Abstract
While the genomes of many Mycoplasma species have been sequenced, there are no collated data on translational start codon usage, and the effects of alternate start codons on gene expression have not been studied. Analysis of the annotated genomes found that ATG was the most prevalent translational start codon among Mycoplasma spp. However in Mycoplasma gallisepticum a GTG start codon is commonly used in the vlhA multigene family, which encodes a highly abundant, phase variable lipoprotein adhesin. Therefore, the effect of this alternate start codon on expression of a reporter PhoA lipoprotein was examined in M. gallisepticum. Mutation of the start codon from ATG to GTG resulted in a 2.5 fold reduction in the level of transcription of the phoA reporter, but the level of PhoA activity in the transformants containing phoA with a GTG start codon was only 63% of that of the transformants with a phoA with an ATG start codon, suggesting that GTG was a more efficient translational initiation codon. The effect of swapping the translational start codon in phoA reporter gene expression was less in M. gallisepticum than has been seen previously in Escherichia coli or Bacillus subtilis, suggesting the process of translational initiation in mycoplasmas may have some significant differences from those used in other bacteria. This is the first study of translational start codon usage in mycoplasmas and the impact of the use of an alternate start codon on expression in these bacteria.
Introduction
Although the number of genes involved in cellular metabolism have been reduced in mycoplasmas, the number of genes involved in transcription and translation are comparable to those of other bacteria [1, 2], and the basic mechanisms of transcription and translation in mycoplasmas are thought to be similar to those of low G+C Gram-positive bacteria. Translation is a complex process that involves four phases—initiation, elongation, termination and ribosome recycling [3]. Translational initiation in bacteria is influenced by a range of factors, including the ribosomal binding site (RBS) [4], mRNA secondary structure near the ribosomal binding site [5], the translational start codon [6] and the sequence downstream of the start codon [7]. In the E. coli K-12 genome, of the 4288 open reading frames annotated, 82% have an ATG start codon, 14.3% a GTG start codon and 3% a TTG start codon [8]. A recent annotation of ten different E. coli strains found that 82.5% of the start codons were ATG, 12.3% were GTG and 5% were TTG, with CTG, ATT and ATC used at lower frequencies [9]. Translational efficiency has been shown to decrease in E. coli when a start codon other than ATG is used, with an eight-fold reduction in translation seen with GTG or TTG start codons [10]. In B. subtilis ATG, TTG and GTG start codons are used in 78%, 13% and 9% of CDSs, respectively [11], with GTG shown to be three- to five-fold less efficient than ATG in translational initiation [12].
Mollicutes generally have genomes with a low G+C content and, as a result, an A or U(T) bias in codon usage, especially in the first and third codon positions. While the coding region of most mycoplasma genes begins with an ATG start codon, alternative start codons, such as GTG and TTG, are used [13]. In the genome of M. gallisepticum Rlow, based on in silico prediction of coding sequences, ATG is the preferred start codon, followed by GTG and TTG (http://services.cbib.u-bordeaux2.fr/molligen/). However, all the genes in the vlhA multigene family, which encodes the abundant phase variable lipoprotein adhesion VlhA, have a GTG translational initiation codon. The high prevalence of an atypical translational initiation codon in gene family encoding a highly expressed lipoprotein in a species with an overall bias against such GC rich codons suggests that this codon may not have the adverse effects on translation of the vlhA genes in M. gallisepticum that are seen in other genes in other bacterial species. In order to explore whether use of a GTG translational initiation codon was more common in mycoplasmas than in other bacterial species, we initially examined the annotated genomes of a number of Mycoplasma species to determine the prevalence of use of alternative translational start codons in predicted coding sequences in this family of bacteria. At present the genomes of 52 Mycoplasma species have been annotated, but there are no collated data on start codon usage in mycoplasmas. We then assessed the effect of the GTG translational start codon on transcription and translation of a reporter lipoprotein gene in M. gallisepticum.
Methods
Bacterial strains and culture conditions
M. gallisepticum strain S6 was grown in mycoplasma broth or on mycoplasma agar at 37°C, with 16 μg gentamicin (Invitrogen)/ml included in the media to select for transformants. E. coli DH5α cells were used as the host for genetic manipulation and cloning of plasmids.
