Abstract
Purpose
To implement a multidirectional motion encoding scheme for magnetic resonance elastography (MRE) of the human brain with reduced acquisition time, and investigate its performance relative to a conventional MRE scheme.
Materials and Methods
The sample interval modulation (SLIM) scheme was implemented in a multishot, variable density spiral MRE sequence. The brains of seven healthy volunteers were investigated with both SLIM-MRE and conventional MRE acquisitions in a single imaging session on a clinical 3 T MRI scanner with 50 Hz vibration. Following extraction of displacement fields, complex shear modulus property maps were estimated for each encoding acquisition.
Results
The SLIM-MRE and conventional MRE acquisitions produced deformation fields that were nearly identical and exhibited an average correlation coefficient of 0.95 (all p < 0.05). Average properties of white matter differed between the two acquisitions by less than 5% for all volunteers, which is better than reproducibility estimates for conventional MRE alone.
Conclusions
The use of SLIM provides very similar quantitative property estimates compared to the conventional MRE encoding scheme. The SLIM acquisition is 2.5 times faster than the conventional acquisition, and may improve the adoption of MRE in clinical settings.
Keywords: magnetic resonance elastography, multidirectional motion encoding, displacement field, brain mechanical properties, multishot spiral
Introduction
In magnetic resonance elastography (MRE), phase contrast-based MRI sequences are synchronized with an applied harmonic vibration to image the resulting mechanical wave motion [1]. Subsequently, tissue mechanical properties are calculated from the measured displacement vector field using inversion algorithms [2]. Pathology affects the mechanical behavior of biological tissue, and quantitative measures of tissue stiffness have demonstrated clinical utility. For example, MRE is becoming widely used for the noninvasive detection and grading of hepatic fibrosis [3,4]. There is a special interest in applying MRE to the brain as, to date, MRE represents the only noninvasive technique capable of determining cerebral mechanical properties in vivo. The technique has revealed a correlation between the stiffness of the brain parenchyma and both demyelination [5] and inflammation [6]. Further, decreased cerebral mechanical properties have been observed in the later stages of neurodegeneration in in vivo human studies using MRE [7,8].
There has been a recent increase in the development of acquisition methods for brain MRE to increase clinical accuracy based on the need for greater spatial resolution [9–11] or the capture of multiple frequencies for rheological tissue parameter assessment [12,13]. In both cases, there exists a complementary need for schemes to reduce the long scan times resulting from increased data acquisition and to improve patient acceptance. Recently, Klatt et al. introduced sample interval modulation (SLIM), which enables the acquisition of all components of the displacement vector in only one temporally resolved, multidirectional experiment through direction-dependent shifting of motion encoding gradients (MEG) relative to the applied vibration [14]. Using conventional, monodirectional motion encoding schemes, each component of the displacement vector is captured in separate, sequential MRE acquisitions. Thus, SLIM-MRE allows for a potential 3x time reduction. Additionally, the new concept results in immediate co-registration of the three displacement components, thus potentially reducing the occurrence of artifacts caused by subject movement during the MRE scan. Of note, the SLIM concept is not restricted to a specific sequence type, but can readily be implemented into either gradient echo or spin echo sequences commonly used in MRE with any readout trajectory, including spirals and echo planar imaging.
Klatt et al. demonstrated in gel phantoms using high field scanners that SLIM encoding does not interfere with the MRE-derived mechanical properties as compared with conventional MRE [14]. Further, Nir et al. have recently proposed to formulate the derivation of wave properties using multidirectional motion encoding as a linear estimation problem and found that the SLIM concept coincides with their least-square solution [15]. While the multidirectional SLIM encoding scheme will not interfere with displacement field estimation, the direction-dependent shifting of the MEG projections involves a necessary increase in the time interval reserved for motion encoding, and thus results in prolonged echo times [14]. The echo time increase can reduce the acquisition signal-to-noise ratio (SNR) and ultimately challenge the practicability of SLIM-MRE for in vivo applications.
In this paper we implement SLIM-MRE with polarity switching of flow-compensated MEGs for minimizing the TE increase. The purpose of the presented work is to show the feasibility of the SLIM encoding concept embedded in a multishot, variable-density spiral sequence for data acquisition on the in vivo human brain. Further, we intend to demonstrate that the wave images acquired using conventional encoding schemes and rapid multidirectional SLIM MRE are highly correlated and produce property measures that agree within the error margins of MRE.
