Abstract
Aberrant behavior and function of neurons are believed to be the primary causes of most neurological diseases and psychiatric disorders. Human postmortem samples have limited availability and, while they provide clues to the state of the brain after a prolonged illness, they offer limited insight into the factors contributing to disease onset. Conversely, animal models cannot recapitulate the polygenic origins of neuropsychiatric disease. Novel methods, such as somatic cell reprogramming, deliver nearly limitless numbers of pathogenic human neurons for the study of the mechanism of neuropsychiatric disease initiation and progression. First, this article reviews the advent of human induced pluripotent stem cell (hiPSC) technology and introduces two major methods, “directed differentiation” and “neuronal induction,” by which it is now possible to generate neurons for modeling neuropsychiatric disease. Second, it discusses the recent applications, and the limitations, of these technologies to in vitro studies of psychiatric disorders.
Keywords: hiPSC, directed differentiation, neuronal induction, iNeuron, modeling neuropsychiatric disease
Introduction
In 2006 and 2007, Takahashi and Yamanaka1 and Takahashi et al.2, respectively, demonstrated that retroviral transduction of OCT4, SOX2, KLF4, and c-MYC (OSKM) was sufficient to reprogram mouse and human fibroblasts to embryonic stem cell (ESC)-like identity, so-called induced pluripotent stem cells (iPSCs). Generation of human iPSCs (hiPSCs) does not require destruction of human embryos, yet the process yields cells with pluripotent characteristics capable of giving rise to every cell type in the body. Moreover, unlike embryonic stem cells (ESCs), hiPSCs can be generated from adult humans after the diagnosis of disease; therefore, these cells can be used as a source with all the known and unknown genetic factors contributing to any complex genetic disease. Thus, hiPSC technology is rapidly being applied to applications such as cell-based therapeutics, disease modeling, and drug development.
Still, Takahashi and Yamanaka’s1 original method for hiPSC generation had significant barriers to translational application. Primarily, retroviral gene integration is a potential risk for tumorigenesis; to address this concern, nonintegrating methods utilizing adenoviruses, Sendai viruses, episomal vectors, synthetic mRNAs, and direct protein delivery reprogramming tools have now been developed.3–8 Owing to their high efficiency and wide commercial availability, the most widely used delivery methods of OSKM at the moment are synthetic mRNA transfection and Sendai viral transduction, the latter being a cytoplasmic RNA virus that does not alter the host genome.7,8
Pluripotency and mechanisms of reprogramming
Pluripotency is defined as a cell’s ability to differentiate into cells of all three germ layers and can be classified into two distinct states in human ESCs (hESCs) and hiPSCs: i) a more primitive “naïve” leukemia inhibitory factor (LIF)-dependent ground state, capable of generating both embryonic and extra-embryonic cell lineages and ii) a basic fibroblast growth factor (bFGF)-dependent “primed” state more reminiscent of “epiblast” identity.9 In both states, pluripotency is maintained through NANOG expression, which together with OCT4 and SOX2, comprises the essential stem cell transcriptional pluripotency network.10 The molecular mechanism of reprogramming remains unclear; particularly, it is uncertain to what extent reprogramming is driven by random molecular “stochastic” events11,12 or by hierarchical “deterministic” events.13,14 In fact, recent studies of the transcriptomic, proteomic, and epigenetic changes characteristic of reprogramming15–17 have identified another alternative stem cell state, dubbed the “F-class” cell, arising from particularly high expression of the reprogramming factors.18 The existence of F-class cells implies that reprogramming may in fact yield a variety of artificial stem cell states. While a number of recent reports have contrasted human “naïve,”19 “primed,”1,2 and “F-class”18 stem cells, the single most relevant detail in the modeling of neuropsychiatric disease is that, although these distinct stem cell populations are cultured differently and have different global transcriptional profiles, all give rise to functional neurons.
hiPSC technology is advancing the study of neurological disease pathology, as hiPSCs can produce nearly limitless numbers of human neurons that retain the genetic information of the donor. Therefore, hiPSC-derived neurons can be used to identify the cellular phenotype(s) and molecular mechanism(s) contributing to disease initiation, progression, and end point.
Generation of Neuronal Subtypes (Relevant to Disease or Disorder)
Neuroscientists first obtained diseased neurons from patient fibroblasts through a slow and labor-intensive process as follows: i) Isolated fibroblasts were expanded from patients and reprogrammed to hiPSCs; ii) hiPSCs were differentiated and neutralized as embryoid bodies (EBs) in suspension culture; iii) EBs were plated and further cultured until neural rosette formation occurred; iv) Neural rosettes were manually picked and dissociated to generate neural progenitor cells (NPCs); v) NPCs were expanded, validated, and subsequently differentiated into functional neurons.20 The total period of time from fibroblast to mature neurons required up to 6 months, and this extended process yielded a population that was a mixture of different neuronal subtypes. To overcome these issues of time and purity, two methods have been established: “directed differentiation” and “induction.”
Directed differentiation
Directed differentiation is an in vitro strategy that mimics in vivo development by applying small molecules and/or morphogens, which mimic the signaling involved in the patterning, specification, and commitment of defined cell types during embryonic development. Treatment of hESCs with Noggin (a bone morphogenetic protein [BMP] inhibitor) and SB431542 (an inhibitor of transforming growth factor-beta [TGF-β]), so-called “dual SMAD inhibition,” directed 80% of hESCs into a population of neural stem cells (NSCs) and neural progenitor cells (NPCs) within 1 week, as assayed by paired box 6 (PAX6) and HES5-eGFP reporter expression.21 Dual SMAD inhibition is a remarkable method for rapidly differentiating neural populations from hiPSCs, expediting the time line and purity of neuronal differentiation protocols.