Construction of plasmids
Plasmid ltufacyphoA (pTAP)
The promoter region of the gene of elongation factor Tu (ltuf) of M. gallisepticum, the leader sequence and acylation sequence of the vlhA1.1 gene from the genomic DNA of M. gallisepticum strain S6 and the E. coli phoA gene, which codes for the E. coli alkaline phosphatase, were ligated into transposon Tn4001 in pISM2062.2, generating the pISM2062.2ltufacyphoA plasmid (pTAP) [14].
Plasmid ltufGTGacyphoA (pTGP)
The ATG translational start codon of the reporter construct in the pTAP plasmid was mutated to GTG by overlap extension PCR. The ltuf promoter and the translational start codon were amplified as a 357 bp product by PCR using the IRF (5′ GGCCGgGATCAAGTCCGTATTATTGTGTAAAAGTgCtaGc 3′) and GTGR (5′ CTTTAAAATGTTTTTTCTCTTCAcTTTTTTAAATATTTCTCC 3′) oligonucleotide primers. Mutation of the first nucleotide of the start codon was achieved by incorporating this change in the GTGR primer.
The translational start codon, the vlhA1.1 signal sequence and the phoA coding sequence were amplified as a product of 1,479 bp from the pTAP plasmid using the oligonucleotide primers GTGF (5′ GGAGAAATATTTAAAAAAgTGAAGAGAAAAAACATTTTAAAG 3′) and PBgR (5′ CCGaGATctaAAAGGACTGttaTATGGCCTTTTTATTTTATTTCAGCCCCAGA 3′). The PCR products were purified after electrophoresis in a 1% agarose gel using the Qiaex gel extraction kit (Qiagen) and joined by overlap extension PCR using the primers IRF and PBgR, resulting in the 1,792 bp ltufGTGacyphoA product.
The resultant PCR product was gel purified and ligated into pGEM-T (Promega) following the manufacturer’s instructions. An E. coli transformant containing a plasmid of the expected size was selected and the insert DNA sequence confirmed using BigDye terminator v3.1 cycle sequencing (Perkin Elmer Applied Biosystems) and the M13 universal primers. The 1,620 bp DNA insert was released from pGEM-T by digestion with the restriction endonucleases NheI and SphI, purified using the Qiaex gel extraction kit and ligated into similarly digested pTAP, resulting in pISM2062.2ltufGTGacyphoA (pTGP).
The plasmid pTGP was introduced into E. coli DH5α by electroporation using a Gene Pulser (Bio-Rad) with settings of 2.5 kV and 25 μF. Transformants were selected for ampicillin resistance and the clones were screened for the presence of the gentamicin resistance gene in the transposon by PCR using the oligonucleotide primers GmF and GmR, which yielded a 223 bp product [14]. Selected clones were cultured in larger volumes and plasmid DNA was extracted using a Midi prep kit (Qiagen) according to the manufacturer’s instructions. The DNA sequence of the pTGP plasmid was confirmed using BigDye terminator v3.1 cycle sequencing (Perkin Elmer Applied Biosystems) and the plasmid was then used to transform M. gallisepticum cells by electroporation and the colonies obtained picked and grown in mycoplasma broth containing gentamicin, as described previously [14]. The presence of the gentamicin resistance gene was confirmed by PCR using the oligonucleotide primers GmF and GmR.
PCR
The thermal cycling conditions for amplification of DNA sequences by PCR and quantitative RT-PCR were as described previously [14].
Insertion points of transposon constructs
To determine the insertion site of the Tn4001 transposon, genomic DNA sequencing was carried out using the ABI Prism BigDye Terminator v3.1 sequencing system (Perkin Elmer Applied Biosystems) and the UBR oligonucleotide primer (5′ GCAGTAATATCGCCCTGAGC 3′) [14].
Detection of PhoA
Immunoblotting, localisation of PhoA (Triton X-114 fractionation, membrane fractionation and trypsin proteolysis) and alkaline phosphatase assays were performed as described previously [14].