Theory
In MRE, the intravoxel coherent motion of an externally introduced mechanical vibration is encoded into the phase ϕ of the MR signal by the interplay between the motion encoding gradient (MEG) G and the displacement u within the time interval of MEG application τ, which corresponds to an integer number of MEG cycles [1]:
| (1) |
Using alternating MEG components with the same period as the harmonic vibration, the solution of the phase integral is given by equation 2:
| (2) |
Herein, and θj are the amplitudes and initial phases of the displacement components. The phase offset ψj corresponds to the difference between the phase of the displacement component at the start time of the respective MEG component and the initial phase of the displacement component. Note that ψj are measured relative to some fixed time, i.e. the initial phase difference of the first sample. The encoding efficiency ξj depends on the specific MEG amplitude and waveform shape, such as sinusoidal or cosinusoidal. Hirsch et al. previously provided a list of ξj for various MEG types [16].
In conventional MRE experiments, ψj components are the same for each direction and are varied in order to sample the temporal evolution of the displacement signal. The sampling interval is generally evenly spaced over a single period of applied vibration. This allows for the components of the complex displacement field to be extracted from a series of sequential acquisitions utilizing different encoding directions.
In SLIM-MRE, each directional component of ψj is spaced evenly over a different number of periods, though with the same number of samples, N. Thus, the sampling interval of the phases is given by,
| (3) |
The discrete form of the MR signal phase ϕn at each sample n = {1,2,…,N} can be obtained by substituting the phases of the MEG components relative to displacement,
| (4) |
into equation 2:
| (5) |
Equation 5 demonstrates that each displacement component is encoded into the MR signal phase ϕn with a different discrete frequency; therefore, the amplitude and phase θj of each component can be derived by applying a discrete Fourier transform to ϕn and extracting from separate bins [14]. Thus, the SLIM encoding concept facilitates the separation of directional displacement components from a single acquisition where MEGs are simultaneously applied on all three gradient axes with varying start phases or onset times. Figure 1 illustrates how the direction-dependent phase offsets evolve across samples.
Figure 1.
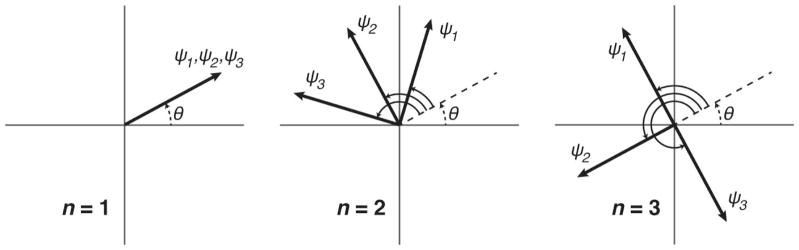
Schematic illustrating the direction-dependent phase offsets of applied MEGs from equation 4. The increment in phase offset between samples is different for each direction, which allows for their relative signal contributions to be separated. Note that the initial phases of the vibration components, θj, are represented as the same for each direction, which is denoted as θ in the figure. However, in principle, these phases can be different.
The phase offsets ψj are defined with respect to an arbitrary origin of the time axis, which is, however, the same for each projection and sample. In the following we define this origin as the onset of the three MEG projections at the first sample. The control of phase offsets is through sequence timing, and the relative start time for each component and sample Sjn is equivalently defined relative to the first MEG projection. Equation 6 converts the phase offsets to start times through the vibration frequency:
| (6) |
Equation 6 includes the modulo operator to reflect that, for a periodic signal, there is no need to shift the start time greater than a single period. Additionally, recognizing that MEGs with inverse polarity have an effective Δψ of π for encoding harmonic vibrations, the range of start times can be further reduced by incorporating MEG polarity reversal for Sjn greater than one half period. Thus, the resulting start times and gradient polarities are provided by equation 7:
| (7a) |
| (7b) |
Equation 7 holds for any excitation frequency, with the specific frequency of interest determining the actual required timings. These equations also hold for the incorporation of fractional encoding [17], or the use of MEGs with a period less than a full cycle of vibration, as SLIM dictates only the phase difference between vibration and MEG components and does not depend on the MEG duration.
Methods
We performed a comparative in vivo human brain study between SLIM-MRE and conventional MRE applied to a cohort of seven male volunteers (mean age 39.3 ± 14.9 years). The Institutional Review Board approved the study and we obtained written informed consent from all subjects. All scanning was performed using a Siemens 3 T Trio whole-body scanner with a 12-channel head coil (Siemens Medical Solutions; Erlangen, Germany).
