Abstract
Rhomboid proteins represent a recently discovered family of intramembrane proteases present in a broad range of organisms and with increasing links to human diseases. The enteric parasite Entamoeba histolytica has evolved multiple mechanisms to adapt to the human host environment and establish infection. Our recent studies identified EhROM1 as a functional E. histolytica rhomboid protease with roles in adhesion to and phagocytosis of host cells. Since those studies were performed in a non-virulent strain, roles in parasite virulence could not be assessed. We focused this study on the comparison and validation of two genetic manipulation techniques: overexpression of a dominant-negative catalytic mutant of EhROM1 and knock down of EhROM1 using a RNAi-based silencing approach followed by functional studies of phenotypic analyses in virulent parasites. Both the EhROM1 catalytic mutant and parasites with EhROM1 downregulation were reduced in cytotoxicity, hemolytic activity, and directional and non-directional transwell migration. Importantly, the role for EhROM1 in cell migration mimics similar roles for rhomboid proteases from mammalian and apicomplexan systems. However, the EhROM1 catalytic mutant and EhROM1 downregulation parasites had different phenotypes for erythrophagocytosis, while complement resistance was not affected in either strain. In summary, in this study we genetically manipulated E. histolytica rhomboid protease EhROM1 by two different approaches and identified similarly attenuated phenotypes by both approaches, suggesting a novel role for EhROM1 in amebic motility.
Keywords: Entamoeba histolytica, rhomboid protease, migration, motility, pathogenicity
Graphical Abstract
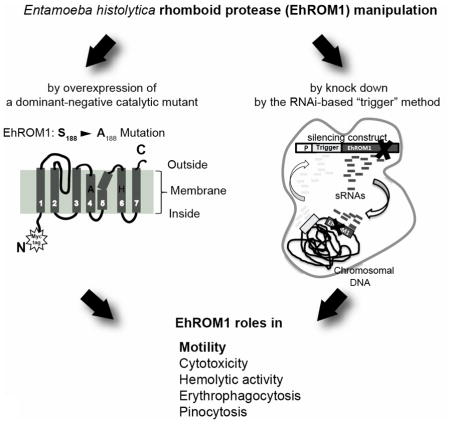
1 Introduction
Intramembrane proteolysis mediated by rhomboid proteases was initially described in Drosophila (1–3). Rhomboid proteases belong to a family of seven transmembrane domain serine proteases harboring serine and histidine as a catalytic dyad (1, 3), which catalyze the release of membrane-bound substrates. Rhomboid proteases (ROMs) are conserved among all kingdoms of life (1, 4) and are known to control a wide range of biologically and medically important processes ranging from insulin resistance and type 2 diabetes by the mitochondrial rhomboid protease PSARL (5), mitochondrial dysfunction and Parkinson’s disease (6, 7), mitochondrial adaptation to stress (7, 8), shedding thrombomodulin and roles in wound healing by RHBDL2 (9), and ER – associated degradation and extraction of misfolded membrane proteins by the endoplasmic reticulum resident rhomboid protease RHBDL4 (10).
In parasitic species multiple rhomboid genes have been identified in Trichomonas vaginalis, Naegleria gruberi, Giardia lamblia, Leishmania major, Cryptosporidium parvum, Babesia bovis, Theileria spp., and Trypanosoma species (11). However, systematic studies to characterize the roles of these proteases have only been performed extensively in Toxoplasma gondii, Plasmodium spp. and Entamoeba histolytica (12–17), where these proteases are found to have multiple important roles. T. gondii contains six rhomboid proteases, one in the mitochondria, and five others that are expressed at different life cycle stages and localized in different cellular compartments. Roles of T. gondii proteases range from intracellular growth, maintenance of micronemal adhesin gradient (thus assuring directional gliding), apical attachment, and efficient host cell invasion (12–15, 18). Human and rodent malaria parasites, P. falciparum and P. bergehi, contain eight rhomboid genes with redundant and multiple roles in parasite development in the asexual blood stage (ROM4, 6, 7 and 8) and in the mosquito or liver stage (ROM1, 3, 9 and 10). Moreover, ROM3 has a vital function in sporogony (19), ROM1 is important for proper formation of the Plasmodium parasitophorous vacuole (20), whereas ROM4 is involved in shedding of various surface adhesins. Shedding of the EBA175 adhesin is an essential event for sialic acid dependent invasion of red blood cells by the merozoite stage (12–14, 21) and removing extracellular adhesive domains of a transmembrane protein TRAP from sporozoite surface is essential for gliding motility and infectivity (22).
Entamoeba histolytica is an intestinal parasitic protozoan that causes colitis and liver abscess. E. histolytica is estimated to cause about 40,000 to 100,000 deaths annually worldwide (23)(24) and is ranked third as a cause of death among parasites (25). E. histolytica possesses several factors that influence tissue invasion including the surface N-acetyl D-galactosamine-inhibitable lectin (Gal/GalNAc lectin) (26–28), cysteine proteases (29–31), pore-forming amoebapores (17, 32, 33) and saposin-like proteins (34). Additionally, phagocytosis (35) and motility are prominent pathological features of invasive amebiasis (36). Motility of E. histolytica pathogenicity is linked to its ability to destroy colonic epithelium and travel to extra-intestinal sites of infection and parasites with reduced motility are less virulent (35, 37, 38).
The genome of E. histolytica contains four genes annotated as rhomboid proteases, with only EhROM1 described to date as an active rhomboid protease (16, 17). Our previous data imply that EhROM1 functions as the sheddase for the amebic Gal/GalNAc lectin on the juxtamembrane site of the host cell (16). EhROM1 knockdown within the genetic background of the E. histolytica G3 strain revealed roles for EhROM1 in parasite adhesion and phagocytosis of host cells (17). However, since the E. histolytica G3 strain is inherently avirulent, studies on cytotoxicity could not be performed (32, 33).
In this work, our main objective was to use two independent approaches for genetic manipulation of ROM1 a virulent parasite strain to allow studies of virulence-associated phenotypes. We overexpressed a catalytic site mutant of EhROM1 (to function in a dominant-negative manner) and achieved RNAi-based downregulation of ROM1 in a virulent E. histolytica strain. We demonstrate that both approaches designed to interfere with EhROM1 activity resulted in parasites with remarkably attenuated transwell migration, significantly reduced cytotoxicity, and defects in pinocytosis. In addition, our study highlights parasite migration as a novel functional property of amebic rhomboid proteases. Thus, EhROM1 has multi-factorial roles in E. histolytica pathogenesis as both motility and migration are prominent pathological features of invasive amebiasis (36).
2 Materials and Methods
2.1 Plasmid construction
The full-length coding region of EhROM1 (EHI_197460) was amplified by PCR from E. histolytica HM-1:IMSS genomic DNA using the following primers: TCCCCCGGGCATTCTCCACCACATAACAATATA (forward primer), GCCGCTCGAGTTAATTGCATTTTCCAACATTGAGTA (reverse primer) containing SmaI and XhoI restriction sites, and cloned into a Topo TA pCR®2.1 vector (Invitrogen, USA). In order to generate the EhROM1 catalytic mutant, the serine at the amino acid position 188 was substituted with alanine using the following primer: GCAACGAACCTGCAGCTCC. Site directed mutagenesis was performed to insert the desired mutations into the DNA motif using the Quikchange site directed mutagenesis protocol (Stratagene). Both, wild type and mutated version of EhROM1 were sub cloned into the pKT-3M vector (39) at the SmaI and XhoI (New England Bio Labs Inc., USA) restriction sites resulting in an N-terminal triple Myc tag fusion. Correct gene insertion was confirmed by sequencing. EhROM1 silencing was achieved by generation of antisense small RNAs by the “trigger” method described by Morf (40). In brief, the first 132bp of the EHI_197520 coding sequence to which large numbers of antisense small RNAs map is designated as a “trigger”-sequence. This trigger was fused with the full-length open reading frame of the EhROM1 gene and resulted in significant downregulation of EhROM1 gene expression.
