Abstract
Fibrosis is the histological manifestation of a progressive usually irreversible process causing chronic and end stage kidney disease. Genome-wide transcriptome studies of a large cohort (n=95) of normal and fibrotic human kidney tubule samples followed by systems and network analyses identified inflammation and metabolism as top dysregulated pathways in diseased kidneys. In particular, we found that humans and mouse models with tubulointerstitial fibrosis had lower expression of key enzymes and regulators of fatty acid oxidation (FAO) and increased intracellular lipid deposition. In vitro experiments indicated that inhibition of fatty acid oxidation in tubule epithelial cells caused ATP depletion, cell death, dedifferentiation and intracellular lipid deposition; a phenotype observed in fibrosis. Restoring fatty acid metabolism by genetic or pharmacological methods protected mice from tubulointerstitial fibrosis. Our results raise the possibility that correcting the metabolic defect may be useful for preventing and treating chronic kidney disease.
Introduction
Fibrosis is the final common pathway and the histological manifestation of chronic kidney disease (CKD)1. While glomerular lesions are specific for the disease etiology, fibrosis shows almost identical manifestation in all progressive CKD2. Fibrosis is characterized by loss of capillary networks, accumulation of fibrillary collagens, activated myofibroblasts and inflammatory cells3,4. In fibrosis, tubular epithelial cells (TECs) are lost due to cell death and the remaining cells dedifferentiate leading to reduced expression of characteristic epithelial markers and increased expression of mesenchymal markers. Increased tubular epithelial Notch, Wnt and Hedgehog signaling can induce dedifferentiation of TECs, a critical characteristic of CKD5–7. While TECs may not be the direct precursors of myofibroblasts, they play an instrumental role in orchestrating fibrosis by multiple mechanisms including secreting different cytokines. Transforming growth factor beta (TGFB1) is a key mediator of tissue fibrosis; it induces secretion of fibrillary collagens, and promotes cell death and dedifferentiation8.
Alteration in cellular metabolism, including changes in fuel source preferences (glucose, fatty acids or ketones) has emerged as an important mechanism of cell differentiation, especially in the context of stem cells and carcinogenesis9. Metabolic reprogramming is a critical constituent of malignant transformation. Many cancers have increased glucose uptake and reduced mitochondrial glucose oxidation, a phenomenon called the “Warburg effect”. Very little is known about the metabolism of renal epithelial cells10,11. Proximal TEC have high levels of baseline energy consumption and a copious supply of mitochondria. Fatty acid oxidation (FAO) is the preferred energy source for highly metabolic cells like cardiac myocytes because it generates more ATP than does oxidation of glucose.
The uptake of long chain fatty acids is facilitated by the long chain fatty acid transporter; cluster of differentiation 36 (CD36)12. Metabolism of fatty acids requires their transport into the mitochondria, which is mediated by carnitine palmitoyl-transferase 1 (CPT1) and this enzyme conjugates fatty acids with carnitine13. CPT1 is considered to be the rate-limiting enzyme in FAO. The peroxisome proliferator-activated receptors (PPAR) and PPARgamma coactivator-1a (PPARGC1A) are the key transcription factors that regulate the expression of proteins involved in fatty acid uptake and oxidation14–16. Normally, fatty acid uptake, oxidation and synthesis are tightly balanced to avoid intracellular lipid accumulation.
Tubule epithelial lipid accumulation has received significant attention especially in the context of acute and diabetic kidney disease17–20. It has been proposed that excess accumulation of triglyceride induce cellular lipotoxicity potentially contributing to fibrosis development17–20. For this reason, we were alerted when our unbiased gene profiling highlighted alterations in cellular metabolism in fibrotic kidneys. We uncovered that enzymes and regulators of FAO were reduced in kidneys from human subjects with CKD and in mouse models of kidney fibrosis. We found that healthy renal TECs primarily rely on FAO as their energy source. Lower FAO by TECs appears to contribute to tubulointerstitial fibrosis development and therefore restoring FAO may be beneficial for treatment of CKD.
RESULTS
The transcriptional landscape of human CKD samples highlighted dyregulation in cellular metabolism
To examine genome wide transcript level changes in human samples with CKD we collected and analyzed a large number (n=95) of microdissected human kidney samples21,22. As the patterns of diabetes- and hypertension-induced tubulointerstitial fibrosis are almost indistinguishable, we performed a combined analysis of tubulointerstitial samples obtained from diabetic and hypertensive CKD samples (Supplementary Table 1). Cases were defined based on their glomerular filtration rate (<l60cc/min/1.72m2) and by the presence of histological lesions of fibrosis. The analysis allowed us to identify common patterns and pathways that characterize CKD in human subjects. We identified 2,497 transcripts with a corrected p-value of 0.05 and at least 30% change in their expression values (Supplementary Fig. 1 and Supplementary Table 2). Diabetic and hypertensive CKD samples showed similar changes.
High-level pattern analysis, using gene ontology tools, highlighted the differential regulation of two key pathways, inflammation and metabolism. Inflammation is known to play an important functional role in fibrosis development (Fig. 1a)23. On the other hand, we also detected a very robust signal indicating the dysregulation of cellular metabolism in CKD (Fig 1a and Supplementary Fig. 2). Gene ontology analysis highlighted specific changes in fatty acid metabolism, beta-oxidation, amino acid catabolism, and carbohydrate metabolism (Fig 1a). Gene set enrichment analysis confirmed the strong enrichment for FAO amongst the differently expressed pathways (Fig. 1b,c and Supplementary Fig. 2). A more detailed interrogation indicated that genes related to fatty acid metabolism and their key transcriptional regulator complex PPARA/PPARGC1A were markedly lower in CKD samples (Fig. 1d–f). There was also a considerable reduction in levels of key regulators of glucose utilization (Fig. 1g). The lower expression of key FAO regulators was associated with higher lipid accumulation in diseased renal TECs (Fig. 1e). Several mitochondrial enzyme and transcription factor mRNA levels were lower in CKD samples, but we found no difference in mitochondrial DNA copy number (Supplementary Fig. 3a,b). In summary, genome wide unbiased transcript analysis of one of the largest collections of human kidney samples identified that enzymes and critical regulators of FAO are lower in kidneys with fibrosis.
Figure 1. The transcriptional landscape of human tubulointerstitial fibrosis.

(a), Gene ontology analysis of human chronic kidney disease (CKD) samples. The graph shows the minus log p-values for the enrichment of a specific pathway. (b), Gene set enrichment analysis highlighted strong enrichment for FAO pathway in CKD human samples. (c), Heatmap analysis of transcripts expression related to fatty acid metabolism. (d), Relative mRNA levels of genes related to FAO in human control (CTL) and CKD samples (CPT1a; carnitine palmitoyl-transferase 1a, CPT2; carnitine palmitoyl-transferase 2, ACOX1; acyl-CoA oxidase 1, ACOX2; acyl-CoA oxidase 2, PPARa; Peroxisome proliferator-activated receptor alpha and PPARGC1a; peroxisome proliferator-activated receptor gamma, coactivator 1 alpha). (e), Representative images of control and CKD human kidney samples stained with periodic acid-schiff (PAS), PPARa and PPARGC1Aa and oil red o (lipid droplets). Scale bar, 20 µm. (f), Quantification of PPARa and PPARGC1a immunostaining in CTL and CKD human kidney samples. (g), Relative transcript levels of enzymes related to carbohydrate metabolism (HK; hexokinase, PFK; phosphofructokinase, GAPDH; Glyceraldehyde 3-phosphate dehydrogenase, PGK1; phosphoglycerate kinase 1, IDH1; isocitrate dehydrogenase 1, PK; pyruvate kinase and GLUT1; Glucose transporter 1). All data are presented as means as ± s.e.m (n=95, 59 of control and 36 of CKD), and * p<0.05 compare to CTL.