Results
Analysis of translational start codons
The data on mycoplasma genomes were downloaded from the NCBI repository of completely sequenced bacterial genomes at ftp://ftp.ncbi.nih.gov/genomes/Bacteria/. The directory on the NCBI site was used to collate the information on start codons. GeneMark 2.5m was used to import the the sequence, G+C content and length of the sequence. The CDS data were obtained from the NC.rpt file. The start codon annotations from Prodigal 2.50 (Prokaryotic Dynamic programming Gene-finding Algorithm) [15] were downloaded and the count of the different start codons compiled. Prodigal detects only the 3 major start codons, AUG, GUG and UUG, with the non-standard codons AUU, AUA and CUG not considered. The tabulated frequencies of use of the different translational initiation codons in mycoplasmas are shown in Table 1. Based on these bioinformatic data, in mycoplasmas ATG was used in 87% of CDSs, GTG in 8% and TTG in 5%. In different M. gallisepticum strains, 85% of CDSs used ATG, 10% used GTG and 5% used TTG. The G+C content of the mycoplasma genomes varied from 24% to 40%. There was a modest correlation between G+C content and use of an ATG start codon (R2 = 0.4).
Table 1. Translational start codon usage in genomes of Mollicutes.
| Species | Proportion (%) of CDSs predicted to use start codon: | % G+C | No. CDSs | Accession No. | ||
|---|---|---|---|---|---|---|
| ATG | GTG | TTG | ||||
| Acholeplasma laidlawii PG-8A | 92.58 | 2.88 | 4.54 | 31.08 | 1380 | NC_010163.1 |
| Mycoplasma agalactiae PG2 | 93.49 | 2.05 | 4.46 | 29.62 | 742 | NC_013948.1 |
| Mycoplasma arthritidis 158L3-1 | 94.32 | 4.10 | 1.58 | 30.71 | 631 | NC_011025.1 |
| Mycoplasma bovis Hubei-1 | 95.14 | 2.24 | 2.62 | 29.29 | 801 | NC_015725.1 |
| Mycoplasma bovis PG45 | 95.45 | 1.67 | 2.87 | 29.31 | 765 | NC_014760.1 |
| Mycoplasma capricolum subsp. capricolum ATCC 27343 | 96.19 | 2.74 | 1.07 | 23.77 | 812 | NC_007633.1 |
| Mycoplasma conjunctivae HRC/581 | 93.46 | 2.67 | 3.86 | 28.49 | 692 | NC_012806.1 |
| Mycoplasma crocodyli MP145 | 97.23 | 0.79 | 1.98 | 26.95 | 689 | NC_014014.1 |
| Mycoplasma fermentans JER | 94.15 | 2.39 | 3.46 | 26.95 | 797 | NC_014552.1 |
| Mycoplasma fermentans M64 | 93.32 | 2.53 | 4.15 | 26.86 | 1050 | NC_014921.1 |
| Mycoplasma gallisepticum F | 89.40 | 7.79 | 2.81 | 31.4 | 756 | NC_017503.1 |
| Mycoplasma gallisepticum R(high) | 83.27 | 11.15 | 5.58 | 31.47 | 766 | NC_017502.1 |
| Mycoplasma gallisepticum R(low) | 83.62 | 10.67 | 5.71 | 31.47 | 763 | NC_004829.2 |
| Mycoplasma genitalium G37 | 86.88 | 9.51 | 3.61 | 31.69 | 475 | NC_000908.2 |
| Mycoplasma haemocanis Illinois | 80.19 | 13.38 | 6.44 | 35.33 | 1156 | NC_016638.1 |
| Mycoplasma haemofelis Ohio2 | 82.78 | 11.15 | 6.07 | 38.81 | 1527 | NC_017520.1 |
| Mycoplasma haemofelis Langford 1 | 83.35 | 11.22 | 5.42 | 38.85 | 1545 | NC_014970.1 |
| Mycoplasma hominis ATCC 23114 | 95.71 | 1.25 | 3.04 | 27.12 | 523 | NC_013511.1 |
| Mycoplasma hyopneumoniae 168 | 92.78 | 4.11 | 3.12 | 28.46 | 693 | NC_017509.1 |
| Mycoplasma hyopneumoniae 232 | 92.79 | 3.68 | 3.53 | 28.56 | 691 | NC_006360.1 |
| Mycoplasma hyopneumoniae 7448 | 93.50 | 3.61 | 2.89 | 28.49 | 657 | NC_007332.1 |
| Mycoplasma hyopneumoniae J | 91.93 | 4.61 | 3.46 | 28.52 | 657 | NC_007295.1 |