2.2 Generation of stable parasite transfectants
Entamoeba histolytica HM-1:IMSS trophozoites were transfected with each construct (“trigger” T-EhROM1-s; “catalytic mutant” EhROM1-SA; or “EhROM1 overexpression” EhROM1-OX) as previously described (17, 39). Briefly, mid-log parasites were seeded into 25 mm Petri dishes, sealed with parafilm and allowed to grow at 37°C for 24 h. Transfection mixture containing 15–20 μg of plasmid DNA and 20 μl of SuperFect (Qiagen, USA) in a total volume of 200 μl of M199 medium (Gibco, USA) was incubated for 10 min at room temperature (RT). Plated amoebae were washed with M199 medium, after which 2 ml of M199 supplemented with 15% heat-inactivated bovine serum (Sigma-Aldrich, USA) was added to the plate. The SuperFect-DNA mixture was pipetted onto the plated amoebae, the dishes sealed and incubated at 37°C for 3 h, iced and transferred to glass tube containing TYI-S-33 medium (41). Parasites were allowed to grow for 48 h before adding 2 μg/ml of G418 (Cellgro, USA). T-EhROM1-s stably transfected parasite line with the corresponding control cell line (trigger fused to a luciferase gene) at 6 μg/ml of G418 were obtained. EhROM1-OX and EhROM1-SA stably transfected parasite lines (with the corresponding control cell line overexpressing luciferase protein) were grown at 12 μg/ml G418 drug selection.
2.3 Western blot analysis
Protein expression was detected using western blot analysis as previously described (17). Lysates were prepared using log phase trophozoites. Trophozoites were iced, pelleted and washed in ice cold PBS, then resuspended in NETN lysis buffer (100 mM NaCl, 20 mM Tris pH 8.0, 1 mM EDTA, 0.2% NP-40) containing 50 μM E-64 and 1 x HALT inhibitor cocktail, incubated 10 min on ice, followed by centrifugation at 14,000 g for 10 min. The remaining pellets were dissolved in 8 M Urea solution. Lysates were applied to a 10–12% SDS-PAGE gel and transferred onto a nitrocellulose membrane. The membrane was blocked with 5% milk in PBS-T (PBS containing 0.1% Tween-20) and incubated with antibodies against Myc (mouse) (1:1000 dilution; Cell Signaling Technology, USA), EhROM1 (rat) (1:20 dilution; custom made by Harlan, USA), EhROM3 (rabbit) (1:500 dilution; custom made by Pierce Biotechnology, USA), and Actin (mouse) (MP Biomedicals 1:1000) followed by incubation with secondary mouse or rat horseradish peroxidase (HRP)-conjugated antibody (1:1,000 dilution; Cell Signaling Technology or Santa Cruz Biotechnology, USA) and developed using enhanced chemiluminescence (ECL Prime) (GE Healthcare, USA). Blots were either scanned on a Kodak Image Station 4000R or imaged on film and developed using a Kodak X-OMAT 2000 processor.
2.4 Adherence assays
Assays to assess adherence to healthy mammalian cells were performed using Chinese hamster ovary (CHO) cells as previously described (17). Briefly, 1×104 parasites and 2×105 CHO cells were mixed together, centrifuged at 150×g for 5 min, and incubated on ice for 1 hr. After incubation, the supernatant was removed, the remaining 100μl media gently resuspended, and a hemocytometer was used to count the parasites. Parasites with three or more attached CHO cells were considered positive for adhesion.
2.5 Cell monolayer destruction assay and hemolytic activity assay
Assays were carried out as previously described (42). Briefly, 5×104 trophozoites were placed on a confluent CHO cell monolayer, centrifuged for 5 min at 50 g and incubated for 2 h at 37°C. Cells were fixed with 4% ultra-pure formaldehyde for 10 min and rinsed twice with PBS, followed by staining with 0.1% methylene blue (OmniPur, USA) diluted in 10 mM borate buffer (pH 8.7) and washed three times with the same buffer. The dye was extracted by adding 1 ml of 0.1 M HCl at 37 C for 30 min. In order to measure the extracted dye, samples were diluted 1:10 with PBS and the absorbance at 650 nm was read in a spectrophotometer. The intact monolayer was set as 0% destruction and a syringe-lysed monolayer was set as 100% destruction and used as a blank.
Hemolytic activity assay was carried out as described (43) with some minor modifications. A total of 1.25×108 human erythrocytes were incubated for 1 h at 37 C with PBS washed 1.25×106 of trophozoites (ratio 100:1) in a final volume of 1mL of PBS. After centrifugation for 5 min at 1000 g, 0.5 ml of the resultant supernatant was used to quantify released hemoglobin by determining the absorbance at 405 nm.
2.6 Transwell migration assays
For transwell migration assays parasites were grown to confluence in glass tubes. In order to monitor migration (without any addition of chemoattractant), cells were iced, pelleted and resuspended in complete TYI-S-33 medium (containing vitamins and serum) at a concentration of 5×105 cells/ml. To assay directed migration properties, cells were washed and resuspended in plain TYI-S-33 medium without addition of vitamins and serum. In both cases parasites (1.5×105) were added to the top of the transwell insert containing 8 μm pores (Costar, USA). Complete TYI-S-33 medium (1 ml) was placed in the bottom of the chamber. The 24 well plate was sealed with parafilm and placed in an anaerobic bag (Becton Dickinson, USA) for 3 h at 37°C. At the desired time point, the transwell inserts were removed and the parasites in the bottom chamber iced, transferred to Eppendorf tubes, centrifuged at 1,000 g for 5min, resuspended in 20 μl of TYI-S-33 and quantified with a hemocytometer.
2.7 Red blood cell phagocytosis assays
Phagocytosis of hRBCs was assayed as previously published (44). Briefly, a total of 1×108 hRBCs were incubated with 1×105 trophozoites in 0.2 ml of TYI-S-33 for 15 min at 37°C. Parasites and hRBCs were pelleted at 1,000 g for 2 min and resuspended twice in ice cold distilled water, ensuring lysis of all extracellular hRBCs. Ingested hRBCs were lysed in concentrated formic acid (88%, Sigma-Aldrich, USA) followed by recording absorbance at 405 nm.
2.8 Pinocytosis assay
To assay fluid phase endocytosis (pinocytosis) trophozoites were iced and washed in 5ml of serum and vitamin free TYI-S-33 medium. A total of 1×105 cells were exposed to 5 mg/ml fluorescein isothiocyanate-dextran (FITC-Dextran; Sigma) in a final volume of 0.5 ml in serum and vitamin free TYI-S-33 for 15 min at 37°C. Parasites were collected by centrifugation at 1000 g for 2 min, washed twice in 1ml ice-cold PBS, and lysed by the addition of 0.2 ml of 2.5% (vol/vol) Triton X-100 in PBS. Total fluorescence of the samples (100 μl) was measured using 96-well plate fluorescence reader (Tecan Spectrafluor Plus) with excitation and emission wavelengths of 485 nm and 528 nm, respectively. Measurements were corrected for autofluorescence by using the zero minute sample.
2.9 Complement resistance assays
To assay resistance of parasites to human complement, a previously published protocol was followed with some modifications (45). Briefly, a total of 1×105 trophozoites were incubated in equal volume with 50 % normal human serum (NHS) in buffer containing: 1x PBS, 0.5 mM MgCl2, 1.25mM CaCl2 for 15–30 min at 37°C. As a control for cell viability, trophozoites were incubated with heat-inactivated NHS. Parasites were centrifuged at 1,000 g for 5 min, resuspended in 50 μl of PBS and stained with 0.2% Trypan blue dye (Gibco) to assess cell viability. Dead cells stain blue, whereas viable cells exclude the dye. A total number of parasites and those that had excluded the dye were quantified. The average number of dead cells that resulted from the incubation with heat-inactivated NHS was subtracted from the average number of dead cells incubated with 50% NHS.
2.10 Statistical analysis
Comparison of data sets between control strains and their corresponding mutant strains were made using Students T-test. Data with a p-value p<0.05 were considered significant.