Genome-wide transcriptome analyses of mouse models of fibrosis indicated differences in FAO
Next, we analyzed genome wide transcriptional changes in mouse models of fibrosis. First, we examined a genetic model of fibrosis created by transgenic overexpression of the Notch1 intracellular domain in TECs by using a doxycycline inducible transgenic system (Tetracycline Responsive Element Intra-Cellular Notch1 [TRE-ICNotch1]) (Supplementary Fig. 4a). Induction of this transgene causes severe tubulointerstitial fibrosis (TIF) (Fig. 2a)5. Transcriptome analysis of control and Pax8rtTA/TREICNotch1 kidneys was accomplished using the Affymetrix ST1.0 platform5. Gene ontology analysis again indicated coordinated differences in metabolic pathways (Fig. 2b). We observed alterations in amino acid, carbohydrate and fatty acid metabolism. Transcript levels of key and rate limiting enzymes of FAO (Cpt, Acox) and glucose utilization (Pgk, Pk) were markedly lower in this mouse fibrosis model (Fig. 2c,d). Immunostaining studies confirmed the lower expression of Ppara; a key regulator of lipid metabolism (Fig 2a).
Figure 2. Dysregulation of FAO in mouse models of tubulointerstitial fibrosis.
(a), Representative images of PAS stained kidney sections of control and Pax8rtTA/TREICNotch1 mice, followed by Ppara immunnostaining images. (b), Gene-ontology analysis of control and Pax8rtTA/TREICNotch1 mice. (c,d), Relative mRNA levels of genes related to FAO (Cpt1a; carnitine palmitoyl-transferase 1a, Cpt2; carnitine palmitoyl-transferase 2, Acox1; acyl-CoA oxidase 1, Acox2; acyl-CoA oxidase 2, Ppara; Peroxisome proliferator-activated receptor alpha and Ppargc1a; peroxisome proliferator-activated receptor gamma, coactivator 1 alpha) and glucose utilization (Hk; hexokinase, Pgk1; phosphoglycerate kinase 1, Gapdh; Glyceraldehyde 3-phosphate dehydrogenase, Idh1; isocitrate dehydrogenase 1, Pk; pyruvate kinase and Glut1; Glucose transporter 1) in control (CTL) and mouse TIF (ICNotch1) samples. All data are presented as means as ± s.e.m (n=6, 3 of control and 3 of Pax8rtTA/TREICNotch1 mice) and * p<0.05 compare to CTL. (e), Relative transcript levels of FAO related enzymes in control (CTL) and FA-induced TIF samples (FAN). All data are presented as means as ± s.e.m (n=12, 6 of control and 6 of FAN mice) and * p<0.05 compare to CTL. (f), Representative images of mouse kidney sections from control and folic acid induced nephropathy (FAN) with PAS staining, immunostaining for Ppara and Ppargc1a and Oil Red O staining. Scale bar, 20 µm. (g), Quantification of Ppara and Ppargc1a images in the CTL and FAN mouse kidney samples. (h), Triglyceride (TG) contents in mouse kidney tissue. N = 10, 4 of CTL and 6 of FAN. * p<0.05 compare to CTL.
Similar changes were observed in kidneys from mice with folic acid-induced kidney fibrosis (FAN). Protein and transcript levels of key FAO enzymes were lower (Fig. 2e). Ppara and Ppargc1a mRNA levels were lower (Fig. 2f,g) and lipid accumulation (Fig. 2f) and triglyceride content (Fig. 2h) were higher in TEC of FAN mice. Mitochondrial copy number was not changed, but the expression of several mitochondrial specific genes was lower in kidneys from mice with FAN (Supplementary Fig. 3c,d). Time-course experiments indicated that the lower expression of FAO in the FAN model preceded the expression of fibrosis markers (Supplementary Fig. 5a–c), potentially suggesting its causal role. Thus, mouse models of kidney fibrosis showed similar metabolic changes to human CKD samples: low fatty acid and carbohydrate metabolism and high intracellular lipid accumulation.
The role of tubule epithelial lipid accumulation in CKD development
Tubule epithelial lipid accumulation was evident both in kidneys from mice and humans with renal fibrosis. It has been proposed that increased lipid accumulation leads to lipotoxicity, contributing to fibrosis development24. Long-chain fatty acids particularly palmitate and stearate mostly enter cells via the long-chain fatty acid transporter (CD36)12. To test this hypothesis we generated a mouse model with kidney specific overexpression of CD36. We placed the human CD36 under a tetracycline responsive element (TRE) and crossed the animals with the Pax8rtTA animals. Upon doxycycline administration (at 4 weeks of age), the expression of human CD36 was induced in TEC (Fig. 3a and Supplementary Fig. 4b). By 8 weeks of age, we observed considerable accumulation of intracellular lipids (Fig. 3b) and triglycerides in the Pax8rtTA/TRECD36 animals compared to controls (Fig. 3c). We performed fatty acid quantification by GC/FID (gas chromatography followed by flame ion diffusion), which indicated that the major fatty acid subtypes in the mouse kidney were stearic, palmitic, linoleic and docosohexaenoic acid (Fig. 3d, Supplementary Table 3). Levels of stearate and palmitate were markedly higher in double transgenic animals compare to WT animals (Supplementary Table 3). The concentration of short chains fatty acids was slightly lower in double transgenic kidney samples. Quantitative analysis of markers of FAO indicated no change or only slightly higher expression of Cpt1a mRNA level by 20 weeks of age (Fig. 3e).
Figure 3. Role of tubular epithelial cell lipid accumulation in tubular interstitial fibrosis development.
(a), Protein expression (Western blot) of human CD36 in control and Pax8rtTA/TRECD36 mouse kidney samples. (b), Representative images of PAS, oil red o and Sirius red stained control and CD36 transgenic animals. Scale bar, 20 µm. (c), Triglyceride (TG) contents in mouse kidney tissue. N = 5 of CTL and n=5 of 20 week old human CD36 transgenic mice. * p<0.05. (d), Fatty acid quantification in the control and CD36 TG mice by GC/FID. (e–g), Quantitative RT-PCR analysis of transcripts of key FAO enzymes and regulators (Cpt1a; carnitine palmitoyl-transferase 1a, Cpt2; carnitine palmitoyl-transferase 2, Acox1; acyl-CoA oxidase 1, Acox2; acyl-CoA oxidase 2 and Ppara; Peroxisome proliferator-activated receptor alpha), markers of fibrosis (Vim; vimentin, Fn; fibronectin, Col1a1; collagen 1a1, Col3a1; collagen 3a1, Col4a1; collagen 4a1 and Acta2; alpha smooth muscle actin) and apoptosis markers (Apaf1; apoptosis inducing factor1, Bax; BCL2 associated X protein, Bcl2; B-cell CLL/ lymphoma 2) in 8 week and 20 week old human CD36 transgenic mice. All data are presented as means as ± s.e.m (n=17, n=5 control, n=5 Pax8rtTA/TRECD36 at 8 weeks, and n=7 of Pax8rtTA/TRECD36 at 20 weeks) and * p<0.05 compared to controls.
Eight-week-old CD36 transgenic animals (at the time when fatty acid accumulation was already evident) showed no evidence of profibrotic marker expression (Fig. 3f), Sirius red staining (Fig. 3b) and change in levels of apoptosis related genes (Fig. 3g). By 20 weeks of age we observed slightly higher expression of Acta2 and Col1a (Fig. 3f). CD36 transgenic animals did not show higher susceptibility to diabetic kidney injury induced by streptozotocin (Supplementary Fig. 6) or folic acid induced fibrosis (data not shown). In summary, CD36 overexpression led to TEC lipid accumulation, but this high lipid (triglyceride and long chain fatty acid) levels alone was not sufficient to induce the full spectrum of fibrosis. We hypothesized that reduced fatty acid utilization (FAO) is the major inciting factor for fibrosis development and that lipid accumulation occurred as a secondary consequence.
