Abstract
Application of next-generation sequencing (NGS) methods for transcriptome analysis (RNA-seq) has become increasingly accessible in recent years and are of great interest to many biological disciplines including, eg, evolutionary biology, ecology, biomedicine, and computational biology. Although virtually any research group can now obtain RNA-seq data, only a few have the bioinformatics knowledge and computation facilities required for transcriptome analysis. Here, we present TRUFA (TRanscriptome User-Friendly Analysis), an open informatics platform offering a web-based interface that generates the outputs commonly used in de novo RNA-seq analysis and comparative transcriptomics. TRUFA provides a comprehensive service that allows performing dynamically raw read cleaning, transcript assembly, annotation, and expression quantification. Due to the computationally intensive nature of such analyses, TRUFA is highly parallelized and benefits from accessing high-performance computing resources. The complete TRUFA pipeline was validated using four previously published transcriptomic data sets. TRUFA’s results for the example datasets showed globally similar results when comparing with the original studies, and performed particularly better when analyzing the green tea dataset. The platform permits analyzing RNA-seq data in a fast, robust, and user-friendly manner. Accounts on TRUFA are provided freely upon request at https://trufa.ifca.es.
Keywords: transcriptomics, RNA-seq, de novo assembly, read cleaning, annotation, expression quantification
Introduction
Since the introduction of the RNA-seq methodology around 2006,1–6 studies based on whole transcriptomes of both model and non-model species have been flourishing. RNA-seq data are widely used for discovering novel transcripts and splice variants, finding candidate genes, or comparing differential gene expression patterns. The applications of this technology in many fields are vast,1,7 including researches on, eg, splicing signatures of breast cancer,8 host–pathogen interactions,9 the evolution of the frog immunome,10 the plasticity of butterfly wing patterns,11 the study of conotoxin diversity in Conus tribblei,12 and the optimization of trimming parameters for de novo assemblies.13
Despite the tremendous decrease in sequencing costs, which allows virtually any laboratory to obtain RNA-seq data, transcriptome analyses are still challenging and remain the main bottleneck for the widespread use of this technology. User-friendly applications are scarce,14 and the post-analysis of generated sequence data demands appropriate bioinformatics know-how and suitable computing infrastructures.
When a reference genome is available, which is normally the case for model system species, a reference-guided assembly is preferable to a de novo assembly. However, an increasing number of RNA-seq studies are performed on non-model organisms with no available reference genome for read mapping (particularly those studies focused on comparative transcriptomics above the species level), and thus require a de novo assembly approach. Moreover, when a reference genome is available, combining both de novo and reference-based approaches can lead to better assemblies.15,16 Analysis pipelines encompassing de novo assemblies are varied, and generally include steps such as cleaning and assembly of the reads, annotation of transcripts, and gene expression quantification.16 A variety of software programs have been developed to perform different steps of the RNA-seq analysis,17–19 but most of them are computationally intensive. The vast majority of these programs run solely with command lines. Processing the data to connect one step to the next in RNA-seq pipelines can be cumbersome in many instances, mainly due to the variety of output formats produced and the postprocessing needed to accept them further as input. Moreover, as soon as a large computing effort is required, interactive execution is usually not feasible and an interface with the underlying batch systems used in clusters or supercomputers is needed. In order to provide users with such a bioinformatics tool that solve the above-mentioned problems, we have developed TRUFA (TRanscriptomes User-Friendly Analysis), an informatics platform for RNA-seq data analysis, which runs on the ALTAMIRA supercomputer at the Instituto de Fisica de Cantabria (IFCA), Spain.20 The platform is highly parallelized both at the pipeline and program level. It can access up to 256 cores per execution instance for certain components of the pipeline. On top of allowing the user to obtain results in a relatively short time thanks to HPC (high-performance computing) resources, TRUFA is an integrative and graphical web tool for performing the main and most computationally demanding steps of a de novo RNA-seq analysis.
