A). These replicon assays measured the ability of 20 μM of each compound in the collection to inhibit Renilla luciferase after cells harboring replicons were exposed to compounds for 72 h (Figure 4B, y-axis). In the second set of assays, the effect of the same compound concentration on BHK cell viability was monitored (Figure 4B, x-axis).
At 20 μM, most of the compounds in the collection had some impact on replicon levels. About half of the compounds with the highest activity against replicons (>65% inhibition) were toxic, meaning that they reduced cell viability and replicon content to a similar degree (upper left quadrant, Figure 4B). Many of the compounds with a modest (20–50%) impact were in the benzothiazole class (ML283 analogues), and most of these were previously reported to have a similarly modest impact on subgenomic HCV replicon levels in Huh7.5 cells.31,36 Only 22 compounds (9%) inhibited the replicon more than 65% and cell viability less than 20% (upper right quadrant, Figure 4B).
The relatively nontoxic replicon inhibitors included compounds in both the benzothiazole and pyrrolone classes. The ability of members of each class to inhibit DENV replicons in a concentration-dependent manner was therefore monitored by applying each active compound to replicon cells in an eight-point two-fold dilution series starting at 100 μM. Compound efficacy was compared with ribavirin and the NS5 inhibitor BG323.8 In these concentration–response assays, only one compound in the benzothiazole class inhibited replicon content more than 90% (compound 24, Table 2). Like the compounds in Table 1, compound 24, also inhibited reactions catalyzed by HCV helicase. For example, 24 inhibited HCV helicase-catalyzed RNA unwinding with an IC50 value of 2.0 ± 0.7 μM. None of the benzothiazoles were tested against authentic virus because of the limited solubility above 50 μM.
All of the pyrrolones tested inhibited the DENV replicon in a concentration-dependent manner (Table 3). The most potent in cells, compound 25, inhibited both DENV helicase-catalyzed ATP hydrolysis and the DENV replicon with half-maximal inhibitory concentrations of 78 ± 23 and 36 ± 6 μM, respectively (Figure 4C). Each pyrrolone tested inhibited cell viability, also, but only at concentrations >50 μM (Figure 4D).
Each of the hits in the pyrrolone series was also tested for its ability to inhibit the replication of West Nile virus (Kunjin strain) in BHK cells by examining plaque titers after incubation with 50 μM of each compound (Figure 4E). Results paralleled those seen with the DENV replicon, with compound 25 lowering plaque-forming units (pfu) to the greatest extent (Figure 4E; Table 3). Effects on WNV were concentration-dependent with 22 μM of compound 25 exerting a half-maximal inhibitory effect (Figure 4F). In similar experiments, exposure of the same WNV-infected cells to 50 μM of BG323 also yielded about 106 pfu of West Nile virus at 72 h,8 suggesting that BG323 and compound 25 yield similar antiviral effects.
DISCUSSION
Our inspiration for developing helicase inhibitors as drugs comes from the fact that helicase inhibitors are now potent antiviral agents that, in some clinical trials, rival many of the drugs traditionally used to treat virus infections.42,43 Boehringer Ingelheim44 and Bayer45 discovered the first antiviral drugs that target the herpes simplex virus helicase. Boehringer Ingelheim identified their inhibitors by screening for compounds that inhibit helicase-catalyzed DNA strand separation. The Bayer compounds were discovered using cytoprotection assays.45 The Bayer thiazolylamide compound BAY 57-1293 inhibits purified helicase-catalyzed ATP hydrolysis (IC50 = 30 nM), can be used to treat HSV in animal models,46,47 and is being tested in humans as the drug pritelivir.48 Astellas Pharma Inc. also developed a HSV helicase inhibitor called amenamevir (ASP2151), which inhibits helicase-catalyzed ATP hydrolysis (IC50 = 78 nM) and shortens the median time for lesion healing.49,50
Helicases are nevertheless challenging drug targets because helicases are difficult to inhibit with small molecules and potent inhibitors are often not specific. Many potent helicase inhibitors interact with the nucleic acid substrate or highly conserved regions of the helicase motor domains.51 Prior high-throughput screens with DENV helicase have yielded inhibitors that were either nucleic acid binding agents2 or nonspecific nucleic acid mimics such as suramin30 and aurintricarboxylic acid (ATA).34 Because previous screens for flavivirus helicase inhibitors used unwinding assays,2,30 we set out to perform screens using an ATPase assay to test the hypothesis that compounds that inhibit helicase-catalyzed ATP hydrolysis might function as antivirals even if they do not inhibit the ability of the protein to unwind RNA.
