Abstract
The Anaphase Promoting Complex/Cyclosome (APC/C) is a massive E3 ligase that controls mitosis by catalyzing ubiquitination of key cell cycle regulatory proteins. The APC/C assembly contains two subcomplexes: the “Platform” centers around a cullin-RING-like E3 ligase catalytic core; the “Arc Lamp” is a hub that mediates transient association with regulators and ubiquitination substrates. The Arc Lamp contains the small subunits APC16, CDC26, and APC13, and tetratricopeptide repeat (TPR) proteins (APC7, APC3, APC6, and APC8) that homodimerize and stack with quasi-twofold symmetry. Within the APC/C complex, APC3 serves as center for regulation. APC3’s TPR motifs recruit substrate-binding coactivators, CDC20 and CDH1, via their C-terminal conserved Ile-Arg (IR) tail sequences. Human APC3 also binds APC16 and APC7, and contains a >200-residue loop that is heavily phosphorylated during mitosis, although the basis for APC3 interactions and whether loop phosphorylation is required for ubiquitination are unclear. Here, we map the basis for human APC3 assembly with APC16 and APC7, report crystal structures of APC3Δloop alone and in complex with the C-terminal domain of APC16, and test roles of APC3’s loop and IR-tail binding surfaces in APC/C-catalyzed ubiquitination. The structures show how one APC16 binds asymmetrically to the symmetric APC3 dimer, and together with biochemistry and prior data explain how APC16 recruits APC7 to APC3, show how APC3’s C-terminal domain is rearranged in the full APC/C assembly, and visualize residues in the IR-tail binding cleft important for coactivator-dependent ubiquitination. Overall, the results provide insights into assembly, regulation, and interactions of TPR proteins and the APC/C.
Keywords: TPR, Anaphase Promoting Complex/Cyclosome, cell cycle, ubiquitin E3 ligase, protein assembly
Introduction
The Anaphase Promoting Complex/Cyclosome (APC/C) is a massive, 1.2 M Da E3 ligase complex that regulates numerous essential eukaryotic processes. APC/C was originally identified for its roles in controlling mitosis1; 2. Indeed, APC/C drives ubiquitin-mediated proteolysis of Cyclin B and Securin, and triggers sister chromatid separation. It is now known that APC/C catalyzes ubiquitination of numerous additional cell cycle regulatory proteins to orchestrate several steps of cell division, spanning from prometaphase, through mitosis, exit from mitosis and G1 3; 4; 5; 6; 7, and APC/C also regulates meiosis 8; 9. Furthermore, a staggering number of cell cycle-independent roles for APC/C-mediated ubiquitination have been identified in neurons 10. These numerous functions depend on the pairing of the core holo-APC/C complex with a coactivator, typically either CDC20 or the homologous CDH1, which bind various motif sequences such as a Lys-Glu-Asn “KEN-box” in substrates and regulatory proteins. Many substrates also display additional APC/C-interaction motifs, including a “D-box” (aka “Destruction-box”) that simultaneously engages a coactivator and the APC/C core subunit APC10 11; 12; 13; 14. Although details of interactions between different substrates and APC/C-coactivator complexes are only beginning to emerge 15; 16; 17, it is clear that ordered APC/C association with different coactivators, together with phosphorylation, regulates the timing of successive steps in the cell division process 4; 5; 6; 7.
The overall architecture of an APC/C-coactivator-substrate complex has been determined by combining information from electron microscopy (EM), biochemistry, and subunit tagging and deletion studies 13; 18; 19; 20; 21; 22; 23; 24; 25; 26. The APC/C core comprises two multiprotein superdomains, which we describe here for human APC/C: the catalytic “Platform”, which contains APC1, APC2, APC4, APC5, APC11, APC15, and the scaffolding “Arc Lamp”, which contains APC8, APC13, APC6, APC12/CDC26, APC3, APC16, APC7, and APC10. Within the Platform, the APC2 and APC11 subunits are homologous to cullin and RBX subunits of cullin-RING E3 ligases, and represent the catalytic core 27; 28. It appears that the major roles of APC1, APC4, APC5, and APC15 in the Platform are to coordinate the placement and conformation of the APC2-APC11 catalytic core for ubiquitin ligation to substrates recruited to domains organized by the Arc Lamp.
The Arc Lamp was named for its curved form resembling an actual arc lamp emanating from the Platform 19. The curved scaffold-like structure is established by layers of homologous homodimeric tetratricopeptide repeat (TPR) proteins - APC8, APC6, APC3, and APC7 in humans - stacked on top of each other. The TPR unit is a ~34-residue degenerate motif, which forms a pair of antiparallel helices 29. Tandem TPR repeats pack in parallel and form superhelical solenoids with various curvatures depending on interactions between adjacent repeats. APC8, APC6, APC3, APC7 are each predicted to contain roughly 14 TPRs, which homodimerize via N-terminal domains in head-to-tail arrangements that generate extended structures with the C-termini of the two protomers on each end 30. Within the holo-APC/C assembly, the border between the Platform and the Arc Lamp is established by the APC8 homodimer (Cdc23p in S. cerevisiae), on which the APC6 homodimer (Cdc16p in S. cerevisiae) is stacked. The APC3 homodimer (Cdc27p in S. cerevisiae) stacks on top of APC6, and in higher eukaryotes, APC7 stacks on the APC3 homodimer to complete the stack of TPR subunits. Also, several small proteins (CDC26, APC13, and APC16) are thought to stabilize the overall assembly of the Arc Lamp. Indeed, disrupting the function of small subunits leads to phenotypes ranging from temperature-sensitive cell cycle arrest in budding yeast (Cdc26) to meiotic and/or mitotic defects upon mutation or knockdown in worms or human cells (APC16) 1; 28; 31; 32; 33; 34; 35; 36; 37.
A major function of the Arc Lamp is to serve as a scaffold for dynamic recruitment of substrates, by APC3 binding to homologous Ile-Arg “IR Tail” sequences displayed at the C-termini of APC10 and coactivators 26; 38; 39. Although there are two protomers of APC3, their context and binding to the different IR tails is asymmetric within the holo-APC/C. One of the two APC3 protomers is constitutively occupied by the IR tail of APC10 26; 39. This interaction is solidified by extensive contacts between APC1 and APC10’s N-terminal domain. Thus, in the context of the holo-APC/C, the APC10 C-terminal IR tail is proximal to only its dedicated partner APC3 protomer 13; 21; 22; 23; 25. This arrangement leaves only the opposite APC3 protomer available to dynamically associate with IR tails from different coactivators at different stages of the cell cycle 7.
Insights into how APC3 assembles into the Arc Lamp and recruits IR-tails were recently provided by a landmark 7.4 Å resolution cryo-EM map and secondary structure definition for an APC/C-CDH1-Substrate complex 23. Nonetheless, residue-specific information is available only for some portions of the Arc Lamp, from crystal structures of a few human components (APC10 N-terminal domain and a subcomplex between human CDC26 and the C-terminal domain of APC6) 39; 40 and homology models based on structures of orthologs (a near full-length complex of S. pombe APC6-CDC26, coactivator complexes with D- and KEN-boxes, and the N-terminal domains of APC8 and APC3 from S. pombe and the parasite E. cuniculi, respectively) 15; 16; 17; 30; 41. Thus, despite importance, our understanding of how an APC3 dimer is integrated into the whole APC complex and recruits coactivators is limited by lack of high-resolution structural data for APC3. Here we address these issues with crystal structures of a human APC3 homodimer, alone and in complex with a fragment of APC16. The structures, along with biochemistry and analyses in light of previous EM data, provide insights into APC/C assembly and regulation.
Results and Discussion
Human APC3 residues 182-453 are dispensable for assembly and activity of recombinant APC/C
To optimize constructs for crystallographic studies, we identified a minimum folded version of human APC3. Several lines of evidence suggested that residues 182-453 are mobile and/or disordered, and not part of the signature TPR folded region of human APC3 (Fig. 1a). First, comparing APC3 sequences across evolution showed that this region is not conserved, and varies from being nonexistent in the parasite E. cuniculi, to spanning more than 300 residues in some organisms 42. Second, secondary structure prediction using Psipred indicated that this region is disordered (Fig. S1) 43. Third, modeling TPR repeats suggested this region is not visible in the APC3 portion of EM maps 19; 20; 21; 23; 44; 45.
