Abstract
The origin of the eukaryotic cell remains one of the most contentious puzzles in modern biology. Recent studies have provided support for the emergence of the eukaryotic host cell from within the archaeal domain of life, but the identity and nature of the putative archaeal ancestor remain a subject of debate. Here we describe the discovery of ‘Lokiarchaeota’, a novel candidate archaeal phylum, which forms a monophyletic group with eukaryotes in phylogenomic analyses, and whose genomes encode an expanded repertoire of eukaryotic signature proteins that are suggestive of sophisticated membrane remodelling capabilities. Our results provide strong support for hypotheses in which the eukaryotic host evolved from a bona fide archaeon, and demonstrate that many components that underpin eukaryote-specific features were already present in that ancestor. This provided the host with a rich genomic ‘starter-kit’ to support the increase in the cellular and genomic complexity that is characteristic of eukaryotes.
Cellular life is currently classified into three domains: Bacteria, Archaea and Eukarya. Whereas the cytological properties of Bacteria and Archaea are relatively simple, eukaryotes are characterized by a high degree of cellular complexity, which is hard to reconcile given that most hypotheses assume a prokaryote-to-eukaryote transition1,2. In this context, it seems particularly difficult to account for the suggested presence of the endomembrane system, the nuclear pores, the spliceosome, the ubiquitin protein degradation system, the RNAi machinery, the cytoskeletal motors and the phagocytotic machinery in the last eukaryotic common ancestor (ref. 3 and references therein). Ever since the recognition of the archaeal domain of life by Carl Woese and co-workers4,5, Archaea have featured prominently in hypotheses for the origin of eukaryotes, as eukaryotes and Archaea represented sister lineages in Woese’s ‘universal tree’5. The evolutionary link between Archaea and eukaryotes was further reinforced through studies of the transcription machinery6 and the first archaeal genomes7, revealing that many genes, including the core of the genetic information-processing machineries of Archaea, were more similar to those of eukaryotes8 rather than to Bacteria. During the early stages of the genomic era, it also became apparent that eukaryotic genomes were chimaeric by nature8,9, comprising genes of both archaeal and bacterial origin, in addition to genes specific to eukaryotes. Yet, whereas many of the bacterial genes could be traced back to the alphaproteobacterial progenitor of mitochondria, the nature of the lineage from which the eukaryotic host evolved remained obscure1,10–13. This lineage might either descend from a common ancestor shared with Archaea (following Woese’s classical three-domains-of-life tree5), or have emerged from within the archaeal domain (so-called archaeal host or eocyte-like scenarios1,14–17). Recent phylogenetic analyses of universal protein data sets have provided increasing support for models in which eukaryotes emerge as sister to or from within the archaeal ‘TACK’ superphylum18–22, a clade originally comprising the archaeal phyla Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota23. In support of this relationship, comparative genomics analyses have revealed several eukaryotic signature proteins (ESPs)24 in TACK lineages, including distant archaeal homologues of actin25 and tubulin26, archaeal cell division proteins related to the eukaryotic endosomal sorting complexes required for transport (ESCRT)-III complex27, and several information-processing proteins involved in transcription and translation2,17,23. These findings suggest an archaeal ancestor of eukaryotes that might have been more complex than the archaeal lineages identified thus far2,23,28. Yet, the absence of missing links in the prokaryote-to-eukaryote transition currently precludes detailed predictions about the nature and timing of events that have driven the process of eukaryogenesis1,2,17,28. Here we describe the discovery of a new archaeal lineage related to the TACK superphylum that represents the nearest relative of eukaryotes in phylogenomic analyses, and intriguingly, its genome encodes many eukaryote-specific features, providing a unique insight in the emergence of cellular complexity in eukaryotes.
Genomic exploration of new TACK archaea
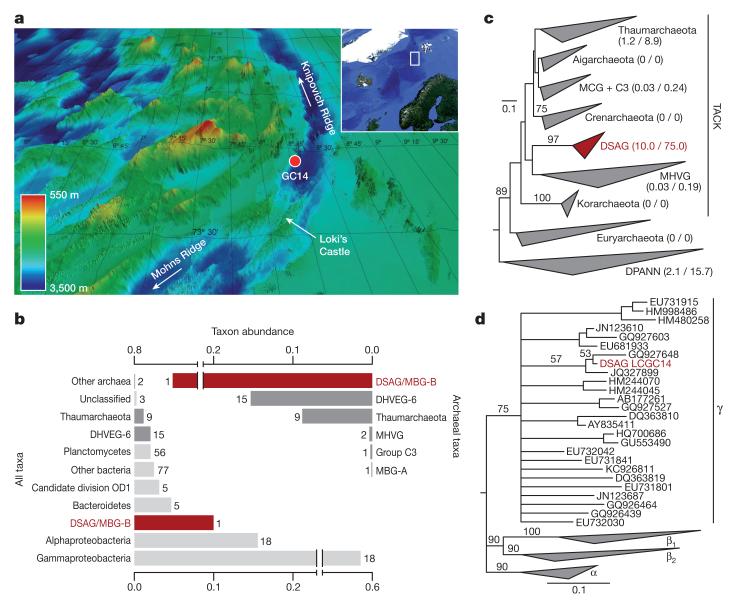
While surveying microbial diversity in deep marine sediments influenced by hydrothermal activity from the Arctic Mid-Ocean Ridge, 16S rRNA gene sequences belonging to uncultivated archaeal candidate lineages were identified in a gravity core (GC14) sampled approximately 15 km north-northwest of the active venting site Loki’s Castle29 at 3283 m below sea level (73.763167 N, 8.464000 E) (Fig. 1a)30,31. Subsequent phylogenetic analyses of these sequences, which comprised ~10% of the obtained 16S reads, revealed that they belonged to the gamma clade of the Deep-Sea Archaeal Group/Marine Benthic Group B (hereafter referred to as DSAG)31–33 (Fig. 1b–d and Supplementary Figs 1 and 2), a clade proposed to be deeply-branching in the TACK superphylum23. DSAG constitutes one of the most abundant and widely distributed archaeal groups in the deep marine biosphere, but so far none of its representatives have been cultured or sequenced31.
Figure 1. Identification of a novel archaeal lineage.
a, Bathymetric map of the sampling site (GC14; red circle) at the Arctic Mid-Ocean Spreading Ridge, located 15 km from Loki’s Castle active vent site. b, 16S rRNA amplicon-based assessment of microbial diversity in GC14. Bars on the left represent the fraction of the respective prokaryotic taxa and bars on the right depict archaeal diversity. Numbers refer to operational taxonomic units for each group. MHVG, Marine Hydrothermal Vent Group; DHVEG-6, Deep-sea Hydrothermal Vent Euryarchaeota Group 6; MBG-A and -B, Marine Benthic Group A and B. c, Maximum-likelihood phylogeny of the archaeal 16S rRNA reads (see b), revealing that DSAG sequences cluster deeply in the TACK superphylum. Numbers between brackets indicate relative abundance (%) of each group relative to total and archaeal reads, respectively. MCG, Miscellaneous Crenarchaeota Group; MHVG, Marine Hydrothermal Vent Group. d, Maximum-likelihood phylogeny of 16S rRNA gene sequences indicating that the DSAG operational taxonomic unit (red font) belongs to the DSAG γ cluster. Bootstrap support values above 50 are shown. c, d, Scale indicates the number of substitutions per site.
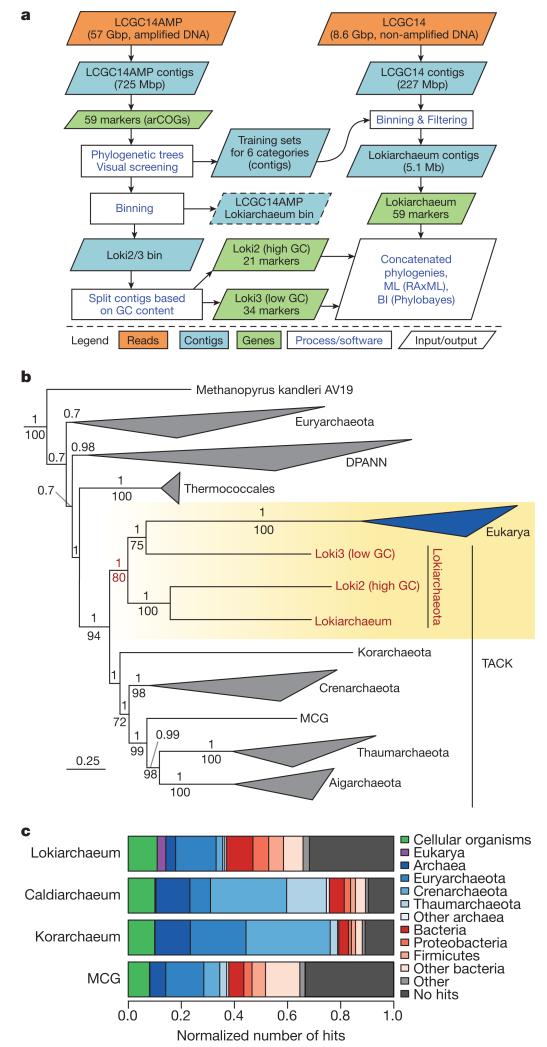
To obtain genomic information for this archaeal lineage, we applied deep metagenomic sequencing to the GC14 sediment sample, resulting in a smaller (LCGC14, 8.6 Gbp) and a larger, multiple-strand displacement amplified (MDA) metagenome data set (LCGC14AMP, 56.6 Gbp; Fig. 2a; Supplementary Fig. 3 and Supplementary Table 1). Given the deeper coverage, the latter data set was used to extract marker genes that carry an evolutionary coherent phylogenetic signal (Supplementary Tables 2 and 3). Using single gene phylogenies of these markers, contigs attributable to either one of the archaeal lineages present in the LCGC14AMP metagenome (DSAG, DSAG-related, DPANN and Thaumarchaeota), could be extracted. These taxon-specific contigs were used as training sets for supervised binning of contigs present in both the LCGC14 and LCGC14AMP metagenomes (Supplementary Fig. 4). This approach resulted in the identification of two DSAG bins (from LCGC14 and LCGC14AMP, respectively) as well as one DSAG-related bin (bin Loki2/3 from LCGC14AMP). We focused on the DSAG bin from the non-amplified data set to avoid potential biases introduced by MDA (see Methods). The analyses of the low-abundant DSAG-related lineages were based on the MDA-amplified LCGC14AMP data set.