Both conventional motion encoding and the SLIM concept of motion encoding were embedded in a multishot, variable-density spiral sequence with six k-space shots [9]. High isotropic spatial resolution of 8 mm3 was achieved across 20 slices with 2 mm thickness in the area of the lateral ventricles using a rectangular field-of-view of 240 mm and a matrix size of 128 × 128. This multishot technique inherently reduces distortions from field inhomogeneity and the oversampled center of each k-space shot is used to correct for motion-induced phase errors [9]. Field inhomogeneity distortions are further corrected during image reconstruction using a fieldmap estimated from an auxiliary scan [18]. Image reconstruction was performed iteratively using nonunifom fast Fourier transforms and linear conjugate gradients with seven time-segments for field inhomogeneity correction [19]. Images were estimated using data from multiple channels using SENSE [20], with coil sensitivity maps also estimated from an auxiliary scan.
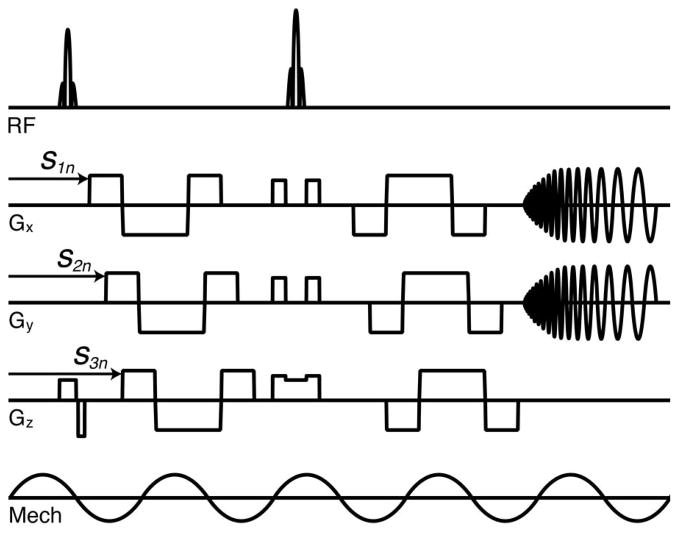
SLIM motion encoding was implemented using cosinusoidal, flow-compensated MEGs with start times and polarities as specified by equation 7 using N = 8 samples (figure 2). Table 1 summarizes the start times and polarities employed. MEGs were applied on both sides of the refocusing pulse while separated by a single vibration period, 20 ms, and with opposite polarity. All MEG components were applied with an amplitude of 18 mT/m, while their durations were matched to the period of the external vibration. The repetition and echo times (TR and TE) were 2400 and 70 ms, respectively. Each of the eight samples was acquired twice with inverted MEGs for the calculation of phase-difference images to remove background phase contributions. This resulted in a total acquisition time of 3 min 51 s. The mutual gradient shift in SLIM-MRE necessitates increased TR and TE relative to the conventional sequence timings of TR/TE = 2000/55 ms. In conventional acquisitions the three displacement components were acquired in consecutive steps, while using the same sequence parameters as in SLIM except for the TR and TE. The conventional acquisition time was 9 min 36 s. Both acquisitions utilized mechanical vibrations at 50 Hz to generate brain deformations, and Figure 3 depicts the use of the Resoundant pneumatic actuation system (Resoundant; Rochester, MN).
Figure 2.
Sequence diagram illustrating SLIM-MRE embedded in a multishot, variable-density spiral sequence. One flow-compensated, cosinusoidal MEG was applied on each side of the refocusing pulse, with their duration and separation matched to the 20 ms period of the 50 Hz vibrations. The start times of the MEG projections are indicated on the gradient axes.
Table 1.
Start times Sjn and polarities (+/−) of MEG components for each time point n (total N = 8).
| n | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| S1n [ms] | 0.0 (+) | 2.5 (+) | 5.0 (+) | 7.5 (+) | 0.0 (−) | 2.5 (−) | 5.0 (−) | 7.5 (−) |
| S2n [ms] | 0.0 (+) | 5.0 (+) | 0.0 (−) | 5.0 (−) | 0.0 (+) | 5.0 (+) | 0.0 (−) | 5.0 (−) |
| S3n [ms] | 0.0 (+) | 7.5 (+) | 5.0 (−) | 2.5 (+) | 0.0 (−) | 7.5 (−) | 5.0 (+) | 2.5 (−) |
Figure 3.
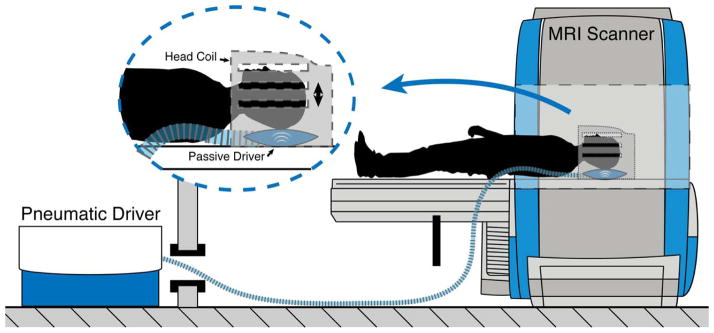
Experimental setup for brain tissue vibration with the Resoundant pneumatic actuator. The subject’s head rests on a pillow-like passive driver that receives vibrations from the active driver located in the control room.