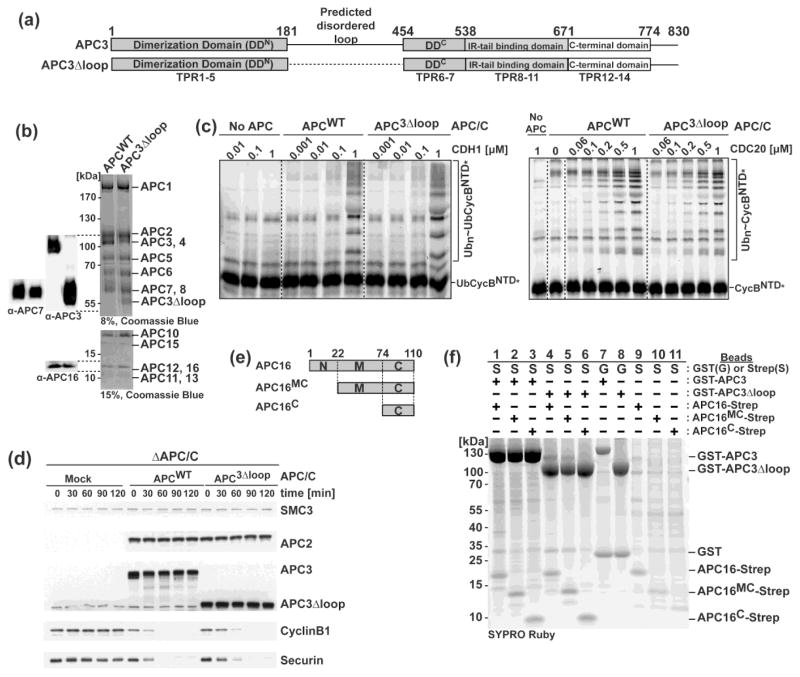
Fig. 1. Characterization of APC3 loop function and identification of a minimal APC3-APC16 complex.
a) Schematic of APC3 and APC3Δloop domains: Dimerization domain (TPRs1-7), Predicted disordered region (residues 182-453), IR tail binding domain (TPRs 8-11), and C-terminal domain (TPRs 12-14).
b) APC/C (Wild-Type and APC3Δloop) complexes used in enzyme assays in c. Right, Coomassie-stained SDS-PAGE. Left panel, immunoblots with α-APC3, α-APC7 and α-APC16.
c) Fluorescent scans showing a role of APC3Δloop in ubiquitination of either Ub-CyclinBNTD* or CyclinBNTD* with APC/C (Wild-Type and APC3 loop), UBCH10, and increasing amounts of CDH1 (left) or CDC20 (right).
d) Role of APC3 loop in APC/C-mediated substrate degradation in X. laevis egg extract. After adding nondegradable Δ90 CyclinB1 for two hours, substrates were added and samples were quenched and analyzed at the indicated times. Western blotting as a function of time shows Cyclin B1 and Securin substrate turnover, APC2 and APC3 (or APC3Δloop) controls for levels of the two versions of APC/C in the assays, and SMC control for comparable levels of extract in each lane.
e) Schematic of APC16. N-terminal domain (N, residues 1-21), Middle domain (M, residues 22-73), and C-terminal domain (C, residues 74-110).
f) Identification of a minimal APC3-APC16 subcomplex. Sypro-ruby stained SDS-PAGE gel after affinity purification of the indicated APC subunits coexpressed in insect cells. Note: Poor expression of APC16C-Strep alone may suggest stabilization upon coexpression with APC3. MW, molecular weight.
To ascertain whether residues 182-453 of human APC3 are required for assembly into the holo-APC/C complex, we used our recombinant system to coexpress in insect cells either wild-type APC3, or a deletion mutant lacking residues 182-453 (referred to hereafter as APC3Δloop), together with all other human APC/C subunits 21; 44; 46. After affinity purification based on a twin-Strep tag on the C-terminus of APC4 46, the complexes were compared by Coomassie-stained SDS-PAGE, and by immunoblotting for APC3, APC7 and APC16. The data showed that APC3’s residues 182-453 are dispensable for assembly into a holo-APC/C complex (Fig. 1b).
We performed two types of assays to test whether the APC3 182-453 loop is required for ubiquitination by APC/C. APC/C uses a two-E2/two-step mechanism for polyubiquitination 47. For humans, APC/C first collaborates with the “initiating” E2 UBCH10 to prime a substrate with monoubiquitin or short ubiquitin chains27; 48; 49. Next, APC/C partners with the distinctive “chain elongating” E2 UBE2S, which generates ubiquitin~ubiquitin linkages to polyubiquitinate the substrate 50; 51; 52. Activity with UBE2S can be assayed by incorporating a priming ubiquitin molecule into the substrate as a linear fusion (Ub-CyclinBNTD*)44; 46; 50; 51; 52. APC/C activity with UBCH10 was assayed toward either this same Ub-CyclinBNTD* substrate, or toward a fluorescent version of Cyclin B’s N-terminal domain without a ubiquitin (CyclinBNTD*) 44; 46. Because a key role of APC3 is to bind a coactivator, we performed assays titrating from sub-saturating to saturating concentrations for either CDH1 or CDC20. In all cases, ubiquitination activity in the entirely purified system was similar for APC/C with wild-type APC3 and with APC3Δloop (Fig. 1c, S2a).
In vivo, phosphorylation of APC/C correlates with APC/CCDC20-dependent ubiquitin-mediated proteolysis during mitosis. A hallmark of phosphorylated APC/C is slower migration of APC3 in a western blot, and previous studies have mapped mitotic phosphorylation sites to APC3’s residues 182-453 53; 54. However, whether this loop is required for degradation of substrates such as Cyclin B1 and Securin has remained unknown. We examined substrate degradation in X. laevis egg extracts. This assay depends on addition of a truncated, nondegradable Cyclin B1 (Δ90) to stimulate CDK1-dependent APC/C phosphorylation and activation (Fig. S2b) 1; 2; 55; 56; 57; 58, as reflected by wild-type APC3 displaying the characteristic phosphorylation-dependent mobility shift (Fig. 1d, S2b). Importantly, the degradation of full-length D-box containing Cyclin B1, and of Securin, depends on APC/C, because this was blocked by immunodepleting APC/C from the extracts, and was rescued by adding back recombinant human APC/C (Fig. 1d, S2b). Adding mutant APC/C harboring APC3Δloop also substantially stimulated substrate degradation, although with a modest delay relative to the rate with wild-type APC/C. Nonetheless, the APC3 loop was not absolutely essential for CDK1-activated APC/C-dependent ubiquitin-mediated proteolysis. Taken together, the data suggested that the APC3Δloop protein would be a suitable model for biochemical and structural studies.
Identification of a minimal APC3-APC16 subcomplex
Although genetic and cell biological studies had previously identified critical roles for APC16 for APC/C-dependent regulation in higher eukaryotes, the functions of APC16 remain incompletely understood 34; 35; 36. Prior EM data suggested that APC3 and APC16 interact with each other, but the resolution of all existing maps precluded identifying the domain of APC16 involved 23; 37. We mapped the interacting regions by coexpressing N-terminally GST-tagged APC3 or APC3Δloop with either wild-type or truncated versions of C-terminally Strep-tagged APC16 (“MC” = middle and C-terminal domains, residues 22-110; “C” = C-terminal domain, residues 74-110), and testing co-association upon affinity purification on Streptactin resin. The deletion mapping identified the APC16C fragment as the minimal region required to bind APC3, and showed that APC3Δloop and APC16C form a stable APC sub-assembly (Fig. 1e, f).
Crystal structure of APC3Δloop reveals homodimer structurally related to but distinct from other APC TPR subunits
To gain insights into APC3 structure and function, we crystallized N-terminally GFP-fused-APC3Δloop alone and in complex with APC16C, and obtained two crystal forms in P65 and P43, which diffracted to 3.3 and 4.25 Å resolution, respectively (Table 1). We determined the structure of the P43 form by the single wavelength anomalous dispersion method for a selenomethionine-containing variant, and subsequently of the P65 form by molecular replacement. Although the GFP moiety was required to obtain the crystals, it was not possible to model GFP into weak density extending from the N-termini of APC3 from either crystal form (Fig. S3a). The density is most consistent with the GFP portions of the fusion proteins adopting multiple positions within the crystal. Also, the electron density maps and crystal packing indicated that the P65 form contained only GFP-APC3Δloop as described in this section, whereas the P43 form contained a complex with APC16C as described in the following section (Fig. S3b–d).
Table 1.