Figure 2. Metagenomic reconstruction and phylogenetic analysis of Lokiarchaeum.
a, Schematic overview of the metagenomics approach. BI, Bayesian inference; ML, maximum likelihood. b, Bayesian phylogeny of concatenated alignments comprising 36 conserved phylogenetic marker proteins using sophisticated models of protein evolution (Methods), showing eukaryotes branching within Lokiarchaeota. Numbers above and below branches refer to Bayesian posterior probability and maximum-likelihood bootstrap support values, respectively. Posterior probability values above 0.7 and bootstrap support values above 70 are shown. Scale indicates the number of substitutions per site. c, Phylogenetic breakdown of the Lokiarchaeum proteome, in comparison with proteomes of Korarchaeota, Aigarchaeota (Caldiarchaeum) and Miscellaneous Crenarchaeota Group (MCG) archaea. Category ‘Other’ contains proteins assigned to the root of cellular organisms, to viruses and to unclassified proteins.
After removal of small (<1 kbp) and low-coverage contigs (Supplementary Fig. 5), reads mapping to the remaining DSAG bin contigs were reassembled into 504 contigs, yielding a 92% complete, 1.4 fold-redundant composite genome (‘Lokiarchaeum’) of 5.1 Mbp, which encodes 5,381 protein coding genes as well as single copies of the 16S and 23S rRNA genes (Supplementary Table 4 and Supplementary Discussion 1). The DSAG-related bin (Loki2/3 from LCGC14AMP) was found to contain two low-abundant, distinct lineages, displaying slight but marked differences in GC content of 32.8 and 29.9%, allowing for separation into two distinct groups (Loki2 and Loki3) (Supplementary Fig. 6). Since these two lineages represent low-abundance community members, only partial genomes could be recovered. The Loki2/3 contigs did not contain 16S rRNA genes, rendering it impossible to attribute them to any of the uncultured archaeal 16S phylotypes identified in the GC14 sediments, such as the low-abundance Marine Hydrothermal Vent Group archaea (abundance ~0.05%; Fig. 1c). However, phylogenetic marker genes were extracted for these lineages as well (21 and 34 markers for Loki2 and Loki3, respectively) since their inclusion was potentially useful in resolving the phylogenetic placement of the Lokiarchaeum lineage.
Lokiarchaeota and Eukarya are monophyletic
To determine the phylogenetic affiliation of Lokiarchaeum and the Loki2/Loki3 lineages, maximum-likelihood and Bayesian inference phylogenetic analyses were performed, using sophisticated models of molecular sequence evolution. By implementing relaxed assumptions of homogeneous amino acid composition across sites or across branches of the tree, these models are less sensitive to long-branch attraction and other phylogenetic artefacts. Both maximum-likelihood and Bayesian inference analyses of concatenated alignments comprising 36 conserved phylogenetic marker proteins20 (Supplementary Tables 2 and 3) revealed that the DSAG and DSAG-related archaea (hereafter referred to as ‘Lokiarchaeota’) represent a monophyletic, deeply branching clade of the TACK superphylum. Loki3 represented the deepest branch of the Lokiarchaeota, and Lokiarchaeum and Loki2 were inferred to be sister lineages with maximum support (Supplementary Fig. 7). Intriguingly, when eukaryotes were included in our phylogenetic analyses, they were confidently positioned within the Lokiarchaeota (posterior probability = 1; bootstrap support = 80; Fig. 2b; Supplementary Figs 8 and 9), as the sister group of the Loki3 lineage (Fig. 2b). Robust assessment of these phylogenetic inferences (Supplementary Figs 10–14 and Supplementary Table 5) revealed strong support for the Lokiarchaeota–Eukarya affiliation (Supplementary Discussion 2).
The proposed naming of the Eukarya-affiliated candidate phylum Lokiarchaeota and the Lokiarchaeum lineage is made in reference to the sampling location, Loki’s Castle29, which in turn was named after the Norse mythology’s shape-shifting deity Loki. Loki has been described as “a staggeringly complex, confusing, and ambivalent figure who has been the catalyst of countless unresolved scholarly controversies”34, in analogy to the ongoing debates on the origin of eukaryotes.
Presence of diverse and abundant ESPs
As our phylogenetic analyses strongly support a common ancestry of Lokiarchaeota and eukaryotes, we investigated the presence of putative ESPs24 in the composite Lokiarchaeum genome. The amount of genomic data obtained for the Loki2/3 lineages was too low to perform detailed gene content analyses. A comparative taxonomic assessment of the Lokiarchaeum composite proteome revealed that a large fraction (32%) of its proteins displayed no significant similarity to any known protein, and that roughly as many proteins display highest similarity to archaeal and bacterial proteins (26% and 29%, respectively; Fig. 2c and Supplementary Fig. 15), which is in accordance with recent findings that suggest major inter-domain gene exchange between Bacteria and Archaea35,36 (Supplementary Discussion 3). Most notably, a significant part of the predicted proteome (175 proteins or 3.3%) was most similar to eukaryotic proteins (Fig. 2c) and revealed a dominance of proteins, which in eukaryotes are involved in membrane deformation and cell shape formation processes, including phagocytosis37 (Extended Data Table 1 and Supplementary Table 6). Several lines of evidence support that the presence of these proteins is not the result of potential contaminating eukaryotic sequence data. First, genes encoding Lokiarchaeum ESPs and other proteins most similar to eukaryotes were always flanked by prokaryotic genes (Supplementary Fig. 16), and most were encoded by contigs that also contained archaeal signature genes. Second, ESP-encoding contigs displayed high (>20×) read coverage, while eukaryotic sequences could not be detected in the LCGC14 data set, and represented only a negligible fraction of the LCGC14AMP metagenome. Furthermore, the amplicon data generated with universal 16S/18S primers did not reveal any 18S rRNA genes of eukaryotic origin (Fig. 1b). Third, phylogenetic analyses of several Lokiarchaeal ESPs revealed their emergence at the base of eukaryotic clades (see below), indicating that these proteins represent archaeal out-groups of the eukaryotic proteins rather than being truly eukaryotic in origin. Fourth, Lokiarchaeum appears to contain bona fide archaeal informational processing machineries (Supplementary Discussion 4 and Supplementary Tables 7–9) and, irrespective of the significant amount of ESPs in its genome, lacks many other key eukaryotic features. Finally, we could also identify highly similar homologues of the Lokiarchaeal ESPs in a recent and independently generated marine sediment metagenome derived from a sediment core sample off the Shimokita Peninsula of Japan, in which DSAG comprises a significant part of the microbial community38. As the function and evolution of the Lokiarchaeal ESPs hold relevance for understanding the origin of the eukaryotic cell, we review some of the key findings in more detail below.
Potential dynamic actin cytoskeleton
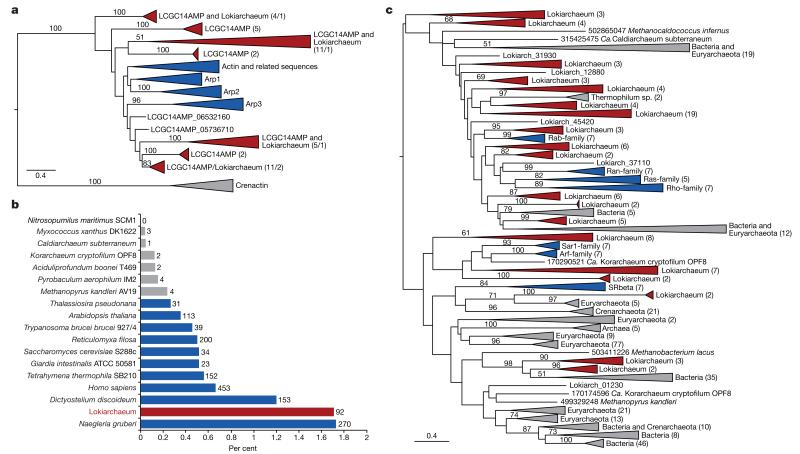
Actins represent key structural proteins of eukaryotic cells and comprise filaments that are crucial for various cellular processes, including cell division, motility, vesicle trafficking and phagocytosis39. The Lokiarchaeum genome encodes five actin homologues that display higher similarity to eukaryotic actins and actin-related proteins (ARPs) than to crenactins, a group of archaeal actin homologues that were recently shown to be involved in cell shape formation25,37,40 (Supplementary Table 6). This observation was confirmed in a phylogenetic analysis of the Lokiarchaeal actins that also included homologues identified in a recently published marine sediment metagenome38 (up to 99% identity) as well as in the LCGC14 and LCGC14AMP metagenomes (Fig. 3a and Supplementary Fig. 17). Lokiarchaeal actins (‘Lokiactins’) comprise several distinct clusters, some of which branch at the base of distinctive eukaryotic actin and ARP clusters, albeit with weak support (Fig. 3a). Despite the poor resolution of several deeper nodes in the actin tree, strong support is provided for a common ancestry of Lokiactins and eukaryotic actins, indicating that the proliferation of actins already occurred in the archaeal ancestor of eukaryotes. Notably, the Lokiarchaeum genome also encodes several hypothetical short proteins containing gelsolin-like domains that so far appear to be absent from bacterial and any other archaeal genomes (Extended Data Table 1, Supplementary Tables 6 and 10 and Supplementary Discussion 5). In eukaryotes, these protein domains are part of the villin/gelsolin superfamily of proteins, which comprise various key regulators of actin filament assembly and disassembly41. Although the function of these hypothetical gelsolin-domain proteins remains to be elucidated, it is tempting to speculate that Lokiarchaeum has a dynamic actin cytoskeleton.