The first step of a de novo RNA-seq analysis consists in assessing data quality and cleaning raw reads. The output of a next-generation sequencing (NGS) reaction contains traces of polymerase chain reaction (PCR) primers and sequencing adapters as well as poor-quality bases/reads. Hence, it is advised to perform read trimming, which has been shown to have a positive effect on the rest of the RNA-seq analysis,21 although parameter values for such trimming have to be optimized.13
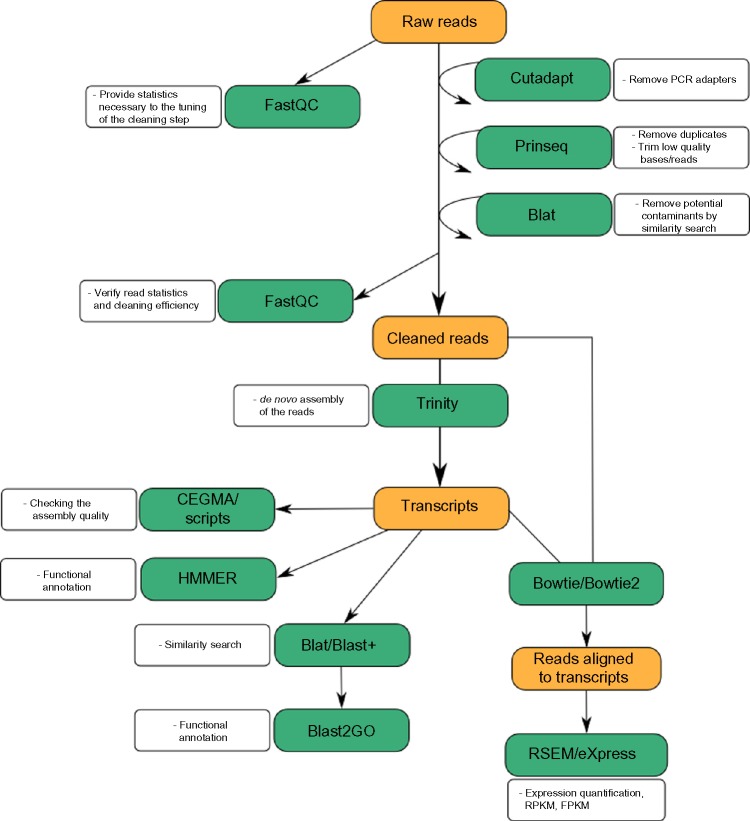
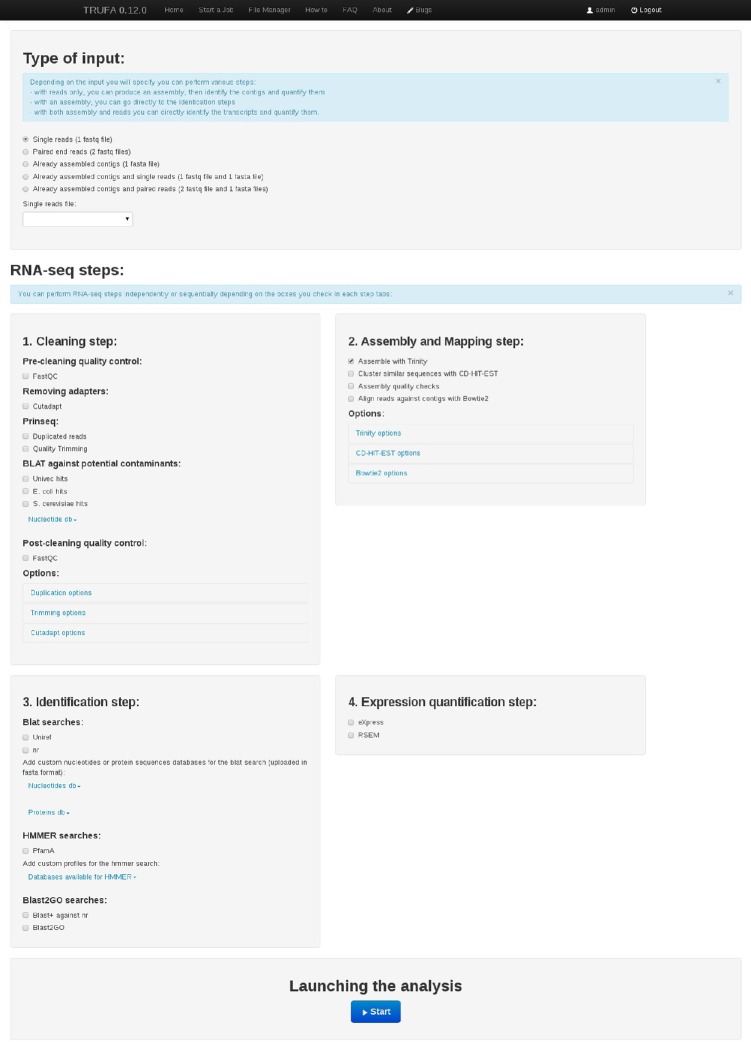
Once reads have been cleaned, they are assembled into transcripts, which are subsequently categorized into functional classes in order to understand their biological meaning. Finally, it is possible to perform expression quantification analyses by estimating the amount of reads sequenced per assembled transcript and taking into account that the number of reads sequenced theoretically correlates with the number of copies of the corresponding mRNA in vivo.6 All the above-mentioned steps in the RNA-seq analysis pipeline are included in TRUFA and correspond to distinct sections in the web-based user interface (see Figs. 1 and 2). For each step, the options available are those that are either critical to the analysis or, to our knowledge, the most widely used in the literature.