The DENV ATPase assay that we have optimized was designed so that it can, in theory, detect compounds that inhibit ATP binding or ATP hydrolysis or that prevent RNA from stimulating helicase-catalyzed ATP hydrolysis. Because RNA is present in the assay at a concentration needed to stimulate ATP hydrolysis to ~85% of its maximal rate, then compounds that either inhibit ATP hydrolysis or RNA binding will be identified as “hits”. The assay is simpler than other colorimetric phosphate assays based on the Fiske–SubbaRow method52 or ammonium molybdate reagents that incorporate the dye malachite green53, 54 because ATP need not be removed and multiple reagents need not be added in a precisely timed procedure.
When the new DENV ATPase HTS assay was used to screen a focused helicase inhibitor library composed of compounds reported to inhibit related helicases such as the HCV NS3 protein,11,35 and more distantly related helicases such as the SV40 Tag protein,55 results showed that the assay was robust (high Z′ factor, Figure 1C) and reproducible (Figure 2A). The ATPase assay was also more sensitive than unwinding assays in detecting helicase inhibitors. As evidence, higher concentrations of compounds were needed to inhibit DENV helicase in RNA unwinding assays than were needed to inhibit ATPase assays to the same extent. Some compounds that inhibited ATP hydrolysis failed to inhibit RNA unwinding even at much higher concentrations (Table 1; Figure 3C).
The most noteworthy of the new DENV helicase inhibitors reported here are a set of pyrrolones, which also inhibited the DENV replicon and WNV replication in cell culture (Figure 4; Table 3). Unlike other helicase inhibitors detected with the assay, the pyrrolones do not inhibit the related HCV helicase under any conditions tested. The pyrrolones were all built around a 4-hydroxy-3-(5-methylfuran-2-carbonyl)-2H-pyrrol-5-one scaffold (see the Supporting Information for synthesis and characterization). Three of the most potent compounds contained a 3,5-dichlorophenyl R1 group with one of three R2 groups. Replacement of the chlorines led to a marked decrease in biological activity. In the R2 position, isoxazole was most active in cells (compound 25), and compounds in which the isoxazole was replaced with either a benzyl (compound 26) or a furanylmethyl (compound 29) were about 3 times more potent inhibitors of DENV helicase-catalyzed ATP hydrolysis in vitro.
The other series of DENV helicase inhibitors that was analyzed in this study was composed of analogues of NIH molecular probe ML283 (compound 13), which was designed to inhibit HCV helicase.31 ML283 is a benzothiazole oligomer that acts by displacing nucleic acids from the HCV helicase.35 Our comparison of the effects of these compounds on the DENV and HCV helicase yielded several noteworthy results. First, lower concentrations of these compounds inhibit HCV helicase-catalyzed unwinding than are needed to inhibit HCV helicase-catalyzed ATP hydrolysis (to the same extent). The reverse is true with DENV helicase. Higher concentrations of the compounds are needed to inhibit DENV helicase-catalyzed unwinding than ATP hydrolysis. Second, some ML283 analogues were better inhibitors of DENV helicase than the HCV helicase. The two most potent, compounds 5 (Table 1) and 24 (Table 2), were >10 times more active against the DENV protein. Although most ML283 analogues had some activity against the DENV replicon when tested at 20 μM, only compound 24 reduced replication of a subgenomic DENV replicon in cells in a concentration-dependent manner (Table 2).