Renal TECs mostly depend of FAO as their energy source
The metabolic derangement observed in patient samples with fibrosis prompted us to examine the metabolic pattern of TECs. To quantitatively analyze metabolism we examined oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of primary TECs and a TEC cell line; HKC-8 (Supplementary Fig. 7, Fig. 4a,b). We found that OCR was markedly higher when palmitic acid was presented to TECs, indicating that TECs efficiently metabolize palmitate. The increase in OCR was sensitive to the Cpt1 inhibitor etoxomir, confirming its specificity (Fig. 4a). OCR was unchanged after glucose injection (Fig. 4b). On the other hand, ECAR was higher following glucose injection, indicating that glucose is mainly used for anaerobic glycolysis (lactate causes acidosis, but does not consume oxygen) (Fig. 4b). FAO is the key contributor to intracellular ATP levels as etoxomir treatment markedly reduced intracellular ATP content in TEC (Fig. 4c). Etomoxir treatment was also associated with higher apoptosis rate, i.e. more cleaved caspase 3 positive cells and lower anti-apoptotic BCL2 transcript levels (Fig. 4d,e). Inhibition of FAO caused dedifferentiation of TECs–including the expression of ACTA2, VIM, COL1A1 and COL3A1 (Fig. 4f)–and higher intracellular lipid droplet accumulation (Fig. 4d). Similar results were observed with ranolazine, a partial FAO inhibitor (Supplementary Fig. 8a–c). In summary, we found that blockade of FAO was associated with higher cell death, dedifferentiation and intracellular lipid accumulation, a phenotype that mimicked what we had observed in human fibrotic kidneys.
Figure 4. Transforming growth factor beta 1 suppresses FAO in renal TEC.
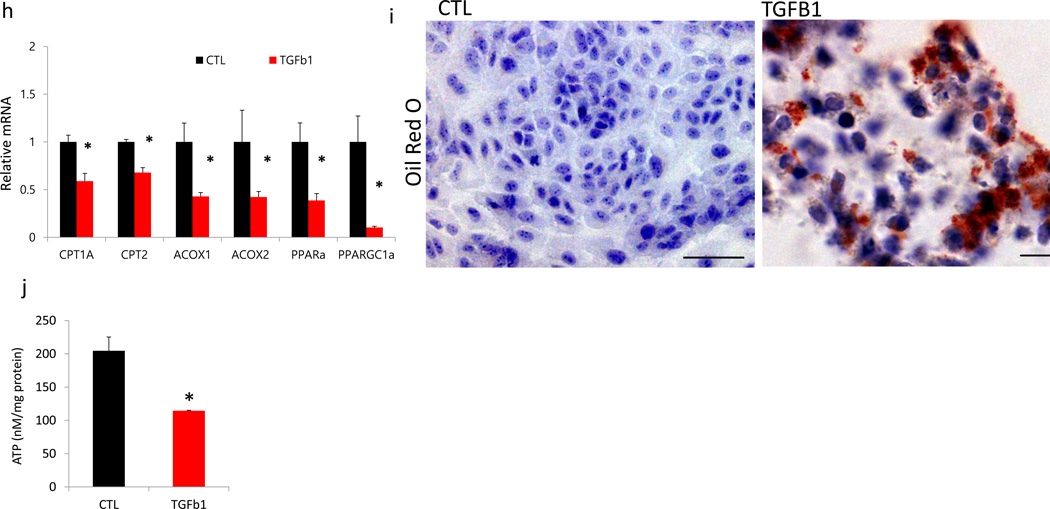
(a), Oxygen consumption rate (OCR) of human tubule epithelial cell line (HKC8); each data-point represents the mean (and S.E.M) of ten independent samples. When indicated palmitate (180 µM), etomoxir (40 µM), and oligomycin (1 µM) was added. (b), OCR and extracellular acidification rate (ECAR) of HKC8 were measured in a Seahorse XF24 analyzer. Each data-point represents the mean of ten independent samples. 10 mM glucose, 2–4 dinitrophenol (2–4 DNP, 100 µM), 2-deoxyglucose (2-DG, 100 mM), and rotenone (1 µM) were injected sequentially at the indicated time points. (c), ATP levels of control (CTL) HKC8 cells, and cells treated with 40 µM etomoxir (CPT1 inhibitor). (d), Bright field, oil red o staining and caspase 3 immunofluorescence images of HKC8 cells, untreated (left) or etomoxir treated (right). (e), Relative transcript amounts of genes related to apoptosis (APAF1; apoptosis inducing factor1, BAX; BCL2 associated X protein, BCL2; B-cell CLL/ lymphoma 2, BCL2L1; BCL2 like 1) of HKC8 cells upon etomoxir treatment (n=6, 3 of control and 3 of etomoxir treated cells). (f), QRT-PCR based relative mRNA levels of markers of cellular dedifferentiation (VIM; vimentin, FN; fibronectin, COL1; collagen1, COL3, collagen3, and ACTA2; alpha smooth muscle actin) of HKC8 cells treated with etomoxir. (g), OCR measurement of HKC8 exposed to 50ng ml−1 TGFβ1 for 48 hrs or controls. Representative traces are shown right, and summary data left analyzed for 16 wells from independent experiments. When indicated palmitate (180 µM), glucose (25mM) and oligomycin (1 µM) were added. (h), Relative mRNA amount of transcripts of FAO enzymes (CPT1a; carnitine palmitoyl-transferase 1a, CPT2; carnitine palmitoyl-transferase 2, ACOX1; acyl-CoA oxidase 1, ACOX2; acyl-CoA oxidase 2, PPARa; Peroxisome proliferator-activated receptor alpha and PPARGC1a; peroxisome proliferator-activated receptor gamma, coactivator 1 alpha) of control and TGFB1 treated cells. All experiments were replicated twice with triplicate repeated measures for each condition. (i), Lipid accumulation was determined by oil red O staining of control and TGFB1 treated cells. Scale bar, 20 µm (j), ATP levels of control (CTL) HKC8 cells, and cells treated with TGFB1. All data are presented as means as ± s.e.m (n=8, 4 of control and 4 of TGFB1) and * p<0.05.
Transforming growth factor beta (TGFB1) suppresses FAO in a SMAD3 and PPARGC1A dependent manner
TGFB1 is one the most powerful profibrotic cytokine; therefore we next examined the effect of TGFB1 on renal epithelial cell metabolism. As described previously, treatment of TEC’s with TGFB1 induced clear profibrotic phenotypic changes characterized by loss of epithelial phenotype and acquisition of a mesenchymal markers (expression of COLa1, COL3a1, VIM, FN and ACTA2 (Supplementary Fig. 8d,e)25. Cells treated with TGFB1 had lower baseline of oxygen consumption levels and they showed a reduction in palmitate-induced elevation in OCR, indicating a low activity of fatty acid metabolism (Fig. 4g). TGFB1 reduced PPARA and PPARGC1A mRNA levels and their downstream targets CPT1 and ACOXs, which was the likely cause for the lower expression of FAO (Fig. 4h). Consistent with the significant metabolic depression, there was an accumulation of oil-red-o positive lipids (Fig. 4i) and lower cellular ATP content (Fig. 4j) in TGB1 treated cells. Levels of glucose metabolism related enzymes (PFK1 and PK) (Supplementary Fig. 8f) were lower in TGFB1 treated cells. Therefore, TGFB1 induces a metabolic reprogramming in TECs, specifically lower FAO and higher lipid accumulation.