| Mycoplasma hyorhinis GDL-1 | 89.51 | 2.92 | 7.57 | 25.91 | 647 | NC_016829.1 |
| Mycoplasma hyorhinis HUB-1 | 89.33 | 3.43 | 7.25 | 25.88 | 658 | NC_014448.1 |
| Mycoplasma hyorhinis MCLD | 89.67 | 3.84 | 6.49 | 25.88 | 778 | NC_017519.1 |
| Mycoplasma leachii PG50 | 94.13 | 2.37 | 3.50 | 23.75 | 882 | NC_014751.1 |
| Mycoplasma mobile 163K | 94.55 | 2.58 | 2.88 | 24.95 | 633 | NC_006908.1 |
| Mycoplasma mycoides subsp. capri LC 95010 | 96.73 | 2.64 | 0.63 | 23.82 | 922 | NC_015431.1 |
| Mycoplasma mycoides subsp. mycoides SC PG1 | 83.90 | 10.62 | 5.48 | 23.97 | 1017 | NC_005364.2 |
| Mycoplasma penetrans HF-2 | 95.86 | 2.79 | 1.35 | 25.72 | 1037 | NC_004432.1 |
| Mycoplasma pneumoniae 309 | 81.82 | 12.65 | 5.53 | 39.98 | 707 | NC_016807.1 |
| Mycoplasma pneumoniae FH | 81.25 | 12.11 | 6.64 | 40.00 | 629 | NC_017504.1 |
| Mycoplasma pneumoniae M129 | 81.41 | 12.30 | 6.28 | 40.01 | 689 | NC_000912.1 |
| Mycoplasma pulmonis UAB CTIP | 92.45 | 4.77 | 2.78 | 26.64 | 782 | NC_002771.1 |
| Mycoplasma putrefaciens KS1 | 92.86 | 5.00 | 2.14 | 26.94 | 650 | NC_015946.1 |
| Mycoplasma suis KI3806 | 65.00 | 10.00 | 25.00 | 31.08 | 794 | NC_015153.1 |
| Mycoplasma suis Illinois | 67.17 | 10.06 | 22.77 | 31.08 | 845 | NC_015155.1 |
| Mycoplasma synoviae 53 | 90.14 | 4.08 | 5.77 | 31.08 | 659 | NC_007294.1 |
| Ureaplasma parvum serovar 3 ATCC 27815 | 92.27 | 3.45 | 4.28 | 31.08 | 609 | NC_010503.1 |
| Ureaplasma parvum serovar 3 ATCC 700970 | 91.92 | 3.39 | 4.68 | 31.08 | 614 | NC_002162.1 |
| Ureaplasma urealyticum serovar 10 ATCC 33699 | 91.82 | 3.48 | 4.70 | 31.08 | 646 | NC_011374.1 |
The CDSs and the %G+C data were obtained from the NCBI site for bacterial genomes (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) and the start codon data compiled using Prodigal 2.50.
Analysis of the codons surrounding the start codon
Analysis of the codons surrounding the start codon in 125 species of bacteria have deetcted a preference for lysine, serine or threonine as the amino acid after the start codon (2nd amino acid). In firmicutes, lysine is the predominant second amino acid [16], and this was the 2nd amino acid in the fusion protein encoded by both pTAP and pTGP. The presence of A/T rich codons around the start codon may reduce formation of secondary structures and favour higher rates of translational initiation [17]. Isoleucine is overrepresented in the fourth to eighth positions in 91% of the prokaryotic genomes that have been analysed, and this was also the case in the fusion protein studied here.
Insertion points of transposon constructs
To confirm that the Tn4001 transposon integrated at random in the genome the site of integration of the transposons from the pTGP plasmid in the M. gallisepticum genome was determined for three of the transformants (TGP2, TGP3, TGP5) by genomic DNA sequencing [14]. The annotated genome sequence of M. gallisepticum strain Rlow (NC_004829.2) was used as the reference genome to determine the location of the transposon. The transposon integrated into the M. gallisepticum genome at random, within lipoprotein (TGP2::MGA_0981), putative bacteriocin/lantibiotic ABC exporter (TGP5::MGA_0022) and hypothetical (TGP3::MGA_0471) genes. The proportion of the coding sequence of these genes upstream of the insertion point of the transposon varied from 40% to 86%.