Figure 1.
Overview of the TRUFA pipeline.
Figure 2.
Snapshot of the TRUFA web page for running RNA-seq analysis.
There are several online platforms already available to perform different parts of a RNA-seq analysis. For example, Galaxy (https://usegalaxy.org/)22 allows analyzing RNA-seq data with a reference genome (using Tophat23 and Cufflinks24), whereas GigaGalaxy (http://galaxy.cbiit.cuhk.edu.hk/) can produce de novo assemblies using SOAPdenovo.25 Another transcriptome analysis package integrated in Galaxy, Oqtans,26 provides numerous features including de novo assembly with Trinity, read mapping, and differential expression. Nonetheless, to our knowledge, GigaGalaxy or Oqtans do not perform de novo annotations. Conversely, Fastannotator27 is a platform specialized in transcript annotations using Blast2GO,28 PRIAM,29 and domain identification pipelines, but does not perform other steps of the RNA-seq analysis.
The TRUFA platform has been designed to be interactive, user-friendly, and to cover a large part of a RNA-seq analysis pipeline. Users can launch the pipeline from raw or cleaned Illumina reads as well as from already assembled transcripts. Each of the implemented programs (Table 1) can be easily integrated into the analysis and tuned depending on the needs of the user. TRUFA provides a comprehensive output, including read quality reports, cleaned read files, assembled transcript files, assembly quality statistics, Blast, Blat, and HMMER search results, read alignment files (BAM files), and expression quantification files (including values of read counts, expected counts, and TPM, ie, transcripts per million30). Some outputs can be directly visualized from the web server, and all outputs can be downloaded in order to locally perform further analyses such as single nucleotide polymorphisms (SNPs) calling and differential expression quantification. The platform is mainly written in Javascript, Python, and Bash. The source code is available at Github (https://github.com/TRUFA-rnaseq). The long-term availability of the TRUFA web server (and further developed versions) is ensured given that it is currently installed in the ALTAMIRA supercomputer, a facility integrated in the Spanish Supercomputing Network (RES). The number of users is currently not limited and accounts are freely provided upon request.
Table 1.
List of available software on TRUFA.
| RNA-SEQ STEPS | AVAILABLE PROGRAMS | VERSIONS |
|---|---|---|
| Read cleaning | PRINSEQ | 0.20.3 |
| CUTADAPT | 1.3 | |
| BLAT | v.35 | |
| Assembly and mapping | Trinity | r2012–06–08 |
| CD-HIT | 4.5.4 | |
| CEGMA | 2.4 | |
| Bowtie | 0.12.8 | |
| Bowtie2 | 2.0.2 | |
| Annotation | BLAT | v.35 |
| HMMER | 3.0 | |
| Blast+ | 2.2.28 | |
| Blast2GO | 2.5.0 | |
| Expression quantification | RSEM | 1.2.8 |
| eXpress | 1.5.1 |
Implementation
The overall workflow of TRUFA is shown in Figure 1. The input, output, and different components of the pipeline are the following:
Input
Currently, the input data accepted by TRUFA includes Illumina read files and/or reads already assembled into contigs. Read files should be in FASTQ format and can be uploaded as gzip compressed files (reducing uploading times). Reads from the NCBI SRA databases can be used but should be first formatted into FASTQ format using, eg, the SRA toolkit.31 Already assembled contigs should be uploaded as FASTA files. Other FASTA files and HMM profiles can be uploaded as well for custom blast-like and protein profile-based transcript annotation steps, respectively. Thus far, no data size limitation is set.