If properly optimized, the two series of compounds reported here could be valuable probes to study the role of the helicase in viral replication, and they might lead to new direct-acting antivirals. Few direct-acting antiflavivirus agents have been reported, and some that were suspected to act via the helicase, such as ivermectin,28 also affect host proteins.56 NS3 is one of the most widely studied antiviral drug targets mainly because of the intense emphasis on HCV drug discovery. However, most HCV drug discovery has focused on the NS3 protease function. NS3 proteins are multifunctional, with the N-terminal domain acting as a protease needed for viral polyprotein processing and the C-terminal domain acting as a helicase. Three NS3 protease inhibitors have recently been approved to treat chronic infection with HCV, which is in the same family (Flaviviridae) as the flaviviruses. Flaviviridae members encode the only proteins known that are both proteases and helicases. The compounds reported here might also prevent flaviviruses from capping their RNA because the same site that cleaves ATP also prepares RNA for capping by removing the 5′ terminal phosphate from the (+) sense RNA genome.25 Regardless of whether or not the new inhibitors here are studied further, the new assays reported here should be valuable tools for discovering additional compounds that inhibit the DENV helicase.
METHODS
Protein Expression and Purification
All oligonucleotides were purchased from Integrated DNA Technologies (Coral-ville, IA, USA). The helicase domain of dengue virus NS3 (NS3 amino acids 171–618) fused at its N-terminus to thioredoxin and a (His)6 tag followed by an enterokinase cleavage site was expressed and purified in Escherichia coli Rosetta (DE3). A plasmid expressing this protein was obtained from Julien Lescar (Singapore),32 and the protein was purified as previously described.57
ATP Hydrolysis (ATPase) Assay
Reactions were performed in 30 μL in clear 384-well microplates (Thermonunc), and they each contained 25 mM Tris, pH 7.5, 1.25 mM MgCl2, 100 μM ATP, 33 μg/mL BSA, 0.07% (v/v) Tween 20, 0.3 mM DTT, 0.6% DMSO, 15 μM poly(U) RNA (expressed as nucleotide concentration), and 3 nM DENV NS3h (or 4 nM HCV NS3h). For automated screening, reactions containing all components except test compounds and MgCl2 were assembled at 1.11 times their final concentration. After 27 μL of this reaction mixture was dispensed in each well, 200 nL of each test compound (6.67 μM final concentration) or DMSO was added (0.3% DMSO v/v, final). Reactions were initiated by adding 3 μL of 12.5 mM MgCl2. After 30 min at 23 °C, 50 μL of Biomol Green reagent (Enzo Life Sciences) was added, and after a final 30 min of incubation at 23 °C, absorbance at 620 nm was read. Percent inhibition was calculated by normalizing the data to assays containing 6.7 μM ATA (100% inhibition) and reactions with DMSO only (0% inhibition).
RNA/DNA Unwinding Assay
Molecular beacon-based DNA and RNA unwinding assays were performed as described previously with HCV helicase.37 For reactions with DENV helicase, the buffer described for ATPase assays (above) and 100 nM DENV was included in each reaction.
DENV Subgenomic Replicon Assay
A total of 258 compounds were screened, 86 in duplicate and 173 in singlicate. Stable baby hamster kidney (BHK-21) DENV-Rluc cells41 were maintained in Dulbecco’s modified Eagle’s medium (DMEM, LifeTech) containing 4.5 g/L D-glucose, L-glutamine, and 110 mg/L sodium pyruvate and supplemented with 10% FBS, 100 U/mL each of penicillin and streptomycin, and 3 μg/mL puromycin to maintain replicon stability. Cells were grown at 37 °C and in atmosphere supplemented with 5% CO2. DENV-Rluc cells were seeded in clear 96-well culture plates (Corning 3599) without puromycin at a concentration of 1000 cells per well. Cells were allowed to adhere to the plate for a minimum of 4 h, but not for more than 24 h. Compounds dissolved in DMSO were first diluted in DMEM and then added to cells such that the final volume in the well was 100 μL, the final DMSO concentration was 1%, and the final compound concentration was 20 μM. Quadruplicate control wells containing DMSO only or 40 μM ribavirin were included on each assay plate. Cells were incubated for 72 h at 37 °C and 5% CO2 atmosphere.