3 Results
3.1 Genetic manipulation of EhROM1 by generation of catalytic mutant and gene downregulation
Our previous studies demonstrated that EhROM1 knockdown (KD) parasites in the avirulent G3 strain had decreased host cell adhesion and erythrophagocytosis (17, 32, 33). Localization of EhROM1 to the parasite surface and relocalization to phagocytic vesicles during phagocytosis (17) suggest that EhROM1 may be involved in other aspects of host-parasite interaction. In order to overcome the limitation of the previous G3-based knock down system and to study biologically relevant phenotypes in a virulent strain, we used two genetic approaches: generation and overexpression of a EhROM1 catalytic mutant, which should serve in a dominant negative fashion and downregulation of EhROM1 in a virulent strain using a newly developed RNAi-based method (40). Since the methods for gene silencing in E. histolytica are hampered by low knockdown efficiency (46), we focused this study on the comparison and validation of two genetic manipulation techniques followed by functional studies of phenotypic analyses.
E. histolytica rhomboid protease 1 possesses the catalytic motif (GASGG), which is highly conserved among rhomboid proteases and also contains a conserved catalytic dyad comprised of serine S188 and histidine H242, which are required for protease function (1). By substituting serine S188 with alanine, we generated a Myc-tagged EhROM1-SA catalytic mutant, which should function in a dominant negative fashion. Importantly, we have previously published that EhROM1 proteolytic activity was abolished with a catalytic serine to alanine mutant (syn-EhR1-SA), in an in vitro cleavage assay against Plasmodium adhesins BAEBL, Rh4, AMA1, and TRAP (16). Furthermore, similar approaches of catalytic mutants serving in a dominant negative manner have been successful in the mammalian mitochondrial rhomboid protease, PARL and in T. gondii (18, 47). E. histolytica wild type EhROM1 gene (EhROM1-OX) and the catalytic mutant EhROM1-SA were both cloned into a vector containing the E. histolytica cysteine synthase promoter, resulting in an N-terminal Myc-tag-protein fusion and ensuring the expression of each protein (39). EhROM1-SA stably transfected parasite line and two control cell lines (one overexpressing the luciferase protein and a second one with EhROM1 overexpression) were obtained at 12 μg/ml of G418 drug selection. Since drug selection can affect the phenotypes tested, the same antibiotic selection was used for all cell lines. In our experience, parasites expressing luciferase largely behave similarly to wild type parasites (2), which encouraged us to use the luciferase overexpressing cell line as our principal control.
Overexpression of Myc-tagged EhROM1 and EhROM1-SA could be detected by Western blot analysis; the endogenous protein level of EhROM1 was low compared to highly overexpressed Myc-tagged EhROM1 and EhROM1-SA proteins (Figure 1A). Knockdown of EhROM1 was achieved according to an RNAi-based method recently developed by our group (40). In brief, the first 132bp of the EHI_197520 coding sequence, to which large numbers of antisense small RNAs map, is designated as a “trigger”-sequence. This trigger was fused with the full-length open reading frame of the EhROM1 gene. Transfection of E. histolytica HM-1:IMSS trophozoites with the silencing construct resulted in parasites with robust gene knock down of EhROM1 with undetectable mRNA as verified by northern blot analysis and RT PCR analysis; these parasites were named Trigger-EhROM1-silenced (T-EhROM1-s) (40). Despite significant reduction of EhROM1 transcript level, expression of the closest homologous gene, EhROM2 (EHI_060330, with 56% sequence identity to EhROM1) was not affected, nor was a recoded EhROM1 targeted for silencing; both controls point to the specificity of EhROM1 silencing (40). T-EhROM1-s stably transfected parasite line and a corresponding control cell line T-luciferase-s (trigger silenced luciferase gene) at 6 μg/ml of G418 were obtained. We confirm a significant reduction of EhROM1 protein level in the T-EhROM1-s cell line (Figure 1B).
Figure 1. Protein levels of endogenous EhROM1 (untransfected), overexpressed Myc-tagged EhROM1, EhROM1-SA, and T-EhROM1-s in E. histolytica HM-1:IMSS trophozoites.
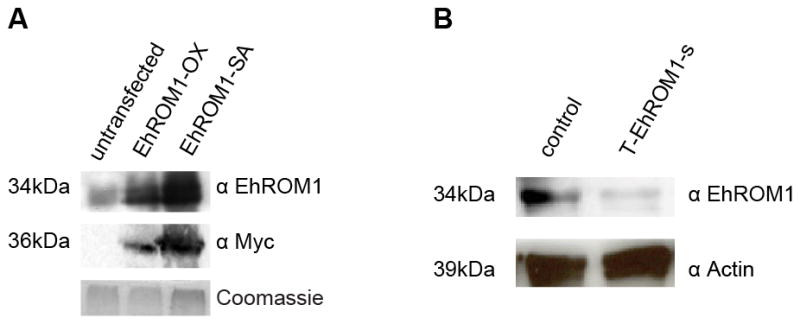
(A) Western blot analysis of wild type (WT) untransfected cells and parasites overexpressing Myc-EhROM1 or Myc-EhROM1-SA with the antibody against EhROM1 resulted in positive signal at the predicted size of 34kDa. Signal against Myc-tag was only present in parasites overexpressing Myc-EhROM1 or Myc-EhROM1-SA protein. Blots were probed with antibodies to EhROM1 (1:20), Myc (1:1000), and EhROM3 (1:500) as a loading control. Coomassie stain was used as an additional loading control. (B) Western blot analysis of EhROM1 resulted in positive signal at the predicted size of 34kDa in the control strain T-luciferase-s and strongly reduced signal in the T-EhROM1-s strain. Blots were probed with antibodies to EhROM1 (1:20) and Actin (1:1000) as a loading control. The control strain is trigger-silencing of the luciferase gene and all parasites are maintained at 6 μg/ml G418 drug concentration.
3.2 Downregulation of EhROM1 results in decreased parasitic cytotoxicity and hemolysis
The cytotoxic activity of E. histolytica is a multistep process and is a major contributor to the virulence phenotype of the parasite (27, 28, 48). The E. histolytica surface carbohydrate-binding lectin is a key virulence factor (26) and is strongly implicated in contact-dependent cytotoxicity and host cell adhesion (27, 28). Our earlier data demonstrate that EhROM1 functions as a sheddase for the amebic Gal/GalNAc lectin (16), suggesting that EhROM1 may play a role in regulation of parasite cytotoxic activity. The previous knockdown of EhROM1 was performed in the avirulent G3 stain and thus cytotoxicity could not be assessed (17). To address the effect of EhROM1 on parasite cytotoxicity, we used the EhROM1-SA mutant and the T-EhROM1-s cell lines. Chinese hamster ovary (CHO) cell monolayer destruction assays and hemolysis of human red blood cells (hRBCs) were performed. Both the EhROM1-SA mutant and the T-EhROM1-s cell lines were significantly attenuated in their ability to destroy mammalian monolayers and to lyse erythrocytes (Figure 2).
Figure 2. Overexpression of EhROM1-SA and downregulation of EhROM1 in E. histolytica trophozoites result in reduced cytotoxicity and hemolytic activity.
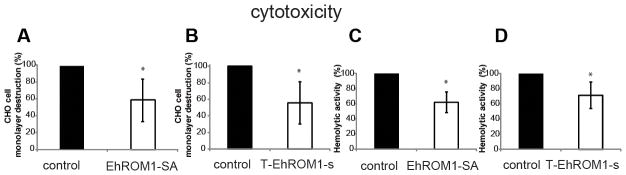
(A and B) Cytotoxicity was measured by placing a total of 0.5×105 trophozoites on a confluent CHO cell monolayer for 2h at 37°C, followed by fixation, staining with 0.1% methylene blue, dye extraction, and spectrophotometric determinations at 650 nm. Overexpression of EhROM1-SA as well as downregulation of EhROM1 resulted in parasites with decreased ability to destroy CHO cell monolayer. *p-value <0.001 control versus EhROM1-SA; *p-value <0.039 control versus T-EhROM1-s. (C and D) Hemolytic activity was measured by incubating human erythrocytes with trophozoites in 100:1 ratio for 1h and released hemoglobin quantified at the absorbance of 405 nm. Overexpression of EhROM1-SA and downregulation of EhROM1 resulted in parasites with decreased hemolytic activity. *p-value <0.0005 control versus EhROM1-SA; *p- value <0.018 control versus T-EhROM1-s. The results represent the mean and standard deviations of three to five independent experiments and are expressed as the percentage of the control strain destruction level or hemolytic activity. The control cell line is one that is overexpressing (A and C) or silencing (B and D) the luciferase gene. The G418 drug concentration in (A and C) is at 12μg/ml and in (B and D) is at 6μg/ml.