Pipeline
Several programs can be called during the cleaning step (Table 1). The program FASTQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc) has been implemented to assess the quality of raw reads and give the statistics necessary to tune cleaning parameters (Fig. 1). After the quality of the data is determined, CUTADAPT32 and PRINSEQ33 allow, among other functionalities, the removal of adapters as well as low quality bases/reads. In particular, PRINSEQ has been chosen for its ability to treat both single and paired-end reads and to perform read quality trimming as well as duplicate removal. Using the BLAT fast similarity search tool, reads can be compared against databases of potential contaminants such as, eg, UniVec (which allows identifying sequences of vector origin; http://www.ncbi.nlm. nih.gov/VecScreen/UniVec.html) or user-specified databases. TRUFA’s scripts will automatically remove those reads, giving hits with such queried databases.
Cleaned reads, after passing an optional second quality control with FASTQC to verify the overall efficiency of the first cleaning step, are ready for assembly. TRUFA implements the software Trinity,34 which is an extensively used de novo assembler and has been shown to perform better than other single k-mer assemblers.35 After the assembly, an in-house script provides basic statistics describing transcripts lengths distribution, total bases incorporated in the assembly, N50, and GC content. In addition, to evaluate the completeness of the assembly, a Blast+36 similarity search is performed against the UniProtKB/Swiss-Prot database, and a Trinity script evaluates whether those assembled transcripts with hits are full-length or nearly full-length. The CEGMA software can also provide a measure of the completeness of the assembly by comparing the transcripts to a set of 248 core eukaryotic genes, which are conserved in highly divergent eukaryotic taxa.37 Both the number of recovered genes from the total of 248 and their completeness have been used for de novo assembly quality assessments.38,39
The newly assembled transcripts can be used as query for similarity searches with BLAT40 or Blast+ against the NCBI nr and UniRef90 databases. In parallel, HMMER41 searches can be performed applying hidden markov models (HMM) against the PFAM-A database. Both analysis can be run as well with user-specified databases or models respectively. Further annotation and assignation of gene ontology (GO) terms can be obtained with Blast2GO28 for the transcripts with blast hits against the nr database.
For expression quantification, Bowtie242 is used to produce alignments of the reads against the assembled transcripts. Alignments are then properly formatted using SAMtools43 and Picard (http://broadinstitute.github.io/picard/).43 Using these alignments, eXpress44 can be used to quantify the expression of all isoforms. Additionally, the script “run_RSEM_align_n_estimate” of the Trinity package implemented in TRUFA uses Bowtie45 and RSEM46 to provide an alternative procedure for expression quantification of both genes and isoforms. Moreover, the percentage of reads mapping back to the assembled transcripts (obtained with Bowtie and Bowtie2) can be used as another indication of the assembly quality.35,38
Output
TRUFA generates a large amount of output information from the different programs used in the customized pipeline. Briefly, a user should be able to download FastQC html reports, FASTQ files with cleaned reads (without duplicated reads and/or trimmed), Trinity-assembled transcripts (FASTA), read alignments against the transcripts (BAM files), GO annotations (.txt and.dat files which can be imported into the Blast2GO java application), and read counts (text files providing read counts and TPM). Various statistics are computed at each step and are reported in text files, such as the percentage of duplicated/trimmed reads, CEGMA completeness report, assembly sequence composition, percentage of mapped reads, and read count distributions.