Dopaminergic neurons
Dopamine (DA) is a neurotransmitter that governs reward-motivated behavior and motor control through the DA system.22 DA neurons in the midbrain (mDA neurons) are associated with distinctive neurological disorders such as Parkinson’s disease (PD), schizo phrenia (SZ), and attention deficit hyperactivity disorder.23 Dual SMAD inhibition followed by Sonic Hedgehog (SHH), brain-derived neurotrophic factor (BDNF), FGF8, and ascorbic acid treatment generates tyrosine hydroxylase (TH)-positive neurons from hiPSCs within 1 month.21 This adherent culture protocol bypasses the EB stage between hiPSC and neural rosette, as well as producing three times more TH-positive neurons (30%) than the conventional EB protocol (10%),20 and recent yields have been reported to be as high as 80%.24 Overexpression of progerin, which facilitates cellular aging, in hiPSC-derived mDA neurons derived from PD patients and healthy controls, reveals distinctive PD phenotypes such as reduction of TH-positive neurons, dendrite degeneration, and enlarged size of mitochondria, specifically in “geriatric” PD hiPSC mDA neurons.25
Glutamatergic neurons
Glutamatergic neurons in the cerebral cortex, generally represented by pyramidal neurons, are projection neurons that relay information to remote areas of the cerebral cortex and other regions of the brain. Aberrant neuronal connectivity and function of the glutamatergic neurons is believed to increase susceptibility to neuropsychiatric disorders such as autism spectrum disorder (ASD) and SZ.26 Synergistic with dual SMAD inhibition, activation of retinoic acid signaling (to restrict dorsal forebrain development) specifies the path for transformation of hESCs and hiPSCs into cortical progenitor cells expressing FOXG1 and EMX1, markers for dorsalized forebrain.27 The cortical stem cell and progenitor cell population generates 70% βIII-TUBULIN-positive neurons by 40 days of differentiation, which express vGLUT1, a glutamate synaptic vesicle marker, with excitatory synaptic property in electrophysiology by 50 days.27 Importantly, neurons with cortical layer-specific makers are generated in a time-dependent manner: deeper layer 6 TBR1+ neurons by day 30, layers 5 and 6 CTIP2+ neurons by day 40, layers 2–4 BRN2+ and CUX1+ neurons by day 50, and SATB2+ neurons by day 80.27 Given that human embryonic cortical neurogenesis also requires roughly 100 days, and that cortical layer formation occurs “inside-out,” in vitro generation of layer-specific neurons from hPSCs appears to recapitulate cortical neurogenesis in the human brain. An alternative methodology, utilizing FGF2 and inhibitors of BMP, WNT/β-CATENIN, and TGF-β/ACTIVIN/NODAL pathways, also induces hiPSCs into NPCs with forebrain fate, which can be further differentiated into presynaptic (SYNAPSIN1+) and postsynaptic (PSD95+) excitatory cortical neurons.28 Moreover, global gene expression profiling reveals the similarity between these in vitro-differentiated glutamatergic neurons and human dorsal telencephalic cells 8–10 weeks postconception. Because cortical glutamatergic neurons have extended projections throughout the brain, one gold standard for assessing hiPSC-derived glutamatergic neurons is to engraft them into mice and observe their integration throughout the host brain. Camacho and colleagues29 differentiated hESCs into cortical progenitor cells and cortical pyramidal neurons and then engrafted them into the cortex of neonatal mice. Engrafted cells sent axonal-like projections to multiple brain regions, including the contralateral cortex, striatum, and thalamus, where they established functional synapses and microcircuits with the host brain, suggesting the functional potential of cortical glutamatergic neurons directly differentiated from hiPSCs.
GABAergic neurons
In the cerebral cortex, GABAergic (producing gamma-aminobutyric acid [GABA]) interneurons are implicated in neuropsychiatric diseases, such as epilepsy, seizure, ASD, and SZ, probably owing to their essential role in fine-tuning and integrating the neural network.30 Cortical GABAergic interneurons initially arise from medial ganglionic eminences (MGEs) of the developing telecephalon and only subsequently migrate into the neocortex.31 To generate MGE NPCs with potential GABAergic identity, the Studer group32 utilized the combinatorial inhibition of dual SMAD (SB431542 and LDN-193189) and WNT (XAV939), together with late activation of SHH, and differentiated hiPSCs into 80% NKX2.1-GFP-positive cells, a marker of ventral cells in the developing forebrain MGE.32 By day 18 of differentiation, these NKX2.1-GFP precursor cells have migratory potential if engrafted in embryonic day (E) 13.5 mouse MGE. By 30 days, NKX2.1-GFP+ cells yield neurons expressing GABA, calbindin, and DLX2, markers for GABAergic identity. If co-cultured with mouse cortical neurons for an additional 30 days, NKX2.1-GFP neurons show physiological activity consistent with GABAergic interneurons, and they further differentiate into somatostatin (SST)-, parvalbumin (PV)-, and calbindin-positive GABAergic interneurons. In parallel, Nicholas et al.33 produced 70% NKX2.1-GFP-positive cells at day 25 by using a similar set of chemical cocktails (dual SMAD inhibition, WNT inhibition [DKK1], and SHH signaling activation). NKX2.1-GFP precursor cells give rise to neurons expressing GABAergic markers with functional synaptic properties consistent with GABAergic neurons. Importantly, upon engraftment into the neonatal mouse cerebral cortex, NKX2.1-GFP precursor cells mature to some subtypes of GABAergic interneurons and functionally integrate into the microcircuitry of the host brain. Recent data suggest that the addition of caudalizing signal by FGF8 enhances the yield of NKX2.1-GFP+ cells more than WNT inhibition and SHH activation alone.34
Generation of morphogen and small molecule directed hiPSC-derived neurons resembles the in vivo differentiation pathways during neural development, though a number of limitations of “directed differentiation” exist. First, the supply of recombinant growth factors may not be economically suitable for methodologies such as massive high-throughput screening. Second, inefficient signaling activation by chemicals can restrain researchers from the precise combinational modulation required for proper differentiation. Third, spatially and temporally impure and heterogeneous neural subtype specification has not been overcome; for instance, though DA neuron differentiation protocols have reached as high as 80% TH+ neurons, the remaining 20% are a mixture of neural and non-neural cells.24 Fourth, directed differentiation yields neurons that are immature relative to those in the human brain, with transcriptional profiles most resembling those of human fetal tissue.28,33,35 Finally, directed differentiation protocols require an extended time course of neuronal differentiation, up to 3 months, leading to slow experiment turnaround. Recently, “neuronal induction” has been shown to be a viable alternative strategy that addresses many of these concerns.