Next, we analyzed the molecular mechanism of the TGFB1-induced FAO blockade. We found that TGFB1’s effects were Smad3 dependent. TGFB1 did not change transcript levels of fatty acid metabolism related enzymes (Cpt, Acox Ppara and Ppargc1a) in Smad3 deficient renal tubule cells (Supplementary Fig. 8g,h). Furthermore using publicly available chromatin immunoprecipitation and sequencing (ChIP-Seq) data we identified that SMAD3 directly binds to an intronic area on the PPARGC1A gene (Fig. 5a) (GSE29422)26. This intronic area is outside of the promoter region of PPARGC1A, and does not overlap with active promoter specific histone tail modifications (H3K4me3) in human kidney TECs27. On the other hand it overlapped with active enhancer specific histone tail modifications (H3K4me1) and this region was a computationally annotated active enhancer (by ChromHMM-based) region in human kidney TECs (Fig. 5a)27. PPARGC1A is a critical transcription factor that regulates mitochondrial biogenesis and the expression of almost all rate-limiting enzymes in the FAO pathway. We did not observe direct SMAD3 binding to PPARA. These results indicate that TGFB1 via SMAD3 can potentially directly regulate PPARGC1A transcript levels.
Figure 5. Transforming growth factor beta (TGFB1) suppresses FAO in a SMAD3 and PPARGC1A dependent manner.
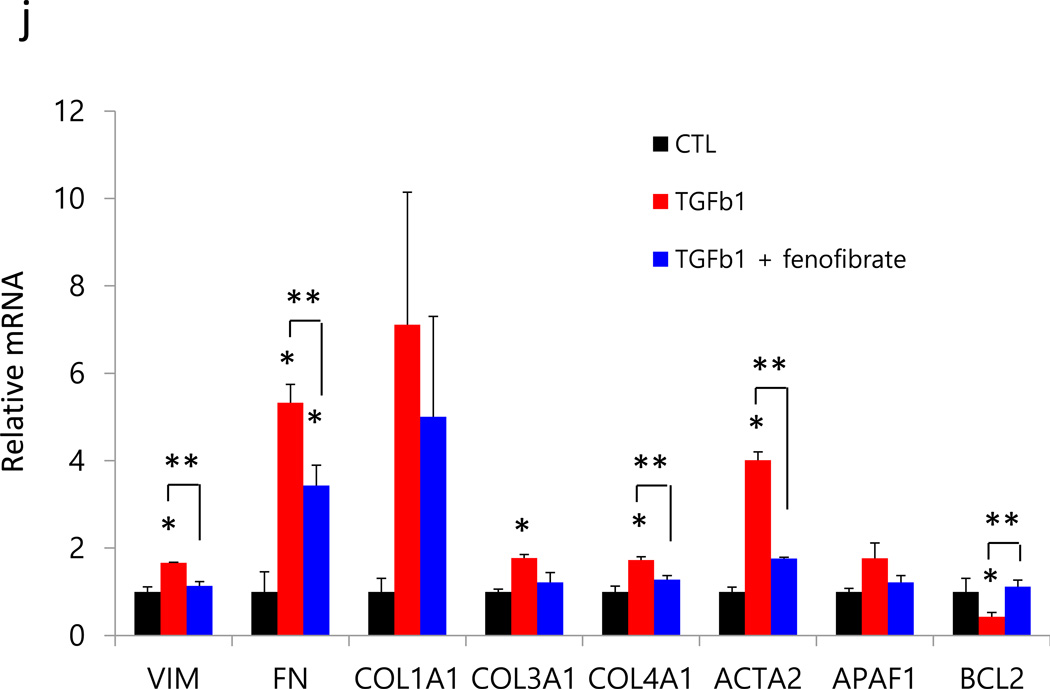
(a) The human PPARGC1A locus followed by SMAD2/3 ChIP-Seq, human kidney H3K4me1 and H3K4me3 ChIPSeq, human kidney specific gene regulatory region annotation (red promoter, yellow enhancer) followed by GERP-based conservation scoring. (b), OCR were measured in primary TEC from wild type (WT) and Pax8rtTA/TREPPARGC1A (TG) mice treated or not with TGFB1. Representative traces are shown right and summary data on left analyzed for 12 wells for independent experiments. All data are presented as means as ± s.e.m and *p<0.05 compare to WT and ** p<0.05 compare to WT-TGF. When indicated palmitate (180 µM), glucose (25mM) and oligomycin (1 µM) were added. (c–d), Expression of key enzymes critically related to FAO and glucose utilization in primary renal tubule epithelial cell from Pax8rtTA/TREPPARGC1A transgenic mice (TG) or control (CTL) exposed with 50ng ml−1 TGFB1 for 48h (Cpt1a; carnitine palmitoyl-transferase 1a, Cpt2; carnitine palmitoyl-transferase 2, Acox1; acyl-CoA oxidase 1, Acox2; acyl-CoA oxidase 2, Ppara; Peroxisome proliferator-activated receptor alpha and Ppargc1a; peroxisome proliferator-activated receptor gamma, coactivator 1 alpha, Hk; hexokinase, G6pc; glucose-6-phosphatase, catalytic subunit, Pfk1; phosphofructokinase 1, Idh2; isocitrate dehydrogenase 2, Pkm2; pyruvate kinase, muscle2, Glut1; glucose transporter 1). (e–f), Protection effects of Ppargc1a transgenic cells from TGFB1 induced fibrosis development including profibrotic, (Vim; vimentin, Fn; fibronectin, Col1a1; collagen 1a1, Col3a1; collagen 3a1, Col4a1; collagen 4a1 and Acta2; alpha smooth muscle actin) and apoptosis related genes (Apaf1; apoptosis inducing factor1 and Bcl2; B-cell CLL/ lymphoma 2). All data are presented as means as ± s.e.m (n=12, 3 of each condition), * p < 0.05 compare to WT and ** p<0.05 compare to WT-TGF. (g), OCR in HKC8 exposed to 50ng ml−1 TGFB1 for 48hrs in the presence or absence of 1 µM fenofibrate. When indicated cells were incubated in palmitate (180 µM), glucose (25mM), and oligomycin (1 µM). (h–i), Relative mRNA expression of transcripts related to FAO, (CPT1a; carnitine palmitoyl-transferase 1a, CPT2; carnitine palmitoyl-transferase 2, ACOX1; acyl-CoA oxidase 1, ACOX2; acyl-CoA oxidase 2, and PPARa; Peroxisome proliferator-activated receptor alpha), fibrosis and apoptosis (VIM; vimentin, FN; fibronectin, COL1A1; collagen1a, COL3A1; collagen3a1, COL4A1; collagen4a1, ACTA2; alpha smooth muscle actin, APAF1; apoptosis inducing factor1, and BCL2; B-cell CLL/ lymphoma 2) in HKC8 cells treated with or without TGFB1 and fenofibrate. All data are presented as means as ± s.e.m (n=9, 3 of control, 3 of TGFB1 and 3 of TGFB1 with fenofibrate treated group), *p<0.05 compare to CTL and ** p<0.05 compare to TGFB1.
We next tested whether the TGFB1-induced metabolic reprogramming was mediated by Ppargc1a using primary murine proximal TECs from mice with tubule specific overexpression of Ppargc1a (Pax8rtTA/TRE-Ppargc1a mice [see description later]). Ppargc1a transgenic cells showed no difference in their palmitate induced oxygen consumption rate after TGFB1 treatment (Fig. 5b). Genetic overexpression of Ppargc1a normalized transcript levels of Cpt and other FAO enzymes (Fig. 5c), however, the defect in glucose utilization related gene expression was not recovered in transgenic cells (Fig. 5d). These results indicate that the TGFB1 induced FAO blockade is mediated by Ppargc1a.