Figure 3. Identification and phylogeny of small GTPases and actin orthologues.
a, Maximum-likelihood phylogeny of 378 aligned amino acid residues of actin homologues identified in Lokiarchaeum and in the LCGC14AMP metagenome, including eukaryotic actins, ARP1–3 homologues and crenactins25. Consecutive numbers in brackets refer to the number of sequences in a respective clade from LCGC14AMP and Lokiarchaeum, respectively. b, Relative amount of small GTPases (assigned to IPR006689 and IPR001806) in the Lokiarchaeum genome in comparison with other eukaryotic, archaeal and bacterial species. Numbers refer to total amount of small GTPases per predicted proteome. c, Maximum-likelihood phylogeny of 150 aligned amino acid residues of small Ras- and Arf-type GTPases (IPR006689 and IPR001806) in all domains of life. Numbers in brackets refer to the number of sequences in the respective clades. a, c, Sequence clusters comprising Lokiarchaeum and/or LCGC14AMP sequences (red), eukaryotes (blue) and Bacteria/Archaea (grey) have been collapsed. Bootstrap values above 50 are shown. Scale indicates the number of substitutions per site.
Genomic expansion of small GTPases
Small GTPases belonging to the Ras superfamily comprise one of the largest protein families in eukaryotes, where they are involved in various regulatory processes, including cytoskeleton remodelling, signal transduction, nucleocytoplasmic transport and vesicular trafficking42. Being key regulators of actin cytoskeleton dynamics, these small GTPases represent essential components for the process of phagocytosis in eukaryotes. Intriguingly, the analysis of Lokiarchaeal ESPs revealed a multitude of Ras-superfamily GTPases, comprising nearly 2% of the Lokiarchaeal proteome (Fig. 3b). The relative amount of small GTPases in the Lokiarchaeum genome is comparable to that observed in several unicellular eukaryotes, only being surpassed by the protist Naegleria gruberi. In contrast, bacterial and archaeal genomes encode only few, if any, small GTPase homologues of the Ras superfamily (Fig. 3b).
Phylogenetic analyses of the Lokiarchaeal small GTPases revealed that these represent several distinct clusters, each of which comprises several GTPase sequences (Fig. 3c and Supplementary Fig. 18). Although phylogenetic analyses failed to resolve most of the deeper nodes, several of the eukaryotic small GTPase families appear to share a common ancestry with Lokiarchaeal GTPases (Fig. 3c), suggesting an archaeal origin of specific subgroups of the eukaryotic small GTPases, followed by independent expansions in eukaryotes and Lokiarchaeota. This scenario contrasts with previous studies that have suggested that eukaryotic small GTPases were acquired from the alphaproteobacterial progenitor of mitochondria37.
Although genes encoding canonical eukaryotic GTPase-activating proteins (GAPs) were absent in Lokiarchaeota, twelve roadblock/LC7-domain-containing proteins were identified (Supplementary Tables 6 and 10). While such proteins have been implicated in dynein organization in eukaryotes, roadblock/LC7 protein MglB of the bacterium Myxococcus xanthus was shown to act as a GAP of the small GTPase MglA43. Hence, the Lokiarchaeal roadblock/LC7 proteins represent possible candidates for alternative GAPs in this archaeon.
Presence of a primordial ESCRT complex
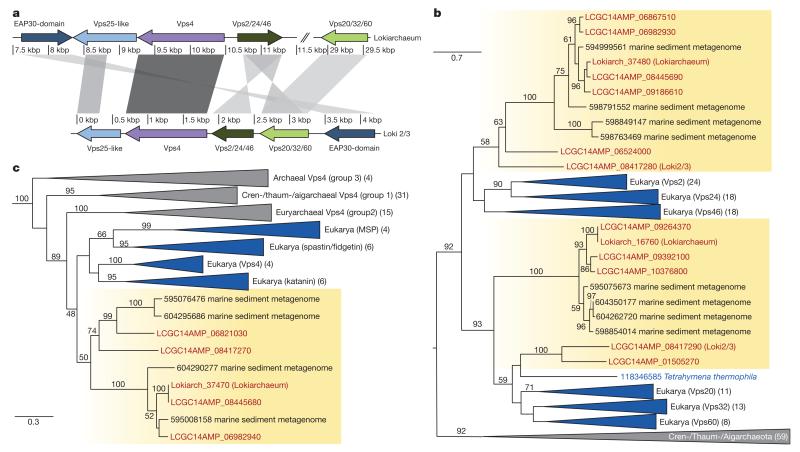
In eukaryotes, the ESCRT machinery represents an essential component of the multivesicular endosome pathway for lysosomal degradation of damaged or superfluous proteins, and it plays a role in several budding processes including cytokinesis, autophagy and viral budding44. The ESCRT machinery generally consists of the ESCRT-I–III subcomplexes, as well as associated subunits45. The analysis of the Lokiarchaeum genome revealed the presence of an ESCRT gene cluster (Fig. 4a), as well as of several additional proteins homologous to components of the eukaryotic multivesicular endosome pathway. For instance, Lokiarchaeum encodes divergent SNF7 domain proteins of the eukaryotic ESCRT-III complex, which appear to represent members of the Vps2/Vps24/Vps46 and Vps20/Vps32/Vps60 families, respectively. A phylogenetic analysis of the Lokiarchaeal SNF7 domain proteins revealed that these branch at the base of these two eukaryotic ESCRT-III families with low bootstrap support (Fig. 4b and Supplementary Fig. 19), not only indicating that they might represent ancestral SNF7 copies, but also suggesting that the last eukaryotic common ancestor already inherited two divergent SNF7-domain-encoding genes from its putative archaeal ancestor rather than a single gene46. Furthermore, the gene cluster encodes an ATPase that displays closest resemblance to eukaryotic VPS4-type ATPases, including katanin, membrane scaffold protein (MSP) and spastin (Fig. 4c and Supplementary Fig. 20) as well as hypothetical proteins that show significant similarity to EAP30-domain-containing proteins (Vps36/22) and Vps25, respectively (Fig. 4a; Supplementary Figs 21 and 22). In eukaryotes, Vps22, Vps25 and Vps36 are components of the ESCRT-II complex, which comprises two to three of these proteins depending on the eukaryotic species46. In addition, a protein domain analysis of the Lokiarchaeum proteome identified a Vps28-like protein, a component of the eukaryotic ESCRT-I subcomplex that links the ubiquitin pathway to vesicular transport and which, apart from Vps28, comprises Vps23 and Vps37 (Extended Data Table 1 and Supplementary Fig. 23). The different subunits of the eukaryotic ESCRT-I complex share similar two-helix core domains and have been suggested to have evolved from a single ancestral sequence47, which we now propose to be of archaeal origin.
Figure 4. Identification of ESCRT components in the Lokiarchaeum genome.
a, Schematic overview of ESCRT gene clusters identified in Lokiarchaeum and Loki2/3. Intensity of shading between homologous sequences is correlated with BLAST bit score. b, Maximum-likelihood phylogeny of 207 aligned amino acid residues of ESCRT-III homologues identified in Lokiarchaeum, LCGC14AMP and other archaeal lineages. Eukaryotic homologues include the two distantly related families Vps2/24/46 and Vps20/32/60. Bootstrap support values above 50 are shown. c, Maximum-likelihood phylogeny of 388 aligned amino acid residues of AAA-type Vps4 ATPases including representatives for each of the four major eukaryotic sub-groups (membrane scaffold protein (MSP), katanin, spastin/fidgetin and Vps4) as well as homologues identified in the Lokiarchaeum genome, in LCGC14AMP and in sequenced archaeal genomes. Bootstrap support values below 45 are not shown. b, c, Scale indicates the number of substitutions per site. Numbers in brackets refer to the number of sequences in the respective clades.