Overall, the level of monolayer destruction by parasites overexpressing the EhROM1-SA catalytic mutant was comparable to trophozoites with silenced EhROM1. EhROM1-SA cannot cleave amebic Gal/GalNAc lectin in an in vitro cleavage assay (16). Thus, the fact that EhROM1-SA affected parasite cytotoxicity, a phenotype regulated by the Gal/GalNAc lectin, is a consistent observation. Although both EhROM1 mutants were also similarly reduced in their hemolytic activity, parasites overexpressing the EhROM1-SA catalytic mutant displayed a greater defect in hemolysis than parasites with silenced EhROM1. Overexpression of wild type EhROM1 did not result in increased cytotoxicity compared to the control strain (data not shown). Reduced cytotoxicity is reminiscent of the severely impaired invasion noted with genetic manipulation of the T. gondii rhomboid protease TgROM4 (15), which is important for shedding of MIC2 a microneme adhesive protein essential for efficient Toxoplasma invasion (49).
3.3 Downregulation of EhROM1 results in parasites with strongly reduced directed and non-directed transwell migration
E. histolytica motility is required to destroy colonic epithelium and parasites with reduced motility are less virulent (35, 37, 38). The decreased cytotoxicity of both EhROM1 mutants prompted us to further investigate whether their motility was also impaired. Therefore we monitored the ability of parasites to migrate through 8μm pores of transwell inserts. In previous studies, no significant defect in transwell migration was noted in ROM (KD) parasites compared to parental G3 strain (17). Those studies used 5-chloromethylfluorescein diacetate (CMFDA) cell tracker dye to label parasites. This dye impairs the general fitness of trophozoites and thus may have negatively impacted motility. In the current study we measured transwell migration directly by counting parasites that had migrated through the pores as has been described in other studies involved in migration analysis of surface proteases (9, 50). Using the direct counting approach, the data indicate that parasites overexpressing catalytic mutant, EhROM1-SA, and trophozoites with silenced EhROM1 resulted in severely (~90%) diminished ability to migrate through the transwell inserts compared to control parasites (Figure 3A and 3B). Precedence for rhomboid proteases with similar roles in migration has been noted. Downregulation of mammalian rhomboid protease RHBDL2 resulted in dramatically impaired transwell migration along with impaired cutaneous wound healing process (9). Conditional knock out of T. gondii TgROM4 resulted in parasites with greater adherence, but impaired motility and cell entry (15), whereas Plasmodium TRAP (the presumptive ROM4 substrate in vivo) rhomboid cleavage site mutants were defective in TRAP shedding and displayed slow, staccato gliding motility and reduced in sporozoite infectivity (22). Similarly the strongly impeded migration phenotype associated with EhROM1 manipulation may be due to impaired cleavage and function of the Gal/GalNAc lectin, which is an EhROM1 substrate (16).
Figure 3. Overexpression of EhROM1-SA and downregulation of EhROM1 in E. histolytica trophozoites result in dramatically decreased motility and chemotaxis.
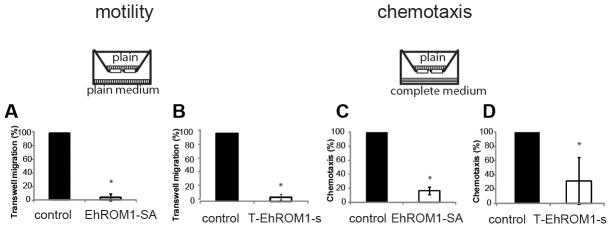
Motility and chemotaxis were measured by assessing the number of parasites that migrated through the transwell chamber. A total of 1.5×105 parasites were added to the upper chamber of a transwell system and allowed to migrate into the lower chamber for 3 h at 37°C. Overexpression of EhROM1-SA and downregulation of EhROM1 resulted in parasites with strongly diminished migration properties (A and B) and chemotaxis (C and D) though the transwell compared to the control. The average of three independent experiments is shown with standard deviation. Data are shown as a percentage of control strain motility or chemotaxis. The control cell line is overexpressing (A and C) or silencing (B and D) the luciferase gene. The G418 drug concentration in (A and C) is at 12 μg/ml and in (B and D) at 6 μg/ml. Motility: *p-value <5×10−6 control versus EhROM1-SA; *p-value <2×10−6 control versus T-EhROM1-s; Chemotaxis: *p-value <1.2 ×10−5 control versus EhROM1-SA; *p-value <0.02 control versus T-EhROM1-s. Cartoon represents transwell migration chamber with vertical bars representing plain TYI-S-33 media; horizontal bars represent TYI-S-33 media completed with serum and vitamins.
In addition to examining non-directional transwell migration, we also assayed directional migration properties towards a nutrient source by placing rich medium on the bottom chamber while the parasites are in a nutrient-poor upper chamber. This approach should provide preliminary insights as to whether rhomboid protease EhROM1 might be involved in regulation of directed movement or chemotaxis. Via its interaction with the Gal/GalNAc lectin, EhROM1 might be involved in cell signaling and chemotaxis. Blazquez et al. previously reported that E. histolytica trophozoites blocked for Gal/GalNAc lectin signaling were inhibited in their oriented movement towards tumor necrosis factor (51). We assayed the directed migration of both EhROM1 mutants by monitoring their ability to move through 8μm pores from plain to rich medium. Both the catalytic mutant and the trigger knockdown EhROM1 strain were strongly diminished in their ability to migrate through the transwell inserts in response to a nutrient gradient compared to control cells (Figure 3C and 3D). Overall, directionally oriented movement was less affected compared to non-oriented movement from plain medium to plain medium. Parasites overexpressing the catalytic EhROM1-SA mutant were almost completely inhibited in non-oriented transwell movement, and were reduced by ~80% in their directed migration towards nutrient rich medium. Trophozoites with silenced EhROM1 were reduced by ~90% attenuation with non-oriented movement, and were reduced ~70% in their directed migration towards nutrient rich medium (Figure 3). There was overall greater migration of wild-type parasites in response to the chemotaxis stimulus (completed medium) compared to migration in plain medium, indicating that the chemotaxis stimulus functioned as expected (data not shown). In summary, parasites with EhROM1 genetically manipulated were strongly impeded in their ability to cross a transwell filter membrane. Reduction in motility was noted in TgROM4 conditional knock out parasites, which were unable to process surface adhesins resulting in disoriented twirling movement and impeded gliding motility (15).
3.4 Overexpression of EhROM1-SA mutant had no effect on erythrophagocytosis in contrast to T-EhROM1-s parasites, which had reduced erythrophagocytosis
Erythrophagocytosis is a phenotype associated with amebic virulence (36). In our previous studies it was shown that G3 parasites with downregulated EhROM1 were reduced in their ability to ingest red blood cells (17). Here we aimed to investigate whether we could confirm this previously observed phenotype, given the earlier observed defect in cytotoxicity. E. histolytica trophozoites were co-incubated with human RBC, and ingested erythrocytes were measured by recording hemoglobin absorbance at 405 nm after lysing the parasites. Parasites overexpressing the catalytic EhROM1-SA mutant (Figure 4A) and parasites overexpressing wild-type EhROM1 (Supplementary Figure 1C and data not shown) did not have any significant changes in their phagocytic ability compared to controls. Mutation of the catalytic serine, which is essential for protease function might thus be dispensable for other EhROM1 functions (i.e. signaling), thus resulting in no impact on erythrophagocytosis. Parasites with silenced EhROM1 displayed a moderate defect (~20%) in erythrophagocytosis similar to previous results in G3 parasites with downregulated EhROM1 (17) (Figure 4B), implying that EhROM1 might only have a secondary or indirect impact on phagocytosis.