Next, we examined whether metabolic reprogramming plays a role in the TGFB1-induced profibrotic phenotype development. We found that Ppargc1a transgenic cells were protected from TGFB1-induced functional changes including the expression of profibrotic (Col3a1, Fn, Acta2, and Col4a1) and apoptosis-associated (Bcl2) gene expression (Fig. 5e,f). These results indicate that metabolic reprogramming plays a critical role in the TGFB1-induced functional changes.
As Ppargc1a acts together with Ppara to regulate FAO, next we examined the effect of the Ppara agonist, fenofibrate. Fenofibrate normalized TGFB1-induced FAO repression (Fig. 5g), Cpt1 and Acox reduction (Fig. 5h), but not the glucose utilization defect (Fig. 5i). It ameliorated TGFB1-induced dedifferentiation and apoptosis (Fig. 5j). Thus, the TGFB1-induced and Smad3-mediated repression of Ppargc1a and FAO are critical components of the TGFB1-induced TECs dedifferentiation and apoptosis.
Genetic or pharmacological improvement of FAO protected animals from kidney fibrosis development in vivo
We next tested whether improving FAO in vivo in mouse models protects animals from tubulointerstitial fibrosis development. We used both transgenic and pharmacological approaches to achieve our goals. First, we analyzed the effect of transgenic overexpression of Ppargc1a. This was achieved by intercrossing the Pax8rtTA animals with the TRE/Ppargc1a animals. Single and double transgenic animals were fed with doxycycline containing food starting at 4 weeks of age. Tubulointerstitial fibrosis was induced by folic acid administration at 6 weeks of age. Double transgenic animals showed improvement in renal histology as shown by the PAS stained kidney sections and QRT-PCR based quantification of profibrotic molecules (Fig. 6a,b). Transgenic expression of Ppargc1a normalized expression of Cpt the rate limiting enzyme in FAO (Fig. 6c). There was lower expression of apoptosis markers in tubules (cleaved caspase3 staining and Bcl2 expression, Fig. 6a,d) of the Ppargc1a transgenic animals. These results indicate that restoring FAO, but not glucose utilization (Fig. 6e) by Ppargc1a ameliorates fibrosis development.
Figure 6. Genetic or pharmacological restoration of FAO protects animals from fibrosis development.
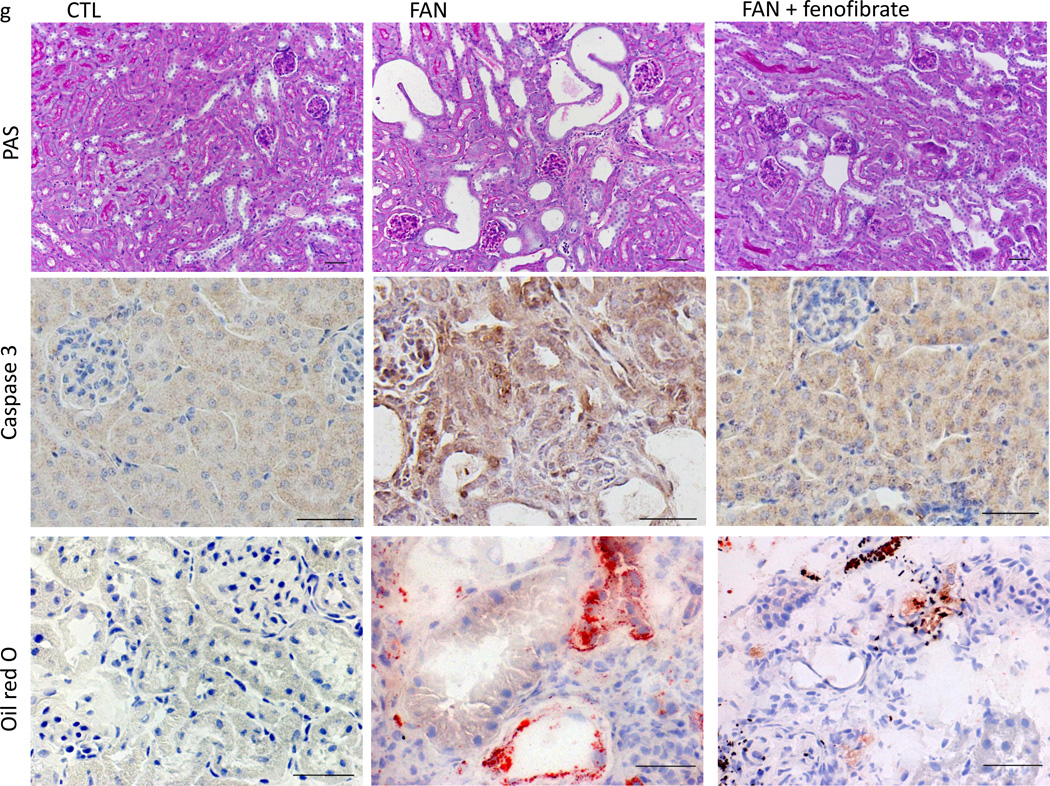
(a), Representative PAS, cleaved caspase 3 and Sirius Red stained kidney sections of control and FAN fibrosis model (day 7). We used wild type and single transgenic animals as controls (CTL, or FAN) and Pax8rtTA/TRE-PPARGC1A transgenic mice with folic acid (Ppargc1a FA). Scale bar, 20 µm. (b), Markedly lower expression of fibrosis related transcripts in PPARGC1A overexpressed mice followed by folic acid administration (Vim; vimentin, Fn; fibronectin, Col1a1; collagen 1a1, Col3a1; collagen 3a1, Col4a1; collagen 4a1, Acta2; alpha smooth muscle actin, and Kim-1; kidney injury molecule-1). (c–e), Relative mRNA levels of FAO enzymes (Cpt1a; carnitine palmitoyl-transferase 1a, Cpt2; carnitine palmitoyl-transferase 2, Acox1; acyl-CoA oxidase 1, Acox2; acyl-CoA oxidase 2 and Ppara; Peroxisome proliferator-activated receptor alpha), apoptosis associated markers (Apaf1; apoptosis inducing factor1, Bcl2; B-cell CLL/ lymphoma 2, and glucose utilization (Hk; hexokinase, G6pc; glucose-6-phosphatase, catalytic subunit, Pfk1; phosphofructokinase 1, Idh2; isocitrate dehydrogenase 2, Pkm2; pyruvate kinase, muscle2, Glut1; glucose transporter 1). All data are presented as means as ± s.e.m (n=18, 6 of control, 6 of FAN and 6 of PPARGC1a FA group), * p<0.05 compare to CTL and ** p<0.05 compare to FAN. (f), Relative mRNA levels of FAO enzymes in control (CTL) folic acid treated (FAN) and folic acid and fenofibrate treated (FAN+fenofibrate) mice. (g), Ppara agonist fenofibrate ameliorated fibrosis development in the FA induced fibrosis model. Representative PAS, cleaved caspase 3, oil red o and sirius red stained kidney sections. Scale bar, 20 µm. (h,i), Relative mRNA levels of fibrosis, apoptosis and glycolysis associated markers. All data are presented as means as ± s.e.m (n=14, 5 of control, 5 of FAN and 4 of FAN with fenofibrate treated group) and * p<0.05. (j), Serum creatinine levels in CTL, FAN and FAN+fenofibrate mice. N = 15, 5 of CTL, 5 of FAN and 5 of FAN with fenofibrate mice. * p<0.05 compare to CTL and **p<0.05 compare to FAN.