Finally, the Lokiarchaeum proteome was found to contain hypothetical proteins containing Longin-like domains, as well as several proteins belonging to the BAR/IMD superfamily (Supplementary Tables 6 and 10), comprising curvature sensing protein families involved in various aspects of vesicle/membrane trafficking or remodelling processes in eukaryotes. These findings suggest that Lokiarchaeum contains a primordial version of a eukaryotic ESCRT vesicle trafficking complex. In eukaryotes, ubiquitylation of target proteins represents a critical step in ESCRT-mediated protein degradation through the multivesicular endosome pathway44,48. The Lokiarchaeum genome contains a gene cluster that encodes several components required for a functional ubiquitin modifier system, including homologues for ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and 26S proteasome regulatory subunit RPN11. In addition, several hypothetical proteins with ubiquitin-like domains were identified in Lokiarchaeum, as well as diverse zinc-finger/RING-domain-containing proteins, some of which might serve as candidates for E3 ubiquitin protein ligases (Supplementary Tables 6 and 10). Several of these components have also been identified in Aigarchaeota49.
A ‘complex’ archaeal ancestor of Eukarya
We have identified and characterized the genome of Lokiarchaeota, a novel, deeply rooting clade of the archaeal TACK superphylum, which in phylogenomic analyses of universal proteins forms a monophyletic group with eukaryotes. While the obtained phylogenomic resolution testifies to a deep archaeal ancestry of eukaryotes, the Lokiarchaeum genome content holds valuable clues about the nature of the archaeal ancestor of eukaryotes, and about the process of eukaryogenesis. Many of the ESPs previously identified in different TACK lineages are united in Lokiarchaeum, indicating that the patchy distribution of ESPs amongst archaea is most likely the result of lineage-specific losses2 (Fig. 5). Moreover, the Lokiarchaeum genome significantly expands the total number of ESPs in Archaea, lending support to the observed phylogenetic affiliation of Lokiarchaeota and eukaryotes. Finally, and importantly, sequence-based functional predictions for these new ESPs indicate a predominance of proteins that play pivotal roles in various membrane remodelling and vesicular trafficking processes in eukaryotes. It is also noteworthy that Lokiarchaeum appears to encode the most ‘eukaryotic-like’ ribosome identified in Archaea thus far (Supplementary Discussion 4), including a putative homologue of eukaryotic ribosomal protein L22e (Fig. 5; Supplementary Fig. 24 and Supplementary Tables 7 and 8).
Figure 5. The complex archaeal ancestry of eukaryotes.
Schematic overview of the distribution of ESPs in major archaeal lineages across the tree of life. Each ESP is depicted as a coloured circle and losses are indicated with a cross. Patchy distribution and absence of a particular ESP in archaeal phyla is indicated by half-shaded and white circles, respectively. aWhile eukaryotes and Lokiarchaeota contain bona fide actins, other archaea encode the more distantly related Crenactins. bOnly few members of the Thaumarchaeota contain distantly related homologs of tubulins (ar-tubulins). cThaum-, Aig- and some Crenarchaeota contain distant homologues of ESCRT-III (SNF7 domain proteins).
Taken together, our data indicate that the archaeal ancestor of eukaryotes was even more complex than previously inferred2 and allow us to speculate on the timing and order of several key events in the process of eukaryogenesis. For example, the identification of archaeal genes involved in membrane remodelling and vesicular trafficking processes indicates that the emergence of cellular complexity was already underway before the acquisition of the mitochondrial endosymbiont, which now appears to be a universal feature of all eukaryotes28,37,50. Indeed, based upon our results it seems plausible that the archaeal ancestor of eukaryotes had a dynamic actin cytoskeleton and potentially endo- and/or phagocytic capabilities, which would have facilitated the invagination of the mitochondrial progenitor.
The present identification and genomic characterization of a novel archaeal group that shares a common ancestry with eukaryotes indicates that the gap between prokaryotes and eukaryotes might, to some extent, be a result of poor sampling of the existing archaeal diversity. Environmental surveys have revealed the existence of a plethora of uncultured archaeal lineages, and some of these likely represent even closer relatives of eukaryotes. Excitingly, the genomic exploration of these archaeal lineages has now come within reach. Such endeavours, combined with prospective studies focusing on uncovering metabolic, chemical and cell biological properties of these lineages, will uncover further details about the identity and nature of the archaeal ancestor of eukaryotes, shedding new light on the evolutionary dark ages of the eukaryotic cell.
METHODS
No statistical methods were used to predetermine sample size.
Sampling site and sample description
A 2-m long gravity core (GC14) was retrieved from the Arctic Mid-Ocean Ridge during summer 2010 (approximately 15 km north-northwest of the active venting site Loki’s Castle; 3283 m below sea level; 73.763167 N, 8.464000 E) (Fig. 1a). Samples for geochemistry and microbiology were collected immediately and either processed on board or frozen for later analysis. Upon port arrival, the core was stored in sealed core liners at 4 °C (core depository facility, University of Bergen, Norway). Comprehensive geochemical and microbial characteristics from this and adjacent sites have been described elsewhere51,52. The core consists of hemipelagic-glaciomarine sediments receiving episodic hydrothermal input. The oxygen penetration depth was estimated to ~50 cm below sea floor (b.s.f.) and the content of organic carbon varied between 0.6 - 1.3%. While no measurable amounts of methane or sulphide could be measured, high and fluctuating levels of dissolved iron were detected. The relative abundance of bacterial and archaeal 16S rRNA gene copy numbers was estimated by quantitative PCR (qPCR) previously52, indicating high abundance of the DSAG in several of the investigated sediment horizons, especially at 75 cm b.s.f. (up to 40% of the total prokaryotic population; 2.7 × 106 copies per gram sediment). Thus, sample material from horizon at 75 cm b.s.f. was used for all downstream analyses including amplicon and metagenome libraries.
DNA extraction and genomic DNA amplification
To obtain sufficient amounts of genomic DNA for sequencing library preparation, new sample material was obtained from the 75-cm-b.s.f. layer of gravity core GC14 in summer 2013. After qPCR-based verification of high DSAG abundance in the re-sampled material, DNA was extracted from 7.5 g sediment using the FastDNA spin kit for soil in conjunction with the FastPrep-24 instrument (MP Biomedicals) following manufacturer’s protocol, except for the addition of polyadenosine as described in ref. 53. The individual extractions were then pooled and concentrated to a final volume of 50 μl using Amicon Ultra-0.5 filters (50.000 NMWL) following the manufacturer’s protocol (Merck Millipore). Due to low yield and presence of inhibitors, 2.73 ng of this genomic DNA was amplified using the REPLI-g ultrafast mini kit (Qiagen) according to the standard protocol for purified genomic DNA.
Amplicon sequencing and analysis of 16S rDNA phylogenetic analyses
To get a better estimate of the microbial diversity of Loki’s Castle sediment core LCGC14, ‘universal’ primer pairs (A519F (5′-CAGCMGCCGCGGTAA-3′) and U1391R (5′-ACGGGCGGTGWGTRC-3′)) were used to amplify a ~900 bp fragment of the 16S rRNA genes present in the non-amplified genomic community DNA (extracted from LCGC14, 75 cm b.s.f.) using the following conditions: 15 min of heat activation of polymerase at 95 °C and 35 cycles of 95 °C (30 s), 54 °C (45 s), 72 °C (60 s), followed by final extension at 72 °C for 7 min. Qiagen HotStar Taq DNA polymerase was used for the PCR reactions. Subsequently, PCR products of the correct size were purified with Qiagen PCR purification kit, and quantified using a Nanodrop ND-3300 fluorospectrometer (Thermo Scientific). Clean PCR products were then used as input materials for library construction using TruSeq DNA LT Sample Prep Kit (Illumina) according to the manufacturer’s instructions and applied to sequencing with an Illumina MiSeq instrument. The Illumina MiSeq run produced two 300-bp paired-end reads. Raw MiSeq fastq sequences were treated with Trimmomatic tool (v0.32)54 using the following options: TRAILING:20, MINLEN:235 and CROP:235, to remove trailing sequences below a phred quality score of 20 and to achieve uniform sequence lengths for downstream clustering processes. Remaining traces of Illumina adaptor sequences were removed by SeqPrep (https://github.com/jstjohn/SeqPrep) and by BLAST55 searches against NCBI Univec database. Quality-filtered MiSeq reads were checked for correct orientation of the 16S rRNA sequence in the paired-end reads and those containing the forward primer sequence (A519F) were extracted for OTU clustering with UPARSE pipeline56, setting a OTU cutoff threshold to 97%. Chimeric sequences were filtered out by the Uchime tool57 integrated in the UPARSE pipeline. Remaining chimeric sequences, if still present, were manually checked and removed. Abundances of each OTU were calculated by mapping the chimaera-filtered OTUs against the quality-filtered reads using the UPARSE pipeline. Using the mothur package (v1.33.2)58, representative sequences for each OTU were aligned together with the Silva NR99 release-11559 alignment file to classify the OTUs.
Phylogenetic analysis of archaeal 16S rRNA gene sequences
Twenty-nine archaeal OTUs identified from the amplicon data were aligned together with 220 sequences representing the major clades in the archaeal 16S rRNA tree according to the study by Durbin and Teske60. A total of 249 sequences were aligned with MAFFT L-INS-i (v7.012b)61, trimmed with TrimAl (v1.4)62, and subjected to a maximum-likelihood phylogeny analysis using RAxML (v8.0.22)63 (GTRGAMMA model of nucleotide substitution and 100 bootstraps). The resulting tree was imported into iTOL online64 to collapse major clades.