For data analysis in SLIM-MRE, a discrete Fourier transform was applied to the temporally resolved phase images. The complex wave images (UX, UY, UZ) captured by motion encoding along the x-, y- and z-directions were taken from the 1st, 2nd and 3rd harmonic, respectively, and scaled to true displacements [14]. Using conventional acquisition schemes, the complex wave images are extracted from the 1st harmonic in each of the three repetitive temporally resolved experiments with varying motion-encoding direction. For noise reduction, a 3D spatial Butterworth low pass filter of order 2 with an eight pixel filter limit (corresponding to 1.6 cm) was applied to each of the three complex wave images [21], as is commonly performed in MRE prior to inversion to improve stability [22]. Subsequently, the curl operator was applied to the wave field for filtering contributions of the compression wave [23], and the complex shear modulus G was calculated by inverting the overdetermined system (Helmholtz equation) in a least square-manner [24]. The storage modulus GS (real part of G) and the loss modulus GL (imaginary part of G) were spatially averaged over the ROI. The ROI was restricted to the white matter region within the eight central brain slices in order to prevent edge errors of the spatial derivative in the inversion process of discrete data. White matter masks were generated by segmenting a separate T1-weighted MPRAGE acquisition (0.9 mm isotropic resolution) in FSL [25], and then registering to the MRE data with a process described previously [26].
We compared the displacement fields measured with both SLIM-MRE and conventional MRE by first calculating the octahedral shear strain (OSS), which is a measure of three-dimensional deformation [27]. We correlated the OSS fields after first co-registering them to the MPRAGE then computing Spearman’s rank correlation coefficients (ρ) using voxel-wise data. We also performed a similar analysis for both GS and GL distributions for each volunteer. Finally, we correlated the property measures spatially averaged over the white matter ROI from each method as these are the most commonly reported measures in brain MRE [7,8].
Results
Consistency was observed between the results from the SLIM-MRE and conventional MRE acquisition schemes. Figure 4a compares full vector displacement fields acquired in one volunteer using both methods. The separation of directional components from the SLIM dataset produced wave images that appear as nearly identical to images acquired in the conventional way, with sequential encoding of directional components. These displacement fields produced very similar property maps after inversion. Figure 4b presents estimated property maps, GS and GL, from the displacements in figure 4a. It is perceptible by visual inspection that the spatial distributions of the complex shear modulus both exhibit common features, including reduced GS in the center of the brain corresponding to the lateral ventricles.
Figure 4.
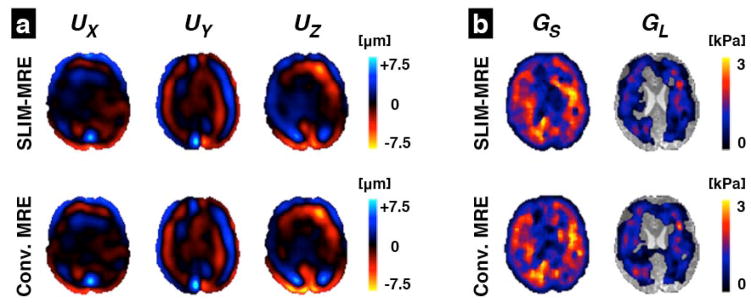
Comparison of SLIM and conventional MRE results in a single axial slice of one volunteer: (a) The complex wave images (real part) corresponding to the three displacement components, UX, UY, and UZ, measured with SLIM-MRE compared with those from conventional MRE; and (b) the property maps, GS and GL, estimated from each displacement field using algebraic Helmholtz inversion. Transparent areas in the loss modulus images indicate discarded outliers.
Table 2 presents the GS and GL values averaged over the white matter ROI as estimated with both methods for each volunteer. Considering conventional MRE as a gold standard, we computed the error in properties estimated by SLIM-MRE, and found that these values were typically less than 4%. Figure 5 illustrates the relationship between the SLIM and conventional MRE, and shows that property measures from both methods are significantly correlated, with Spearman correlation coefficients of 0.85 and 0.88 for GS and GL, respectively (p < 0.05).
Table 2.