Crystallographic data and refinement statistics
| Native GFP-APC3Δloop/APC16C |
SeMet GFP-APC3ΔloopΔC20/APC16C |
Native GFP-APC3Δloop |
|
|---|---|---|---|
| Data Collection | |||
| Beam line | NECAT 24-ID-C | NECAT 24-ID-E | NECAT 24-ID-C |
| Space Group | P43 | P43 | P65 |
| Unit cell parameters | |||
| a, b, c (Å) | 116.8, 116.8, 185.1 | 116.0, 116.0, 184.3 | 118.4, 118.4, 273.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) (Highest shell) | 50 – 3.3 (3.42 – 3.3) | 50 – 3.25 (3.39 – 3.25) | 50 – 4.25 (4.40 - 4.25) |
| Wavelength (Å) | 1.07160 | 0.97918 | 0.97920 |
| Number of measured reflections | 254827 | 162277 | 89519 |
| Number of unique refections | 37171 | 38356 | 15458 |
| Overall Rsym | 0.100 (0.829) | 0.125 (0.760) | 0.119 (0.826) |
| Completeness (%) | 100.0 (100.0) | 99.9 (100.0) | 99.9 (100.0) |
| Overall I/σI | 23.7 (3.5) | 12.8 (2.3) | 17.2 (2.5) |
| Multiplicity | 6.9 | 4.2 | 5.8 |
| Refinement | |||
| Resolution (Å) | 50 - 3.3 | 49.94 – 3.25 | 49.67 – 4.25 |
| Rwork/Rfree | 0.2033/0.2441 | 0.2097/0.2485 | 0.2548/0.2823 |
| rmsd bond lengths (Å) | 0.006 | 0.006 | 0.005 |
| rmsd bond angles (°) | 1.039 | 1.065 | 1.243 |
| Number of atoms | |||
| Proteins | 7598 | 7542 | 6066 |
| Ramachandran statistics | |||
| Preferred (%) | 93.9 | 93.6 | 94.2 |
| Allowed (%) | 6.1 | 6.4 | 5.6 |
| Disallowed (%) | 0 | 0 | 0.2 |
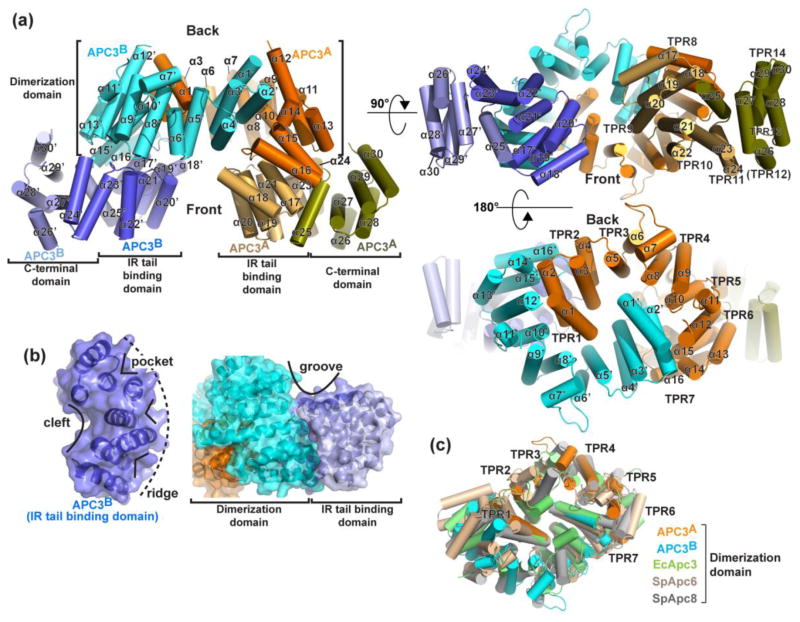
The crystal structure of APC3Δloop alone showed a symmetric homodimer (Fig. 2a). Helices from 13 of the 14 predicted TPRs (TPRs 1-11 and 13-14) are visible in the electron density, and form a superhelical structure. The homodimeric arrangement leads to an overall V-shaped structure, with the N-terminal dimerization domain in the center, and the C-termini at the two tips of the “V”.
Fig. 2. Crystal structure of APC3.
a) Three views of human APC3Δloop crystal structure, with domains (Dimerization, IR-tail binding, and C-terminal), TPRs (1-11 and 13-14) and helices (α1-α30, from N-to C-terminus) labeled. Note TPR12 is not properly folded.
b) Four distinctive types of surface from arc-shaped TPR domains are shown: 1) cleft, 2) pocket, 3) ridge and 4) groove.
c) Superimposition of dimerization domain from human APC3 (protomers in orange and blue) with that from Apc3 of E. cuniculi (EcApc3, PDB ID: 3KAE, lime, 2.9 Å rmsd for 205 Cα), of Apc6 from S. pombe (SpApc6, PDB ID: 2XPI, wheat, 3.3 Å rmsd for 225 Cα) and Apc8 from S. pombe (SpApc8, PDB ID: 3ZN3, gray, 3.3 A rmsd for 205 Cα) 30; 41.
Each half of the “V” can be viewed as consisting of 3 arc-shaped subdomains. For a given arc, the functional properties rely on interactions mediated by three distinctive types of surfaces established by the tandem arrays of TPRs: (1) the concave side of an arc forms a bowl-shaped cleft that can accommodate binding partners, (2) the convex side of an arc can contain pockets either between the helices or between TPRs that hold relatively small surfaces of partners, and (3) the convex side also forms a ridge that itself can fit into complementary clefts or grooves (Fig. 2b). Also, upon packing together in full-length APC3, the arcs generate nearly two complete turns of superhelical structure, which establish a fourth type of interacting surface – grooves between the arcs (Fig. 2b).
Each of APC3’s arc-shaped subdomains has important functions. The N-terminal arcs from the two APC3 protomers interact with each other to form the homodimerization domain (Fig. 2a). The residues involved in the dimer interface are shown in Fig. S4. For each molecule of APC3 in the homodimer, this domain encompasses residues 1-181 that adopt five TPRs preceding the start of the deletion, and the subsequent residues 454-537 that form two additional TPRs. The seven dimerization domain TPRs from an APC3 protomer adopt a continuous superhelix. Homodimerization is mediated by one protomer packing in the cleft from the opposite protomer in a manner that resembles a pair of clasped hands: the two N-termini form interacting “fingers” that pack in the “palms” formed by TPRs 3-7 of the opposite hand. The N-terminal 5 TPRs of the dimerization domain adopt a relatively flat ovoid structure, while the 6th and 7th TPR “wrists” begin to wrap around the oval due to their concave clefts accommodating the convex ridges from TPRs 1-2 from the opposite protomer. Overall, the superhelical structure projects the terminal TPRs of the dimer domain toward one face of the oval that we refer to here as the “front”. This would correspond to the top if viewing the entire APC3 homodimer as a “V”. In agreement with predictions based on sequence alignments 41; 42, the human APC3 dimer domain superimposes with that from the parasite E. cuniculi with an RMSD of 2.9 Å, and with the dimer domains of APC8 and APC6 from S. pombe with RMSDs of 3.3 and 3.3 Å, respectively (Fig. 2c). The data are consistent with overall similar modes of dimerization for Arc Lamp TPR subunits despite only 15%–21% sequence identity.
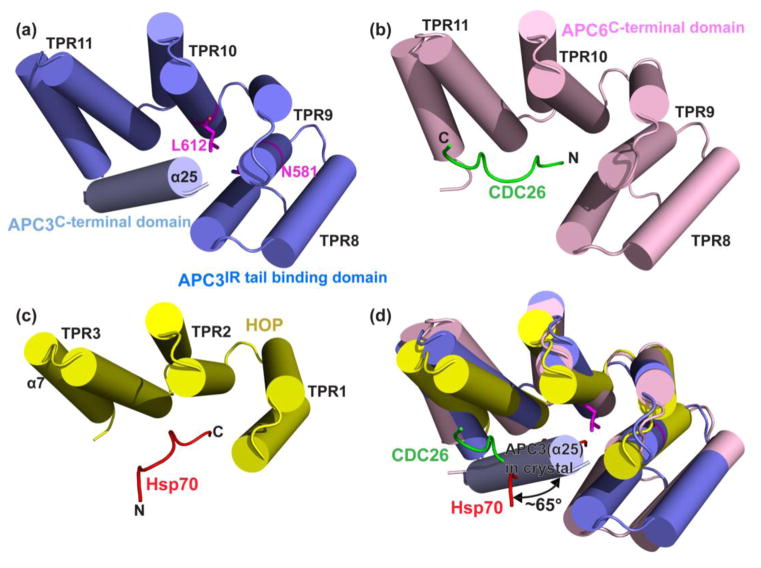
The central arc comprises TPRs 8-11, which for each APC3 monomer forms a side of the “V” by extending away from the front of the dimerization domain (Fig. 2a, 3a). In the context of a full APC3 protomer, the N-terminal and central arcs together form a continuous, roughly one-and-a-half turn superhelix comprising TPRs 1-11, with both the cleft and ridge sides of the central arc exposed to mediate interactions. The ridged side of APC3’s central domain binds APC16C as discussed below, whereas the cleft side represents a typical TPR domain-peptide interaction surface. Indeed, a search for similar structures with the DALI server showed homology to other peptide-binding TPR proteins 59. Interestingly, high scoring homologs include a complex between APC6’s central and C-terminal domains bound to CDC26, and the HOP-Hsp70 structure (Z-scores of 18.4 and 12.4 respectively) (Fig. 3b, c) 40; 41; 60; 61. Notably, superimposing the TPRs from APC3’s central arc on the corresponding regions of the APC6-CDC26 or HOP-Hsp70 complexes show a potential peptide-binding surface on the concave cleft from APC3’s central domain (Fig. 3d). In the crystal, this APC3 cleft binds a rearranged α25-helix as discussed below. However, in the setting of a holo-APC/C complex, this central arc corresponds to the IR-tail binding domain 23; 38.