Results and Discussion
We have built an informatics platform that performs a nearly complete de novo RNA-seq analysis in a user-friendly manner (amenable to the nonexpert user, avoiding command lines, and providing a lightweight visual interface), and tested its performance using four publicly available transcriptome datasets. A small dataset of the fission yeast, Schizosaccharomyces pombe, which is provided in a published Trinity tutorial,47 was used to test the correct functioning of the assembly process on TRUFA. Two previously well-characterized datasets from the green tea, Camelia sinensis (SRX020193), and the fruit fly, Drosophila melanogaster (SRR023199, SRR023502, SRR023504, SRR023538, SRR023539, SRR023540, SRR023600, SRR023602, SRR023604, SRR027109, SRR027110, SRR027114 and SRR035403), were used to compare assembly and read mapping statistics with the results from Zhao et al.35 Finally, TRUFA was tested using a rice (Oryza sativa) dataset48,49 (SRX017630, SRX017631, SRX017632, SRX017633). When applicable, reads corresponding to each end of a pair-ended reaction were concatenated separately into two files, and all files were compressed with gzip before uploading to the platform. Each of the compressed read files was uploaded to TRUFA in less than a day (typical uploading times from a personal computer anywhere ranging from 30 seconds to 12 hours for files ranging from 200 MB to 12 GB, ie, between 0.25 and 25 Gbp).
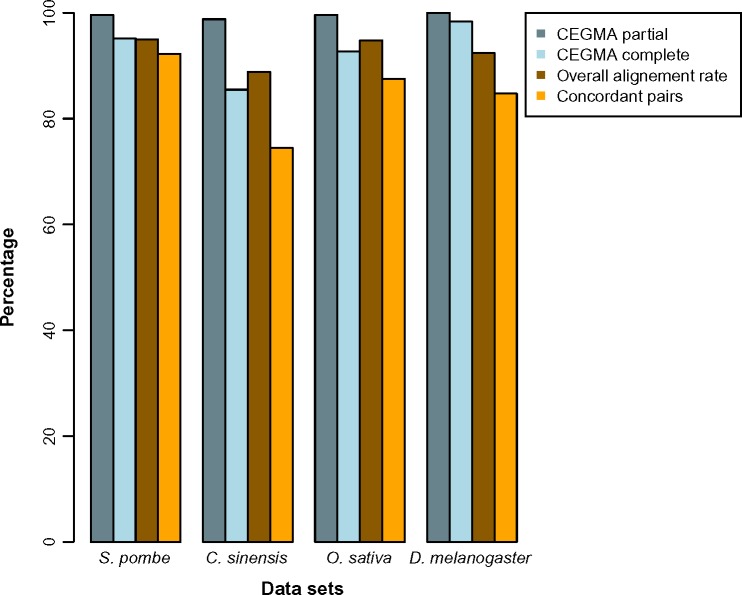
The results of a first run performing only a FASTQC analysis were used to set the parameters (see Supplementary Table 1) for the cleaning process, except for the yeast dataset, which was assembled without preprocessing. Read cleaning, assembly, mapping, and annotation statistics are shown in Tables 2 and 3. The yeast dataset showed highly similar results to the original analysis, validating the TRUFA assembly. The difference observed in the number of transcripts is most likely due to the not fully deterministic nature of the Trinity algorithm.47 However, the percentage of reads mapped back to the transcripts was slightly higher in the original study.47 For the other three datasets, TRUFA showed globally comparable results. Except for the mean transcript length for the C. sinensis assembly, all other statistics for both C. sinensis and D. melanogaster assemblies were higher in the present analyses with respect to the original ones (Table 2). Remarkably, the percentage of reads mapping back to the transcripts was significantly higher for the green tea dataset using TRUFA. This could be due to a more efficient read-cleaning step or to differences between Bowtie2 (used in TRUFA) and Bowtie used by Zhao et al (2011) mappings. CEGMA analysis showed that more than 80% (range 85.5%–98.39%) of the core eukaryotic genes are fully recovered and more than 98% (range 98.8%–100%) are partially recovered in all dataset assemblies (Fig. 3). This indicates an overall high completeness of the assemblies performed herein with TRUFA. In addition to the assembly and the mapping of the reads, TRUFA was able to annotate de novo 25%–42% of the transcripts using the Blat, Blast+, and Blast2GO pipeline with an e-value of <10−6 (Table 3). HMMER searches identified 17%–60% of the transcripts with at least one hit with an e-value <10−6. The expression of each transcript was quantified using RSEM and eXpress, although no data were available for comparison with the original studies.
Table 2.