Neuronal induction
Patient-derived somatic cells can now be rapidly and directly converted from differentiated cells into neurons. The manipulation of key transcription factors is sufficient to change cell fate from one committed cell type to another. For example, expression of MyoD converts mouse fibroblast into muscle myoblast cells, and Pax5-deleted mature lymphocyte B cells from mouse dedifferentiate back to lymphoid progenitors, which can be differentiated into T cells.36,37 From these two discoveries came the idea that cell types can be “induced” to form NSCs/NPCs or directly change to neurons.
Induced NSCs/NPCs
Brief expression of Oct4 at the initial stage of reprogramming, together with constitutive induction of Sox2, Klf4, and c-Myc, converts mouse fibroblasts into “induced neural stem cells (iNSCs)” that exhibit similar gene expression to primary NSCs and can give rise to functional neural lineage cells (neuron, astrocyte, and oligodendrocyte).38 Transduction of five factors, including Pou3f2, Nr2e1, Sox2, c-Myc, and Hes5, induces formation of NPCs from fibroblasts, which display chemotactic behavior similar to primary NPCs.39 Moreover, Sox2 expression alone is sufficient to generate iNSCs in 30 days, which differentiate into multiple neural cells and integrate into the host brain.40 Unlike iPSCs, iNSCs do not produce tumors in engraftment assays, suggesting that these cells may be a more desirable source for cell replacement therapeutics for neurodegenerative disease.
iNeurons
During neurogenesis, a series of proneuronal transcription factors orchestrate the global gene expression network required for cell fate specification, driving the cellular transition from NSCs/NPCs to mature neurons. Expression of key neurogenic regulators is sufficient to induce donor fibroblasts into neurons (iNeurons). In 2009, the Wernig group41 demonstrated that three proneuronal transcription factors – Ascl1, Brn2, and Myt1l (BAM) – directly converted mouse fibroblasts into heterogeneous but functional neurons in just 20 days. In humans, combining NeuroD1 with these three BAM factors induced neurons from human fibroblasts, even though these iNeurons only formed fully functional excitatory synapses when co-cultured with mouse primary cortical neurons.42 The molecular mechanism of BAM neuronal induction begins with the opening of the chromatin complex by Ascl1, followed by transcriptional activation of key neuronal genes by Brn2 and Myt1l.43 Recently, Chanda et al.44 reported that ASCL1 alone is sufficient to induce excitatory neurons from both human fibroblasts and ESCs. Interestingly, expression of two microRNAs (miR-9 and miR-124) with ASCL1, MYT1L, and NEUROD2 improved neuronal induction efficiency, yielding iNeurons with electrical synaptic property independent of primary neuron co-culture.45 These studies opened the possibility that combinatorial activation of BAM factors with lineage-specific regulators may facilitate the conversion of donor cells into precise neuronal subtypes.
Induced motor neurons
Along with the BAM factors, two sets of motor neuron progenitor and committed-motor neuronal factors (Sox1, Pax6, Nkx6.1, Olig2 or Ngn2, Lhx3, Lsl1, Hb9) can convert fibroblasts into functional spinal motor neurons (induced motor neurons [iMNs]) within 35 days. These iMNs show both gene expression patterns and electrophysiological activity analogous to embryonic motor neurons, elicit rhythmic contraction of C2C12 muscle cell line in vitro, and integrate into the developing chick spinal cord.46 These iMNs are sensitive to the degenerative stimuli from glial cells harboring a SOD1 mutation, known to cause amyotrophic lateral sclerosis (ALS), demonstrating their value in modeling complex neurological disease.
Induced glutamatergic neurons
Expression of Neurogenin 2 (Ngn2), a dorsal telencephalic fate determinant, transdifferentiates cortical astroglial cells into glutamatergic neurons with functional synapses in vitro47,48 and hiPSCs into functional iNeurons.49 When combined with selection for Ngn2 expression, >90% of cells express MAP2, a dendritic marker, within 14 days and elicit electrical characteristics of excitatory synaptic function, when co-cultured with mouse cortical neurons, within 21 days. Furthermore, Ngn2-iNeurons express glutamatergic synaptic proteins vGLUT2, PSD95, and SYNAPSIN1, as well as successfully integrating when transplanted into a mouse brain.
Induced dopaminergic neurons
Combinatorial transduction of BAM factors, together with LMX1A and FOXA2, fate determinants of mDA neurons, yields mDA-like cells that express TH but show poor functionality.50 This may reflect the fact that BRN2 (one of the BAM factors) is enriched in the pyramidal neurons of layers 2–4,51 indicating its putative role in cortical neurogenesis rather than in midbrain specification. Accordingly, Addis et al.52 transduced just Ascl1, Lmx1b, and Nurr1 in mouse astrocytes, yielding TH+ neurons that secrete DA. Similar sets of transcription factors (Ascl1, Lmx1a, and Nurr1) induced functional mDA neurons (iDA neurons) from both mouse and human fibroblasts, which expressed DA machinery components such as VMAT2, DAT, ALDH1A1, and CALBINDIN.53 These iDA neurons also fired action potentials and released DA following K+ stimulation. Using Pix3-eGFP reporter mouse ESC lines, Kim et al.54 identified the six factors (Ascl1, Pitx3, Lmx1a, Nurr1, Foxa2, and EN1) that most enriched conversion efficiency of fibroblasts into functional mDA neurons; these imDA neurons, when transplanted into the striatum, improved the behavioral phenotypes in a PD mouse model.54 Recently, transduction of ASCL1, LMX1A, and NURR1 (ALN) yielded populations of 60% pure DA neurons (TH+, βIII-TUBULIN+ double neurons) from hiPSCs within 14 days.55
Induced GABAergic neurons
Induction into GABAergic neuronal fate has only just begun to be explored. Expression of Ascl1 together with Dlx2, a factor essential for GABAergic neuronal differentiation, is sufficient to transdifferentiate mouse astroglial cells to synapse-forming neurons within 21 days, which are positive with GABAergic neuronal markers, including GAD67, calretinin and vGAT.48 More recently, coexpression of miR-9 and miR-124, together with MYT1L and three transcription factors enriched in the developing striatum, BCL11B (also known as CTIP2), DLX1, and DLX2. induced human fibroblasts into a population analogous to striatal medium spiny neurons.56 These induced GABAergic neurons fire action potential trains with a long delay to initial spike, and if transplanted into the mouse brain, extend projections to the anatomical targets of medium spiny neurons.