The effect of compounds on DENV replication was assessed by measuring Renilla luciferase reporter gene activity using a Renilla luciferase assay kit (Promega, Madison, WI, USA). Medium was aspirated and cells were washed with PBS before being lysed with 70 μL of Renilla luciferase lysis buffer. Cells were rocked for 1 h at room temperature, and then 50 μL of lysate was transferred to a black 96-well luminescence plate (Nunc 9502867). Luciferase activity was measured in a FluoSTAR Omega (BMG Labtech, Germany) after the injection of 25 μL of luciferase substrate and reading for 10 s. Percent inhibition was normalized to DMSO-only controls.
Compounds that inhibited at least 50% and reduced cell viability less than 20% were tested again in confirmatory “cherry pick” assays, and those that inhibited confirmatory assays at least 50% were assayed in eight-point two-fold concentration–response (from 200 μM to 1.6 μM final concentrations) to obtain IC50 values.
Cell Viability Assays
To determine compound toxicity, BHK DenV-Rluc cells were plated and treated as above, and cell viability was assessed using the CellTiter-Glo luminescent cell viability kit (Promega). At the end of 72 h, the medium was aspirated and an equal volume of CellTiter-Glo reagent and medium was added to each well. After incubation for 30 min at room temperature, luminescence was read for 5 s using a FLUOstar Omega (BMG Labtech, Germany) and converted to percentage viability by normalizing readings to those obtained from cells treated with DMSO only (i.e., negative controls).
West Nile Virus (Kunjin Subtype) Assay
Effects of compounds on West Nile virus were measured as previously described.8
Data Analysis
Z′ factors were calculated as described by Zhang et al.33 Half-maximal inhibitory concentrations were calculated from concentration–response curves using nonlinear regression to fit data to a log(inhibitor) versus normalized response equation with variable slope. Therapeutic index (TI) was defined as CC50/EC50.
Instant JChem 6.0 (2013) was used for structure database management, search, and prediction (ChemAxon (http://www.chemaxon.com).
Supplementary Material
Acknowledgments
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI088001 (to D.N.F.) and U54 HG005031 to Jeffrey Aubé, (Molecular Libraries Probe Production Centers Network, University of Kansas Specialized Chemistry Center), a Bradley Catalyst Grant from the UW– Milwaukee Research Foundation (to D.N.F.), and grants 5U54AI065357 (Sub#6142) and 1R01AI114675 to B.J.G.
ABBREVIATIONS
- ATA
aurintricarboxylic acid
- BHK
baby hamster kidney
- HCV
hepatitis C virus
- HTS
high-throughput screen
- DENV
Dengue virus
- NS3h
nonstructural protein 2 lacking the protease domain
- WNV
West Nile virus
Footnotes
The following file is available free of charge on the ACS Publications website at DOI: 10.1021/id5000458.
Experimental and characterization details for the synthesis of compounds 25–30 (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim SP, Wang QY, Noble CG, Chen YL, Dong H, Zou B, Yokokawa F, Nilar S, Smith P, Beer D, Lescar J, Shi PY. Ten years of dengue drug discovery: progress and prospects. Antiviral Res. 2013;100:500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Scaturro P, Trist IM, Paul D, Kumar A, Acosta EG, Byrd CM, Jordan R, Brancale A, Bartenschlager R. Characterization of the mode of action of a potent dengue virus capsid inhibitor. J Virol. 2014;88:11540–11555. doi: 10.1128/JVI.01745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niyomrattanakit P, Chen YL, Dong H, Yin Z, Qing M, Glickman JF, Lin K, Mueller D, Voshol H, Lim JY, Nilar S, Keller TH, Shi PY. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J Virol. 2010;84:5678–5686. doi: 10.1128/JVI.02451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson SM, Watowich SJ. Anthracene-based inhibitors of dengue virus NS2B-NS3 protease. Antiviral Res. 2011;89:127–135. doi: 10.1016/j.antiviral.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng J, Li N, Liu H, Zuo Z, Liew OW, Xu W, Chen G, Tong X, Tang W, Zhu J, Zuo J, Jiang H, Yang CG, Li J, Zhu W. Discovery of novel small molecule inhibitors of dengue viral NS2B-NS3 protease using virtual screening and scaffold hopping. J Med Chem. 2012;55:6278–6293. doi: 10.1021/jm300146f. [DOI] [PubMed] [Google Scholar]