Comparison of outputs between original and TRUFA analyses.
| NO. OF RAW BASES | S. pombe | C. sinensis | O. sativa | D. melanogaster | ||||
|---|---|---|---|---|---|---|---|---|
| PESS | PE | PE | PE | |||||
| 544M | 2320M | 5983M | 24740M | |||||
| Pipeline | Trufa | Haas et al (2013) | Trufa | Zhao et al (2011) | Trufa | Xie et al (2014) | Trufa | Zhao et al (2011) |
| No. of bases after cleaning | No cleaning | No cleaning | 2,017M | NA | 5,342M | NA | 5,028M | NA |
| No. of transcripts | 9,370 | 9,299 | 201,892 | 188,950 | 166,512 | 170,880 | 80,999 | 70,906 |
| Mean transcript length | 1,014 | NA | 319 | 332 | 480 | 552 | 847 | 751 |
| No. of bases in the assembly | 9M | NA | 64M | 63M | 80M | 94M | 69M | 53M |
| N50 | 1,585 | 1,585 | 542 | 525 | 1,205 | 1,392 | 2,960 | 2,499 |
| No. of transcripts >1000 nt | 3,680 | NA | 13,276 | 12,495 | 22,317 | 28,578 | 17,251 | 12,511 |
| Total alignment rate | 94.98% | 99.93% | 88.84% | 61.04% | 94.76% | NA | 92.39% | 89.9 |
| Concordant pairs | 92.21% | 93.12% | 74.45% | NA | 87.51% | NA | 84.73% | NA |
Note: Concordant pairs are considered when they report at least one alignment.
Abbreviations: PE, Paired-end; SS, strand-specific; M, million; NA, data not available.
Table 3.
Summary of the de novo annotation step for the four assembled transcriptomes.
| S. pombe | C. sinensis | O. sativa | D. melanogaster | |
|---|---|---|---|---|
| # transcripts | 9,370 | 201,892 | 166,512 | 80,999 |
| # Blast Hits | 8,257 | 72,559 | 66,129 | 29,924 |
| # Annotations | 3,922 | 51,272 | 50,721 | 22,534 |
| % of annotated transcripts | 42% | 25% | 30% | 28% |
| # HMMER hits | 5,588 | 34,689 | 28,736 | 16,552 |
| User time | 11 h | 3 d 19 h | 6 d 8 h | 4 d 15 h |
Notes: # Transcripts, number of transcripts assembled by Trinity; # Blast hits, number of transcripts with at least one hit against the NCBI nr database (e-value <10−6); # Annotation, number of transcripts with at least one annotation after Blast2GO analysis; # HMMER hits, number of transcripts with at least one hit against the Pfam A database (e-value <10−6); User time, time needed to perform the complete pipeline (cleaning, assembly, annotation, and expression quantification).
Figure 3.
Measures of completeness and read usage for the assemblies produced with TRUFA. CEGMA results represent the percentage of completely and partially recovered genes in the assemblies for a subset of 248 highly conserved core eukaryotic genes. Overall alignment rate and concordant pairs (providing at least one alignment) were computed with Bowtie2.
Considering the entire pipeline, each testing dataset was analyzed by TRUFA in less than a week (Table 3), confirming a good time efficiency of the platform. According to Macmanes13 on the effect of read trimming for RNA-seq analysis, optimizing trimming parameters leads to better assembly results. This optimization should take no longer than 3 days of computation for datasets such as the ones used here and can be easily done with TRUFA by producing in parallel various assemblies and their quality statistics with different sets of trimming parameters and parameter values.
In Prospect
To complete the RNA-seq analysis pipeline available in TRUFA, we plan to expand the platform by incorporating programs for differential expression analysis and SNP calling. Other programs, especially for assembly (eg, SOAPdenovo-Trans, Velvet-Oases) and visualization (eg, GBrowse) of the data, are planned to be also included in the future. In addition, integrating GO terms for each annotated transcripts would permit the user to browse sequences of interest directly from the web server without the need to download large quantities of output. We also plan to complete the platform by providing features for read mapping against a reference genome (such as, eg, STAR,50 Tophat, and Cufflinks). A cloud version of TRUFA, which would increase considerably its global capabilities, is also envisioned to be run in the EGI.eu Federated Cloud (see https://www.egi.eu/infrastructure/cloud/) in the near future.