Spatially averaged storage and loss moduli of white matter from both SLIM and conventional MRE acquisitions for each volunteer.
| Volunteer # | I | II | III | IV | V | VI | VII |
|---|---|---|---|---|---|---|---|
| Age [years] | 36 | 42 | 26 | 26 | 67 | 29 | 49 |
| GS (SLIM) [kPa] | 1.63 | 1.66 | 1.58 | 1.88 | 1.50 | 1.66 | 1.56 |
| GS (MRE) [kPa] | 1.68 | 1.67 | 1.60 | 1.91 | 1.57 | 1.66 | 1.62 |
| GL (SLIM) [kPa] | 0.89 | 0.98 | 0.89 | 0.91 | 0.84 | 0.94 | 0.82 |
| GL (MRE) [kPa] | 0.91 | 0.99 | 0.92 | 0.90 | 0.86 | 0.95 | 0.82 |
Figure 5.
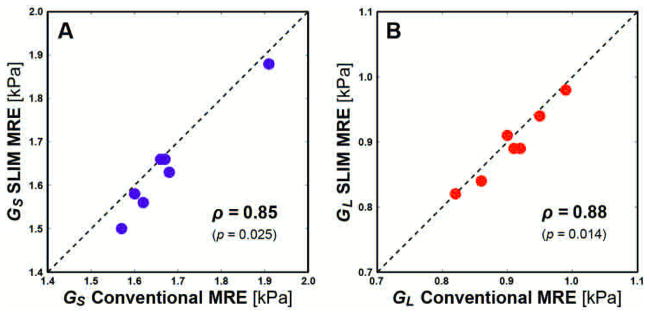
Correlation of spatially averaged values of the (A) storage modulus GS and (B) loss modulus GL for both methods. Spearman’s rank correlation coefficient (ρ) is provided for each relationship with corresponding p-value. SLIM and conventional MRE measures were significantly correlated in for both GS and GL (p < 0.05). Line with unity slope is included for illustration only.
Further, table 3 presents the voxel-wise correlations for both displacement and property fields as estimated from the two methods for each volunteer. The OSS deformations captured by SLIM and MRE were highly correlated, with an average coefficient of 0.94 over a range from 0.88 to 0.98 (all significant at p < 0.05). Similarly, the two methods produced highly correlated measures for both GS and GL, with an average correlation coefficient of 0.73 (range: 0.63 to 0.88).
Table 3.
Spatial correlation coefficients (ρ) relating the deformation fields (OSS) and property maps (GS and GL) from the SLIM acquisition with the conventional MRE acquisition for each volunteer (all p < 0.05).
| Volunteer # | I | II | III | IV | V | VI | VII |
|---|---|---|---|---|---|---|---|
| OSS | 0.94 | 0.97 | 0.91 | 0.98 | 0.97 | 0.96 | 0.88 |
| GS | 0.69 | 0.70 | 0.66 | 0.87 | 0.79 | 0.76 | 0.64 |
| GL | 0.67 | 0.71 | 0.64 | 0.88 | 0.79 | 0.77 | 0.63 |
Discussion
We have demonstrated that the simultaneous acquisition of the full vector displacement field in MRE can be achieved in vivo without compromising the quality of the resulting properties. Previously, research groups have described how the simultaneous application of gradients on all three axes can reduce MRE acquisition times in simulation and in phantoms [14,15,28]. However, for in vivo studies on clinical MRI scanners, the adoption of these approaches is non-trivial. The use of lower vibration frequencies in human applications causes the TE increase from mutual gradient shifting to be more significant. Additionally, the increasing use of spin-echo sequences in MRE, including in liver MRE to account for iron overload [29], amplifies the TE increase.
The incorporation of the SLIM encoding scheme required modifications to sequence timings relative to the conventional encoding scheme, including both TE and TR (by 15 and 400 ms, respectively). By implementing SLIM with varying gradient polarity, we were able to minimize the required TE. Additionally, we further mitigated the increase by also shifting the vibration trigger time relative to the sequence by the lowest start time at each sample. In this sense, the maximum relative gradient shift is only 5 ms, and we were able to achieve a TE that is only 15 ms greater than the conventional scheme while accounting for the spin-echo preparation.
However, the TE increase also lengthened each slice acquisition block and necessitated an increased TR. We were able to achieve a TR of 2400 ms in our implementation of SLIM-MRE, compared to a TR of 2000 ms in the conventional scheme, while maintaining the timing necessary for preserving phase relative to the continuous tissue vibration [9,30]. Thus, the total acquisition time was 3 min 51 s, which represents a 2.5x decrease in time relative to the acquisition with conventional encoding.
SLIM-MRE produced very similar estimates of brain mechanical properties to those acquired using conventional MRE. The storage and loss modulus maps exhibited high correlation in property topology between the two methods, resulting in nearly identical measures across all subjects. The mean disagreement was approximately 2% for each measure, with the greatest error being less than 5%, which is better than previously reported reproducibility estimates for a similar acquisition [26].