Figure 4. Downregulation of EhROM1 results in differing phenotypes in erythrophagocytosis between EhROM1-SA and EhROM1 knock down but comparable defects of both EhROM1 mutants in pinocytosis with no significant effect on complement resistance.
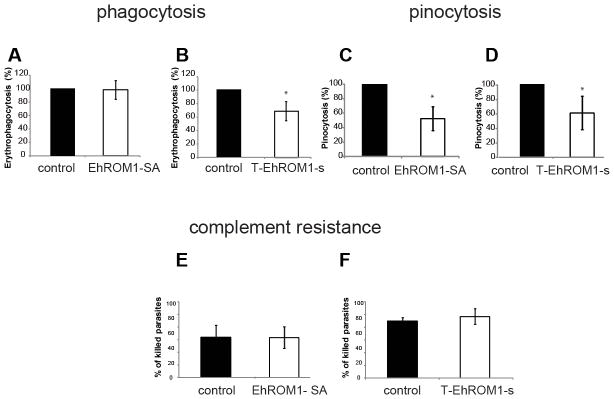
Erythrophagocytosis was measured by incubation 1×105 trophozoites with 1×108 hRBC for 15 min at 37°C, followed by lysis of extracellular hRBC and measurement of ingested erythrocytes at 405 nm. No significant changes in phagocytosis of hRBC noted in E. histolytica strains that overexpressed mutant EhROM1-SA compared to control (A), but parasite cell line T-EhROM1-s displayed a moderate defect in erythrophagocytosis (B) *p-value <0.005 control versus T-EhROM1-s parasites; The results represent the mean and standard deviations of three to four independent experiments and are expressed as the percentage of the control strain erythrophagocytosis level. Pinocytosis (C and D) is assayed by incorporation of fluorescein isothiocyanate-dextran by 1×105 trophozoites and total fluorescence with wavelengths of 485 nm and 528 nm is measured. Parasites overexpressing mutant EhROM1-SA protein (C) and EhROM1 knock down mutant (D) were reduced in their ability to incorporate dextran.*p-value <0.007 control versus EhROM1-SA parasites; *p-value <0.01 control versus T-EhROM1-s parasites; The average of three independent experiments is shown with standard deviation. Data are shown as a percentage of control strain pinocytosis. Complement resistance was measured by incubation of 1 × 105 trophozoites in equal volume with 50 % normal human serum (NHS) 15–30 min at 37°C (E and F). As a control for cell viability, trophozoites were incubated with heat-inactivated NHS and stained with 0.2% Trypan blue dye to assess cell viability. The average number of dead cells resulting from the incubation with heat-inactivated NHS was subtracted from the average number of dead cells incubated with 50% NHS. No significant changes in complement resistance in parasite cell lines EhROM1-SA (E) and T-EhROM1-s (F) were noted compared to control. The average of three independent experiments is shown with standard deviation. Data are shown as a percentage of killed parasites. The control cell line is overexpressing (A, C, E) or silencing (B, D, F) the luciferase gene. The G418 drug concentration in (A, C, E) is at 12 μg/ml and in (B, D, F) is at 6 μg/ml.
3.5 Downregulation of EhROM1 results in parasites with significantly reduced pinocytosis
Fluid-phase endocytosis or pinocytosis is important for amebic nutrient acquisition (52) and has been described as one of E. histolytica virulence functions (53, 54). Since the ability to ingest human RBCs by parasites overexpressing catalytic EhROM1-SA mutant differed from that of parasites with downregulated EhROM1, we aimed to examine whether there were any differences in fluid-phase endocytosis. Trophozoites were exposed to fluorescein isothiocyanate-conjugated dextran as a fluid-phase marker and the fluorescence of the ingested dextran was assessed. Both independently generated EhROM1 mutants were similarly diminished in their ability to ingest dextran. Parasites overexpressing mutant EhROM1-SA were reduced by ~50% in their pinocytosis ability and trophozoites with downregulated EhROM1 displayed a moderate but significant defect of ~40% compared to the control cell line (Figure 4C and 4D). This implies that a direct involvement of EhROM1 protease activity is required for function within the endocytosis pathways in contrast to phagocytosis.
3.6 Downregulation of EhROM1 has no significant effect on complement resistance
E. histolytica is known to employ numerous strategies to avoid lysis by the host complement system (55–59). One of these processes is surface receptor capping, which is induced by the presence of antibodies against parasite surface proteins and results in their polarization and release from the parasite (45, 56). The amebic Gal/GalNAc lectin is one of the proteins targeted to the cap for release (45, 56). In our previous work we observed relocalization of EhROM1 during surface receptor capping to the cap, suggesting that EhROM1 may facilitate E. histolytica escape from the host cellular immune response. However G3-ROM (KD) parasites formed caps that were morphologically indistinguishable from G3 parasites and no change in complement resistance was noted (17). We assessed the newly generated EhROM1 mutants and their susceptibility to human complement. Complement resistance was measured by incubating EhROM1-SA mutant and T-EhROM1 silenced parasites with human serum for 15 or 30 min followed by trypan blue staining to assess parasite viability. No significant changes in parasite viability compared to control cell lines were noted (Figure 4E and 4F). The general viability of EhROM1-SA parasites was higher (~50% survival) compared to T-EhROM1-s parasites (~20% survival).
4 Discussion
Intramembrane proteolysis mediated by rhomboid proteases is conserved in a range of organisms from bacteria to mammals with roles in bacterial quorum sensing, initiation of cell signaling events, regulation of mitochondrial homeostasis, and dismantling of adhesion junctions of parasitic protozoa (1, 4). We used two methods of genetic manipulation of the E. histolytica rhomboid protease gene, EhROM1, to demonstrate that this protein has effects on multiple aspects of amebic virulence. Overexpression of a catalytically inactive EhROM1 mutant mimicked the phenotype obtained by downregulation of EhROM1. Both parasite strains were similarly attenuated in their ability to destroy mammalian cell monolayer, hemolyze erythrocytes, ingest a fluid-phase marker and migrate through transwell chambers (summarized in Table 1). The diversity of defects identified using these approaches implies that EhROM1 may not be restricted to one substrate and that substrate profiling should be pursued in future studies.
Table 1.
Summary of phenotypes observed in Entamoeba histolytica HM-1: IMSS parasites with silenced EhROM1 expression (T-EhROM1-s) versus parasites over-expressing the EhROM1-SA gene (EhROM1-SA).
| Phenotype | EhROM1-SA | T-EhROM1-s |
|---|---|---|
| Cytotoxicity | ++ | ++ |
| Hemolytic activity | ++ | ++ |
| Erythrophagocytosis | − | + |
| Pinocytosis | ++ | + |
| Motility | +++ | +++ |
| Chemotaxis | +++ | ++ |
| Complement resistance | − | − |
+++ strong defect, ++ considerable defect, + moderate defect, − no change
Overall, the data suggest complex roles of rhomboid protease EhROM1 in ameba-host interactions. Similarly complex phenotypes were described for EhMSP-1, a surface metalloprotease involved in regulation of amebic adherence, with additional effects on cell motility, monolayer destruction, and phagocytosis (50). In a previous study the knock down of EhROM1 in the G3 strain resulted in decreased adherence to healthy mammalian cells but not towards apoptotic cells (17), suggesting that EhROM1 may directly or indirectly impact signaling during the adhesion process. The cytoplasmic domain of the Gal/GalNAc lectin contains an integrin-like motif, which regulates signaling and plays roles in adherence and virulence (60). Downregulation of EhROM1 results in decreased adherence (17), which might be due to impaired cleavage and function of the Gal/GalNAc lectin leading to reduced cytotoxicity towards cell monolayers and also in strongly impeded transwell migration. A conditional knock out of T. gondii rhomboid protease TgROM4 results in parasites unable to process surface adhesins (e.g. AMA1), displaying disoriented twirling movement and impeded gliding motility (15). Robust gliding motility and efficient invasion requires constant disengagement of Plasmodium TRAP from the sporozoite surface and extracellular matrix (or host cell receptors) by a rhomboid protease (22). Interestingly, in COS cell assays TRAP is effectively cleaved by wild type EhROM1 but not by a catalytic mutant (16), implying EhROM1 may have a role in regulating motility. Apicomplexan motility is a substrate-dependent form of locomotion, which occurs by gliding and that does not involve significant change in cell shape, in contrast to the crawling motility of ameba (61). However both modes of motility are similarly engaged to move across substrates and to actively invade host cells, since non-motile Plasmodium parasites fail to invade host cells (22) whereas amebic trophozoites are less virulent (35, 37, 38).