Phylogenetic analysis of DSAG-related OTU’s
All 16S rRNA gene sequences classified as DSAG by Jørgensen et al.52 were used as queries in a BLAST search (E < 10−5, identity > 83%) against all archaeal entries in the SILVA database (release 119) that met the following criteria: sequence length > 900 bp, alignment identity > 70, alignment quality > 75 and pintail quality > 75 and the quality of recovered sequences was checked (for example, using ‘cut-head’ and ‘cut-tail’ information). The number of sequences in the data set was reduced while keeping maximum diversity as follows. First, the retained 16S rRNA sequences were aligned with SINA (v1.2.11)65, using all archaea in the SILVA database as reference. The alignment was manually curated with Seaview (v4)66. Upon removal of gaps, sequences were used to create OTUs with UCLUST (v1.2.22)67 (94% identity cut-off and the ‘–optimal’ option). All sequences that corresponded to OTU seeds were selected to represent full DSAG genetic diversity and, upon adding archaeal outgroup sequences and the single amplicon OTU, classified as DSAG, the final data set was aligned with SINA (v1.2.11) as described above, trimmed with TrimAl (v1.4) (gap threshold of 50%) and subjected to RAxML phylogenetic analyses (v7.2.8; GTRGAMMA substitution model, 100 rapid bootstraps). All internal branches with ≤40 bootstrap support were collapsed with Newick-Utilities (v1.6)68. The resulting tree was then imported into iTOL online64 to collapse major clades.
Metagenome sequencing and assembly
Library preparation and shotgun sequencing
Nextera libraries (Illumina) were prepared according to the manufacturer’s instructions, using unamplified LCGC14 (20 ng) and amplified LCGC14AMP (50 ng) as input DNA. Since less starting material was used for the generation of the unamplified library, a total of eight amplification cycles were used in the PCR step during which the Illumina barcodes and adapters (NextEra Index kit) were fused, rather than the default five cycles. The LCGC14 and LCGC14AMP NextEra libraries were sequenced with three and two lanes, respectively, of HiSeq2500 (Illumina), using rapid mode setting, generating two 150-bp paired reads. These runs yielded 8.6 Gbp and 56.6 Gbp of data with an average insert size of 620 and 350 bp for the LCGC14 and LCGC14AMP NextEra libraries, respectively.
Read preprocessing
SeqPrep (v.b5efabc5f7, https://github.com/jstjohn/SeqPrep) was used to merge overlapping paired-end reads and to trim adapters, with default settings. Merged reads and non-merged pairs were trimmed with Sickle (v.1.210, https://github.com/najoshi/sickle), using “se” and “pe” options, respectively, and default settings.
Metagenomic assembly
Pre-processed paired-end reads and single reads were assembled with SPAdes v. 3.0.069 in single-cell mode, to take into account the widely varying coverage of metagenomics contigs as well as to try to assemble contigs with low coverage. The read correction tool was turned on and kmers 21, 33, 55 and 77 were used. Mismatch correction was not performed on the LCGC14AMP data set. Contigs shorter than 1 kbp were discarded.
Gene predictions
Protein coding genes (CDS) were identified with prodigal v. 2.6070, using the ‘meta’ option for metagenomes. Ribosomal RNA (rRNA) genes were called with rnammer v.1.271, using the archaeal model and searching for all three rRNA subunits. Transfer RNA genes (tRNA) were identified with tRNAscan-SE v.1.2372, using the ‘–G’ option for metagenomes and ‘–A’ option for the Lokiarchaeum composite genome (see subsequent paragraphs). For the latter, the analysis was also run with SPLITSX (no version number available; source code downloaded on 14 August 2014)73 to detect tRNA genes that are split or that have multiple introns.
Protein clustering
Archaea-specific clusters of orthologous genes (arCOGs)74, based on 120 archaeal proteomes (hereafter called arCOGs2012), were extended with proteomes from 45 recently sequenced organisms, including 31 single-cell amplified genomes (SAGs) (Supplementary Table 1). First, existing arCOGs were attributed to the new proteomes: protein sequences in each of the 10,323 arCOGs2012 were aligned with MAFFT L-INS-i v.7.130b61. Each alignment was used as a query (-in_msa) to search the new proteomes using PSI-BLAST55, ignoring the master sequence, using 10−4 as an E-value cut-off, fixing the database size to 108, gathering at most 1,000 sequences, and not using composition-based statistics. Hits were then sorted per subject protein and, for each subject, the highest-scoring query alignment was deemed the main arCOG. Whenever applicable, the next-highest, non-overlapping query alignment was deemed the secondary arCOG. Second, proteins without arCOG attribution (singletons) in both the original and extended set of proteomes were gathered, and new arCOGs (arCOGs2014) were created from symmetrical best hits, using the tools available in COG software suite, release 201204 (ref. 75). PSI-BLAST searches were performed according to the COG software instructions. Lineage-specific expansions were identified with COGlse, using a job-description file containing all possible pairs of organisms that do not belong to the same phylum. COGtriangles was run with default settings, and yielded 3,570 new arCOGs. Of the 325,405 proteins in the combined data sets (165 proteomes), 29,249 (9%) had no arCOG attribution.
Attribution of arCOGs to metagenomes or composite genomes in this study was performed with PSI-BLAST as described above, using the arCOGs2014 as queries.
Phylogenetic analyses of ‘taxonomic marker’ proteins for binning and concatenated protein trees
Phylogenetic inference
Maximum-likelihood phylogenies were inferred with RAxML 8.0.963, calculating 100 non-parametric bootstraps. PROTGAMMALG and GTRGAMMA were used for amino acid and nucleotide alignments, respectively, unless otherwise stated. Bayesian inference phylogenies were calculated with PhyloBayes MPI 1.5a76, using the CAT model and a GTR substitution matrix. Four chains were run, and runs were checked for convergence. Whenever convergence was not reached, the topology of individual chains was compared. Consensus trees were obtained with bpcomp, using all four chains and a burn-in of at least half the generations. To add bootstrap support values to the Bayesian phylogenies, sumtrees.py (DendroPy package77) was used, with default settings, taking the Bayesian inference tree as a guide tree and the 100 bootstraps as input. For concatenated phylogenies, amino-acid sequences were aligned again with MAFFT L-INS-i individually for each cluster. Positions with >50% gaps were trimmed and alignments were concatenated.
Amino acid bias filtering
To assess the effect of amino acid bias on the phylogenies, a χ2 filtering analysis was performed on the concatenated alignment. For a complete description, see refs 78 and 79. In brief, a global χ2 score is calculated for the concatenated alignment, by summing, for each amino acid and each sequence, the normalized squared difference between the expected and observed frequency of the amino acid in this particular sequence and its frequency expected from the whole alignment. Each position in the alignment is individually trimmed and the difference (Δχ2) between the global χ2 score and the χ2 score calculated on the trimmed alignment provides an estimation of the relative contribution of each position to the global amino acid composition heterogeneity. Positions are then ranked by their Δχ2 values, and the most or least biased sites up to a threshold are removed.
Tree topology tests
To compare how well different trees explained the aligned sequence data, approximately unbiased tests80 were performed on concatenated as well as single-gene alignments. Two maximum-likelihood hypothesis trees were tested against the alignments. The first one, showing Lokiarchaeota grouping with eukaryotes, was obtained from the concatenation of 36 markers, shown in Fig. 2b. The second was obtained from the concatenation of the 21 ribosomal proteins present in the previous set, and shows Korarchaeota grouping with eukaryotes. For individual gene trees, the taxa missing in the alignment were also pruned from the hypothesis trees using the utility nw_prune from the Newick Utilities package68. For each alignment tested, per-site maximum likelihood was calculated for both hypothesis trees with RaxML 8.0.9, using the option ‘–f G’, and the PROTGAMMALG model. CONSEL 0.2081 was then used to perform approximately unbiased tests, using default settings.
Identification of taxonomic markers
A reference set of 59 highly conserved, low- or single-copy genes were used both as taxonomic markers in the binning process and for concatenated phylogenies (Supplementary Table 2). Fifty-seven of these, which were shown to be prone to very few or no horizontal gene transfers were taken from ref. 79. Two further arCOGs (arCOG04256 and arCOG04267, subunits A″ and A′ of the DNA-directed RNA polymerase, respectively) were added to the set (see Supplementary Information and Supplementary Table 2 for a list over which arCOG is included in each phylogeny).
Unless otherwise stated, all trees included the same set of 101 reference genomes: 58 archaeal genomes selected79 from the 120 analysed by Wolf et al.74; 21 selected from the 45 newly sequenced organisms that were also used for clustering, some of them already analysed in Guy et al.82; two groups of three closely related SAGs were pooled to provide more complete proteomes; ten bacteria and ten eukaryotes, as in Guy et al.82 (Supplementary Table 1). To remove paralogues and obtain sets with at most one homologue per genome, members of each of the selected arCOGs were aligned with MAFFT L-INS-i and a maximum-likelihood phylogeny was inferred with RAxML, under a PROTCATLG model with 100 slow bootstraps. Previously removed paralogues79 were not included. Trees were then visually inspected and paralogues removed using the same guidelines as in ref. 79. This set, including at most one copy of each of the 59 reference arCOGs in 101 genomes, is hereafter referred to as ‘59ref’.
Binning
Training set
After arCOG attribution (see above), genes from LCGC14AMP belonging to the respective arCOGs were added to the 59ref set. Sequences were aligned and individual trees were built for each arCOG, as described above. Trees were then visually inspected and sequences from LCGC14AMP were classified in the following categories: Lokiarchaeum, Loki2/3 (distant Lokiarchaeaum-related clades), Thaumarchaeota, DPANN, Diapherotrites, Mimivirus, Bacteria or unknown. Classification was based on phylogenetic placement. In some cases where the phylogenetic placement was inconclusive, presence on the same contig of another gene already classified was used to aid classification. The fact that Lokiarchaeum is the only clade for which four to six distinct but closely related strains are present in LCGC14AMP greatly aided classification. In a minority of cases, some genes were classified in a category but marked as ‘putative’, as their attribution was slightly ambiguous.