Levels of transcription of phoA in transformants
Three pTAP transformants (pTAP3, 4, 9) and three pTGP transformants (pTGP2, 3, 5) were selected and mRNA extracted to compare levels of transcription of the phoA gene by quantitative RT-PCR. The phoA mRNA abundance relative to GAPDH mRNA in each transformant was determined in triplicate, and the mean levels of transcription of phoA in the pTAP and pTGP transformants were compared. The levels of phoA mRNA (mean ± SEM) were determined in pTGP2 (4.8 ± 0.8), pTGP3 (6.4 ± 1.1), pTGP5 (3.5 ± 0.5), pTAP3 (12.5 ± 1.5), pTAP4 (10.9 ± 1.4) and pTAP9 (13.4 ± 1.5). Thus the relative abundances of phoA mRNA transcripts were similar in the clones transformed with the same construct and the mean level of phoA transcription in pTAP transformants (12.1 ± 0.7) was significantly higher than in pTGP transformants (4.9 ± 0.8) (P = 0.003, Student’s t-test).
Alkaline phosphatase activity of pTGP-transformed M. gallisepticum
The level of alkaline phosphatase activity was determined in five randomly selected pTAP and pTGP transformants in triplicate as described previously [14]. The mean level (± SEM) of alkaline phosphatase activity for pTAP transformants was 190 ± 8 U/mg total cell protein, which was significantly higher than in pTGP transformants (119 ± 11 U/mg total cell protein) (P = 0.03, Student’s t-test).
Efficiency of translational initiation codon
The relative efficiency of translation was determined by dividing alkaline phosphate activity by the relative phoA mRNA concentrations. The relative levels of translation were determined in the transformants pTGP2, pTGP3, pTGP5, pTAP3, pTAP4 and pTAP9 as shown in Table 2. The relative efficiency of PhoA translation in pTGP transformants (26.6 ± 1.4) was significantly higher than in pTAP transformants (16.7 ± 2.6) (P = 0.045, Student’s t-test).
Table 2. Efficiency of transcription and translation of phoA constructs with a GTG (TGP) or ATG (TAP) translational start codon in M. gallisepticum transformants.
| Transformant | Alkaline phosphatase (mean units/mg ± SEM) | mRNA (mean ± SEM) | Relative efficiency of translation (mean ± SEM) |
|---|---|---|---|
| TGP2 | 118.7 ± 11.3 | 4.8 ± 0.8 | 27.5 ± 8.5 |
| TGP3 | 142.4 ± 18.2 | 6.4 ± 1.1 | 23.8 ± 4.8 |
| TGP5 | 94.5 ± 3.9 | 3.5 ± 0.5 | 28.4 ± 5.7 |
| TAP3 | 259.4 ± 5.9 | 12.5 ± 1.5 | 21.4 ± 2.9 |
| TAP4 | 172.2 ± 7.8 | 10.9 ± 1.4 | 16.3 ± 1.9 |
| TAP9 | 160.5 ± 1.2 | 13.4 ± 1.5 | 12.3 ± 1.4 |
Localisation of PhoA fusion protein
Whole cell proteins of the pTGP3 transformant were subjected to Triton X-114 fractionation and the protein fractions were separated by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes and immunostained using a monoclonal antibody against alkaline phosphatase [14]. A band of 47 kDa, corresponding to the predicted molecular weight of expressed PhoA fusion protein, was detected in pTGP3-transformed M. gallisepticum whole cell proteins (Fig 1A, lane W) and in the hydrophobic fraction (Fig 1A, lane H), but not in the aqueous fraction (Fig 1A, lane A).
Fig 1. Localisation of PhoA proteins in M. gallisepticum cells.
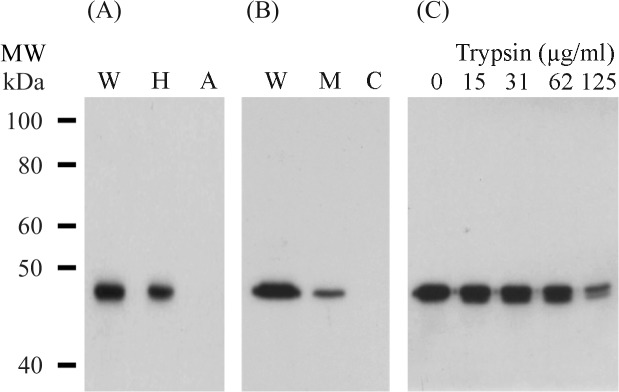
(A) Proteins of the pTGP3 transformant were separated into hydrophobic and aqueous fractions by Triton X-114 partitioning. Equivalent amounts of the fractions were separated by SDS-PAGE in 10% polyacrylamide gels, Western transferred and probed with a monoclonal antibody against alkaline phosphatase. Lanes: W, whole cells; H, hydrophobic fraction; A, aqueous fraction. The molecular weight (MW) markers were the biotinylated protein ladder from Cell Signaling Technology. (B) The membrane and cytosolic fractions of transformant pTGP3 were separated by SDS-PAGE in a 10% polyacrylamide gel and Western transferred. Immunostaining with a monoclonal antibody to alkaline phosphatase demonstrated the presence of PhoA in the whole cells (lane W) and the membrane fraction (lane M), but not in the cytosolic fraction (lane C). (C) Whole cells of the pTGP3 transformant were treated with increasing concentrations of trypsin. The cells in lane 0 were treated with TS buffer only. Proteins were separated by SDS-PAGE in 10% polyacrylamide gels and immunostained with a monoclonal antibody against alkaline phosphatase.