There is also growing body of evidence indicating that mammalian rhomboid proteases are involved in cell migration with implications for wound healing and cancer (9, 62). Downregulation of mammalian rhomboid protease RHBDL2 resulted in dramatically impaired transwell migration of keratinocyte cell lines (9). Moreover, the fact that oriented movement was strongly impeded in trophozoites inhibited for Gal/GalNAc function and also that genes encoding Gal/GalNAc lectin were upregulated during chemotaxis and involved in cytoskeleton dynamics (51) matches our previous results as Gal/GalNAc lectin being a substrate of EhROM1 (16).
Our data represent the first study of a rhomboid protease in an extracellular human parasite. Although intriguing functional diversity has been described for rhomboid proteases across evolution (63), to date little is known about roles of rhomboid proteases in phagocytosis or pinocytosis. However mutants of mitochondrial rhomboid protease PARL are impaired in mitophagy, a process known as autophagic removal of damaged mitochondria, suggesting that defects in mitophagy may be an underlying mechanism of Parkinson’s disease (47). Interestingly, endocytic events that are important for the nutrient uptake rely on cholesterol containing lipid raft membranes enriched with heavy and light subunits of Gal/GalNAc lectin (52, 53). Cholesterol depletion results in reduced pinocytosis and adhesion to host cells as a likely consequence of lectin loss (52, 53) suggesting that EhROM1 might be transiently targeted to lipid rafts where it interacts with the Gal/GalNAc lectin.
Parasites with silenced EhROM1 match the phenotype of reduced erythrophagocytosis noted previously in the G3 strain (17). The difference in phenotype between the EhROM1-SA mutant and downregulated EhROM1 in erythrophagocytosis could be explained by the fact that the intact endogenous copy of EhROM1 is sufficient to overcome or compensate for the dominant negative suppression. Alternatively, rhomboid proteases often “share” substrates with other proteases such as metalloproteases (64) such that only inhibition of background cleavage would reveal the target of rhomboid protease. According to our data, mutation of the catalytic serine, which is absolutely essential for the protease function such as various substrate shedding, is most likely dispensable for EhROM1 signaling function. Catalytically-inactive rhomboid -like proteins (called iRhoms) still retain their important biological roles despite their lack of protease activity in such diseases as inherited condition tylosis with esophageal cancer (62) or indirectly impact tumor necrosis factor signaling by direct interaction with a disintegrin and metalloprotease called ADAM (64, 65)
The genome of E. histolytica contains four genes annotated as rhomboid proteases (17). In addition to EhROM1, EhROM3 (EHI_029220) has the catalytically important serine (S133) and histidine (H179) residues and thus might share substrate redundancy and functionally compensate for down regulation of EhROM1. Although neither ROM2 (EHI_060330) or ROM4 (EHI_128190) possesses active site residues required for protease function, we cannot rule out that they also contribute to signaling events, as data for catalytically inactive rhomboid proteases are emerging (62, 66).
5 Conclusion
In this study we genetically manipulated E. histolytica rhomboid protease EhROM1 by two different approaches and identified similarly attenuated phenotypes by both approaches. Our work provides confirmation of a new and effective RNAi-based gene silencing technique that overcomes the limitation of low knockdown efficiency in E. histolytica. Additionally, our data shed light on the complexity and importance of intramembrane proteolysis for multiple aspects of virulence in a medically important parasite. The intriguing diversity of phenotypes implies that EhROM1 may not be restricted to only one substrate target but may be similar to other rhomboid proteases, which possess multiple or overlapping targets. In summary, we highlight the unexpected role for EhROM1 in cell motility noted. Further studies are needed to fully identify the rhomboid proteases targets that impact parasite motility and chemotaxis.
Supplementary Material
(A) Cytotoxicity was measured by placing a total of 0.5×105 trophozoites on a confluent CHO cell monolayer for 2h at 37°C, followed by fixation, staining with 0.1% methylene blue, dye extraction, and spectrophotometric determinations at 650 nm. Overexpression of mutant EhROM1-SA protein resulted in parasites with decreased ability to destroy CHO cell monolayer *p-value <0.02 control versus EhROM1-SA (A). The results represent the mean and standard deviations of three independent experiments and are expressed as the percentage of the control strain destruction level. (B) Adhesion was measured with a Chinese hamster ovary (CHO) cell rosette assay. Parasites were mixed with CHO cells at 4°C for 2 hr, after which parasites with three or more CHO cells attached were counted as positive. Data shown is an average of 3 experiments each with two replicates. Standard deviation is shown. *p-value <0.006. Erythrophagocytosis (C) was measured by incubating 1×105 trophozoites with 1 ×108 hRBC for 15 min at 37°C, followed by lysis of extracellular hRBC and measurement of ingested erythrocytes at 405 nm. No significant changes in phagocytosis of hRBC were noted in E. histolytica strain that overexpressed EhROM1-SA compared to control (C). The results are expressed as the percentage of the control strain erythrophagocytosis level. The average of three independent experiments is shown with standard deviation. Motility (D) was measured by assessing the number of parasites that migrated through the transwell chamber. A total of 1.5×105 parasites were added to the upper chamber of a transwell system and allowed to migrate into the lower chamber for 3 h at 37°C. Overexpression of mutant EhROM1-SA protein resulted in parasites with reduced migration properties *p-value <0.02 control versus EhROM1-SA (D). The average of three independent experiments is shown with standard deviation. Data are shown as a percentage of control strain motility. Cartoon represents transwell migration chamber with vertical bars representing plain TYI-S-33 media. The control cell line is overexpressing wild type EhROM1 gene. The G418 drug concentration is at 12μg/ml.
HIGHLIGHTS.