Quality control of the training set
Contigs containing markers classified in the first six categories mentioned above were extracted from the assembly, and their tetranucleotide frequencies (TNF) were calculated. To then assess the reliability of the classification, linear discriminant analysis (LDA) was performed in R83 with package MASS84, using GC content and TNF as input data: half of the contigs belonging to each of the six selected categories were randomly selected (excluding the contigs marked as ‘putative’), and used to calculate LDA (function ‘lda’ in MASS) (Supplementary Fig. 4). Based on this, classification was predicted using the MASS function predict.lda. Incorrect predictions (that is, when the prediction based on LDA was not congruent with the classification based on the phylogenetic trees) were recorded. The procedure was repeated 100 times, and contigs that were attributed to the wrong category 30 times or more were manually reviewed and eventually discarded from the training set (Supplementary Fig. 4a). Contigs marked as putative were attributed to the category if the prediction was congruent with the putative classification 90 times or more, or discarded otherwise. A further cycle of LDA calculation and prediction was performed, with no contigs classified as ‘putative’ this time (Supplementary Fig. 4b). To further investigate the robustness of the method, we randomized the categories of the input and performed the same LDA calculation and prediction as above, and assessed the number of incorrect predictions in each case (Supplementary Fig. 4c). This test confirmed that classifications based on trees were generally congruent with predictions based on LDA, significantly more often than just by chance (Supplementary Fig. 4d–f). The final set of contigs was used as a training set for phymmBL85(see below), and comprised 839 kbp for Lokiarchaeum, 544 kbp for Loki2/3, 521 kbp for Thaumarchaeota, 646 kbp for DPANN, 43 kbp for Diapherotrites and 21 kbp for Mimivirus.
Binning using PhymmBL
PhymmBL version 4.085 was run separately for binning the contigs larger than 1 kbp from both LCGC14AMP and LCGC14. As training sets, all prokaryotic genomes published in GenBank (retrieved on 2014-03-04, 2716 genomes) were complemented with the 60 newly sequenced genomes used to constitute the arCOG set that was absent from GenBank (Supplementary Table 1), and the six training sets (Lokiarchaeum, Loki2/3, Thaumarchaeota, DPANN, Diapherotrites and Mimivirus) obtained from LCGC14AMP as described above.
Reassembly of Lokiarchaeum bin
In the LCGC14 assembly, 3,165 contigs (18.6 Mbp in total) were predicted to belong to the Lokiarchaeum genus, indicating a large degree of microdiversity. In order to reduce redundancy, contig sets were constituted, with increasing low-coverage cut-offs (from 1 to 100×, with a 1× increment). The completeness and redundancy of each set was then estimated using the micomplete script (manuscript in preparation). In brief, micomplete bases its predictions on the presence or absence of a set of single-copy panorthologs, in this case 162 markers defined in ref. 86. To avoid overemphasizing the presence of markers that are often very close to each other (for example, ribosomal proteins), each marker receives a weight coefficient based on the distance between this marker and its closest neighbours both upstream and downstream, averaged over a representative set of 70 Archaea (set described in ref. 79). Completeness is the fraction of weighted markers present, and is thus constrained between 0 (no marker present) and 1 (all markers present). Redundancy is calculated as the total number of copies of weighted markers present divided by the number of weighted markers present, and is thus always greater than one, where one would mean that all markers present are single copy. These two numbers were calculated for each contig set, and a cut-off of 24× represented the best compromise between completeness (0.89) and redundancy (1.67) (Loki24× set, Supplementary Fig. 5a).
To obtain a better assembly with longer contigs with only reads from Lokiarchaeum, reads belonging to Lokiarchaeum contigs were reassembled as follows. Reads from the LCGC14 data set, corrected by SPAdes, were mapped against the whole LCGC14 assembly with bwa-mem87, and reads that matched contigs in the Loki24× set were extracted. For paired-end reads, both reads were retained if at least one read matched the Loki24× set. These extracted reads were assembled with SPAdes as above, but without the single-cell mode and without read correction. Again, completeness and coverage were assessed for sets of contigs with increasing low-coverage cut-offs, and a threshold of 20× coverage was found to give the best compromise between completeness (0.92) and redundancy (1.44) (Supplementary Fig. 5b). The selected 504 contigs, hereafter referred to as ‘Lokiarchaeum’, represented 5.14 Mbp of sequence. The N50 and N90 of this assembly were 15.4 and 5 kbp, respectively.
Annotation and contamination assessment of Lokiarchaeum genome bin
Annotation of all predicted open reading frames of the Lokiarchaeum genome bin was done using prokka88, using a concatenation of the three kingdom-specific protein databases shipped with prokka as the main database, predicting tRNA and rRNA as above. Furthermore, proteins were compared to sequences in NCBI’s non-redundant database and RefSeq using BLAST55 and results were inspected using MEGAN89. Additionally, an InterProScan 590 (which integrates a collection of protein signature databases such as BlastProDom, FPrintScan, HMMPIR, HMMPfam, HMMSmart, HMMTigr, ProfileScan, HAMAP, PatternScan, SuperFamily, SignalPHMM, TMHMM, HMMPanther, Gene3D, Phobius and Coils) was performed and the genome was viewed and analysed in MAGE91. Selected genes of interest for the evolution of the eukaryotic cell and/or subjected to phylogenetic analyses were checked manually and annotated according to their protein domains/signatures based on PSI-BLAST55 results, arCOG attributions (Supplementary Tables 6–10) as well as protein structure predictions using Phyre292. To check for the presence of particular genes of interest, such as specific eukaryotic ribosomal proteins, or eukaryotic ribosomal protein L41e which has been detected in several Euryarchaeota93, existing alignments from arCOGs and/or KOGs were downloaded from eggNOG94 and used in PSI-BLAST searches as query against the Lokiarchaeal composite genome.
Several controls were performed to confirm the absence of obvious contaminants in the final Lokiarchaeum bin. Most importantly, all contigs containing ESPs discussed in the manuscript were manually inspected to verify that these actually belong to Lokiarchaeum, by: (1) inspecting neighbouring genes for the presence of archaeal markers; (2) by querying all proteins present on contigs containing ESPs against the LCGC17 metagenome to check whether highly similar homologues could be found several times in the sample (generally between 3–7 copies) accounting for the different, highly related Lokiarchaeota strains; and (3) by querying the same proteins against environmental metagenomes publicly available at NCBI, controlling that most of them had highly similar homologues in an ocean sediment metagenome95, but not in any other metagenome. This last check was based on our finding that all ESPs of Lokiarchaeum had highly similar homologues in this marine sediment metagenome (for example, up to 98% for Lokiactins) indicating that closely related genomes of members of Lokiarchaeota are present, which is in accordance with the finding that DSAG represents an abundant group in these sub-seafloor sediments96.
Finally, proteins comprising informational processing machineries were also investigated using MEGAN89. The absence of bacterial informational processing proteins indicated that there is no bacterial contamination in the final bin (see Supplementary Discussion 3).
Identification of taxonomic markers in the bins
For Lokiarchaeum, arCOG attribution was performed as described above, and taxonomic markers were identified by their arCOG attribution. Whenever there were two copies of the same marker, the copy located on the contig with the highest coverage was selected.
For Loki2/3, the category had two copies of 19 out of 36 markers present, with divergent phylogenetic placement. A clear GC content difference could also be observed between the copies, and, with a single exception, the two sets of copies were not overlapping (Supplementary Fig. 6). The exception was discarded and the remaining two-copy markers were divided into two bins, Loki2 (high GC, ranging between 32.2–37.3%) and Loki3 (low GC, ranging between 27.7–30.7%). Single-copy markers with a GC content falling into the range of either of Loki2 or Loki3 were attributed to the corresponding bin, the other copies were discarded. Loki2 (high GC, average 32.8%) consisted of 21 markers and Loki3 (low GC, average 29.9%) of 34 markers.
Taxonomic affiliation of the Lokiarchaeum proteome
To estimate how Lokiarchaeum relates to its closest relatives, its proteome was aligned to NCBI’s non-redundant database using blastp, with an E threshold of 0.001. To provide a way to compare results, the complete proteomes of ‘Candidatus Korarchaeum cryptofilum’ OPF8, ‘Candidatus Caldiarchaeum subterraneum’ and the incomplete proteome of SCGC AB-539-E09, sole representative of the Miscellaneous Crenarchaeotal group (MCG) were similarly analysed. The results of the blasts were filtered to remove self-hits and hits to organisms belonging to the same phylum. In the case of the MCG representative, only self-hits were removed. Filtered results were then analysed with MEGAN 5.4.0. Last common ancestor parameters were set as follows: Min Score, 50; Max Expected, 0.01; Top Percent, 5; Min Support, 1; Min Complexity, 0.0. For each result, branches were uncollapsed at the level below super-kingdom. Profiles were compared using Absolute counts, and the results were exported and further analysed in R. Categories to which less than 100 hits were attributed in Lokiarchaeum were grouped under the ‘Other Archaea’ or ‘Other Bacteria’ categories. Hits to ‘root’, viruses, unclassified sequences and hits not assigned were grouped under the ‘Other’ category. Results are shown in Fig. 2b. Using the same parameters, functional COG categories were assigned to the Lokiarchaeal proteome to get insights into the functional and taxonomic affiliation of the Lokiarchaeal proteome (Supplementary Fig. 15).