One of the major concerns about the “neuronal induction” is whether forced expression of neuronal transcription factors will overcome disease-specific deficits in neuronal patterning and/or maturation. An important proof of concept was the demonstration that iNeurons recapitulate the expected AMPAR-mediated excitation deficits when generated from mice with Neuroligin-3 mutations, reminiscent of the neuronal phenotypes observed in primary neurons from these same mutant mice.57 This strongly supports the utility of iNeurons for disease modeling. In contrast to directed differentiation, overexpression of transcription factor facilitates rapid conversion of somatic cells and iPSCs into a variety of functional neuronal subtypes in a dramatically shorter period of time. Furthermore, fate regulator-mediated induction orchestrates a more uniform conversion process in donor cells, giving rise to relatively homogeneous populations of neuronal subtypes.49,55 However, the necessity of co-culture with other neurons or glial cells to enable synaptic maturation remains an important challenge when considering application of iNeurons to high-throughput screening for drug development.
Modeling Neuropsychiatric Disease with hiPSCs
Neuropsychiatric disease is typically associated with dysregulated neuronal connectivity between diverse neuronal populations.58 Genetic studies have unraveled many linkages between neuropsychiatric disease and genetic variations, including common single-nucleotide polymorphism and copy number variation (CNV), primarily involving genes responsible for neuronal functions. Heritable neuropsychiatric disorders, including SZ, ASD, and bipolar disorder (BD), are highly penetrant, with a heritability approaching 80%–90%; however, the multifactorial genetic etiology hinders the research with animal models, which best recapitulate monogenic defects. Though neuronal phenotypes have been studied in postmortem human brain, this tissue is in limited supply, and also only informs on the end-stage disease, providing limited insight into disease onset.
hiPSC-based studies of ASD
Since the discovery of hiPSCs, neuroscientists have pursued the potential of hiPSCs to model neuropsychiatric disease because this methodology provides theoretically limitless numbers of neurons that retain the genetic information of the donor somatic cells. One of the earliest demonstrations was the generation of hiPSCs from Rett Syndrome patients with mutation in MECP2.59 Rett hiPSC-derived neurons had signatures of excitatory synaptic deficits: reduced spine number and vGlut1 puncta, weaker postsynaptic current, and less frequent Ca2+ transients.59 hiPSC neurons derived from patients with another rare ASD, Timothy Syndrome, caused by a mutation on L-type calcium channel Cav1.2, revealed poor Ca2+ signaling and increased production of dopamine and norepinephrine, phenotypes that were ameliorated by treatment with the L-type channel inhibitor roscovitine.60 Together, these two studies indicated that it might be possible for hiPSC-based studies to model multifactorial psychiatric disorders.
hiPSC-based studies of SZ
There is reduced spine density in postmortem prefrontal cortex and disrupted synaptic maturation and function in mouse models of SZ.61 In 2011, Brennand et al.62 generated hiPSCs from genetically heterogeneous SZ patient fibroblasts and differentiated these hiPSCs into neurons, observing reduced levels of neuronal connectivity, neurite outgrowth, and synaptic protein such as PSD95, which could be rescued by loxapine treatment, an antipsychotic medication. Moreover, they identified altered expression of genes involved in glutamate, c-AMP, and WNT signaling pathways in SZ hiPSC neurons, consistent with synaptic dysregulation. Meanwhile, from an independent SZ patient cohort, hair follicle-derived SZ hiPSCs were found to i) poorly pattern into DA neurons and ii) show deficits in synaptic maturation of glutamatergic neurons, potentially owing to mitochondrial dysfunction.63 Interestingly, gene expression patterns consistent with aberrant neuronal differentiation, migration, and synaptic function signaling pathways are already specified in SZ hiPSC-derived NPCs, before full neuronal differentiation.35 Other studies also reported that SZ NPCs display imbalance of Zn2+ and K+,64 increased oxidative stress, and mitochondrial damage.63,65 Moreover, SZ NPCs exhibit increased cell-to-cell variability in heat shock protein 70 (HSP70) mRNA levels following sublethal environmental challenges of oxidative stress35 or ethanol treatment,66 supporting the idea that environmental challenges increase the risk to the onset of neuropsychiatric disorder. Aberrant hippocampal structure and function have been linked to SZ (reviewed in61). To study this, Yu et al.67 differentiated SZ hiPSCs into hippocampal neurons and observed reduced neuronal activity and neurotransmitter release. Recently, SZ patients with defined mutations68 in SZ risk genes69 have been modeled with hiP-SCs. 15q11.2 hiPSC-derived NPCs are defective in apical polarity and adherence junction, probably owing to haploinsufficiency of CYFIP1, a component of the WAVE complex regulating cytoskeletal dynamics.68 hiPSC-derived forebrain neurons derived from patients with mutant DISC1 have deficits in synaptic vesicle release, which can be ameliorated following genetic correction of the locus.69
hiPSC-based modeling of neuropsychiatric disease, particularly SZ, has recently demonstrated the proof-of-concept ability to model the “cause and effect” between neuronal dysfunction and disease occurrence. However, current methods of obtaining diseased neuronal subtype require further refinement, and difficulties in recapitulating neuronal networks composed of specific neuronal subtypes in vitro remain a critical roadblock in understanding the etiology of neuropsychiatric disease.
Accelerated aging of hiPSC neurons
Although gene expression and phenotypic analysis of hiPSC-derived neurons indicate that they most resemble human fetal brain tissue,28,35 there has at least been good concordance between hiPSC studies and reports of aberrant migration,68,70 reduced neurite outgrowth,71,72 and impaired synaptic activity73–78 in mouse models of both SZ and the monogenic ASD Rett Syndrome. This may reflect the fact that while ASD, SZ, and BD typically present clinically through childhood (ASD) and late adolescence (SZ, BD), all three are now considered to be neurodevelopmental conditions that result from abnormal neurodevelopmental processes initiated in utero and/or early childhood.79,80 Given the common genetic risk underlying these disorders,81,82 we speculate that phenotypes observed through current hiPSC-based modeling of ASD, SZ, and BD may appear convergent, reflecting the lack of incorporation of neuronal circuitry, activity, and plasticity, as well as inflammation and other environmental effects, into the existing models. To date, because hiPSC neurons mimic the molecular and cellular states existing before symptom onset, it is most accurate to state that hiPSC-based studies are more suitable for studying genetic predisposition rather than the disease state itself.