- 7.Xie X, Wang QY, Xu HY, Qing M, Kramer L, Yuan Z, Shi PY. Inhibition of dengue virus by targeting viral NS4B protein. J Virol. 2011;85:11183–11195. doi: 10.1128/JVI.05468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stahla-Beek HJ, April DG, Saeedi BJ, Hannah AM, Keenan SM, Geiss BJ. Identification of a novel antiviral inhibitor of the flavivirus guanylyltransferase enzyme. J Virol. 2012;86:8730–8739. doi: 10.1128/JVI.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Clercq E. Current race in the development of DAAs (direct-acting antivirals) against HCV. Biochem Pharmacol. 2014 doi: 10.1016/j.bcp.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Frick DN. The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr Issues Mol Biol. 2007;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee S, Hanson AM, Shadrick WR, Ndjomou J, Sweeney NL, Hernandez JJ, Bartczak D, Li K, Frankowski KJ, Heck JA, Arnold LA, Schoenen FJ, Frick DN. Identification and analysis of hepatitis C virus NS3 helicase inhibitors using nucleic acid binding assays. Nucleic Acids Res. 2012;40:8607–8621. doi: 10.1093/nar/gks623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ndjomou J, Kolli R, Mukherjee S, Shadrick WR, Hanson AM, Sweeney NL, Bartczak D, Li K, Frankowski KJ, Schoenen FJ, Frick DN. Fluorescent primuline derivatives inhibit hepatitis C virus NS3-catalyzed RNA unwinding, peptide hydrolysis and viral replicase formation. Antiviral Res. 2012;96:245–255. doi: 10.1016/j.antiviral.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee S, Weiner WS, Schroeder CE, Simpson DS, Hanson AM, Sweeney NL, Marvin RK, Ndjomou J, Kolli R, Isailovic D, Schoenen FJ, Frick DN. Ebselen inhibits hepatitis C virus NS3 helicase binding to nucleic acid and prevents viral replication. ACS Chem Biol. 2014;9:2393–2403. doi: 10.1021/cb500512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saalau-Bethell SM, Woodhead AJ, Chessari G, Carr MG, Coyle J, Graham B, Hiscock SD, Murray CW, Pathuri P, Rich SJ, Richardson CJ, Williams PA, Jhoti H. Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function. Nat Chem Biol. 2012;8:920–925. doi: 10.1038/nchembio.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 19.Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assenberg R, Mastrangelo E, Walter TS, Verma A, Milani M, Owens RJ, Stuart DI, Grimes JM, Mancini EJ. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol. 2009;83:12895–12906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 22.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aydin C, Mukherjee S, Hanson AM, Frick DN, Schiffer CA. The interdomain interface in bifunctional enzyme protein 3/4A (NS3/4A) regulates protease and helicase activities. Protein Sci. 2013;22:1786–1798. doi: 10.1002/pro.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, Davidson AD, Kotaka M, Lescar J, Vasudevan SG. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benarroch D, Selisko B, Locatelli GA, Maga G, Romette JL, Canard B. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004;328:208–218. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Bera AK, Kuhn RJ, Smith JL. Structure of the flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J Virol. 2005;79:10268–10277. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borowski P, Niebuhr A, Schmitz H, Hosmane RS, Bretner M, Siwecka MA, Kulikowski T. NTPase/helicase of Flaviviridae: inhibitors and inhibition of the enzyme. Acta Biochim Polym. 2002;49:597–614. [PubMed] [Google Scholar]