Next, we examined whether pharmacological activation of Ppara can also protect mice from tubule epithelial fibrosis development using the folic acid-induced injury model. Animals were injected with fenofibrate starting one day prior to folic acid injection. Fenofibrate administration strongly induced the expression of FAO enzymes Cpt1, 2 and Acox1, 2 (Fig. 6f). Folate-treated mice injected with fenofibrate had improved renal histology, lower expression of cleaved caspase 3 (Fig. 6g), reduced fibrosis, kidney injury, apoptosis rate (Fig. 6h), and improved renal function (Fig. 6i) compared to sham injected animals. On the other hand, enzymes level related to glucose utilization were not recovered by fenofibrate treatment (Fig. 6j). These observations were not limited to the folic acid induced kidney fibrosis model and similar results were observed upon analyzing the unilateral ureteral obstruction-(UUO) induced kidney fibrosis model. There was low expression of FAO-related genes in the UUO model (Supplementary Fig. 9). Treatment with fenofibrate restored FAO related enzyme expression, lower expression of epithelial injury (Kim-1) and fibrosis markers (collagens) (Supplementary Fig. 9).
Next, to provide additional evidence that kidney fibrosis is specifically linked to a defect in FAO, we tested the effect of etoxomir; a specific inhibitor Cpt, the key enzyme in the FAO pathway. Mice treated with etomoxir developed more severe kidney injury after folic acid injection; they showed markedly higher expression of fibrosis markers and higher serum creatinine (Supplementary Fig. 10). Etomoxir inhibited Cpt and shifted metabolism to more glycolysis, evident by higher Glut1 and Pfk1 expression. However, the glycolytic phenotype did not improve (rather worsened) the fibrosis development.
In addition, we also tested the effect of C75, a synthetic compound that increases Cpt1 activity and blocks fatty acid synthase28. C75 treatment restored expression of Cpt1 and Acox1, and markedly lowered apoptosis rate. C75 treatment improved kidney fibrosis and function (Supplementary Fig. 11) without restoring the defect in glucose utilization, indicating again the critical role of FAO in fibrosis development. In summary, our results indicate that improving FAO by genetic or pharmacological activation of Ppargc1a, Ppara or Cpt1 protects animals from fibrosis development.
Discussion
In this study we aimed to identify and examine common mechanisms of kidney fibrosis. Here we present genome-wide transcript data from a large human CKD kidney sample cohort. Our non-biased studies indicated consistent differences in two gene groups; inflammation and metabolism. Several metabolic pathways were dysregulated in kidney fibrosis. Carbohydrate, amino acid and lipid metabolism were markedly depressed in patient samples with fibrosis.
Research into lipid metabolism in the kidney has been limited29,30. Clinical observations indicate a potential association between lipid levels and CKD development31. Most prior observations focused at higher lipid accumulation in TECs in CKD kidneys12,18. Higher intracellular lipid accumulation was proposed to play a key role in fibrosis development by inducing lipotoxicity12,20,32. Our results, however, indicate that higher intracellular triglyceride accumulation alone was not sufficient to induce the full spectrum of fibrosis in our CD36 transgenic mouse model and substrate (fatty acid) availability does not seem to be the limiting factor in kidney fibrosis. We also found that depressed FAO was associated with intracellular lipid accumulation (Supplementary Fig. 12). Our data therefore suggest a modest role for triglyceride accumulation in kidney fibrosis development.
We found that renal TEC critically depend on fatty acid oxidation as their energy source. This observation is in line with (almost half a century old) studies showing that kidneys preferentially take up and oxidize fatty acids and they are not a major contributor to circulating glucose catabolism33. As TECs are one of the highest energy demanding cells in the body, this carbon preference is quite logical. Other high-energy demanding organs (for example the heart) also depend on FAO24.
Increased glucose utilization is the key metabolic phenotype of renal cell cancer and polycystic kidney disease11,34,35. It is important to note that in fibrosis, tubule cells did not significantly revert to glucose oxidation. Additional genetic mutations or signaling mechanisms are likely needed to make this metabolic switch and simply loosing the capacity for FAO does not induce a change in energy substrate preference. Upon inhibition of FAO in TEC, cellular ATP level decreased, likely contributing to the increased apoptosis rate and dedifferentiation. The key observation of the current work is that the decreased FAO alone was sufficient to reprogram TEC cells into a profibrotic phenotype, while improving FAO restored cell differentiation.
We also found that TGFB1 appears to be an important upstream modulator of fatty acid metabolism in renal TECs. In TECs TGFB1 acted upstream of Ppargc1a. Chromatin immunoprecipitation studies indicate that Smad3 can directly bind to the Ppargc1a regulatory region36. A similar mechanism has already been described in adipocytes; a TGFB1-driven Smad3-dependent pathway appears to control Pgc1a and downstream fat deposition36. There might be other factors and cytokines that regulate Ppargc1a and FAO in kidney fibrosis (Supplementary Fig. 12), for example Lkb1 and Ampk could be important controllers of FAO in TECs37 (Drs. Han and Susztak unpublished observation). In our models and human samples, we did not observe a significant change in mitochondrial copy number, on the other hand the expression of several important mitochondrial genes was lower in fibrotic kidney samples. As beta-oxidation also takes places in the mitochondria we cannot exclude that the reduced FAO is just one manifestation of a broader mitochondrial dysfunction and/or oxidative phosphorylation defect. Mitochondrial dysfunction proposed to play important role in diabetic kidney disease development. Future studies shall determine the role of FAO and mitochondria in fibrosis development. Performing metabolic studies in glomerular cells and glomerular injury models will also be important.
Our studies could have several important clinical implications. FAO inhibitors have received significant attention recently, when it was found that these drugs significantly sensitize leukemia cells for cell death because leukemia cells use fatty acid as their key energy substrate38. Short-term treatment of animals with FAO inhibitors did not induce kidney fibrosis, however mice treated with FAO inhibitors showed increased fibrosis following injury. Future studies shall determine the long-term renal consequences of fatty oxidation inhibitors.
On the other hand our experiments indicate that drugs that enhance FAO might be of therapeutic benefit for patients with kidney fibrosis. For example the Ppara agonist fenofibrate might confer therapeutic benefit for patients with chronic kidney disease, as fenofibrate is widely used clinically for the treatment of hyperlipidemia. Indeed, some long-term studies indicate reduction in albuminuria in patients with type 2 diabetes39–41. Unfortunately, the effect of fibrates on renal function is less obvious as it can be associated with a rise in serum creatinine. Our results indicate that Ppargc1 or Cpt1 agonists might have a better therapeutic profile.
In summary, our studies show that decreased tubule epithelial FAO causes metabolic reprogramming of TECs inducing increased apoptosis and dedifferentiation. Restoring FAO could offer new therapeutic approaches for the treatment and prevention of kidney fibrosis.
Online Methods
Antibodies and reagents
PPARa (IHC, Abcam, #ab8934), PPARGC1A (IHC, Abcam, #ab54481), and Cleaved caspase 3 (IF, IHC, Cell signaling, #9662), RPMI 1640 (Cellgro), CD36 (WB, Santa Cruz #sc-7309), FBS (Atlanta Biologicals), EGF (Peprotech), bFGF (Peprotech), Penicilin Streptomycin (Cellgro), ITS (Cellgro), hydrocortisone (Sigma), TGFβ1 (Peprotech), fenofibrate (Sigma), etomoxir (Sigma), ranolazine (Sigma), RNAeasy mini columns (Qiagen, Valencia, CA), collagenase I (Worthington Biochemical Product), 100 µm mesh (Fisherbrand), doxycycline-containing chow (Bioserv S3888), oil red o (EMD chemical), palmitate conjugated BSA (Seahorse Bioscience), oligomycin (Seahorse Bioscience), DNP (Sigma), 2-DG (Sigma), and rotenone (Sigma).