The immaturity of hiPSC-derived neurons relative to the adult human brain poses a significant challenge for the use of hiPSC-based models for late-onset neurodegenerative disorders such as Alzheimer’s disease (AD), PD, and ALS, as well as adult-onset psychiatric disorders such as SZ, BD, and addiction.62,83 Unlike hiPSC-derived cells from young and old controls, hiPSC-derived fibroblasts and mDA neurons derived from patients with Hutchinson–Gilford progeria syndrome (HGPS), a disease characterized by accelerated aging, show evidence of age-associated phenotypes.25 Furthermore, expression of progerin, a protein expressed from the mutated gene that causes HGPS, in hiPSC-derived DA neurons from PD and control patients revealed neurodegeneration-related phenotypes unique to PD neurons.25 Although it remains to be determined how applicable progerin-induced “aging” is to hiPSC-based models of psychiatric disorders such as SZ, BD, and addiction, this novel strategy promises to revolutionize current hiPSC-based models for neurodegenerative disorders, including PD, AD, and ALS.
Caveats to modeling neuropsychiatric disease with hiPSC-derived neurons
In designing hiPSC-based studies, it is important to be aware that genetic mutations and epigenetic faulty remodeling can occur during the reprogram ming process. First, both CNVs84–86 and somatic coding mutations87 will change the donor DNA. Importantly, more CNVs are present in early-passage hiPSCs than in higher-passage hiPSCs, implying that most novel CNVs generated during the reprogramming process are lost before the time that any neuronal differentiation would occur.88 Across 22 hiPSC lines reprogrammed using five different methods, each contained an average of five protein-coding point mutations, though at least half of these reprogramming-associated mutations preexisted in fibroblast progenitors at low frequencies.87 Second, at the epigenetic level, evidence now demonstrates that aberrant DNA methylation remodeling89–91 and an erosion of X chromosome inactivation91,92 can occur in hiPSCs. Consistent with this, evidence suggests that donor cell type can influence the epigenome and differentiation potential of hiPSCs.93,94 These genetic and epigenetic effects contribute to the “intraindividual variation” observed in hiPSC-based studies, which exists because each hiPSC line generated from a given person will show subtle differences in gene expression and propensity toward neural differentiation.
Conversely, “interindividual variation” represents biological differences among individuals and can be addressed by studying ever-larger cohorts of patients and controls, better capturing the heterogeneity among individuals. Unpublished data obtained by other researchers and us suggest that intrapatient variability is less than interpatient variability. Therefore, well-designed and controlled experiments are critical to ensure that researchers can draw meaningful conclusions from hiPSC-based studies of psychiatric disorders; we recommend that at least three hiPSC lines should be compared per individual, to reduce the likelihood that a rare genetic or epigenetic mutation might affect disease-specific hiPSC lines in a meaningfully different way than its effect on control hiPSC lines.
When investigating the effect of a single disease-associated allele, whether in the context of a simple Mendelian disorder or a complex genetic disease, an alternative to increasing cohort size is to instead compare isogenic hiPSC lines. In fact, a burst of recent hiPSC-based studies has used isogenic controls to demonstrate the precise effects of a single gene on neural phenotypes or gene expression.69,95,96 For example, in order to confirm that the synaptic defects observed in two psychiatric patients were due to the identified DISC1 frameshift mutation, Wen et al.69 produced isogenic hiPSC lines, by both engineering the DISC1 mutation into a control hiPSC line and also repairing the mutation in a DISC1 patient hiPSC line. In this way, they showed precisely that mutant DISC1 causes synaptic vesicle release deficits and also dysregulates expression of many genes related to synapses and psychiatric disorders in hiPSC-derived forebrain neurons.69
Perhaps most critically, while recent studies have reported the importance of heritable genetic factors in neuropsychiatric disease and have modeled the correlation between these risk factors and disease phenotype, it is still challenging to unravel the causality of environmental risk factors such as stressful life events, social anxiety, and neurotrauma,97 which remain infeasible to recapitulate in existing cell-based systems in vitro. Moreover, the question remains as to how to link relatively simple cellular phenotypes from hiPSC-derived neurons with complex behavioral phenotypes of neuropsychiatric patients, encompassing delusions, hallucinations, negative affect, and impaired cognition. One strategy will be to build increasing complexity into hiPSC-based models. Future models will necessarily incorporate neuronal circuits comprising at least two distinct neuronal cell types, synapsed in a defined orientation, together with oligodendrocytes – to provide myelination – and astrocytes and microglia, to incorporate critical aspects of inflammation and synaptic pruning. Circuits will need to be stimulated repeatedly to establish plasticity and to be exposed to meaningful levels of stress hormones and other environmental factors. A second strategy will be to transplant each of these relevant human cell types (neurons, astrocytes, oligodendrocytes, and microglia) into mouse models of disease, yielding increasingly humanized platforms for study. The ultimate solution, of course, will be to pursue all of the strategies we have discussed in tandem: larger cohorts, isogenic controls, improved patterning and maturation of a variety of human neural cell types, cultured either as artificial circuits or transplanted into mice. While models, by definition, must always lack the intricacies of human disease, the goal of hiPSC scientists should always be toward ever-increasing complexity of their models.
Conclusion
Somatic cell reprogramming confers the ability to model human neural development and complex neurological diseases in a dish, using human patient-derived neural cells. To generate neurons, two major methods have been widely adopted: “directed differentiation,” which modulates key neural patterning pathways through treatment with morphogens and small molecules, and “neuronal induction,” which converts donor cells into functional neurons by expression of proneuronal transcription factors. Both approaches have limitations concerning length of differentiation, subtype-specific neuronal purity, and neuronal maturity, which remain to be addressed so as to yield functional neurons with high efficiency and efficacy. Recently, several neuropsychiatric diseases have been modeled using these strategies, with particular focus to date on disorders involving single genetic mutations. Given that most neuropsychiatric disorders have multifactorial genetic origins, probably leading to defects in patterning, maturation, and/or synaptic function of one or more diverse neuronal subtypes, advances in neuronal differentiation and/or induction will facilitate mechanistic studies and high-throughput drug screening of neuropsychiatric diseases, to one day identify novel therapeutics for these common but devastating disorders.