Fig. 3. Structural feature of APC3 IR tail binding domain.
a) Structure of APC3 IR tail binding domain, also showing the mis-arranged α25 from the C-terminal domain in the crystal. N581 and L612, which correspond to N548 and L579 that when mutated in the S. cerevisiae ortholog Cdc27 disrupt CDH1 binding are shown in magenta sticks 38.
b) Corresponding region of human APC6TPR-CDC26N (PDB ID: 3HYM) 40.
c) Corresponding region of human HOP-Hsp70 (PDB ID: 3ESK) 61.
d) Superposition of the structure of APC3s IR tail binding domain and corresponding regions of APC6-CDC26 (1.6 Å rmsd for 132 Cα) and HOP-Hsp70 (2.3 Å rmsd for 108 Cα).
The C-terminal subdomain (residues 672-830) of human APC3 is predicted to contain 9-helices, with 6 of them arranged as tandem TPRs (Fig. S1b). In the crystal, helices 27-30 pack as the predicted TPRs 13-14, and the subsequent residues 775-830 are not visible in the electron density and are presumably disordered (Fig. 2a). However, the predicted TPR 12 that corresponds to helices 25 and 26 is disrupted with helix 26 partially disordered. Furthermore, unexpectedly, in the crystal, helix 25 is not part of TPR12, but instead packs inside the groove of the IR-tail binding domain (Fig. 3a, d). This may be an artifact of crystallization as described below. Nonetheless, the crystal packing does demonstrate propensity for the IR-tail binding domain to make protein-protein interactions – albeit by mediating non-native contacts in the absence of other APC/C subunits.
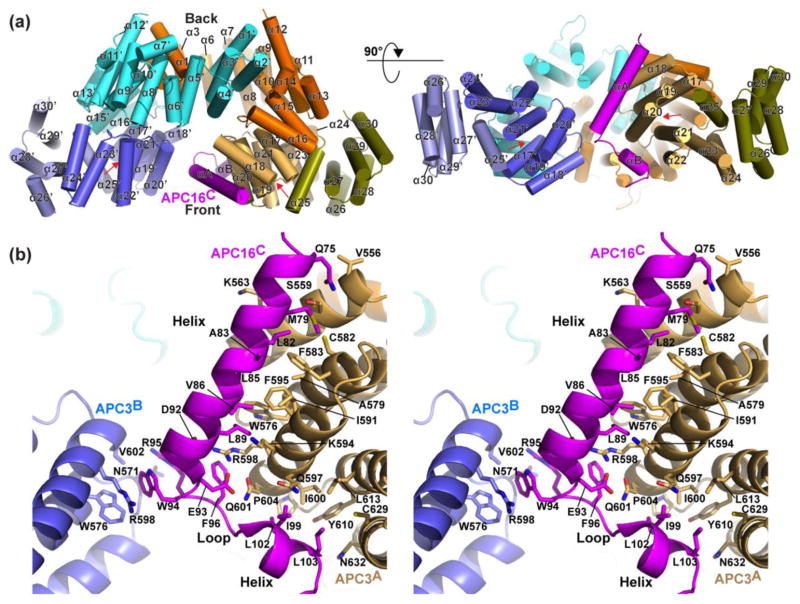
Structure of 1:2 APC16C:APC3Δloop complex: asymmetric interactions with a symmetric interface
In the complex with APC16C, the crystal structure of the APC3Δloop dimer is essentially identical to that crystallized alone (Fig. 4a, S5). The APC16C binding site is located centrally in front of the APC3 dimerization domain. The two molecules of APC3 together cradle a single molecule of APC16C. From APC3, interactions are mediated by pockets in the ridged sides of the IR-tail binding domains.
Fig. 4. Crystal structure of APC3-APC16: asymmetric interactions in a symmetric binding pocket.
a) Overall structure of APC3Δloop homodimer complex with APC16C. The APC3 domains are colored as in Fig. 2a. APC16 is colored magenta. Red arrow indicates the IR tail binding cleft.
b) Stereo view of close-up of the APC3-APC16 interface, showing side-chains mediating contacts.
APC16C adopts a helix-loop-helix structure, with the first helix, loop, and second helix zig-zagging to contact alternating protomers in the APC3 homodimer (Fig. 4b, S6). This establishes the asymmetric 1:2 APC16:APC3 stoichiometry. The first element encompasses APC16 residues 74-93, which form a kinked helix from which side-chains along one face pack in a cleft between TPR8 and TPR9 from one APC3 protomer, which we refer to as “A”. Here, contacts center around hydrophobic interactions between APC16’s Met79, Leu82, Ala83, Leu85, Val86, and Leu89 and the aliphatic portions of APC3’s Ser559 and Lys563 from TPR8, and Trp576, Ala579, Cys582, Phe583, Ile591, Phe595, Lys594, and Arg598 from TPR9. The two ends of this interface are anchored by electrostatic interactions between APC16’s Gln75 and the backbone from APC3’s Val556 backbone and side chain from Ser559, and between a cluster of hydrogen bonds involving APC16’s Leu89, Asp92, and Glu93 and APC3’s Arg598 and Gln601. Interactions from the APC16C loop involve Trp94 and Arg95 inserting into a cleft in the opposite APC3 protomer (here referred to as “B”) involving Asn571, Trp576, Arg598, and Val602 from TPR9. Finally, APC16 crosses back to TPR9 and TPR10 from the opposite APC3 protomer (“A”), where APC16’s Phe96, Ile99, Leu102, and Leu103 pack against APC3’s Gln597, Arg598, Ile600, Gln601, Pro604, Tyr610, Leu613, Cys629, and Asn632.
Insights into APC3 structure in holo-APC from docking APC3-APC16 into prior cryo-EM density map
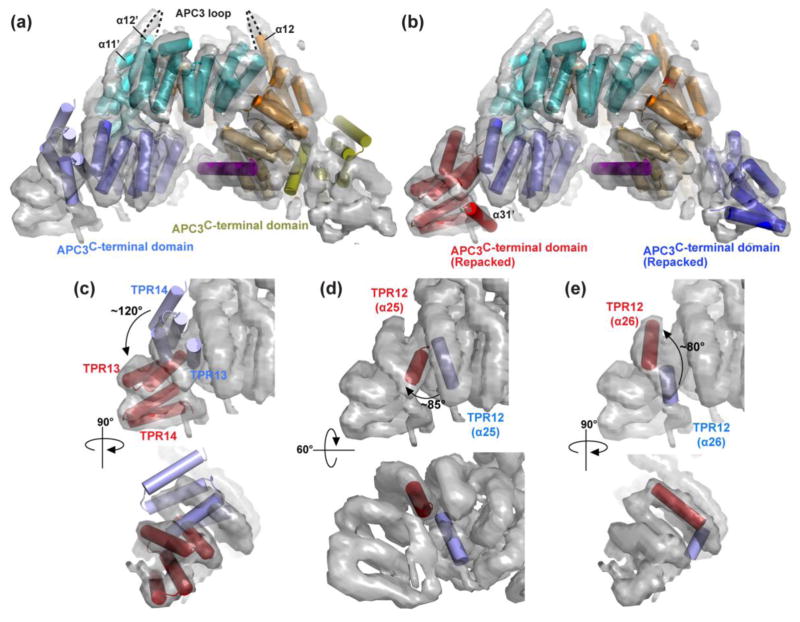
It was possible to assess how the helices in the APC3-APC16 subcomplex are arranged in the holo-APC/C complex by docking domains from the crystal structure into the recent cryo-EM map of an APC/C-CDH1-Substrate complex 23. Although the resolution of the cryo-EM data allowed clear visualization of secondary structures, the amino acid sequences could not be resolved from the EM density alone. Obtaining high-resolution information requires a hybrid method in which either structures or homology models are docked into the map.
Using Chimera, we readily fit the portion of the crystal structure encompassing APC16C and APC3’s dimerization and IR-tail binding domains into the map as a single unit, with minimal manual readjusting in Coot (Fig. 5a) 23; 62; 63; 64. However, APC3’s C-terminal domain is substantially rearranged in the holo-APC/C complex in comparison to the crystal (Fig. 5b). We approximated this region of the holo-APC/C by subdividing the crystal structure and rearranging elements as follows. One unit consisting of TPRs 13-14 was placed by ~120° rotation relative to the arrangement in the crystal (Fig. 5c). Placing TPR12 between the IR-tail binding domain and TPRs 13-14 required: (1) extracting helix 25 from its crystallographic packing in the concave surface of the IR-tail binding domain and repositioning by an ~85° rotation (Fig. 5d); and (2) separately rotating the subsequent helix 26 and extending this as an idealized helix (Fig. 5e). Also, another helix 31 was observed in the EM density map following helix 30/TPR14 of APC3 B. Thus, it is possible to generate a full model of APC3Δloop-APC16C in the context of the holo-APC/C, albeit with uncertainties in the detailed structure of TPR12 and the C-terminus. Notably, the modeling is consistent with the 182-453 loop being disordered in the context of an APC/C-CDH1-Substrate complex.