Human kidney tissue handling and microdissection
Tissue was placed into RNALater and manually microdissected at 4°C for glomerular and tubular compartments as described earlier. Dissected tissue was homogenized and RNA was prepared using RNAeasy mini columns according to manufacturer’s instruction. RNA quality and quantity was determined using Lab-on-Chip Total RNA PicoKit (Agilent BioAnalyzer). Only samples without evidence of degradation were used.
Gene expression analysis using Affymetrix Arrays
Transcript levels were analyzed by Affymetrix U133A and 1.0ST arrays. Probes were prepared using Affymetrix 3’ IVT kit. After hybridization and scanning, raw data files were imported into Genespring GX software (Agilent Technologies, USA). Raw expression levels were normalized using RMA16 summarization algorithm. Genespring GX software was then used for statistical analysis; the data was above the 20th percentile when filtered by expression. We used a Benjamini-Hochberg multiple testing correction with a p-value<0.05. Heatmap of gene expression data were generated using unsupervised hierarchical clustering method calculated by squared Euclidean distances. The Database for Annotation, Visualization and Integrated Discovery13 bioinformatics package was also used for gene ontology and pathway analysis. In addition, the Ingenuity Pathway Analysis (IPA, Redwood City, CA) was used to generate networks.
Primary culture of human and mouse renal tubule cells
Kidneys were collected from the non-cancerous part of nephrectomies. Cell were isolated by 2 mg ml−1 collagenase I digestion for 30 mins at 37 °C with gentle stirring and then filtered through the 100 µm mesh to collect single cells. Cell were cultured in RPMI 1640 supplement with 10% FBS, 20 ng ml−1 EGF, 20ng ml−1 bFGF and 1 % Penicilin Streptomycin at 5% CO2, 37 °C. HKC8 cells were cultured in DMEM/F12 supplemented with 5 % FBS, 1X ITS, 0.5 ug ml−1 hydrocortisone and 1 % Penicilin Streptomycin at 5% CO2, 37 °C. At 80% confluence, cells were starved in 0.5% FBS (RPMI) overnight and treated with 50 ng ml−1 TGFB1 with or without 1µM fenofibrate, 40 µM etomoxir or 200 µM ranolazine for 48h.
Animals
Male FvB mice (~25 gr, 6 weeks old), Pax8-rtTA transgenic mice and TRE-Pgc1a animals were purchased from Jackson Laboratory (Stock numbers 007176, 012387, respectively). Transgenic mice carrying the TRE-ICNotch 1 transgene were obtained from Dr D Melton (Harvard Medical School). Mice with TRE-hCD36 transgene will be described later (SH Ahn and Susztak unpublished observation). Briefly the human CD36 cDNA was cloned into the pTRE Tight vector (Clontech) and the construct was injected into FvB oocytes at the Einstein Transgenic facility. Transgenic mice were identified by genomic PCR analysis. Several founder lines were obtained and two lines with the highest expression of CD36 were kept for further analysis. We used human CD36 specific QRT-PCR primers and antibodies to distinguish endogenous (mouse) and transgene (human) expression. Mice were placed on doxycycline-containing chow starting at 3 weeks of age. Only male mice were used in the study. Mice (3–6 weeks of age) were injected with FA (250 mg kg−1, dissolved in 300 mM NaHCO3) or streptozotocin (50mg kg−1 for 5 days) intraperitoneally. One day before the FA injection, Ppara agonist fenofibrate (50mg kg−1 for 3 days and 100mg kg−1 for 5 days) was administered by oral gavage. Mice were randomly allocated to FA injected (FAN, n=5), FA with fenofibrate injected (FAN + fenofibrate, n=5) and sham-treated group (n=5). C75 (15mg kg−1) was injected i.p one day before FA injection. Etomoxir (30mg kg−1 for 2 days) was injected i.p two day before the FA injection. For UUO experiments, fenofibrate (125mg kg−1) was administered for 10 days. The treatment was started 3 days before the surgery and ended on day 7. Under sterile conditions, animals were anaesthetized with a mixture of isoflurane and oxygen. UUO was performed by complete ligation of the left ureters with 6-0 non-absorbable suture. Animal studies were approved by the Animal Care Committee of the University of Pennsylvania.
QRT-PCR
RNA was isolated from harvested cells and kidneys using RNAeasy Mini kit. 1 µg RNA was reverse transcribed using cDNA archival kit (Life Technology) and QRT-PCR was run in the ViiA 7 System (Life Technology) machine using SYBRGreen Master Mix and gene specific primers. The data was normalized and analyzed using the delta delta CT method. Primers used are listed in Supplementary Table I.
Histology and immunostaining
We used formalin-fixed, paraffin-embedded kidney sections stained with periodic acid Schiff (PAS) and OCT embedded frozen sections for oil red o staining. We performed immunocyto- and -histochemistry on paraformaldehyde fixed cells and formalin- fixed, paraffin-embedded kidney sections. We used the following primary antibodies: PPARa (1:1,000), PPARGC1A (1:500), and Cleaved caspase 3 (1:1,000). Staining was visualized using peroxidase-conjugated antibodies to mouse immunoglobulin using the Vectastain Elite kit and 3,3-diaminobenzidine (DAB) (Vector Labs).
Gas chromatography followed by flame ion diffusion (GC/FID)
Analysis of fatty acid profiles of control (CTL) and hCD36 transgenic (TG) mice was done at the Metabolomics Core of the Diabetes Center of the Albert Einstein College of Medicine.
Measurement of oxygen consumption rate and extracellular acidification rate
A Seahorse Bioscience X24 extracellular flux analyzer was used to measure the rate change of dissolved O2 in medium immediately surrounding adherent cells cultured in an XF24 V7 cell culture microplate (Seahorse Bioscience). Human and mouse primary tubular epithelial cells and HKC8 cultured in RPMI 1640 supplement with 0.5 % FBS with or without 50ng ml−1 TGFB1 or 1uM fenofibrate were seeded in XF24 V7 cell culture microplate at 1.0 × 104 cells per well. Oxygen consumption rates (pmol min−1) and extracellular acidification rate were assessed at baseline and after the after the addition of palmitate conjugated BSA or 10 mM glucose followed by the addiction of the carnitine palmitoyltransferase-1 (CPT1) inhibitor etomoxir or the mitochondrial upcoupler, 2-4 DNP or glycolysis inhibitor 2-DG. Final state was determined after the addition of the ATP synthase inhibitor oligomycin or rotenone.
ATP measurement
ATP content was determined using the ATP Colorimetric Assay kit (Biovision), following the manufacturer's instructions, on HKC8 cells treated with etomoxir or TGFB1.
Mitochondrial copy number determination
Mitochondrial copy number was determined using NovaQUANT™ Mitochondrial to Nuclear DNA Ratio Kit (Novagen) following the manufacturer's instructions using human and mouse kidney genomic DNA.
Triglyceride measurement
Kidney triglyceride was measured using quantification kits (Cayman Chemical) according to the manufacturer’s instructions.
Statistical analyses
The data were presented as mean ± s.e.m. Unpaired student’s t-test was used for comparisons between two groups. When needed multiple testing correction was performed by one way ANOVA with Tukey’s post hoc tests. P value < 0.05 was considered to be significant.