- 28.Mastrangelo E, Pezzullo M, De Burghgraeve T, Kaptein S, Pastorino B, Dallmeier K, de Lamballerie X, Neyts J, Hanson AM, Frick DN, Bolognesi M, Milani M. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J Antimicrob Chemother. 2012;67:1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrd CM, Grosenbach DW, Berhanu A, Dai D, Jones KF, Cardwell KB, Schneider C, Yang G, Tyavanagimatt S, Harver C, Wineinger KA, Page J, Stavale E, Stone MA, Fuller KP, Lovejoy C, Leeds JM, Hruby DE, Jordan R. Novel benzoxazole inhibitor of dengue virus replication that targets the NS3 helicase. Antimicrob Agents Chemother. 2013;57:1902–1912. doi: 10.1128/AAC.02251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basavannacharya C, Vasudevan SG. Suramin inhibits helicase activity of NS3 protein of dengue virus in a fluorescence-based high throughput assay format. Biochem Biophys Res Commun. 2014 doi: 10.1016/j.bbrc.2014.09.113. [DOI] [PubMed] [Google Scholar]
- 31.Li K, Frankowski KJ, Hanson AM, Ndjomou J, Shanahan MA, Mukherjee S, Kolli R, Shadrick WR, Sweeney NL, Belon CA, Neuenswander B, Ferguson J, Aube J, Schoenen FJ, Blagg BSJ, Frick DN. Hepatitis C virus NS3 helicase inhibitor discovery. Probe Reports from the NIH Molecular Libraries Program. 2011 available from http://www.ncbi.nlm.nih.gov/books/NBK143540/ [PubMed]
- 32.Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J. Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J Virol. 2005;79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 34.Shadrick WR, Mukherjee S, Hanson AM, Sweeney NL, Frick DN. Aurintricarboxylic acid modulates the affinity of hepatitis C virus NS3 helicase for both nucleic acid and ATP. Biochemistry. 2013;52:6151–6159. doi: 10.1021/bi4006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweeney NL, Shadrick WR, Mukherjee S, Li K, Frankowski KJ, Schoenen FJ, Frick DN. Primuline derivatives that mimic RNA to stimulate hepatitis C virus NS3 helicase-catalyzed ATP hydrolysis. J Biol Chem. 2013;288:19949–19957. doi: 10.1074/jbc.M113.463166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K, Frankowski KJ, Belon CA, Neuenswander B, Ndjomou J, Hanson AM, Shanahan MA, Schoenen FJ, Blagg BS, Aube J, Frick DN. Optimization of potent hepatitis C virus NS3 helicase inhibitors isolated from the yellow dyes thioflavine S and primuline. J Med Chem. 2012;55:3319–3330. doi: 10.1021/jm300021v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanson AM, Hernandez JJ, Shadrick WR, Frick DN. Identification and analysis of inhibitors targeting the hepatitis C virus NS3 helicase. Methods Enzymol. 2012;511:463–483. doi: 10.1016/B978-0-12-396546-2.00021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam AM, Rypma RS, Frick DN. Enhanced nucleic acid binding to ATP-bound hepatitis C virus NS3 helicase at low pH activates RNA unwinding. Nucleic Acids Res. 2004;32:4060–4070. doi: 10.1093/nar/gkh743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura GT, Costa EC, Capaccia AM, Mohana-Borges R. pH-dependent conformational changes in the HCV NS3 protein modulate its ATPase and helicase activities. PLoS One. 2014;9(e115941) doi: 10.1371/journal.pone.0115941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Annu Rep Comput Chem. 2008;4:217–241. [Google Scholar]
- 41.Whitby K, Pierson TC, Geiss B, Lane K, Engle M, Zhou Y, Doms RW, Diamond MS. Castanospermine, a potent inhibitor of dengue virus infection in vitro and in vivo. J Virol. 2005;79:8698–8706. doi: 10.1128/JVI.79.14.8698-8706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleymann G. Helicase primase: targeting the Achilles heel of herpes simplex viruses. Antivir Chem Chemother. 2004;15:135–140. doi: 10.1177/095632020401500303. [DOI] [PubMed] [Google Scholar]