Fig. 5. Comparison of APC3 C-terminal domain structure in crystal and in APC/C complex.
a) Superimposing APC3Δloop-APC16C structure (colored as in Fig. 4) with cryo-EM map (grey) of APC/C-CDH1-Substrate (EMD-2651) using Chimera 23; 79. Dashed line shows the location of the deleted APC3 “loop” spanning residues 182-453 between α11 and α12.
b) APC3Δloop-APC16C with APC3 C-terminal domain helices arranged as in the holo APC/C complex 23. The C-terminal domains are colored blue for APC3 protomer “A” and red for “B”.
c–e) Comparison of overall arrangements of helices from APC3 C-terminal domain in crystal and in the holo APC/C complex 23, showing TPR13-14s rotated by ~120° in c, rotation of helix 25 of TPR12 ~85° in d, and helix 26 of TPR12 rotated by ~80° in e.
Comparing APC3-APC16 and APC6-CDC26 subcomplex structures reveals distinct features with potential implications for APC3 assembly into the Arc Lamp
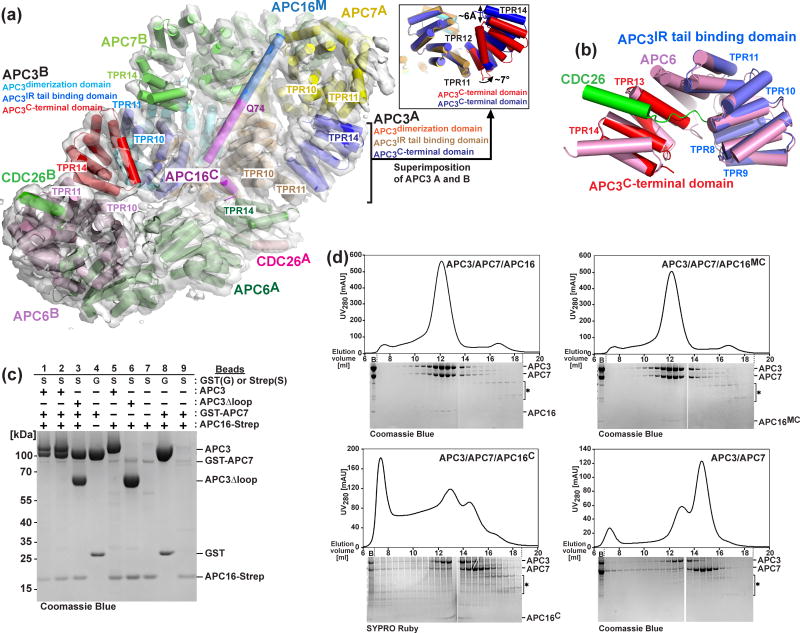
Comparing features of the APC3Δloop-APC16C subcomplex with those described previously for APC6-CDC26 40; 41 reveals interesting similarities and differences that might relate to APC3 assembly into the Arc Lamp. Interestingly, like APC3, the C-terminal region of APC6 is also not properly folded on its own 40. However, unlike APC3, APC6’s central and C-terminal arcs are structurally stabilized through forming a 1:1 complex with CDC26 40. CDC26 forms a rod-like linchpin support for the APC6 TPR superhelix (Fig. 6a, b) 40; 41. Interestingly, when extracted out of the APC3Δloop crystal structure, the separate IR-tail binding subdomain and TPR13-14 unit superimpose well on the corresponding regions of the APC6-CDC26 complex (Fig. 6b). However, without a corresponding stabilizing “linchpin”, proper folding of the APC3 C-terminal domain may rely on interactions with other APC/C subunits.
Fig. 6. Insights into APC3-APC16 assembly into Arc Lamp and role of APC16 in stabilizing APC7 binding to APC3.
a) Secondary structures of Arc Lamp subunits APC6-CDC26-APC3-APC16-APC7, as identified by 23, are shown in their cryo-EM map of APC/C-CDH1-Substrate complex, with APC3 colored as in Fig. 5b, and the two molecules each of APC7, APC6 and CDC26 colored yellow and lime, forest and lightpink, salmon and green, respectively. APC16M is colored aquamarine. Q74 from the crystal structure of of APC3Δloop-APC16C is denoted to show the boundary between APC16M and APC16C. The asymmetric arrangement of helices from the two APC3 protomers is shown in inset.
b) APC3 IR tail binding domain and C-terminal domains are shown individually superimposed on the corresponding regions of human APC6TPR-CDC26N 40.
c) Coomassie-stained SDS-PAGE gels showing role of APC3 in complex formation between APC7 and APC16 after glutathione (G) or Strep (S) affinity purification of the indicated APC/C subunits coexpressed in insect cells. Note: Lanes 1 and 2 are from the same pulldown, but loading half the volume in lane 1 is in lane 2. Molecular weights are indicated.
d) Role of APC16 in complex formation between APC3 and APC7 was tested by gel filtration chromatography of the indicated versions of APC3-APC7-APC16 complexes. Elution profiles and Coomassie or SYPRO-Ruby-stained SDS-PAGE gels of corresponding gel filtration chromatography fractions are shown for APC3-APC7-APC16 (top left), APC3-APC7-APC16MC (top right), APC3-APC7-APC16C (bottom left), and APC3-APC7 (bottom right). ‘B’ is the sample before injection to size exclusion column, after insect cell coexpression of GST-APC7 and APC3 in the absence or presence of indicated versions of Strep-tagged APC16, affinity purification, and TEV treatment.
The model for APC3Δloop-APC16C within the context of the holo-APC/C complex provides insights into how proper folding of the two APC3 C-terminal domains might be achieved. Within the Arc Lamp, the APC8, APC6, APC3, and APC7 homodimers stack through complementary placement of the ridges from the convex side of TPR arcs from one subunit into the grooves between superhelical turns from a neighboring subunit in the stack 30. At lower resolution this arrangement was thought to be uniform across an entire TPR protein dimer. However, the hybrid structural modeling enables visualizing localized asymmetry between the two APC3 protomers in the holo-APC/C (Fig. 6a). Whereas the dimerization and IR-tail binding domains are symmetrically arranged, the angles between TPRs 11 and 12 differ by roughly 7° for the two APC3 molecules (Fig. 6a). This leads to a relative 6 Å displacement of TPR14 for the two APC3 protomers, which make different interactions. The C-terminal domain of one APC3 protomer (“B”) docks exclusively into a groove from APC6. By contrast, the opposite APC6 molecule and one molecule of APC7 embrace the C-terminal domain from the other APC3 protomer (“A”). Thus, it is conceivable that the propensity for APC3’s C-terminal domain to rearrange facilitates assymetric assembly into the Arc Lamp (Fig. 5b, 6a).
APC16 stabilizes interactions between APC3 and APC7
Through docking APC3Δloop-APC16C into the EM density map for holo-APC/C, we noticed that a relatively N-terminal portion of APC16 forms a helix that extends away from APC3, and packs between ridges in the APC7 dimerization domain (Fig. 6a). Thus, we investigated interactions between APC3, APC7, and APC16 by coexpressing in insect cells Strep-tagged APC16, GST-tagged APC7, and/or untagged APC3 or APC3Δloop. Affinity purification on either Streptactin or Glutathione sepharose showed that the three proteins form a subcomplex (Fig. 6c, lanes 1-3). In experiments testing binary APC16 interactions with individual subunits, binding was observed with APC3 or APC3Δloop, in agreement with the crystal structure (Fig. 6c, lanes 5–6). However, APC7 and APC16 did not co-associate in the absence of APC3 (Fig. 6c, lanes 8–9).
To better understand formation of the APC3-APC7-APC16 subcomplex, we coexpressed GST-APC7 and untagged APC3, alone or with various truncated versions of Strep-tagged APC16. After Streptactin or glutathione affinity purification, the complexes were treated with TEV protease to liberate the GST-tag from APC7 and the Strep-tag from APC16, and the products were subjected to gel filtration chromatography. In the absence of APC16, the APC3 that initially co-purified with GST-APC7 separated during gel filtration (Fig. 6d), and the APC3-APC16C complex and APC7 also did not copurify (Fig. 6d). However, the ternary complex was stable both with full-length APC16, and a truncated version (APC16MC) that encompasses APC16’s residues 22 through the C-terminus. Thus, the capacity of APC16 to interact with both APC3 and APC7 promotes assembly (Fig. 6d).