Conclusion
We presented TRUFA, a bioinformatics platform offering a web interface for de novo RNA-seq analysis. It is intended for scientists analyzing transcriptome data who do not have either bioinformatics skills or access to fast computing services (or both). TRUFA is essentially a wrapper of various widely used RNA-seq analysis tools, allowing the generation of RNA-seq outputs in an efficient, consistent, and user-friendly manner, based on a pipeline approach.
The trimming and assembly steps are guided by the integration of widely used quality control programs toward the optimization of the assembly process. Moreover, the implementation of HMMER, BLAST+, and Blast2GO to the platform for de novo annotation is, to our knowledge, a feature not available in other RNA-seq analysis web servers such as GigaGalaxy or Oqtans. This step is the most computationally demanding among all RNA-seq analysis steps (including SNPs calling and differential expression), and TRUFA uses highly parallelized steps to obtain annotations in a relatively short time frame. Although annotations can be performed in other platforms such as FastAnnotator, having all these steps from cleaning to annotations and expression quantification in the same pipeline reduces unnecessary transfer of large outputs and provides an advantage to the nonexpert user.
Data Accessibility
TRUFA platform, user manual, example data sets and tutorial videos are accessible at the web page https://trufa.ifca.es/web. Accession numbers to the read files used in this study are provided in the Results and Discussion section and can be obtained from http://www.ncbi.nlm.nih.gov/sra/.
Supplementary Material
Supplementary Table 1. List of the main command lines used for the analysis of each data sets. Datasets: 1, S. pombe; 2, C. sinensis; 3, O. sativa; 4, D. melanogaster.
Acknowledgments
We would like to thank Beatríz Ranz for her help during the validation process and Iván Cabrillo for his help managing the accounts on the Altamira supercomputer. We are grateful to Federico Abascal for his input in the review process, his suggestions, and his help during the beta testing. Thanks to all other beta testers: Anna María Addamo, Carlos Canchaya, Michel Domínguez, Iván Gómez-Mestre, Iker Irisarri, Nathan Kenny, Diushi Keri, David Osca, Snæbjörn Pálsson, Sara Rocha, Diego San Mauro, Maria Torres, Juan E. Uribe, Ignacio Varela, and Joel Vizueta. We thank five anonymous reviewers for their valuable feedback on a previous version of the manuscript. The platform was developed thanks to the use of bootstrap (http://getbootstrap.com/), jquery (http://jquery.com/), sqlite (https://sqlite.org/), webpy (http://webpy.org/), and filemanager (https://github.com/simogeo/Filemanager). The Altamira supercomputer is member of the Spanish Supercomputing Network.
Glossary
Abbreviations
- TRUFA
transcriptome user-friendly analysis
- TPM
transcripts per million
- SNP
single nucleotide polymorphism
- HPC
high performance computing
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
FUNDING: This work was partially funded with Spanish Ministry of Science and Innovation grants CGL2010–18216 and CGL2013–45211-C2–2-P to RZ and CGL2013–45211-C2–1-P to JR. JR was partially supported by ICREA Academia (Generalitat de Catalunya; Spain). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived the study: RZ, EK. Constructed the pipeline: EK. Performed testing runs: EK, CFL. Implemented the web version: LC, FA, EK, JM. Tuned parts of the pipeline: CFL, JR. All authors contributed to the writing and improving of the manuscript, and read and approved the final version.