The pTGP3 transformant was separated into membrane and cytosolic fractions by differential ultracentrifugation [14]. The proteins were separated by SDS-PAGE, transferred to PVDF membranes and immunostained. The PhoA fusion protein was detected in the membrane fraction (Fig 1A, lane M), and also in whole cells (Fig 1A, lane W), but not in the cytosolic fraction (Fig 1A, lane C).
The surface exposure of PhoA in the pTGP3 transformant was examined by trypsin proteolysis [14]. Immunostaining of trypsin-treated cells with a monoclonal antibody against alkaline phosphatase demonstrated a gradual loss of reactivity with increasing concentrations of trypsin (Fig 1C), indicating surface exposure of the PhoA fusion protein.
Discussion
There was a 2.5 fold decrease in transcription of the ltufacyphoA gene when the GTG start codon was used. As the nucleotides in the untranslated leader region (UTR) were not mutated, a possible factor influencing transcription could be the mRNA secondary structure. The minimal free energy for optimal secondary structure was higher with the ATG codon (-2.5 kCal mol-1), which could have reduced the stability of the mRNA secondary structure and increased transcriptional efficiency. A significantly higher free energy for genes with an ATG initiation codon has been reported in B. subtilis and it is possible that start codon preference is partly influenced by mRNA structure [11]. In B. subtilis, different start codons affect the stability of the ΔermC mRNA, with mutation of the start codon from ATG to GTG resulting in a significant decrease in the mRNA half-life, from 8.2 min to 6.5 min, which has been putatively attributed to an effect of ribosomal binding and ternary complex formation on stability [18]. In E. coli mRNA levels are highly correlated with folding energy near the 5’ end of the transcript. The secondary structure can obstruct the binding of the ribosomal subunit and thus translational initiation. When there is reduced ribosomal binding, mRNA is also exposed to nuclease digestion [19]. Mazin et al have recently described an experimental genome-wide study of regulation of transcription in M. gallisepticum strain S6, and detected transcriptional changes under different stress conditions. The A/T content, the first nucleotide of the transcript and the spacer distance appeared to correlate with variations in transcription [20].
It has been suggested that when GTG replaces the ATG codon there is less efficient pairing with the fMet-tRNA, which can reduce the rate of translational initiation [21]. A 3 to 8 fold reduction in protein expression is seen with a GTG start codon in B. subtilis [12, 22] and E. coli [10, 12, 23, 24]. While alteration of the start codon of the phoA gene in the pTAP plasmid from ATG to GTG resulted in reduced levels of expression of alkaline phosphatase in M. gallisepticum, the reduction in expression was considerably less than the reduction in transcription of the gene.
The relative efficiency of translation was determined for the ATG and GTG initiation codons. The GTG codon appeared to result in a significantly higher relative efficiency of translation (26.6 ± 1.4) than the ATG codon (16.7 ± 2.6), suggesting that it was a more efficient translational initiation codon. While a relatively small number of transformants generated with each construct were evaluated, the within-transformant variation was similar to the between-transformant variation for the three pTGP and three pTAP transformants evaluated. Furthermore, there was a significant difference in levels of transcription, expression and translational efficiency between the transformants with the different start codons in the construct. Therefore the small number of transformants was sufficient for effective analysis.
The phoA reporter gene used in this study was fused to the lipoprotein signal sequence of a vlhA gene, that we have previously shown results in translocation of the protein through the cytoplasmic membrane, acylation of the protein and exposure on the surface of the mycoplasma cell. In order to confirm that there was not a differential effect of the two alternative start codons on the processing of the lipoprotein reporter, we examined the localization of the expression products of the two constructs. Partitioning of cellular proteins into the hydrophobic and hydrophilic fractions with Triton X-114 demonstrated that with both constructs the PhoA fusion protein was within the hydrophobic fraction, and cell surface proteolysis showed that it was also sensitive to trypsin. This confirmed that the PhoA fusion protein was exported to the membrane and surface exposed, suggesting that lipoprotein processing and export was similar with both ATG [14] and GTG start codons.