Supplementary Material
Acknowledgement
Financial support for this work provided by the NIDDK; R01DK087635, R01DK076077 to K. Susztak and Diabetic Complications Consortium (DiaComp, www.diacomp.org), grant DK076169 to K Susztak and E Bottinger. I. Goldberg is supported by grants HL45095 and HL73029 and K. Sharma is supported by NIH DP3 DK 094352. H. Kang is supported by a post-doctoral fellowship from the Juvenile Diabetes Research Foundation (3-2013-182). We thank the Einstein (P60-DK020541) and Penn Diabetes Research Centers (P30-DK19525) for GC/FID and Seahorse measurements, respectively and the Einstein Analytical imaging facility for the EM studies. Part of the work was presented at the Annual Nephrology Society Meeting in 2011 and 2013.
Footnotes
Author’s contribution
HMK performed all in vitro and most in vivo experiments, SHA and KSu generated and analyzed the CD36 transgenic animals, SHH, FC and JT helped with the animal experiments, PC, AESP, MP, JP were involved in collecting and analyzing human kidney tissue samples, YAK generated and analyzed the ChiP-seq data, EPB, KSh and IJG provided help with conceptual design, HMK and KSu wrote the paper and IJG assisted with editing the manuscript.
References
- 1.Brosius FC, 3rd, et al. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2009;20:2503–2512. doi: 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nath KA, Salahudeen AK, Clark EC, Hostetter MK, Hostetter TH. Role of cellular metabolites in progressive renal injury. Kidney Int Suppl. 1992;38:S109–S113. [PubMed] [Google Scholar]
- 3.Humphreys BD, et al. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeisberg M, et al. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 5.Bielesz B, et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 2010;120:4040–4054. doi: 10.1172/JCI43025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato H, et al. Wnt/beta-catenin pathway in podocytes integrates cell adhesion, differentiation, and survival. The Journal of biological chemistry. 2011;286:26003–26015. doi: 10.1074/jbc.M111.223164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabian SL, et al. Hedgehog-Gli pathway activation during kidney fibrosis. Am J Pathol. 2012;180:1441–1453. doi: 10.1016/j.ajpath.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottinger EP, Bitzer M. TGF-beta signaling in renal disease. J Am Soc Nephrol. 2002;13:2600–2610. doi: 10.1097/01.asn.0000033611.79556.ae. [DOI] [PubMed] [Google Scholar]
- 9.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldfield MD, et al. Advanced glycation end products cause epithelial-myofibroblast transdifferentiation via the receptor for advanced glycation end products (RAGE) The Journal of clinical investigation. 2001;108:1853–1863. doi: 10.1172/JCI11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowe I, et al. Defective glucose metabolism in polycystic kidney disease identifies a new therapeutic strategy. Nat Med. 2013;19:488–493. doi: 10.1038/nm.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Susztak K, Ciccone E, McCue P, Sharma K, Bottinger EP. Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med. 2005;2:e45. doi: 10.1371/journal.pmed.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran M, et al. PGC-1alpha promotes recovery after acute kidney injury during systemic inflammation in mice. J Clin Invest. 2011;121:4003–4014. doi: 10.1172/JCI58662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proctor G, et al. Regulation of renal fatty acid and cholesterol metabolism, inflammation, and fibrosis in Akita and OVE26 mice with type 1 diabetes. Diabetes. 2006;55:2502–2509. doi: 10.2337/db05-0603. [DOI] [PubMed] [Google Scholar]
- 16.Chau BN, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Science translational medicine. 2012;4:121ra118. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Portilla D, et al. Metabolomic study of cisplatin-induced nephrotoxicity. Kidney international. 2006;69:2194–2204. doi: 10.1038/sj.ki.5000433. [DOI] [PubMed] [Google Scholar]
- 18.Herman-Edelstein M, Scherzer P, Tobar A, Levi M, Gafter U. Altered Renal Lipid Metabolism and Renal Lipid Accumulation in Human Diabetic Nephropathy. Journal of lipid research. 2013 doi: 10.1194/jlr.P040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, et al. Transgenic expression of proximal tubule peroxisome proliferator-activated receptor-alpha in mice confers protection during acute kidney injury. Kidney international. 2009;76:1049–1062. doi: 10.1038/ki.2009.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decleves AE, et al. Regulation of lipid accumulation by AMP-activated kinase [corrected] in high fat diet-induced kidney injury. Kidney international. 2014;85:611–623. doi: 10.1038/ki.2013.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woroniecka KI, et al. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60:2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si H, et al. Human and murine kidneys show gender- and species-specific gene expression differences in response to injury. PloS one. 2009;4:e4802. doi: 10.1371/journal.pone.0004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vielhauer V, et al. CCR1 blockade reduces interstitial inflammation and fibrosis in mice with glomerulosclerosis and nephrotic syndrome. Kidney Int. 2004;66:2264–2278. doi: 10.1111/j.1523-1755.2004.66038.x. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg IJ, Trent CM, Schulze PC. Lipid metabolism and toxicity in the heart. Cell Metab. 2012;15:805–812. doi: 10.1016/j.cmet.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SW, et al. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Developmental biology. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Ko YA, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14:R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhajda FP, et al. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proceedings of the National Academy of Sciences. 2000;97:3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wanner C, Drechsler C, Krane V. Lipid metabolism in chronic kidney disease: the role of statins in cardiovascular risk. J Ren Nutr. 2007;17:75–78. doi: 10.1053/j.jrn.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Wanner C, Drechsler C, Krane V. Lipid metabolism in chronic kidney disease: the role of statins in cardiovascular risk. Journal of renal nutrition : the official journal of the Council on Renal Nutrition of the National Kidney Foundation. 2007;17:75–78. doi: 10.1053/j.jrn.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Kovesdy CP, Anderson JE, Kalantar-Zadeh K. Inverse association between lipid levels and mortality in men with chronic kidney disease who are not yet on dialysis: effects of case mix and the malnutrition-inflammation-cachexia syndrome. J Am Soc Nephrol. 2007;18:304–311. doi: 10.1681/ASN.2006060674. [DOI] [PubMed] [Google Scholar]
- 32.Khan S, et al. Lipotoxic disruption of NHE1 interaction with PI(4,5)P2 expedites proximal tubule apoptosis. The Journal of clinical investigation. 2014 doi: 10.1172/JCI71863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer C, Nadkarni V, Stumvoll M, Gerich J. Human kidney free fatty acid and glucose uptake: evidence for a renal glucose-fatty acid cycle. Am J Physiol. 1997;273:E650–E654. doi: 10.1152/ajpendo.1997.273.3.E650. [DOI] [PubMed] [Google Scholar]
- 34.Li B, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014 doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yadav H, et al. Protection from obesity and diabetes by blockade of TGF-beta/Smad3 signaling. Cell metabolism. 2011;14:67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dugan LL, et al. AMPK dysregulation promotes diabetes-related reduction of superoxide and mitochondrial function. The Journal of clinical investigation. 2013;123:4888–4899. doi: 10.1172/JCI66218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samudio I, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. The Journal of clinical investigation. 2010;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jun M, et al. Effects of fibrates in kidney disease: a systematic review and meta-analysis. J Am Coll Cardiol. 2012;60:2061–2071. doi: 10.1016/j.jacc.2012.07.049. [DOI] [PubMed] [Google Scholar]
- 40.Davis TM, et al. Effects of fenofibrate on renal function in patients with type 2 diabetes mellitus: the Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) Study. Diabetologia. 2011;54:280–290. doi: 10.1007/s00125-010-1951-1. [DOI] [PubMed] [Google Scholar]
- 41.Ansquer JC, et al. Fenofibrate reduces progression to microalbuminuria over 3 years in a placebo-controlled study in type 2 diabetes: results from the Diabetes Atherosclerosis Intervention Study (DAIS) Am J Kidney Dis. 2005;45:486–493. doi: 10.1053/j.ajkd.2004.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.