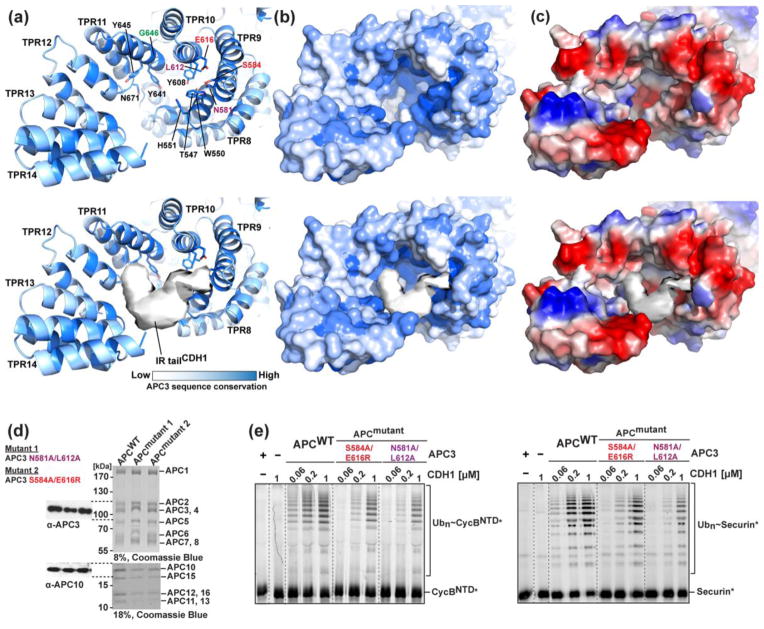
Insights into CDH1-binding groove of APC3
In the hybrid structural model, EM density for CDH1’s IR-tail fills the cleft of the IR-tail binding domain from one APC3 protomer (“B”) (Fig. 7a–c). The corresponding region of the opposite APC3 protomer (“A”) binds the IR-tail from the core subunit APC10 (not shown). In the APC3Δloop crystal structure, this cleft is lined by numerous hydrophobic side-chains that could potentially contact the hydrophobic “Ile” side chain, as well as aromatic, polar and acidic side chains that may interact with the “Arg” in the “IR” motif (Fig. 7a, c). Also, APC3’s superhelical structure places the cleft from its C-terminal domain in position to seal the IR-tail in place (Fig. 7a). Sequence analysis of APC3 suggests that these interactions with IR tails are conserved across eukaryotes, as the side chains lining the cleft of the IR-tail binding are among the most conserved (Fig. 7a, c). In addition, human APC3 Gly645, which corresponds to the site of a temperature sensitive mutant of S. cerevisiae 18, is invariant and located proximal to IR tail binding pocket (Fig. 7a).
Fig. 7. The IR tail binding domain of APC3.
a) Close-up of IR-tail binding domain from APC3Δloop-APC16C (top), and superimposed with EM density for CDH1 IR tail extracted from APC/C-CDH1-Substrate complex (bottom). APC3 residues were shaded by sequence conservation using the program ProtSkin 78; 82, with dark blue indicating high conservation and white not conserved. Conserved hydrophobic residues lining the IR-tail binding groove are shown as sticks with residues tested for function in e highlighted (S584 and E616 in red; N581 and L612 in purple). Gly646, which corresponds to a ts mutant allele in S. cerevisiae Cdc27, is green18.
b) Surface representation of a.
c) Representation of b, with surface colored by electrostatic potential.
d) Coomassie stained SDS-PAGE gels of APC/C complexes used in enzyme assays in e. The presence of APC3 (WT and mutants) and APC10 were confirmed by immunoblotting using α-APC3 and α-APC10 (left).
e) Effects of APC3 mutations in the IR-tail binding domain on CDH1-dependent APC/C-mediated ubiquitination. Fluorescence scans are shown for SDS-PAGE gels monitoring ubiquitination of fluorescent CyclinBNTD* (left) or Securin* (right), with recombinant APC/C harboring either wild-type (WT) APC3 or the S584A/E616R or N581A/L612A mutants, UBCH10, and the indicated concentrations of CDH1.
We wished to experimentally validate the importance of residues within the IR-tail binding cleft in human APC/C. We were able to generate two variants of recombinant APC/C, each containing doubly mutated versions of APC3. One APC/C mutant has APC3’s Ser584 and Glu616 simultaneously replaced by alanine and arginine, and the other has Ala substitutions in place of Asn581 and Leu612. This latter mutant corresponds to the N548A/L579A double-mutant version of S. cerevisiae APC3 that was previously shown to assemble into APC/C and yet to display defects in IR-tail dependent coactivator functions 38. Similarly, for human APC/C, both mutant versions of APC3 did not cause obvious defects in APC/C assembly (Fig. 7d), and they allowed stoichiometric incorporation of the core subunit APC10 presumably via extensive interactions in the APC10-APC1 interface 23. Although CDH1 also interacts with APC/C through multiple interfaces, we could test effects of mutations on CDH1-dependent activity by monitoring ubiquitination as a function of the concentration of CDH1 added to reactions. Relative to wild-type APC/C, the mutants in the APC3 IR-tail binding cleft required higher CDH1 concentrations to ubiquitinate substrates (Fig. 7e). These results confirm the importance of the structurally-observed IR-tail binding groove for human APC/C.
Concluding Remarks
At 1.2 MDa, with numerous subunits and transiently associated regulatory subunits, the APC/C is a dynamic and complicated molecular machine 1; 2. Although structural insights from crystallographic data for some subunits and from EM maps of APC/C complexes are emerging, our knowledge of detailed features of many APC/C subunits has remained relatively rudimentary. This has included APC3, despite its identification as a core APC/C subunit almost 20 years ago 1. Structural characterization of APC3 and its complex with APC16 and biochemical studies described herein shed light on both regulation and assembly of APC/C.
Unexpectedly, general enzymatic properties of APC/C were maintained even upon deleting the nearly one-third of the APC3 protein encompassing residues 182-453, which are presumably disordered at least in some holo-APC/C assemblies (Fig. 1, 5a, S1). APC3’s 182-453 loop has long been known to be phosphorylated, and this correlates with ubiquitin-mediated proteolysis of APC/C substrates 53. Nonetheless, the role of this loop has been called into question for the S. cerevisiae ortholog Cdc27, where substituting CDK sites with alanines supported CDH1-dependent functions and yeast growth at all temperatures tested, although the mutations did cause delays in mitosis and CDC20-dependent substrate degradation65. Accordingly, we found that the APC3 loop is dispensable for human APC/C-mediated ubiquitination in vitro using purified recombinant components. Nonetheless, the APC3 loop does contribute to, but is not absolutely required for, Cyclin B1-CDK1-dependent substrate turnover in mitotic extracts (Fig. 1, S2). Our results underscore the complexities of phosphoregulation of APC/C. It will be interesting in the future to understand both how the APC3 loop contributes to APC/C activity, and to identify regions of APC/C that are responsible for CDK1-dependent activation of APC/C lacking the APC3 loop.
Although at this point we do not know how APC/C assembles in vivo, our identifying binding partners for the isolated APC3Δloop construct enabled determining the crystal structure in complex with APC16C, modeling of APC3-APC16 in the context of the holo-APC/C, pinpointing APC16’s critical role in stabilizing interactions with APC7, and contrasting and comparing features of APC3 with other TPR subunits. It seems likely that the different properties of APC6-CDC26, APC3-APC16C and APC7-APC16M, established in part by distinctive roles of the small subunits CDC26 an APC16, play key roles in APC/C assembly. Whereas the APC6-CDC26 complex is properly folded due to the CDC26 “linchpin”, it seems likely that proper folding of APC3’s C-terminal domain is coupled to association with other Arc Lamp subunits. Our finding that incorporation of APC7 into stable subcomplexes required both APC3 and APC16 raises the possibilities that the APC3-APC16-APC7 subcomplex assembles into the Arc Lamp as a unit, or that incorporation of APC7 follows that of APC3 and APC16. During APC/C assembly, APC3-APC16 or APC3-APC16-APC7 subcomplexes may bind APC6-CDC26, perhaps in the context of the rest of the Arc Lamp or APC/C. Although the propensity for APC3’s C-terminal domain to adopt different structures may reflect the requirement for forming two orientations in the holo-APC/C (Fig. 6a), alternative conformations of the C-terminal domain of unassembled APC3 could also potentially help protect the IR-tail binding domain from mediating wayward protein-protein interactions (Fig. 3a, 5a, 5b).
Despite their potential to form symmetric homodimeric structures (Fig. 2 and 30), the TPR subunits of the Arc Lamp make asymmetric interactions in the context of the holo-APC/C. The APC3Δloop-APC16C complex shows how the sequence and structure of APC16’s C-terminal domain breaks symmetry, with different elements forming distinct interactions by zig-zagging between the two symmetrically arranged APC3 protomers in the dimer domain (Fig. 6a). Interestingly, examination of EM data raises the possibility that APC16’s middle domain mediates similar asymmetric interactions with APC7 (Fig. 6), and that another small subunit, APC13, likewise makes asymmetric contacts with APC8 and APC6 23. Furthermore, in the context of the holo-APC/C, the two APC3 protomers associate asymmetrically with adjacent TPR homodimers of APC6 and APC7. Thus, the overall architecture establishes distinctive environments for each APC3 subunit, and by extension also for APC10 and coactivators that bind the two APC3 IR-tail binding clefts. Although APC10 appears to constitutively occupy one APC3 protomer, the other APC3 molecule’s IR-tail binding cleft interacts transiently with different coactivators and regulators. Notably, this latter cleft has also been implicated as a target for the small molecule APC/C inhibitor, Tosyl-L-arginine methyl ester (TAME) 66; 67; 68; 69: TAME has a similar structure to an Ile-Arg dipeptide, and competes with the IR tail of CDC20 for binding to APC3 67. The visualization of side-chains lining the IR-tail binding cleft in the APC3Δloop-APC16C crystal structure may enable future efforts to generate higher affinity TAME analogs for use as efficient APC/C and cell cycle inhibitors.