Phylogenetic analyses of selected eukaryotic signature proteins (ESPs)
Selection of ESCRT-III homologues
For the ESCRT-III phylogeny, eukaryotic ESCRT-III homologues as described in Makarova et al.97 (comprising the families Vps60/Vps20/Vps32 and Vps46/2/24), as well as archaeal ESCRT-III homologues belonging to arCOG00452, arCOG00453 and arCOG00454 families present in Crenarchaeota, Thaumarchaeota and Aigarchaeota were extracted from GenBank. The more distantly related SNF7-like arCOG families (arCOG09747, arCOG09749 and arCOG07402)97 present in a few euryarchaeal species were not included in the alignment. Subsequently, respective arCOGs were retrieved from both the LCGC14AMP metagenome and Lokiarchaeum final bin (see section on arCOG attribution). The ESCRT operon present on a Loki2/3 contig revealed the presence of an additional ESCRT-III homologue (most similar to eukaryotic Vps20/32/60 sequences), which was not attributed to an archaeal COG. This homologue was used as an additional query to retrieve highly similar sequences from the LCGC14AMP metagenome as well as the Lokiarchaeum final bin using blastp. Finally, each of the two different SNF7-family proteins, which are part of the ESCRT operons of Lokiarchaeum and Loki2/3, respectively, were used as queries to search published metagenomes (NCBI) with blastp. Highly similar sequences (coverage > 70%; identity > 40%) were retrieved and included in the phylogeny as well.
Selection of Vps4 homologues
Archaeal sequences assigned to arCOG01307 (cell division ATPase of the AAA+ class, ESCRT system component) as well as eukaryotic Vps4 homologues, including a few proteins of the cdc48 subfamily, were retrieved from GenBank. The latter protein family served as outgroup, as described in Makarova et al.97 Sequences assigned to arCOG01307 were also extracted from LCGC14AMP metagenome as well as from the Lokiarchaeum bin, and sequences highly similar to the Vps4 of Lokiarchaeum were retrieved from published metagenomes (coverage > 60%; identity > 50%). The LCGC14AMP metagenome contained a large amount of sequences assigned to arCOG01307, including hits to Vps4 homologues of Thaumarchaeota. However, ATPases that, based on phylogenetic analyses, turned out to be unrelated to Vps4 were removed from the analysis. Based on the initial phylogeny that included all of these sequences, only those LCGC14AMP Vps4 homologues that clustered with the Vps4 homologue of the Lokiarchaeum bin were selected to avoid the inclusion of false positives.
Selection of EAP30-domain (Vps22/36-like) and Vps25 homologues
EAP30 and Vps25 homologues have so far not been detected in Archaea and thus the respective sequences present in Lokiarchaeum (Extended Data Table 1 and Supplementary Table 6) have not been assigned to an arCOG family. Thus, only Lokiarchaeum homologues, as well as selected representative eukaryotic sequences spanning the eukaryotic diversity that were retrieved from the GenBank database were included in these phylogenetic reconstructions. Putative EAP30- and Vps25-like homologues were discovered in the Lokiarchaeum genome since they are part of the ESCRT operon present on contig119. These sequences were used as queries to also retrieve homologues from the LCGC14AMP metagenome (E cut-off, 0.1; q coverage, 85) as well as from metagenomes deposited at NCBI.
Selection of small GTPase family homologues (IPR006689 and IPR001806)
The investigation of the Lokiarchaeum proteome revealed large numbers of proteins homologous to small GTPases of the Ras and Arf families. In order to reliably identify all putative small GTPases in the Lokiarchaeum bin, an InterPro scan90,98 was performed and all proteins assigned to IPR006689 (Ras type of small GTPases) and IPR001806 (Arf/Sar type of small GTPases) were extracted. Subsequently, archaeal reference sequences belonging to these IPR families were retrieved from GenBank. Eukaryotic and bacterial reference sequences were selected based on a previous study by Dong et al.99 that investigated the phylogenetic relationships of members of the Ras superfamily. Due to the large number of GTPase homologues in the Lokiarchaeum bin, and the difficulty assigning these proteins to a particular taxon, it was decided not to analyse all GTPase homologues present in metagenomes. Upon inspection of the MAFFT L-INS-i alignment, partial sequences and extremely divergent homologues were removed.
Selection of actin homologues
So far, the only actin-related proteins detected in a few members of the archaea belong to arCOG05583 and have been referred to as crenactins100. Three proteins encoded by the Lokiarchaeum genome were assigned to this arCOG, and a blastp search against RefSeq revealed that these proteins are more closely related to bona fide actins of Eukaryotes than to archaeal crenactins. In order to identify additional full-length actin homologues, blastp (E-value cut-off <10−10) searches were performed against the Lokiarchaeum genome as well as the LCGC14AMP metagenome, using this Lokiarchaeum actin homologue as query. Finally, a total of five and 42 full-length (>180 amino acids) actin-related proteins were retrieved from the Lokiarchaeum bin and from the LCGC14AMP metagenome, respectively. These sequences were merged with the archaeal protein sequences belonging to arCOG05583 as well as with major eukaryotic actin families (actins and ARP1–3 (refs 101, 102)). We also assessed the phylogenetic position of the bacterial actin-related protein (BARP) of the bacterium Haliangium ochraceum103 in light of the new Lokiarchaeal actin homologues, and concluded that the Haliangium BARP was most likely acquired via horizontal gene transfer from eukaryotes.
Phylogenetic reconstructions
For all of these ESPs, the selected sequences were aligned using MAFFT L-INS-i61 and trimmed with TrimAl62 to retain only those columns present in at least 50% (for ESCRT-III; Vps4; actin homologues), 40% (EAP30-domain and Vps25 homologues) and 80% (small GTPases) of the sequences. Alignments were visually inspected and manually edited whenever necessary and subsequently subjected to maximum-likelihood phylogenetic analyses using RAxML (8.0.22, PROTGAMMALG) with the slow bootstrap option (100 bootstraps).
Contig maps
The contig maps displayed in Fig. 4a were drawn with the software genoPlotR v.0.8.2 (ref. 104).
Extended Data
Extended Data Table 1.
Overview of Lokiarchaeal ESPs
| Suggested function | Product | Locus tag | IPR-domains | Comment |
|---|---|---|---|---|
| Putative ESCRT-III proteins | Vps2/24/46-like protein* | Lokiarch_37480 | IPR005024 Snf7 | More distant homologs also present in several other members of the TACK superphylum. |
| Vps20/32/60-like protein* | Lokiarch_16760 | IPR005024 Snf7 | ||
| Putative ESCRT-II proteins | EAP30 domain protein (Vps22/36-like)* | Lokiarch_37450 | IPR007286 EAP30 | Previously not found in Archaea. |
| Vps25-like protein* | Lokiarch_37460 | IPR014041 ESCRT-II complex, Vps25 subunit, N-terminal Winged helix; IPR008570 ESCRT-II complex, Vps25 subunit; IPR011991 Winged helix-turnhelix DNA-binding domain | ||
| Putative ESCRT-I protein | Hypothetical protein with Vps28-like domain† | Lokiarch_10170 | IPR007143 Vacuolar protein sortingassociated, Vps28 | Vps28 is part of ESCRT-I, potential interacting protein Lokiarch_16740 (see Table S6). |
| Putative ESCRT-associated protein | Vps4 ATPase* | Lokiarch_37470 | IPR003959 ATPase, AAA-type, core; IPR027417 P-loop containing nucleoside triphosphate hydrolase; IPR003593 AAA+ ATPase domain; IPR007330 MIT-domain | Also present in other members of the Archaea. |
| Putative vesicular trafficking machinery associated proteins | Hypothetical proteins vacuolar fusion domain MON1‡ | Lokiarch_21780 Lokiarch_01670 Lokiarch_15160 |
IPR004353 Vacuolar fusion protein MON1 | Previously not found in other prokaryotic organisms (see Table S6 and Table S10 for more details) |
| Hypothetical proteins with longin-like domains | Lokiarch_01890 Lokiarch_13110 Lokiarch_03280 Lokiarch_22790 Lokiarch_04850 |
IPR011012 Longin-like domain; IPR010908 Longin domain | ||
| BAR/IMD domain-like superfamily protein‡ |
Lokiarch_46220
Lokiarch_08900 |
IPR004148 BAR domain; IPR009602 FAM92 protein | Includes various protein families that bind membranes and detect membrane curvature. | |
| Cell division/cytoskeleton related proteins | Actin and related proteins* |
Lokiarch_44920
Lokiarch_36250 Lokiarch_10650 Lokiarch_09100 Lokiarch_41030 |
IPR004000, Actin-related protein; IPR020902 Actin/actin-like conserved site | Some Cren- Kor- and Aigarchaeota encode crenactins 25 (arCOG05583) |
| Hypothetical proteins with gelsolin-like domains‡ | 12 proteins, Suppl Table S6 | IPR007122 Villin/Gelsolin; IPR029006 ADF-H/Gelsolin-like domain; IPR007123 Gelsolin-like domain | Previously not found in Archaea. Serve as candidates for potential actin-binding proteins. | |
| Small GTP-binding domain proteins with Ran-/Ras- /Rab-/Rho- and Arf-domain signatures*§ | 92 proteins, see Suppl Table S6 | IPR001806 Small GTPase superfamily; IPR003579 Small GTPase superfamily, Rab type; IPR027417 P-loop containing nucleoside triphosphate hydrolase; IPR020849 Small GTPase superfamily, Ras type; IPR002041 Ran GTPase; IPR003578 Small GTPase superfamily, Rho type; IPR005225 Small GTP-binding protein domain; IPR024156 Small GTPase superfamily, ARF type | Extreme proliferation of small GTP-binding proteins in Lokiarchaeum (92 proteins in composite genome, see Fig. 3b and c); in addition Lokiarchaeum encodes 12 Roadblock/LC7 domain proteins, which might serve as GTPase activating enzyme (see Suppl Table S6). | |
| Ubiquitin modifier system related proteins | Ubiquitin-like proteins‡ |
Lokiarch_29280
Lokiarch_29310 Lokiarch_37670 |
IPR029071 Ubiquitin-related domain; IPR000626 Ubiquitin-like | Ubiquitin modifier system was previously identified in Aigarchaeota49; Canonical E3 ubiquitin ligases are not present in Caldiarchaeum subterraneum and Lokiarchaeum. However, both archaeal genomes contain RING-domain proteins‡ that could serve as candidates for E3 ligases, e.g. Lokiarch_34010 (see Suppl. Table S6). |