Figure 1.
Generation of subtype-specific human iNeurons. (A) Ngn2 overexpression yields pure populations of glutamatergic neurons. (Adapted with permission from Zhang et al, 201349) (B) Overexpression of miR-9/9* and miR-124, together with CTIP2, DLX1, DLX2, and MYT1L, yields 80% pure populations of striatal medium spiny (GABAergic) neurons. (Adapted with permission from Victor et al, 201456) (C). Overexpression of Ascl1, Lmx1a, and Nurr1 yields 3% pure populations of dopaminergic neurons. (Adapted with permission from Caiazzo et al, 201153).
Figure 2.
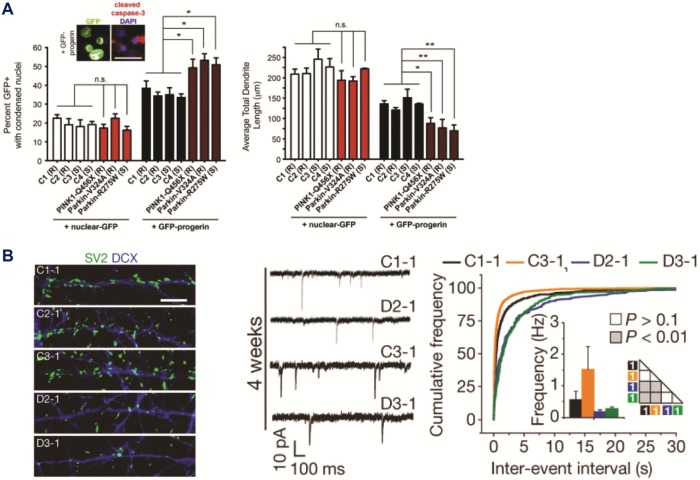
Representative cellular phenotypes recently identified through cell-based modeling of neurological diseases. (A) hiPSC-based model of Parkinson’s disease (PD) shows patient-specific cell loss, only in the condition of accelerated aging via progerin overexpression. Left: Analysis of PD patient and control hiPSC DA neurons, with and without progerin overexpression, reveals elevated cell death specifically in progerin-expressing PD neurons. Right: Quantification of total dendrite lengths, with and without progerin overexpression, shows dendrite shortening in PD hiPSC DA neurons compared to apparently healthy controls (C1–4). (Adapted with permission from Miller et al, 201425) (B) hiPSC-based models of schizophrenia. Left: Decreased synaptic density of SV2+ puncta in hiPSC neurons differentiated from hiPSC lines derived from schizophrenia patients (D2 and D3) carrying the DISC1 mutation compared to control lines. Right: Defects in the frequency of glutamatergic synaptic transmission in DISC1-mutant hiPSC neurons. (Adapted with permission from Wen et al, 201469).
Table 1.
Summary of methods of neural differentiation and neural induction in human and mouse fibroblasts and iPSCs.
| NEURONAL TYPE (REGIONAL PATTERNING) | CELL SOURCE | FACTORS INVOLVED | TIME TO EXPRESS REPRESENTATIVE NEURONAL MARKER | YIELD OF REPRESENTATIVE NEURONAL MARKER | IN VITRO VALIDATION OF ELECTROPHYSIOLOGICAL MATURITY, *INDICATES SPONTANEOUS SYNAPTIC ACTIVITY (TIME POINT OF EARLIEST DEMONSTRATION) | CO-CULTURE REQUIRED FOR IN VITRO MATURATION | IN VIVO VALIDATION BY TRANSPLANTATION | OTHER VALIDATIONS | REFERENCE |
|---|---|---|---|---|---|---|---|---|---|
| Excitatory and inhibitory neuron (majority cortical) | Mouse fibroblast | Brn2, Ascl1, Myt1l | 2 weeks (Tuj1/MAP2) | 19% | *yes (ND) | Mouse cortical neuron or glial cells | N/A | Immunostaining | Vierbuchen et al.41 |
| Excitatory neuron (18% cortical, 21% PNS) | Human fibroblast | Brn2, Ascl1, Myt1l NeuroD1 | 8 days (MAP2) | ND | *Yes (5 weeks) | Mouse cortical neuron or glial cells | N/A | Immunostaining Fluidigm single cell analysis | Pang et al.42 |
| Excitatory and inhibitory neuron (majority cortical) | Human fibroblast | miR-9/9*, miR-124 NEUROD, ASCL1, MYT1L | 3wks (MAP2) | 80% | *Yes (6wks) | N/A | N/A | Immunostaining qRT-PCRCa2+ imaging (Fluo-2 AM) | Yoo et al.45 |
| Excitatory and inhibitory neuron (majority cortical) | Human ESCs | Ascl1 | 4wks (Tuj1) | ND | *Yes (ND) | Mouse cortical neuron or glial cells | N/A | Immunostaining Fluidigm single cell analysis | Chanda et al.44 |
| Glutamatergic neuron (majority cortical) | Human PSCs | Neurogenin2 | 2wks (MAP2) | 90% | *Yes (3wks) | Mouse cortical neuron or glial cells | Mouse striatum | Immunostaining Fluidigm single cell analysis Ca2+ imaging (GCaMP) | Zhang et al.49 |
| Motor neuron | Mouse fibroblast | Ascl1, Brn2, Myt1l, Lhx3, Hb9, Isl1, Ngn2 | 10 days (Hb9-eGFP) | 5% | *Yes (ND) - neuromuscular junction | N/A | Chick spinal cord | Immunostaining Transcriptional profiling | Son et al.46 |
| Dopaminergic neuron (midbrain) | Mouse astrocyte | ASCL1, LMX1B, NURR1 | 19 days (TH)* | 18% | ND | N/A | N/A | Immunostaining Transcriptional profiling Ca2+ imaging (GCaMP) DA quantification | Addis et al.52 |
| Dopaminergic neuron (midbrain) | Mouse fibroblast Human fibroblast | Ascl1, Lmx1a, Nurr1 | 16 days (Tuj1/TH)* 16 days (Tuj1/TH)* |
15% 3% |
Yes (16 days) | N/A | Mouse brain (ventricle) | Immunostaining Transcriptional profiling Promoter methylation assay Ca2+ imaging (FM 4–64 dye) DA quantification | Caiazzo et al.53 |
| Dopaminergic neuron (midbrain) | Mouse fibroblast | Ascl1, Pitx3, Lmx1a, Nurr1, Foxa2, EN1 | 18 days (Pitx3-eGFP) | 9% | Yes (15 days) | N/A | Mouse brain (striatum of Parkinson’s disease mouse model) | Immunostaining Transcriptional profiling DA quantification | Kim et al.54 |
| Dopaminergic neuron (midbrain) | hiPSCs | ASCL1, LMX1A NURR1 | 2wks (Tuj1/TH) | 60% | Yes (3wks) | N/A | N/A | Immunostaining DA quantification | Theka et al.55 |
| GABAergic neuron | Mouse astroglial cell | Dlx2, Ascl1 | 3wks (GAD67) | ND | *Yes (4wks) | N/A | N/A | Immunostaining | Heinrich et al.48 |
| GABAergic neuron (striatum) | Human fibroblast | miR-9/9* miR-124 CTIP2, DLX1, DLX2, MYT1L | 5wks (MAP2/GABA)* | 80% | *Yes (12wks) | Rat glial cell | Mouse striatum | Immunostaining Transcriptional profiling | Victor et al.56 |
Abbreviation: ND, not determined.