Catalytic mutant and gene downregulation of Entamoeba rhomboid protease 1
ROM1 has roles in numerous aspects of parasite biology, including virulence
New roles in parasite migration and motility noted for ROM1
ROM1 role in cell migration mimics roles in mammalian and apicomplexan systems
Acknowledgments
We thank all members of the Singh lab for helpful discussion and invaluable input. We especially thank Leigh Baxt and Audrie Lin for the initial cloning of the EhROM1-SA mutant. This work was supported by Stanford University Dean’s fellowship to ER, SwissNational Foundation to LM (PBZHP3-130988 and PBZHP3_141513), and grants from the NIH (AI085178 and AI053724) to US.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 2.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci U S A. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 5.Walder K, Kerr-Bayles L, Civitarese A, Jowett J, Curran J, Elliott K, Trevaskis J, Bishara N, Zimmet P, Mandarino L, Ravussin E, Blangero J, Kissebah A, Collier GR. The mitochondrial rhomboid protease PSARL is a new candidate gene for type 2 diabetes. Diabetologia. 2005;48:459–468. doi: 10.1007/s00125-005-1675-9. [DOI] [PubMed] [Google Scholar]
- 6.Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- 7.Greene JG. Current status and future directions of gene expression profiling in Parkinson’s disease. Neurobiol Dis. 2012;45:76–82. doi: 10.1016/j.nbd.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjuan Szklarz LK, Scorrano L. The antiapoptotic OPA1/Parl couple participates in mitochondrial adaptation to heat shock. Biochim Biophys Acta. 2012;1817:1886–1893. doi: 10.1016/j.bbabio.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng TL, Wu YT, Lin HY, Hsu FC, Liu SK, Chang BI, Chen WS, Lai CH, Shi GY, Wu HL. Functions of rhomboid family protease RHBDL2 and thrombomodulin in wound healing. J Invest Dermatol. 2011;131:2486–2494. doi: 10.1038/jid.2011.230. [DOI] [PubMed] [Google Scholar]
- 10.Greenblatt EJ, Olzmann JA, Kopito RR. Making the cut: intramembrane cleavage by a rhomboid protease promotes ERAD. Nat Struct Mol Biol. 2012;19:979–981. doi: 10.1038/nsmb.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos MJ, Graindorge A, Soldati-Favre D. New insights into parasite rhomboid proteases. Mol Biochem Parasitol. 2012;182:27–36. doi: 10.1016/j.molbiopara.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc Natl Acad Sci U S A. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brossier F, Starnes GL, Beatty WL, Sibley LD. Microneme rhomboid protease TgROM1 is required for efficient intracellular growth of Toxoplasma gondii. Eukaryot Cell. 2008;7:664–674. doi: 10.1128/EC.00331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowse TJ, Koussis K, Blackman MJ, Soldati-Favre D. Roles of proteases during invasion and egress by Plasmodium and Toxoplasma. Subcell Biochem. 2008;47:121–139. doi: 10.1007/978-0-387-78267-6_10. [DOI] [PubMed] [Google Scholar]
- 15.Buguliskis JS, Brossier F, Shuman J, Sibley LD. Rhomboid 4 (ROM4) affects the processing of surface adhesins and facilitates host cell invasion by Toxoplasma gondii. PLoS Pathog. 2010;6:e1000858. doi: 10.1371/journal.ppat.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baxt LA, Baker RP, Singh U, Urban S. An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev. 2008;22:1636–1646. doi: 10.1101/gad.1667708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baxt LA, Rastew E, Bracha R, Mirelman D, Singh U. Downregulation of an Entamoeba histolytica rhomboid protease reveals roles in regulating parasite adhesion and phagocytosis. Eukaryot Cell. 2010;9:1283–1293. doi: 10.1128/EC.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos JM, Ferguson DJ, Blackman MJ, Soldati-Favre D. Intramembrane cleavage of AMA1 triggers Toxoplasma to switch from an invasive to a replicative mode. Science. 2011;331:473–477. doi: 10.1126/science.1199284. [DOI] [PubMed] [Google Scholar]
- 19.Lin JW, Meireles P, Prudencio M, Engelmann S, Annoura T, Sajid M, Chevalley-Maurel S, Ramesar J, Nahar C, Avramut CM, Koster AJ, Matuschewski K, Waters AP, Janse CJ, Mair GR, Khan SM. Loss-of-function analyses defines vital and redundant functions of the Plasmodium rhomboid protease family. Mol Microbiol. 2013;88:318–338. doi: 10.1111/mmi.12187. [DOI] [PubMed] [Google Scholar]
- 20.Vera IM, Beatty WL, Sinnis P, Kim K. Plasmodium protease ROM1 is important for proper formation of the parasitophorous vacuole. PLoS Pathog. 2011;7:e1002197. doi: 10.1371/journal.ppat.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger TW, Blackman MJ. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J Cell Biol. 2006;174:1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ejigiri I, Ragheb DR, Pino P, Coppi A, Bennett BL, Soldati-Favre D, Sinnis P. Shedding of TRAP by a rhomboid protease from the malaria sporozoite surface is essential for gliding motility and sporozoite infectivity. PLoS Pathog. 2012;8:e1002725. doi: 10.1371/journal.ppat.1002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heiman FL, Wertheim PHaJPW. Atlas of Human Infectious Diseases. 1. Blackwell Publishing Ltd; 2012. [Google Scholar]
- 24.Stauffer W, Ravdin JI. Entamoeba histolytica: an update. Curr Opin Infect Dis. 2003;16:479–485. doi: 10.1097/00001432-200310000-00016. [DOI] [PubMed] [Google Scholar]
- 25.Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N Engl J Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 26.Ravdin JI, Murphy CF, Salata RA, Guerrant RL, Hewlett EL. N-Acetyl-D-galactosamine-inhibitable adherence lectin of Entamoeba histolytica. I. Partial purification and relation to amoebic virulence in vitro. J Infect Dis. 1985;151:804–815. doi: 10.1093/infdis/151.5.804. [DOI] [PubMed] [Google Scholar]
- 27.Petri WA, Jr, Smith RD, Schlesinger PH, Murphy CF, Ravdin JI. Isolation of the galactose-binding lectin that mediates the in vitro adherence of Entamoeba histolytica. J Clin Invest. 1987;80:1238–1244. doi: 10.1172/JCI113198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffer LD, Petri WA., Jr Role of the galactose lectin of Entamoeba histolytica in adherence-dependent killing of mammalian cells. Infect Immun. 1991;59:4681–4683. doi: 10.1128/iai.59.12.4681-4683.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moncada D, Keller K, Ankri S, Mirelman D, Chadee K. Antisense inhibition of Entamoeba histolytica cysteine proteases inhibits colonic mucus degradation. Gastroenterology. 2006;130:721–730. doi: 10.1053/j.gastro.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Tillack M, Nowak N, Lotter H, Bracha R, Mirelman D, Tannich E, Bruchhaus I. Increased expression of the major cysteine proteinases by stable episomal transfection underlines the important role of EhCP5 for the pathogenicity of Entamoeba histolytica. Mol Biochem Parasitol. 2006;149:58–64. doi: 10.1016/j.molbiopara.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Bracha R, Nuchamowitz Y, Anbar M, Mirelman D. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2006;2:e48. doi: 10.1371/journal.ppat.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracha R, Nuchamowitz Y, Mirelman D. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot Cell. 2003;2:295–305. doi: 10.1128/EC.2.2.295-305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bujanover S, Katz U, Bracha R, Mirelman D. A virulence attenuated amoebapore-less mutant of Entamoeba histolytica and its interaction with host cells. Int J Parasitol. 2003;33:1655–1663. doi: 10.1016/s0020-7519(03)00268-6. [DOI] [PubMed] [Google Scholar]
- 34.Winkelmann J, Leippe M, Bruhn H. A novel saposin-like protein of Entamoeba histolytica with membrane-fusogenic activity. Mol Biochem Parasitol. 2006;147:85–94. doi: 10.1016/j.molbiopara.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Labruyere E, Guillen N. Host tissue invasion by Entamoeba histolytica is powered by motility and phagocytosis. Arch Med Res. 2006;37:253–258. doi: 10.1016/j.arcmed.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Griffin JL. Human amebic dysentery. Electron microscopy of Entamoeba histolytica contacting, ingesting, and digesting inflammatory cells. Am J Trop Med Hyg. 1972;21:895–906. [PubMed] [Google Scholar]
- 37.Voigt H, Olivo JC, Sansonetti P, Guillen N. Myosin IB from Entamoeba histolytica is involved in phagocytosis of human erythrocytes. J Cell Sci. 1999;112 (Pt 8):1191–1201. doi: 10.1242/jcs.112.8.1191. [DOI] [PubMed] [Google Scholar]