Materials and Methods
Protein purification
All proteins used in this study are human. For crystallography, N-terminally GST-tagged GFP-APC3Δloop (residues 1-181, 454-830) and GFP-APC3ΔloopΔC20 (1-181, 454-810) were either expressed alone or co-expressed in a baculovirus expression system with C-terminally twin-Strep-tagged APC16C (74-C). To obtain phases for crystal structure determination, selenomethionine (SeMet)-labeled GFP-APC3ΔloopΔC20 (deleted for the C-terminal 20 residues)-APC16C was expressed as described 70. Complexes with APC16 and variants were purified by streptactin affinity chromatography followed by glutathione affinity chromatography. The GST and Strep tags were cleaved off their fusion partners by TEV protease, and the resultant GFP-APC3Δloop-APC16C complexes were further purified by ion exchange chromatography and gel filtration chromatography.
For pulldown assays comparing interactions of deletion mutants, APC3, APC16, and/or APC7 were co-expressed by using the indicated baculoviruses, and affinity purified using either Streptactin or glutathione beads. For experiments monitoring stable complex persistence throughout gel filtration chromatography, GST-APC7 and/or untagged APC3 were co-expressed with C-terminally twin-Strep-tagged APC16 and/or deletion mutants. Complexes were purified by Streptactin affinity chromatography and treated with TEV protease. To examine complex formation in the absence of APC16, APC3 was co-expressed in insect cells with GST-APC7, and the complex was initially purified by glutathione affinity chromatography and subsequently treated with TEV protease. Gel filtration chromatography was performed with a Superose 6, 10/300 GL column (GE healthcare) in 20 mM Tris pH 7.6, 300 mM NaCl, 1 mM dithiothreitol. All fractions were analyzed by SDS-PAGE followed by staining with Coomassie blue or SYPRO Ruby.
For ubiquitination assays, recombinant APC/C, UBA1, UBCH10, UBE2S, N-terminally 3xMyc-His6-CDH1, CycBNTD, and Ub-CycBNTD were purified, and CycBNTD* and Ub-CycBNTD* were fluorescently labeled as described 21; 44; 46. Securin was expressed with an N-terminal GST-tag, with its two native cysteines (C197 and C198) replaced with alanines, and a Cys-His6 tag introduced at the C-terminus. The Securin substrate was purified by glutathione affinity chromatography, treated with TEV protease, and the resultant GST-free substrate was purified by nickel-affinity chromatography. After labeling with Fluorescein-5-maleimide as described previously 46, Securin* was purified by size-exclusion chromatography. APC/C harboring the APC3Δloop mutant was generated by coexpressing in insect cells all APC/C subunits but with an APC3Δloop baculovirus instead of WT APC3. WT APC/C and APC/C harboring the APC3Δloop mutant were affinity purified by Streptactin affinity chromatography based on the C-terminal twin Strep tag on APC4, and further purified by size exclusion chromatography. Stoichiometry of APC/C complexes was confirmed by comparing Coomassie-stained SDS-APGE and western blotting with anti-APC3 (sc-9972, Santa Cruz Biotechnology), anti-APC7, anti-APC10 (sc-20989, Santa Cruz Biotechnology) and anti-APC16 (sc-135452, Santa Cruz Biotechnology) antibodies.
Enzyme assays
The APC/C-mediated ubiquitination assays were performed as described 46. Proteins were purified or desalted into 20 mM HEPES pH 8.0, 200 mM NaCl, 1 mM DTT, and 30 nM recombinant APC/C (wild-type or with APC3Δloop) were mixed on ice for 30 minutes with 0.1 μM UBA1, 0.2 μM E2 (UBCH10 or UBE2S), 0.25 mg/ml BSA, substrate (0.25 μM CyclinBNTD* or 0.5 μM Ub-CyclinBNTD* or 0.125 μM Securin*, as indicated), coactivator (CDH1 concentrations ranging from 0.001 – 1 μM, or CDC20 ranging from 0.06 – 1 μM), 5 mM MgCl2 and 5 mM ATP. The mixes were equilibrated to room temperature for 10 minutes, and the reactions were initiated by adding 125 μM ubiquitin. The samples were quenched by mixing with SDS sample loading buffer, separated by SDS-PAGE, and were analyzed based on a fluorescein signal of CyclinBNTD*, Ub-CyclinBNTD* or Securin* using a Typhoon FLA 9500 Phosphoimager (GE Healthcare).
Recombinant human APC/C-dependent degradation in Xenopus egg extracts depleted of endogenous APC/C
Interphase egg extracts were prepared as described 46, except that eggs were activated with Ca2+-ionophore A23187 (Calbiochem) at 0.6 μg/ml in MMR for 3 min. Cycloheximide was added to 50 μg/ml to arrest the extract in interphase. To deplete APC/C, 70 μl of interphase extract was mixed with 2.5 μg of anti-APC3 antibody coupled to 10.5 μl of Affiprep Protein A beads and incubated at 4 °C for 40 min, twice. Approximately 1.05 μg of recombinant APC/C complex was added to 15 μl of APC/C- depleted extract. Interphase extract was induced to enter mitosis by addition of non-degradable cyclin B (Δ90) at 300 nM for 120 min prior to assay. Reactions were incubated at 22 °C for the indicated times after recombinant human securin and cyclin B1/Cdk1 addition, and the reactions were quenched with SDS-PAGE sample buffer and boiled for 3 min.
Crystallization and structure determination
For crystallization, GFP-APC3Δloop-APC16C, GFP-APC3ΔloopΔC20-APC16C and GFP-APC3Δloop complexes were concentrated to 2 mg/ml, mixed with reservoir solutions at 1:1 volume ratio for crystallization by the hanging drop vapor diffusion method. Reservoir solutions were with ranges of 0.1M MES pH 6.0–6.2, 0.16–0.32 M MgCl2, and 6–8 % PEG6000. After initial crystallization, quality was improved by streak-seeding. Crystals were cryoprotected in reservoir solution supplemented with 8% glycerol, 8% ethylene glycol and 8% xylitol, and flash frozen in liquid nitrogen prior to data collection. Diffraction data were processed with HKL2000 for GFP-APC3Δloop-APC16C and GFP-APC3Δloop, and with RAPD (https://rapd.nec.aps.anl.gov/rapd) for SeMet-labeled GFP-APC3ΔloopΔC20/APC16C. The initial electron density map was calculated by SeMet-SAD using RAPD. The structure of GFP-APC3Δloop-APC16C was determined by molecular replacement using Phaser71 with GFP-APC3ΔloopΔC20/APC16C as a search model. The structure of GFP-APC3Δloop was determined by molecular replacement using Phaser71 with the structure of GFP-APC3Δloop from the complex as a search model. Model construction and rebuilding was performed using COOT62. The structures of GFP-APC3Δloop-APC16C and GFP-APC3ΔloopΔC20-APC16C were refined using Refmac572 in the CCP4 software suite73, and that of GFP-APC3Δloop was refined using Phenix74. Diffraction data and refinement statistics are provided in Table 1. Although the same protein prep yielded the crystals for GFP-APC3Δloop-APC16C and GFP-APC3Δloop, there was no electron density for APC16C in the P65 crystal form (Fig. S3b, c). Moreover, the P65 crystal packing is different from that of P43, and precludes APC16 binding to APC3 (Fig. S3d).
Structure analysis
The disorder prediction and the secondary structure prediction were performed using the PSIPRED server43. TPR prediction was done by TPRpred75. All structural figures were generated by PyMOL76. Structural comparisons were performed by the DALI search engine77. To map the protein sequence conservation onto APC3, ProtSkin program was used78.
For the figures, the cryo EM map for the structure of the APC/C-CDH1-Hsl1 complex (7.4Å resolution, EMDB accession code: EMD-2651 23) was segmented using Chimera79. To fit the structures or the models into the cryo-EM map, rigid-body fitting with a command, fit in map, was applied in Chimera, and then minimal manual readjustment was done in COOT62. The models of APC7 and APC6-CDC26 were made by i-Tasser80 and Phyre81 as described 23.
Supplementary Material
Highlights.