A major benefit of the SLIM motion encoding concept is that it allows for a reduction in acquisition time without a theoretical loss of signal-to-noise ratio (SNR). Typically, the acquisition of more time points (samples) will increase the SNR of the displacement field, since there is a greater number of discrete frequency bins for equal distribution of white Gaussian noise. Since both SLIM and conventional MRE utilize the same number of time points, each frequency bin experiences the same amount of noise. The difference is that SLIM utilizes three bins in a single acquisition, while each direction of a conventional MRE acquisition uses only one bin. Thus, the theoretical SNR between the two acquisition schemes is identical, provided all other sequence parameters are shared.
Our implementation of SLIM and its mutual gradient shifting requirements necessitated a difference in TE and TR relative to the conventional acquisition, and thus resulted in a difference in SNR due to tissue relaxation. Brain white matter at 3 T has T1 and T2 relaxation times of 1084 and 69 ms, respectively [31]. Accounting for the increased TE and TR of the SLIM acquisition, it should have approximately 85% of the SNR of the conventional acquisition. As has been previously noted [32], average property estimates from direct inversion decrease with SNR level, which likely explains why the SLIM-MRE property estimates were lower for nearly all measures. Despite this decrease in SNR, the magnitude of the disagreement in property measures is well within normal levels of uncertainty in MRE.
The SLIM concept also allows for an implementation with a further reduced TE, and thus improved SNR, by incorporating MEGs with both sinusoidal and cosinusoidal profiles. This would allow additional quarter period timing opportunities in addition to those explained in table 1, and thus the maximum mutual shift would be only 2.5 ms. Theoretically, both MEG shapes have the same encoding efficiency, but sinusoidal MEGs do not have the property of first-order nulling, or flow-compensation, as cosinusoidal MEGs do. Flow-compensation greatly reduces signal contributions from low-frequency vibrations, such as from brain pulsation [33], which can corrupt the usable MRE motion estimates. Thus, we chose to adopt the SLIM scheme using only cosinusoidal MEGs with flow-compensation to avoid significant artifacts at the expense of slightly reduced SNR.
Despite the strong agreement found between SLIM-MRE and conventional MRE acquisitions, there are some limitations to this analysis. First, we compared the two techniques as only applied to the in vivo human brain and not to other tissues commonly studied with MRE, such as the liver [29]. While, in principle, the benefits of SLIM should apply equivalently to abdominal organs, the added complexity stemming from the need for breathholds may make the comparison less clear. Additionally, shorter T2 relaxation times [31] can amplify the SNR loss due to lengthened echo times from mutual gradient shifting, thus reducing the benefit margin from SLIM. Finally, no gold standard exists for in vivo MRE measurements, and the comparison with the conventional MRE approach is therefore limited. However, we expect that SLIM should be superior given its short acquisition time and co-registration of displacement components, and thus a comparison with a true gold standard may reveal better agreement than even the conventional approach.
The adoption of MRE in clinical settings has resulted in increased demand for fast acquisition methods that provide robust and reliable tissue property maps. In this work we implemented a multidirectional motion encoding scheme into a brain MRE sequence and demonstrated that it returns virtually unchanged property estimates in a greatly reduced total scan time. This faster acquisition can aid in the adoption of MRE into routine clinical protocols, but also provide an avenue for capturing additional displacement data without increasing scan time. Additionally, the shorter acquisition reduces the opportunities for intra-scan subject motion to corrupt resulting property estimates, and SLIM-MRE measures may have a greater reproducibility.
Acknowledgments
Grant Support: NIH/NIBIB R01-EB001981, R01-EB007537
The authors acknowledge Mr. Joseph L. Holtrop for assistance in collecting data. Portions of this work were presented at the 22nd Annual Meeting of the International Society for Magnetic Resonance in Medicine.
Klatt D, Johnson CL, Yasar TK, Holtrop JL, Sutton BP, Royston TJ, Magin RL. Simultaneous Acquisition of the 3D Displacement Vector in Magnetic Resonance Elastography of the In Vivo Human Brain. In: Proceedings of the 22nd Annual Meeting of ISMRM, Milan, Italy, 2014 (abstract 1685).