- 38.Guillen N. Role of signalling and cytoskeletal rearrangements in the pathogenesis of Entamoeba histolytica. Trends Microbiol. 1996;4:191–197. doi: 10.1016/0966-842x(96)10033-0. [DOI] [PubMed] [Google Scholar]
- 39.Saito-Nakano Y, Yasuda T, Nakada-Tsukui K, Leippe M, Nozaki T. Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica. J Biol Chem. 2004;279:49497–49507. doi: 10.1074/jbc.M403987200. [DOI] [PubMed] [Google Scholar]
- 40.Morf LPJM, Pearson RJ, Wang AS, Singh U. Robust gene silencing mediated by antisense small RNAs in the pathogenic protist Entamoeba histolytica. Nucleic Acids Research. 2013;41:9424–9437. doi: 10.1093/nar/gkt717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 42.Hellberg A, Nickel R, Lotter H, Tannich E, Bruchhaus I. Overexpression of cysteine proteinase 2 in Entamoeba histolytica or Entamoeba dispar increases amoeba-induced monolayer destruction in vitro but does not augment amoebic liver abscess formation in gerbils. Cell Microbiol. 2001;3:13–20. doi: 10.1046/j.1462-5822.2001.00086.x. [DOI] [PubMed] [Google Scholar]
- 43.Biller L, Schmidt H, Krause E, Gelhaus C, Matthiesen J, Handal G, Lotter H, Janssen O, Tannich E, Bruchhaus I. Comparison of two genetically related Entamoeba histolytica cell lines derived from the same isolate with different pathogenic properties. Proteomics. 2009;9:4107–4120. doi: 10.1002/pmic.200900022. [DOI] [PubMed] [Google Scholar]
- 44.Mora-Galindo J, Gutierrez-Lozano M, Anaya-Velazquez F. Entamoeba histolytica: kinetics of hemolytic activity, erythrophagocytosis and digestion of erythrocytes. Arch Med Res. 1997;28(Spec No):200–201. [PubMed] [Google Scholar]
- 45.Calderon J, Tovar R. Loss of susceptibility to complement lysis in Entamoeba histolytica HM1 by treatment with human serum. Immunology. 1986;58:467–471. [PMC free article] [PubMed] [Google Scholar]
- 46.MacFarlane RC, Singh U. Loss of dsRNA-based gene silencing in Entamoeba histolytica: implications for approaches to genetic analysis. Exp Parasitol. 2008;119:296–300. doi: 10.1016/j.exppara.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, Ross OA, Farrer M, McQuibban GA, Bulman DE. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum Mol Genet. 2011;20:1966–1974. doi: 10.1093/hmg/ddr077. [DOI] [PubMed] [Google Scholar]
- 48.Huston CD, Houpt ER, Mann BJ, Hahn CS, Petri WA., Jr Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell Microbiol. 2000;2:617–625. doi: 10.1046/j.1462-5822.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 49.Wan KL, Carruthers VB, Sibley LD, Ajioka JW. Molecular characterisation of an expressed sequence tag locus of Toxoplasma gondii encoding the micronemal protein MIC2. Mol Biochem Parasitol. 1997;84:203–214. doi: 10.1016/s0166-6851(96)02796-x. [DOI] [PubMed] [Google Scholar]
- 50.Teixeira JE, Sateriale A, Bessoff KE, Huston CD. Control of Entamoeba histolytica adherence involves metallosurface protease 1, an M8 family surface metalloprotease with homology to leishmanolysin. Infect Immun. 2012;80:2165–2176. doi: 10.1128/IAI.06389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blazquez S, Guigon G, Weber C, Syan S, Sismeiro O, Coppee JY, Labruyere E, Guillen N. Chemotaxis of Entamoeba histolytica towards the pro-inflammatory cytokine TNF is based on PI3K signalling, cytoskeleton reorganization and the Galactose/N-acetylgalactosamine lectin activity. Cell Microbiol. 2008;10:1676–1686. doi: 10.1111/j.1462-5822.2008.01158.x. [DOI] [PubMed] [Google Scholar]
- 52.Goldston AM, Powell RR, Temesvari LA. Sink or swim: lipid rafts in parasite pathogenesis. Trends Parasitol. 2012;28:417–426. doi: 10.1016/j.pt.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laughlin RC, McGugan GC, Powell RR, Welter BH, Temesvari LA. Involvement of raft-like plasma membrane domains of Entamoeba histolytica in pinocytosis and adhesion. Infect Immun. 2004;72:5349–5357. doi: 10.1128/IAI.72.9.5349-5357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimmer S, van Deurs B, Sandvig K. Membrane ruffling and macropinocytosis in A431 cells require cholesterol. J Cell Sci. 2002;115:2953–2962. doi: 10.1242/jcs.115.14.2953. [DOI] [PubMed] [Google Scholar]
- 55.Reed SL, Ember JA, Herdman DS, DiScipio RG, Hugli TE, Gigli I. The extracellular neutral cysteine proteinase of Entamoeba histolytica degrades anaphylatoxins C3a and C5a. J Immunol. 1995;155:266–274. [PubMed] [Google Scholar]
- 56.Calderon J. Dynamic changes on the surface of Entamoeba induced by antibodies. Arch Invest Med (Mex) 1980;11:55–61. [PubMed] [Google Scholar]
- 57.Espinosa-Cantellano M, Martinez-Palomo A. Entamoeba histolytica: mechanism of surface receptor capping. Exp Parasitol. 1994;79:424–435. doi: 10.1006/expr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 58.Hamelmann C, Foerster B, Burchard GD, Shetty N, Horstmann RD. Induction of complement resistance in cloned pathogenic Entamoeba histolytica. Parasite Immunol. 1993;15:223–228. doi: 10.1111/j.1365-3024.1993.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 59.Silva PP, Martinez-Palomo A, Gonzalez-Robles A. Membrane structure and surface coat of Entamoeba histolytica. Topochemistry and dynamics of the cell surface: cap formation and microexudate. J Cell Biol. 1975;64:538–550. doi: 10.1083/jcb.64.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vines RR, Ramakrishnan G, Rogers JB, Lockhart LA, Mann BJ, Petri WA., Jr Regulation of adherence and virulence by the Entamoeba histolytica lectin cytoplasmic domain, which contains a beta2 integrin motif. Mol Biol Cell. 1998;9:2069–2079. doi: 10.1091/mbc.9.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchison TJCL. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- 62.Blaydon DC, Etheridge SL, Risk JM, Hennies HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr NK, Bishop DT, Ellis A, Jankowski J, Field JK, Leigh IM, South AP, Kelsell DP. RHBDF2 mutations are associated with tylosis, a familial esophageal cancer syndrome. Am J Hum Genet. 2012;90:340–346. doi: 10.1016/j.ajhg.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freeman M. Rhomboids: 7 years of a new protease family. Semin Cell Dev Biol. 2009;20:231–239. doi: 10.1016/j.semcdb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, Lang KS, Haussinger D, Wakeham A, Itie-Youten A, Khokha R, Ohashi PS, Blobel CP, Mak TW. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Cytotoxicity was measured by placing a total of 0.5×105 trophozoites on a confluent CHO cell monolayer for 2h at 37°C, followed by fixation, staining with 0.1% methylene blue, dye extraction, and spectrophotometric determinations at 650 nm. Overexpression of mutant EhROM1-SA protein resulted in parasites with decreased ability to destroy CHO cell monolayer *p-value <0.02 control versus EhROM1-SA (A). The results represent the mean and standard deviations of three independent experiments and are expressed as the percentage of the control strain destruction level. (B) Adhesion was measured with a Chinese hamster ovary (CHO) cell rosette assay. Parasites were mixed with CHO cells at 4°C for 2 hr, after which parasites with three or more CHO cells attached were counted as positive. Data shown is an average of 3 experiments each with two replicates. Standard deviation is shown. *p-value <0.006. Erythrophagocytosis (C) was measured by incubating 1×105 trophozoites with 1 ×108 hRBC for 15 min at 37°C, followed by lysis of extracellular hRBC and measurement of ingested erythrocytes at 405 nm. No significant changes in phagocytosis of hRBC were noted in E. histolytica strain that overexpressed EhROM1-SA compared to control (C). The results are expressed as the percentage of the control strain erythrophagocytosis level. The average of three independent experiments is shown with standard deviation. Motility (D) was measured by assessing the number of parasites that migrated through the transwell chamber. A total of 1.5×105 parasites were added to the upper chamber of a transwell system and allowed to migrate into the lower chamber for 3 h at 37°C. Overexpression of mutant EhROM1-SA protein resulted in parasites with reduced migration properties *p-value <0.02 control versus EhROM1-SA (D). The average of three independent experiments is shown with standard deviation. Data are shown as a percentage of control strain motility. Cartoon represents transwell migration chamber with vertical bars representing plain TYI-S-33 media. The control cell line is overexpressing wild type EhROM1 gene. The G418 drug concentration is at 12μg/ml.