APC/C is a multisubunit E3 ligase that regulates mitosis, meiosis, and neurons
The APC3 subunit is phosphorylated, binds APC7 and APC16, and recruits coactivators
Structure shows symmetric APC3 TPR homodimer binding asymmetrically to one APC16
Mutations inform APC3-APC16-APC7 assembly, and roles of APC3 in APC/C E3 activity
Insights into assembly, regulation, and interactions of TPR proteins and the APC/C
Acknowledgments
We thank J. Wang and G. Petzold for advice and insights about APC3, K. Perry for assistance with synchrotron data collection, H. Stark and P. Neumann for helpful discussions regarding EM data, I. Davidson for assistance with APC/C depletion and addback assays using Xenopus egg extracts, and S. Bozeman, D.W. Miller, and J. Bollinger for support. For funding, we thank the Jane Coffin Childs Foundation (N.G.B.); Boehringer Ingelheim, the Laura Bassi Centre for Optimized Structural Studies, EUFP7 grant no. 227764 MitoSys, and the Austrian Research Fund (J.-M.P.); ALSAC, NIH R37GM065930 and P30CA021765, and HHMI (B.A.S.). NECAT and APS where synchtron data were collected were supported by NIH P41 GM103403 and DOE Contract No. DE-AC02-06CH11357.
Abbreviations
- APC/C
Anaphase Promoting Complex/Cyclosome
- TPR
tetratricopeptide repeat
- PDB
Protein Data Bank
- EM
Electron Microscopy
- EMDB
Electron Microscopy Data Bank
Footnotes
Accession numbers
Atomic coordinates and structure factors of GFP-APC3Δloop-APC16C (native), GFP-APC3ΔloopΔC20-APC16C (SeMet) and GFP-APC3Δloop have been deposited in the RCSB Protein Data Bank with PDB IDs: 4RG6, 4RG9 and 4RG7, respectively.
Author statement
The authors have no competing financial interests in this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–88. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 2.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–97. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16:2179–206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 4.McLean JR, Chaix D, Ohi MD, Gould KL. State of the APC/C: organization, function, and structure. Crit Rev Biochem Mol Biol. 2011;46:118–36. doi: 10.3109/10409238.2010.541420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–38. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 6.Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. J Cell Biol. 2013;201:177–89. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 8.Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–99. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper KF, Strich R. Meiotic control of the APC/C: similarities & differences from mitosis. Cell Div. 2011;6:16. doi: 10.1186/1747-1028-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puram SV, Bonni A. Novel functions for the anaphase-promoting complex in neurobiology. Semin Cell Dev Biol. 2011;22:586–94. doi: 10.1016/j.semcdb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–95. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18:543–53. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 13.Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–8. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–13. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 16.Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc Natl Acad Sci U S A. 2012;109:18419–24. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, Chao WC, Zhang Z, Yang J, Cronin N, Barford D. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol Cell. 2013;50:649–60. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb JR, Michaud WA, Sikorski RS, Hieter PA. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–8. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dube P, Herzog F, Gieffers C, Sander B, Riedel D, Muller SA, Engel A, Peters JM, Stark H. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20:867–79. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–81. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, Mechtler K, Hyman AA, Stark H, Schulman BA, Peters JM. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19:1116–23. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–32. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 23.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–93. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohi MD, Feoktistova A, Ren L, Yip C, Cheng Y, Chen JS, Yoon HJ, Wall JS, Huang Z, Penczek PA, Gould KL, Walz T. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol Cell. 2007;28:871–85. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–60. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–68. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–66. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 28.Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–9. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 29.Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, Chang L, Yang J, Conin N, Kulkarni K, Barford D. The four canonical tpr subunits of human APC/C form related homo-dimeric structures and stack in parallel to form a TPR suprahelix. J Mol Biol. 2013;425:4236–48. doi: 10.1016/j.jmb.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Araki H, Awane K, Ogawa N, Oshima Y. The CDC26 gene of Saccharomyces cerevisiae is required for cell growth only at high temperature. Mol Gen Genet. 1992;231:329–31. doi: 10.1007/BF00279807. [DOI] [PubMed] [Google Scholar]
- 32.Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–4. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- 33.Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973;74:267–86. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kops GJ, van der Voet M, Manak MS, van Osch MH, Naini SM, Brear A, McLeod IX, Hentschel DM, Yates JR, 3rd, van den Heuvel S, Shah JV. APC16 is a conserved subunit of the anaphase-promoting complex/cyclosome. J Cell Sci. 2010;123:1623–33. doi: 10.1242/jcs.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shakes DC, Allen AK, Albert KM, Golden A. emb-1 encodes the APC16 subunit of the Caenorhabditis elegans anaphase-promoting complex. Genetics. 2011;189:549–60. doi: 10.1534/genetics.111.131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters JM, Hyman AA, Durbin R, Pepperkok R, Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–7. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A, Lawo S, Pelletier L, Stark H, Nasmyth K, Ellenberg J, Durbin R, Buchholz F, Mechtler K, Hyman AA, Peters JM. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–9. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wendt KS, Vodermaier HC, Jacob U, Gieffers C, Gmachl M, Peters JM, Huber R, Sondermann P. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8:784–8. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Dye BT, Rajashankar KR, Kurinov I, Schulman BA. Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure. Nat Struct Mol Biol. 2009;16:987–9. doi: 10.1038/nsmb.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. Embo J. 2010;29:3733–44. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Roe SM, Diogon M, Kong E, El Alaoui H, Barford D. Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. J Mol Biol. 2010;397:1316–28. doi: 10.1016/j.jmb.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 43.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frye JJ, Brown NG, Petzold G, Watson ER, Grace CR, Nourse A, Jarvis MA, Kriwacki RW, Peters JM, Stark H, Schulman BA. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol. 2013;20:827–35. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Yang J, Kong EH, Chao WC, Morris EP, da Fonseca PC, Barford D. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) Biochem J. 2013;449:365–71. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 46.Brown NG, Watson ER, Weissmann F, Jarvis MA, VanderLinden R, Grace CR, Frye JJ, Qiao R, Dube P, Petzold G, Cho SE, Alsharif O, Bao J, Davidson IF, Zheng JJ, Nourse A, Kurinov I, Peters JM, Stark H, Schulman BA. Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly. Mol Cell. 2014;56:246–260. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–39. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 48.Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman JV. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci U S A. 1996;93:4294–9. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–76. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106:18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–9. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–60. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. Embo J. 2003;22:6598–609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hegemann B, Hutchins JR, Hudecz O, Novatchkova M, Rameseder J, Sykora MM, Liu S, Mazanek M, Lenart P, Heriche JK, Poser I, Kraut N, Hyman AA, Yaffe MB, Mechtler K, Peters JM. Systematic phosphorylation analysis of human mitotic protein complexes. Sci Signal. 2011;4:rs12. doi: 10.1126/scisignal.2001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shteinberg M, Protopopov Y, Listovsky T, Brandeis M, Hershko A. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem Biophys Res Commun. 1999;260:193–8. doi: 10.1006/bbrc.1999.0884. [DOI] [PubMed] [Google Scholar]
- 56.Lahav-Baratz S, Sudakin V, Ruderman JV, Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci U S A. 1995;92:9303–7. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Felix MA, Labbe JC, Doree M, Hunt T, Karsenti E. Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature. 1990;346:379–82. doi: 10.1038/346379a0. [DOI] [PubMed] [Google Scholar]
- 58.Patra D, Dunphy WG. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase- promoting complex at mitosis. Genes Dev. 1998;12:2549–59. doi: 10.1101/gad.12.16.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 61.Kajander T, Sachs JN, Goldman A, Regan L. Electrostatic interactions of Hsp-organizing protein tetratricopeptide domains with Hsp70 and Hsp90: computational analysis and protein engineering. J Biol Chem. 2009;284:25364–74. doi: 10.1074/jbc.M109.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol. 2000;149:1377–90. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verma R, Peters NR, D’Onofrio M, Tochtrop GP, Sakamoto KM, Varadan R, Zhang M, Coffino P, Fushman D, Deshaies RJ, King RW. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–20. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 67.Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–95. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng X, King RW. An APC/C inhibitor stabilizes cyclin B1 by prematurely terminating ubiquitination. Nat Chem Biol. 2012;8:383–92. doi: 10.1038/nchembio.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sackton KL, Dimova N, Zeng X, Tian W, Zhang M, Sackton TB, Meaders J, Pfaff KL, Sigoillot F, Yu H, Luo X, King RW. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514:646–9. doi: 10.1038/nature13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cronin CN, Lim KB, Rogers J. Production of selenomethionyl-derivatized proteins in baculovirus-infected insect cells. Protein Sci. 2007;16:2023–9. doi: 10.1110/ps.072931407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 73.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karpenahalli MR, Lupas AN, Soding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- 77.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–1. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deprez C, Lloubes R, Gavioli M, Marion D, Guerlesquin F, Blanchard L. Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J Mol Biol. 2005;346:1047–57. doi: 10.1016/j.jmb.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 79.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 80.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 82.Ritter B, Denisov AY, Philie J, Deprez C, Tung EC, Gehring K, McPherson PS. Two WXXF-based motifs in NECAPs define the specificity of accessory protein binding to AP-1 and AP-2. EMBO J. 2004;23:3701–10. doi: 10.1038/sj.emboj.7600378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.