Footnotes
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
FUNDING: The Brennand Laboratory is supported by a Brain and Behavior Young Investigator grant, National Institutes of Health grant R01 MH101454, and the New York Stem Cell Foundation. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: KJB is a New York Stem Cell Foundation - Robertson Investigator. Other authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the first draft of the manuscript: SH, AT, KJB. Contributed to the writing of the manuscript: SH, AT, KJB. Agree with manuscript results and conclusions: SH, AT, KJB. Jointly developed the structure and arguments for the paper: SH, AT, KJB. Made critical revisions and approved final version: SH, AT, KJB. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–53. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–6. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–62. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ban H, Nishishita N, Fusaki N, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci USA. 2011;108:14234–9. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–63. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buecker C, Geijsen N. Different flavors of pluripotency, molecular mechanisms, and practical implications. Cell Stem Cell. 2010;7:559–64. doi: 10.1016/j.stem.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Buganim Y, Faddah DA, Cheng AW, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14(6):427–39. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502(7469):65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 14.Ross J, Busch J, Mintz E, et al. A rare human syndrome provides genetic evidence that WNT signaling Is required for reprogramming of fibroblasts to induced pluripotent stem cells. Cell Reports. 2014;9(5):1770–80. doi: 10.1016/j.celrep.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee DS, Shin JY, Tonge PD, Puri MC, Lee S. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat Commun. 2014;5:5619. doi: 10.1038/ncomms6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clancy JL, Patel HR, Hussein S, Tonge PD. Small RNA changes en route to distinct cellular states of induced pluripotency. Nat Commun. 2014;5:5522. doi: 10.1038/ncomms6522. [DOI] [PubMed] [Google Scholar]
- 17.Benevento M, Tonge PD, Puri MC, Hussein S. Proteome adaptation in cell reprogramming proceeds via distinct transcriptional networks. Nat Commun. 2014;5:5613. doi: 10.1038/ncomms6613. [DOI] [PubMed] [Google Scholar]
- 18.Tonge PD, Corso AJ, Monetti C, Hussein S, Puri MC. Divergent reprogramming routes lead to alternative stem-cell states. Nature. 2014;516(7530):192–7. doi: 10.1038/nature14047. [DOI] [PubMed] [Google Scholar]
- 19.Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–6. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 20.Boyer L, Campbell B, Larkin S, Mu Y, Gage FH. Dopaminergic differentiation of human pluripotent cells. Curr Protoc Stem Cell Biol. 2012:6. doi: 10.1002/9780470151808.sc01h06s22. Chapter 1:Unit1H. [DOI] [PubMed] [Google Scholar]
- 21.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krügel U, Kittner H, Franke H, Illes P. Stimulation of P2 receptors in the ventral tegmental area enhances dopaminergic mechanisms in vivo. Neuropharmacology. 2001;40:1084–93. doi: 10.1016/s0028-3908(01)00033-8. [DOI] [PubMed] [Google Scholar]
- 23.Sillitoe RV, Vogel MW. Desire, disease, and the origins of the dopaminergic system. Schizophr Bull. 2008;34:212–9. doi: 10.1093/schbul/sbm170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kriks S, Shim JW, Piao J, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JD, Ganat YM, Kishinevsky S, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Bartolomeis A, Latte G, Tomasetti C, Iasevoli F. Glutamatergic postsynaptic density protein dysfunctions in synaptic plasticity and dendritic spines morphology: relevance to schizophrenia and other behavioral disorders pathophysiology, and implications for novel therapeutic approaches. Mol Neurobiol. 2014;49:484–511. doi: 10.1007/s12035-013-8534-3. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15(477–86):S1. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mariani J, Simonini MV, Palejev D, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770–5. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Espuny-Camacho I, Michelsen KA, Gall D, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–56. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Marín O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13(2):107–20. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 31.Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–63. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- 32.Maroof AM, Keros S, Tyson JA, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–72. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholas CR, Chen J, Tang Y, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–86. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TG, Yao R, Monnell T, et al. Efficient specification of interneurons from human pluripotent stem cells by dorsoventral and rostrocaudal modulation. Stem Cells. 2014;32:1789–804. doi: 10.1002/stem.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennand K, Savas JN, Kim Y, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015 Mar;20(3):361–8. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 37.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449(7161):473–7. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 38.Thier M, Wörsdörfer P, Lakes YB, et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–9. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Tian C, Ambroz RJ, Sun L, et al. Direct conversion of dermal fibroblasts into neural progenitor cells by a novel cocktail of defined factors. Curr Mol Med. 2012;12:126137. doi: 10.2174/156652412798889018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ring KL, Tong LM, Balestra ME, et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–9. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pang ZP, Yang N, Vierbuchen T, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–3. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wapinski OL, Vierbuchen T, Qu K, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–35. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chanda S, Ang CE, Davila J, et al. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports. 2014;3(2):282–96. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo AS, Sun AX, Li L, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Son EY, Ichida JK, Wainger BJ, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–18. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berninger B, Costa MR, Koch U, et al. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J Neurosci. 2007;27(32):8654–64. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinrich C, Blum R, Gascón S, et al. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Pak C, Han Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–98. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfisterer U, Kirkeby A, Torper O, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108:10343–8. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23(11):2632–43. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Addis RC, Hsu FC, Wright RL, Dichter MA, Coulter DA, Gearhart JD. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One. 2011;6:e28719. doi: 10.1371/journal.pone.0028719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caiazzo M, Dell’Anno MT, Dvoretskova E, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–7. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 54.Kim J, Su SC, Wang H, et al. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413–9. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theka I, Caiazzo M, Dvoretskova E, et al. Rapid generation of functional dopaminergic neurons from human induced pluripotent stem cells through a single-step procedure using cell lineage transcription factors. Stem Cells Trans Med. 2013;2:473–9. doi: 10.5966/sctm.2012-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Victor MB, Richner M, Hermanstyne TO, et al. Generation of human striatal neurons by MicroRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84:311–23. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chanda S, Marro S, Wernig M, Südhof TC. Neurons generated by direct conversion of fibroblasts reproduce synaptic phenotype caused by autism-associated neuroligin-3 mutation. Proc Natl Acad Sci USA. 2013;110:16622–7. doi: 10.1073/pnas.1316240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spellman T, Gordon J. Synchrony in schizophrenia: a window into circuit-level pathophysiology. Curr Opin Neurobiol. 2015;30:1723. doi: 10.1016/j.conb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchetto MC, Carromeu C, Acab A, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–39. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paşca SP, Portmann T, Voineagu I, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–62. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brennand KJ, Gage FH. Concise review: the promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem Cells. 2011;29:1915–22. doi: 10.1002/stem.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brennand KJ, Simone A, Jou J, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robicsek O, Karry R, Petit I, et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013;18:1067–76. doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- 64.Paulsen Bda S, Cardoso SC, Stelling MP, Cadilhe DV, Rehen SK. Valproate reverts zinc and potassium imbalance in schizophrenia-derived reprogrammed cells. Schizophr Res. 2014;154:3035. doi: 10.1016/j.schres.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 65.Paulsen Bda S, de Moraes Maciel R, Galina A, et al. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant. 2012;21:1547–59. doi: 10.3727/096368911X600957. [DOI] [PubMed] [Google Scholar]
- 66.Hashimoto-Torii K, Torii M, Fujimoto M, et al. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron. 2014;82:560572. doi: 10.1016/j.neuron.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu DX, Di Giorgio FP, Yao J, et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports. 2014;2:295–310. doi: 10.1016/j.stemcr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon KJ, Nguyen HN, Ursini G, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wen Z, Nguyen HN, Guo Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515(7527):414–8. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meechan DW, Tucker ES, Maynard TM, LaMantia AS. Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc Natl Acad Sci USA. 2009;106:16434–45. doi: 10.1073/pnas.0905696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kvajo M, McKellar H, Arguello PA, et al. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci USA. 2008;105:7076–81. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krivosheya D, Tapia L, Levinson JN, et al. ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J Biol Chem. 2008;283:32944–56. doi: 10.1074/jbc.M800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwon OB, Longart M, Vullhorst D, Hoffman DA, Buonanno A. Neuregulin-1 reverses long-term potentiation at CA1 hippocampal synapses. J Neurosci. 2005;25:9378–83. doi: 10.1523/JNEUROSCI.2100-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woo RS, Li XM, Tao Y, et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54:599–610. doi: 10.1016/j.neuron.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 75.Chun S, Westmoreland JJ, Bayazitov IT, et al. Specific disruption of thalamic inputs to the auditory cortex in schizophrenia models. Science. 2014;344:1178–82. doi: 10.1126/science.1253895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maher BJ, LoTurco JJ. Disrupted-in-schizophrenia (DISC1) functions presynaptically at glutamatergic synapses. PLoS One. 2012;7:e34053. doi: 10.1371/journal.pone.0034053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Niwa M, Kamiya A, Murai R, et al. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2014;65:480–9. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tropea D, Giacometti E, Wilson NR, et al. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci USA. 2009;106:2029–34. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–9. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 80.Marin O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13:107–20. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 81.Network, Pathway Analysis Subgroup of the Psychiatric Genomics C, International Inflammatory Bowel Disease Genetics C, International Inflammatory Bowel Disease Genetics Consortium I Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen HM, DeLong CJ, Bame M, Rajapakse I. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiatry. 2014;4:e375. doi: 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laurent LC, Ulitsky I, Slavin I, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–18. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu P, Kaplan A, Yuan B, Hanna JH, Lupski JR. Passage number is a major contributor to genomic structural variations in mouse iPSCs. Stem Cells. 2014;32(10):2657–67. doi: 10.1002/stem.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu J, Li H, Hu M, et al. The distribution of genomic variations in human iPSCs is related to replication-timing reorganization during reprogramming. Cell Rep. 2014;7:70–8. doi: 10.1016/j.celrep.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gore A, Li Z, Fung HL, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–7. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hussein SM, Batada NN, Vuoristo S, Ching RW. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471(7336):58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 89.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma H, Morey R, O’Neil RC, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–83. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nazor KL, Altun G, Lynch C, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–34. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nature BIotechnol. 2011;29(12):1117–9. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bar-Nur O, Russ H, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 95.Chen H, Qian K, Du Z, et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament Balance in motor neurons. Cell Stem Cell. 2014;14:796–809. doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu GH, Qu J, Suzuki K, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–7. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Howes OD, McDonald C, Cannon M, et al. Pathways to schizophrenia: the impact of environmental. Neuropsychopharm. 2004;7(suppl 1):S7–13. doi: 10.1017/S1461145704004122. [DOI] [PubMed] [Google Scholar]