REFERENCES
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marguerat S, Wilhelm BT, Bähler J. Next-generation sequencing: applications beyond genomes. Biochem Soc Trans. 2008;36(pt 5):1091–6. doi: 10.1042/BST0361091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol. 2009;12(2):107–18. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48(3):249–57. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Bainbridge MN, Warren RL, Hirst M, et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 7.Marguerat S, Bähler J. RNA-seq: from technology to biology. Cell Mol Life Sci. 2010;67(4):569–79. doi: 10.1007/s00018-009-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eswaran J, Horvath A, Godbole S, et al. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep. 2013;3:1689. doi: 10.1038/srep01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nat Rev Microbiol. 2012;10(9):618–30. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- 10.Savage AE, Kiemnec-Tyburczy KM, Ellison AR, Fleischer RC, Zamudio KR. Conservation and divergence in the frog immunome: pyrosequencing and de novo assembly of immune tissue transcriptomes. Gene. 2014;542(2):98–108. doi: 10.1016/j.gene.2014.03.051. [DOI] [PubMed] [Google Scholar]
- 11.Daniels EV, Murad R, Mortazavi A, Reed RD. Extensive transcriptional response associated with seasonal plasticity of butterfly wing patterns. Mol Ecol. 2014;23(24):6123–34. doi: 10.1111/mec.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barghi N, Concepcion GP, Olivera BM, Lluisma AO. High conopeptide diversity in Conus tribblei revealed through analysis of venom duct transcriptome using two high-throughput sequencing platforms. Mar Biotechnol. 2014;17(1):81–98. doi: 10.1007/s10126-014-9595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macmanes MD. On the optimal trimming of high-throughput mRNA sequence data. Front Genet. 2014;5:13. doi: 10.3389/fgene.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DR. The battle for user-friendly bioinformatics. Front Genet. 2013;4:187. doi: 10.3389/fgene.2013.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain P, Krishnan NM, Panda B. Augmenting transcriptome assembly by combining de novo and genome-guided tools. Peer J. 2013;1:e133. doi: 10.7717/peerj.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12(10):671–82. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 17.Bao S, Jiang R, Kwan W, Wang B, Ma X, Song YQ. Evaluation of next-generation sequencing software in mapping and assembly. J Hum Genet. 2011;56(6):406–14. doi: 10.1038/jhg.2011.43. [DOI] [PubMed] [Google Scholar]
- 18.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8(6):469–77. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 19.Lee HC, Lai K, Lorenc MT, Imelfort M, Duran C, Edwards D. Bioinformatics tools and databases for analysis of next-generation sequence data. Brief Funct Genomics. 2012;11(1):12–24. doi: 10.1093/bfgp/elr037. [DOI] [PubMed] [Google Scholar]
- 20.Cabrillo I, Cabellos L, Marco J, Fernandez J, Gonzalez I. Direct exploitation of a top 500 supercomputer for analysis of CMS data. J Phys Conf Ser. 2014;513(3):032014. [Google Scholar]
- 21.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One. 2013;8(12):e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreedharan VT, Schultheiss SJ, Jean G, et al. Oqtans: the RNA-seq workbench in the cloud for complete and reproducible quantitative transcriptome analysis. Bioinformatics. 2014;30(9):1300–1. doi: 10.1093/bioinformatics/btt731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen TW, Gan RC, Wu TH, et al. FastAnnotator – an efficient transcript annotation web tool. BMC Genomics. 2012;13(suppl 7):S9. doi: 10.1186/1471-2164-13-S7-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 29.Claudel-Renard C. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31(22):6633–9. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131(4):281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 31.Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39(Database issue):D19–21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10. [Google Scholar]
- 33.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–4. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 2011;12(suppl 14):S2. doi: 10.1186/1471-2105-12-S14-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–7. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 38.Moreton J, Dunham SP, Emes RD. A consensus approach to vertebrate de novo transcriptome assembly from RNA-seq data: assembly of the duck (Anas platyrhynchos) transcriptome. Front Genet. 2014;5:190. doi: 10.3389/fgene.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One. 2013;8(3):e59534. doi: 10.1371/journal.pone.0059534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ. BLAT – The BLAST-like alignment tool. Genome Res. 2002;12(4):656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10(1):71–3. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory- efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haas BJ, Papanicolaou A, Yassour M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Y, Wu G, Tang J, et al. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 2014;30(12):1660–6. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 49.Zhang G, Guo G, Hu X, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010;20(5):646–54. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. List of the main command lines used for the analysis of each data sets. Datasets: 1, S. pombe; 2, C. sinensis; 3, O. sativa; 4, D. melanogaster.
Data Availability Statement
TRUFA platform, user manual, example data sets and tutorial videos are accessible at the web page https://trufa.ifca.es/web. Accession numbers to the read files used in this study are provided in the Results and Discussion section and can be obtained from http://www.ncbi.nlm.nih.gov/sra/.