The native tuf gene has an ATG start codon, while the native vlhA 1.1 gene has a GTG start codon. It is not clear if the sequence of the promoter region may have played a role in the higher levels of transcription seen with the ATG codon in the pTAP transformants. It is possible that the reduced translational efficiency of the gene containing the GTG codon may have directly influenced the efficiency of transcription of the gene and it is also possible that there is not a linear correlation between transcript abundance and translation.
The mean usage of the three major initiation codons across 620 bacterial chromosomes is 80.1% for ATG, 11.6% for GTG and 7.8% for TTG, with low G+C genomes showing a greater bias towards use of the ATG codon [9]. Mycoplasmas deviate from the universal genetic code in using TGA as a codon for tryptophan [25, 26], which is presumed to be a result of directional mutational pressure during evolution towards an A+T rich genome. The predominant translational initiation codon based on in silico prediction of coding sequences in Mycoplasma spp. is ATG (Table 1), as might be expected in organisms with a low genomic G+C content. However, the order of preference of ATG > GTG > TTG is more similar to the pattern seen in E. coli than to that seen in B. subtilis [11], and does not appear to concord with the strong bias in mycoplasmas towards codons with a low G+C content. These results are primarily based on genome annotation and could change with experimental analysis. In M. pneumoniae using proteomic approaches two novel genes were identified and one translational start codon was corrected [27], while in Mycoplasma mobile both genomic and proteogenomic mapping by mass spectrometry were combined to guide annotation of the genome [28]. In future such combinations of proteogenomic approaches may lead to correction of genome annotation errors, and identification and validation of additional or alternative translational start codons in the genome [29].
The transcriptional and translational regulation mechanisms and signals in mycoplasmas are considered similar, but not identical, to those seen in other bacteria [30]. In M. pneumoniae there is only a modest correlation between mRNA and protein abundance, and it has been suggested that translational regulation may be more important in mycoplasmas than regulation of transcription [31]. While the much smaller difference in the effect of a differing start codon on the efficiency of expression in M. gallisepticum compared to that seen in other bacteria can explain why there is not a strong selection against use of a GTG start codon, it is not clear why there is such a strong preference for its use in a specific gene family. Further studies on regulation of transcription and translation in mycoplasmas will be necessary to establish whether the GTG start codon plays a role in translational regulation of protein expression that was not elucidated in the studies we have described here.
In conclusion the ATG start codon is the preferred start codon for translation in most mycoplasma species. In this study, using the pTAP construct, GTG was shown to be a more efficient translational initiation codon in M. gallisepticum, although less efficient transcription in constructs using this start codon resulted in lower overall levels of expression. This finding contrasts with observations of reduced protein expression when ATG start codons were replaced with GTG in E. coli and B. subtilis, suggesting there may be significant differences in the mechanisms involved in translational initiation and regulation in mycoplasmas.
Data Availability
All relevant data are within the paper.