References
- 1.Muthupillai R, Lomas DJ, Rossman PJ, Greenleaf JF, Manduca A, Ehman RL. Magnetic Resonance Elastography by Direct Visualization of Propagating Acoustic Strain Waves. Science. 1995;269:1854–1857. doi: 10.1126/science.7569924. [DOI] [PubMed] [Google Scholar]
- 2.Glaser KJ, Manduca A, Ehman RL. Review of MR Elastography Applications and Recent Developments. J Magn Reson Imaging. 2012;36:757–774. doi: 10.1002/jmri.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asbach P, Klatt D, Schlosser B, Biermer M, Muche M, Rieger A, Loddenkemper C, Somasundaram R, Berg T, Hamm B, Braun J, Sack I. Viscoelasticity-Based Staging of Hepatic Fibrosis with Multifrequency MR Elastography. Radiology. 2010;257:80–86. doi: 10.1148/radiol.10092489. [DOI] [PubMed] [Google Scholar]
- 4.Yin M, Talwalkar JA, Glaser KJ, Manduca A, Grimm RC, Rossman PJ, Fidler JL, Ehman RL. Assessment of Hepatic Fibrosis with Magnetic Resonance Elastography. Clin Gastroenterol H. 2007;5:1207–1213. doi: 10.1016/j.cgh.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schregel K, Wuerfel E, Garteiser P, Gemeinhardt I, Prozorovski T, Aktas O, Merz H, Petersen D, Wuerfel J, Sinkus R. Demyelination Reduces Brain Parenchymal Stiffness Quantified in Vivo by Magnetic Resonance Elastography. P Natl Acad Sci USA. 2012;109:6650–6655. doi: 10.1073/pnas.1200151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riek K, Millward JM, Hamann I, Mueller S, Pfueller CF, Paul F, Braun J, Infante-Duarte C, Sack I. Magnetic Resonance Elastography Reveals Altered Brain Viscoelasticity in Experimental Autoimmune Encephalomyelitis. NeuroImage Clin. 2012;1:81–90. doi: 10.1016/j.nicl.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy MC, Huston J, Jack CR, Glaser KJ, Manduca A, Felmlee JP, Ehman RL. Decreased Brain Stiffness in Alzheimer’s Disease Determined by Magnetic Resonance Elastography. J Magn Reson Imaging. 2011;34:494–498. doi: 10.1002/jmri.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wuerfel J, Paul F, Beierbach B, Hamhaber U, Klatt D, Papazoglou S, Zipp F, Martus P, Braun J, Sack I. MR-Elastography Reveals Degradation of Tissue Integrity in Multiple Sclerosis. NeuroImage. 2010;49:2520–2525. doi: 10.1016/j.neuroimage.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Johnson CL, McGarry MDJ, Van Houten EEW, Weaver JB, Paulsen KD, Sutton BP, Georgiadis JG. Magnetic Resonance Elastography of the Brain Using Multishot Spiral Readouts with Self-Navigated Motion Correction. Magn Reson Med. 2013;70:404–412. doi: 10.1002/mrm.24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson CL, Holtrop JL, McGarry MDJ, Weaver JB, Paulsen KD, Georgiadis JG, Sutton BP. 3D Multislab, Multishot Acquisition for Fast, Whole-Brain MR Elastography with High Signal-to-Noise Efficiency. Magn Reson Med. 2014;71:477–485. doi: 10.1002/mrm.25065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun J, Guo J, Lutzkendorf R, Papazoglou S, Hirsch S, Sack I, Bernarding J. High-Resolution Mechanical Imaging of the Human Brain by Three-Dimensional Multifrequency Magnetic Resonance Elastography at 7T. NeuroImage. 2014;90:308–314. doi: 10.1016/j.neuroimage.2013.12.032. [DOI] [PubMed] [Google Scholar]
- 12.Klatt D, Hamhaber U, Asbach P, Braun J, Sack I. Noninvasive Assessment of the Rheological Behavior of Human Organs Using Multifrequency MR Elastography: a Study of Brain and Liver Viscoelasticity. 2007;52:7281–7294. doi: 10.1088/0031-9155/52/24/006. [DOI] [PubMed] [Google Scholar]
- 13.Papazoglou S, Hirsch S, Braun J, Sack I. Multifrequency Inversion in Magnetic Resonance Elastography. 2012;57:2329–2346. doi: 10.1088/0031-9155/57/8/2329. [DOI] [PubMed] [Google Scholar]
- 14.Klatt D, Yasar TK, Royston TJ, Magin RL. Sample Interval Modulation for the Simultaneous Acquisition of Displacement Vector Data in Magnetic Resonance Elastography: Theory and Application. 2013;58:8663–8675. doi: 10.1088/0031-9155/58/24/8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nir G, Sahebjavaher RS, Sinkus R, Salcudean SE. A Framework for Optimization-Based Design of Motion Encoding in Magnetic Resonance Elastography. Magn Reson Med. 2014 doi: 10.1002/mrm.25280. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch S, Klatt D, Freimann FB, Scheel M, Braun J, Sack I. In Vivo Measurement of Volumetric Strain in the Human Brain Induced by Arterial Pulsation and Harmonic Waves. Magn Reson Med. 2013;70:671–683. doi: 10.1002/mrm.24499. [DOI] [PubMed] [Google Scholar]