- 43.Field HJ, Mickleburgh I. The helicase-primase complex as a target for effective herpesvirus antivirals. Adv Exp Med Biol. 2013;767:145–159. doi: 10.1007/978-1-4614-5037-5_7. [DOI] [PubMed] [Google Scholar]
- 44.Crute JJ, Grygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, Kibler P, Liuzzi M, Cordingley MG. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 45.Kleymann G, Fischer R, Betz UA, Hendrix M, Bender W, Schneider U, Handke G, Eckenberg P, Hewlett G, Pevzner V, Baumeister J, Weber O, Henninger K, Keldenich J, Jensen A, Kolb J, Bach U, Popp A, Maben J, Frappa I, Haebich D, Lockhoff O, Rubsamen-Waigmann H. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 46.Baumeister J, Fischer R, Eckenberg P, Henninger K, Ruebsamen-Waigmann H, Kleymann G. Superior efficacy of helicase-primase inhibitor BAY 57-1293 for herpes infection and latency in the guinea pig model of human genital herpes disease. Antivir Chem Chemother. 2007;18:35–48. doi: 10.1177/095632020701800104. [DOI] [PubMed] [Google Scholar]
- 47.Kaufman HE, Varnell ED, Gebhardt BM, Thompson HW, Atwal E, Rubsamen-Waigmann H, Kleymann G. Efficacy of a helicase-primase inhibitor in animal models of ocular herpes simplex virus type 1 infection. J Ocul Pharmacol Ther. 2008;24:34–42. doi: 10.1089/jop.2007.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wald A, Corey L, Timmler B, Magaret A, Warren T, Tyring S, Johnston C, Kriesel J, Fife K, Galitz L, Stoelben S, Huang ML, Selke S, Stobernack HP, Ruebsamen-Schaeff H, Birkmann A. Helicase-primase inhibitor pritelivir for HSV-2 infection. N Engl J Med. 2014;370:201–210. doi: 10.1056/NEJMoa1301150. [DOI] [PubMed] [Google Scholar]
- 49.Tyring S, Wald A, Zadeikis N, Dhadda S, Takenouchi K, Rorig R. ASP2151 for the treatment of genital herpes: a randomized, double-blind, placebo- and valacyclovir-controlled, dose-finding study. J Infect Dis. 2012;205:1100–1110. doi: 10.1093/infdis/jis019. [DOI] [PubMed] [Google Scholar]
- 50.Katsumata K, Weinberg A, Chono K, Takakura S, Kontani T, Suzuki H. Susceptibility of herpes simplex virus isolated from genital herpes lesions to ASP2151, a novel helicase-primase inhibitor. Antimicrob Agents Chemother. 2012;56:3587–3591. doi: 10.1128/AAC.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shadrick WR, Ndjomou J, Kolli R, Mukherjee S, Hanson AM, Frick DN. Discovering new medicines targeting helicases: challenges and recent progress. J Biomol Screen. 2013;18:761–781. doi: 10.1177/1087057113482586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simoni RD, Hill RL, Vaughan M. The determination of phosphorus and the discovery of phosphocreatine and ATP: the work of Fiske and SubbaRow. J Biol Chem. 2002;277:21e. [PubMed] [Google Scholar]
- 53.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 54.Frick DN, Banik S, Rypma RS. Role of divalent metal cations in ATP hydrolysis catalyzed by the hepatitis C virus NS3 helicase: magnesium provides a bridge for ATP to fuel unwinding. J Mol Biol. 2007;365:1017–1032. doi: 10.1016/j.jmb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seguin SP, Evans CW, Nebane-Akah M, McKellip S, Ananthan S, Tower NA, Sosa M, Rasmussen L, White EL, Maki BE, Matharu DS, Golden JE, Aube J, Brodsky JL, Noah JW. High-throughput screening identifies a bisphenol inhibitor of SV40 large T antigen ATPase activity. J Biomol Screen. 2012;17:194–203. doi: 10.1177/1087057111421630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belon CA, High YD, Lin TI, Pauwels F, Frick DN. Mechanism and specificity of a symmetrical benzimidazole-phenylcarboxamide helicase inhibitor. Biochemistry. 2010;49:1822–1832. doi: 10.1021/bi901974a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.