Funding Statement
ISP was in receipt of an International Postgraduate Research Scholarship from the Commonwealth of Australia. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Glass JI, Lefkowitz EJ, Glass JS, Heiner CR, Chen EY, Cassell GH. The complete sequence of the mucosal pathogen Ureaplasma urealyticum . Nature. 2000;407(6805):757–762. . [DOI] [PubMed] [Google Scholar]
- 2. Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, et al. The minimal gene complement of Mycoplasma genitalium . Science. 1995;270(5235):397–403. . [DOI] [PubMed] [Google Scholar]
- 3. Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69(1):101–123. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stenstrom CM, Jin H, Major LL, Tate WP, Isaksson LA. Codon bias at the 3'-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli . Gene. 2001;263(1–2):273–284. . [DOI] [PubMed] [Google Scholar]
- 5. de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87(19):7668–7672. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234(2):187–208. . [DOI] [PubMed] [Google Scholar]
- 7. Guillerez J, Gazeau M, Dreyfus M. In the Escherichia coli lacZ gene the spacing between the translating ribosomes is insensitive to the efficiency of translation initiation. Nucleic Acids Res. 1991;19(24):6743–6750. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. . [DOI] [PubMed] [Google Scholar]
- 9. Villegas A, Kropinski AM. An analysis of initiation codon utilisation in the Domain Bacteria—concerns about the quality of bacterial genome annotation. Microbiology. 2008;154(9):2559–2661. 10.1099/mic.0.2008/021360-0 [DOI] [PubMed] [Google Scholar]
- 10. Sussman JK, Simons EL, Simons RW. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo . Mol Microbiol. 1996;21(2):347–360. . [DOI] [PubMed] [Google Scholar]
- 11. Rocha EP, Danchin A, Viari A. Translation in Bacillus subtilis: roles and trends of initiation and termination, insights from a genome analysis. Nucleic Acids Res. 1999;27(17):3567–3576. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo . Mol Microbiol. 1992;6(9):1105–1114. . [DOI] [PubMed] [Google Scholar]
- 13. Dybvig K, Voelker LL. Molecular biology of mycoplasmas. Annu Rev Microbiol. 1996;50:25–57. . [DOI] [PubMed] [Google Scholar]
- 14. Panicker IS, Kanci A, Chiu CJ, Veith PD, Glew MD, Browning GF, et al. A novel transposon construct expressing PhoA with potential for studying protein expression and translocation in Mycoplasma gallisepticum . BMC Microbiol. 2012;12(1):138 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li W, Zou H, Tao M. Sequences downstream of the start codon and their relations to G+C content and optimal growth temperature in prokaryotic genomes. Antonie Van Leeuwenhoek. 2007;92(4):417–427. . [DOI] [PubMed] [Google Scholar]
- 17. Zalucki YM, Beacham IR, Jennings MP. Biased codon usage in signal peptides: a role in protein export. Trends Microbiol. 2009;17(4):146–150. 10.1016/j.tim.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 18. Sharp JS, Bechhofer DH. Effect of translational signals on mRNA decay in Bacillus subtilis . J Bacteriol. 2003;185(18):5372–5379. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli . Science. 2009;324(5924):255–258. 10.1126/science.1170160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mazin PV, Fisunov GY, Gorbachev AY, Kapitskaya KY, Altukhov IA, Semashko TA, et al. Transcriptome analysis reveals novel regulatory mechanisms in a genome-reduced bacterium. Nucleic Acids Res. 2014;42(21):13254–13268. 10.1093/nar/gku976 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. . [DOI] [PubMed] [Google Scholar]
- 22. Yeh CM, Chang HK, Hsieh HM, Yoda K, Yamasaki M, Tsai YC. Improved translational efficiency of subtilisin YaB gene with different initiation codons in Bacillus subtilis and alkalophilic Bacillus YaB. J Appl Microbiol. 1997;83(6):758–763. . [DOI] [PubMed] [Google Scholar]
- 23. Reddy P, Peterkofsky A, McKenney K. Translational efficiency of the Escherichia coli adenylate cyclase gene: mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc Natl Acad Sci USA. 1985;82(17):5656–5660. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Looman AC, van Knippenberg PH. Effects of GUG and AUG initiation codons on the expression of lacZ in Escherichia coli . FEBS Lett. 1986;197(1–2):315–320. . [DOI] [PubMed] [Google Scholar]
- 25. Yamao F, Muto A, Kawauchi Y, Iwami M, Iwagami S, Azumi Y, et al. UGA is read as tryptophan in Mycoplasma capricolum . Proc Natl Acad Sci USA. 1985;82(8):2306–2309. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inamine JM, Ho KC, Loechel S, Hu PC. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum . J Bacteriol. 1990;172(1):504–506. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, et al. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 2000;28(17):3278–3288. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, et al. The complete genome and proteome of Mycoplasma mobile . Genome Res. 2004;14(8):1447–1461. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Armengaud J. A perfect genome annotation is within reach with the proteomics and genomics alliance. Curr Opin Microbiol. 2009;12(3):292–300. 10.1016/j.mib.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 30. Muto A, Ushida C. Transcription and translation In: Razin S, Herrmann R, editors. Molecular Biology and Pathogenicity of Mycoplasmas. New York: Kluwer Academic / Plenum Publishers; 2002. p. 323–345. [Google Scholar]
- 31. Maier T, Schmidt A, Guell M, Kuhner S, Gavin AC, Aebersold R, et al. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol. 2011;7:511 10.1038/msb.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