- 17.Rump J, Klatt D, Braun J, Warmuth C, Sack I. Fractional Encoding of Harmonic Motions in MR Elastography. Magn Reson Med. 2007;57:388–395. doi: 10.1002/mrm.21152. [DOI] [PubMed] [Google Scholar]
- 18.Funai AK, Fessler JA, Yeo DTB, Olafsson VT, Noll DC. Regularized Field Map Estimation in MRI. IEEE T Med Imaging. 2008;27:1484–1494. doi: 10.1109/TMI.2008.923956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutton BP, Noll DC, Fessler JA. Fast, Iterative Image Reconstruction for MRI in the Presence of Field Inhomogeneities. IEEE T Med Imaging. 2003;22:178–188. doi: 10.1109/tmi.2002.808360. [DOI] [PubMed] [Google Scholar]
- 20.Pruessmann KP, Weiger M, Börnert P, Boesiger P. Advances in Sensitivity Encoding with Arbitrary K-Space Trajectories. Magn Reson Med. 2001;46:638–651. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 21.Buttkus B. Spectral Analysis and Filter Theory in Applied Geophysics. Berlin: Springer; 1991. [Google Scholar]
- 22.Sack I, Beierbach B, Hamhaber U, Klatt D, Braun J. Non-Invasive Measurement of Brain Viscoelasticity Using Magnetic Resonance Elastography. NMR Biomed. 2008;21:265–271. doi: 10.1002/nbm.1189. [DOI] [PubMed] [Google Scholar]
- 23.Sinkus R, Tanter M, Xydeas T, Catheline S, Bercoff J, Fink M. Viscoelastic Shear Properties of in Vivo Breast Lesions Measured by MR Elastography. Magn Reson Imaging. 2005;23:159–165. doi: 10.1016/j.mri.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 24.Manduca A, Oliphant TE, Dresner MA, Mahowald JL, Kruse SA, Amromin E, Felmlee JP, Greenleaf JF, Ehman RL. Magnetic Resonance Elastography: Non-Invasive Mapping of Tissue Elasticity. Med Image Anal. 2001;5:237–254. doi: 10.1016/s1361-8415(00)00039-6. [DOI] [PubMed] [Google Scholar]
- 25.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Johnson CL, McGarry MDJ, Gharibans AA, Weaver JB, Paulsen KD, Wang H, Olivero WC, Sutton BP, Georgiadis JG. Local Mechanical Properties of White Matter Structures in the Human Brain. NeuroImage. 2013;79:145–152. doi: 10.1016/j.neuroimage.2013.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGarry MDJ, Van Houten EEW, Perriñez PR, Pattison AJ, Weaver JB, Paulsen KD. An Octahedral Shear Strain-Based Measure of SNR for 3D MR Elastography. 2011;56:N153–N164. doi: 10.1088/0031-9155/56/13/N02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasar TK, Klatt D, Magin RL, Royston TJ. Selective Spectral Displacement Projection for Multifrequency MRE. 2013;58:5771–5781. doi: 10.1088/0031-9155/58/16/5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesh SK, Yin M, Ehman RL. Magnetic Resonance Elastography of Liver: Technique, Analysis, and Clinical Applications. J Magn Reson Imaging. 2013;37:544–555. doi: 10.1002/jmri.23731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinkus R, Lorenzen J, Schrader D, Lorenzen M, Dargatz M, Holz D. High-Resolution Tensor MR Elastography for Breast Tumour Detection. 2000;45:1649–1664. doi: 10.1088/0031-9155/45/6/317. [DOI] [PubMed] [Google Scholar]
- 31.Stanisz GJ, Odrobina EE, Pun J, Escaravage M, Graham SJ, Bronskill MJ, Henkelman RM. T1, T2 Relaxation and Magnetization Transfer in Tissue at 3T. Magn Reson Med. 2005;54:507–512. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MC, Huston J, Jack CR, Glaser KJ, Senjem ML, Chen J, Manduca A, Felmlee JP, Ehman RL. Measuring the Characteristic Topography of Brain Stiffness with Magnetic Resonance Elastography. PLoS One. 2013;8:e81668. doi: 10.1371/journal.pone.0081668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver JB, Pattison AJ, McGarry MDJ, Perreard IM, Swienckowski JG, Eskey CJ, Lollis SS, Paulsen KD. Brain Mechanical Property Measurement Using MRE with Intrinsic Activation. 2012;57:7275–7287. doi: 10.1088/0031-9155/57/22/7275. [DOI] [PMC free article] [PubMed] [Google Scholar]