| Putative E1-like ubiquitin activating protein | Lokiarch_15900 Lokiarch_29320 |
IPR023280 Ubiquitin-like 1 activating enzyme, catalytic cysteine domain; IPR019572 Ubiquitin-activating enzyme (see SOM for more details) | ||
| Putative E2-like ubiquitin conjugating protein |
Lokiarch_10330
Lokiarch_41760 Lokiarch_29330 |
IPR016135 Ubiquitin-conjugating enzyme/RWD-like; IPR000608 Ubiquitin-conjugating enzyme, E2 | ||
| Hypothetical proteins with JAB1/MPN/MOV34 metalloenzyme domain |
Lokiarch_29340
Lokiarch_43590 Lokiarch_26830 Lokiarch_08140 |
IPR000555 JAB1/MPN/MOV34 metalloenzyme domain | ||
| Eukaryotic ribosomal protein | Putative homolog of eukaryotic ribosomal protein L22e† | Lokiarch_30160 | - | Previously not found in Archaea. Best blast hit: gb|EPR78232.1| 60S ribosomal protein L22 [Spraguea lophii 42_110] - Expect = 0.21 |
| Oligosaccharyl transferase complex proteins | Ribophorin 1 superfamily protein | Lokiarch_43710 | IPR007676 Ribophorin I | Previously not found in Archaea |
| Putative oligosaccharyl transferase complex, subunit OST3/OST6 |
Lokiarch_24040
Lokiarch_25040 |
IPR021149 Oligosaccharyl transferase complex, subunit OST3/OST6 | Previously not found in Archaea. | |
| Putative oligosaccharyl transferase STT3 subunit | Lokiarch_28460 | IPR003674 Oligosaccharyl transferase, STT3 subunit | Homologs also present in some other Archaea. |
Locus tags that are highlighted in bold indicate a significant top blast hit of the respective protein of Lokiarchaeum to a eukaryotic sequence (see Supplementary Table 6 for further details).
Phylogenetic analyses have been performed.
Alignments shown in Supplementary figures.
Protein domain assignments for these proteins listed in Supplementary Table 10.
While most small GTPases encoded by Lokiarchaeum have highest similarity to eukaryotic homologues, approximately 10% are most similar to Archaea and/or Bacteria (see Supplementary Table 6 for more details).
Supplementary Material
Acknowledgements
We dedicate this paper to the memory of Rolf Bernander. We thank P. Offre and I. de Bruijn for technical advice and useful discussions, and A. Denny for image processing. We also acknowledge the help from chief scientist R. B. Pedersen, the scientific party and the entire crew on board the Norwegian research vessel G.O. Sars during the summer 2010 expedition. All sequencing was performed by the National Genomics Infrastructure sequencing platforms at the Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VR-RFI) and the Knut and Alice Wallenberg Foundation. We thank the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) at Uppsala University and the Swedish National Infrastructure for Computing (SNIC) at the PDC Center for High-Performance Computing for providing computational resources. This work was supported by grants of the Swedish Research Council (VR grant 621-2009-4813), the European Research Council (ERC Starting grant 310039-PUZZLE_CELL) and the Swedish Foundation for Strategic Research (SSF-FFL5) to T.J.G.E., and by grants of the Carl Tryggers Stiftelse för Vetenskaplig Forskning (to A.S.), the Wenner-Gren Stiftelserna (to J.H.S.), and by Marie Curie IIF (331291 to J.H.S.) and IEF (625521 to A.S.) grants by the European Union. S.L.J received financial support from the H2DEEP project through the EuroMARC program, and by the Research Council of Norway through the Centre for Geobiology, University of Bergen. C.S. is supported by the Austrian Science Fund (FWF grant P27017).
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Information Sequence data have been deposited to the NCBI Sequence Read Archive under study number SRP045692, which includes 16 rRNA reads (experiment number SRX872366). Protein sequencesof Loki2/3 were deposited to GenBank under accession numbers KP869578–KP869724. The Lokiarchaeum genome bin and the LCGC14 metagenome projects have been deposited at DDBJ/EMBL/GenBank under the accessions JYIM00000000 and LAZR00000000, respectively. The versions described in this paper are versions JYIM01000000 and LAZR01000000. The authors declare no competing financial interests.
References
- 1.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Yutin N. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb. Perspect. Biol. 2014;6:a016188. doi: 10.1101/cshperspect.a016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koumandou VL, et al. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit. Rev. Biochem. Mol. Biol. 2013;48:373–396. doi: 10.3109/10409238.2013.821444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pühler G, et al. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc. Natl Acad. Sci. USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult CJ, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc. Natl Acad. Sci. USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McInerney JO, O’Connell MJ, Pisani D. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nature Rev. Microbiol. 2014;12:449–455. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- 10.Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nature Rev. Microbiol. 2010;8:743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- 11.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol. Biol. Evol. 2008;25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rochette NC, Brochier-Armanet C, Gouy M. Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol. Biol. Evol. 2014;31:832–845. doi: 10.1093/molbev/mst272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiergart T, Landan G, Schenk M, Dagan T, Martin WF. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol. Evol. 2012;4:466–485. doi: 10.1093/gbe/evs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson E, et al. A new ribosome structure. Science. 1984;225:510–512. doi: 10.1126/science.6429855. [DOI] [PubMed] [Google Scholar]
- 15.Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake JA. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988;331:184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- 17.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 18.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster PG, Cox CJ, Embley TM. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Phil. Trans. R. Soc. Lond. B. 2009;364:2197–2207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy L, Saw JH, Ettema TJ. The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 2014;6:a016022. doi: 10.1101/cshperspect.a016022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasek-Nesselquist E, Gogarten JP. The effects of model choice and mitigating bias on the ribosomal tree of life. Mol. Phylogenet. Evol. 2013;69:17–38. doi: 10.1016/j.ympev.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc. R. Soc. Lond. B. 2012;279:4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guy L, Ettema TJ. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Hartman H, Fedorov A. The origin of the eukaryotic cell:a genomic investigation. Proc. Natl Acad. Sci. USA. 2002;99:1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ettema TJ, Lindås A-C, Bernander R. An actin-based cytoskeleton in archaea. Mol. Microbiol. 2011;80:1052–1061. doi: 10.1111/j.1365-2958.2011.07635.x. [DOI] [PubMed] [Google Scholar]
- 26.Yutin N, Koonin EV. Archaeal origin of tubulin. Biol. Direct. 2012;7:10. doi: 10.1186/1745-6150-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindås A-C, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc. Natl Acad. Sci. USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martijn J, Ettema TJ. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem. Soc. Trans. 2013;41:451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen RB, et al. Discovery of a black smoker vent field and vent fauna at the Arctic Mid-Ocean Ridge. Nat. Commun. 2010;1:126. doi: 10.1038/ncomms1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jørgensen SL, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl Acad. Sci. USA. 2012;109:E2846–E2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jørgensen SL, Thorseth IH, Pedersen RB, Baumberger T, Schleper C. Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge. Front. Microbiol. 2013;4:299. doi: 10.3389/fmicb.2013.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inagaki F, et al. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 2003;69:7224–7235. doi: 10.1128/AEM.69.12.7224-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Von Schnurbein S. The function of Loki in Snorri Sturluson’s Edda. Hist. Relig. 2000;40:109–124. [Google Scholar]
- 35.Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, Lopez-Garcia P. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic thaumarchaeota and euryarchaeota. Genome Biol. Evol. 2014;6:1549–1563. doi: 10.1093/gbe/evu127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson-Sathi S, et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biol. Direct. 2009;4:9. doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai M, et al. High frequency of phylogenetically diverse reductive dehalogenase-homologous genes in deep subseafloor sedimentary metagenomes. Front. Microbiol. 2014;5:80. doi: 10.3389/fmicb.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernander R, Lind AE, Ettema TJ. An archaeal origin for the actin cytoskeleton: Implications for eukaryogenesis. Commun. Integr. Biol. 2011;4:664–667. doi: 10.4161/cib.16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 42.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Franco M, Ducret A, Mignot T. A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol. 2010;8:e1000430. doi: 10.1371/journal.pbio.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurley JH. The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Field MC, Dacks JB. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr. Opin. Cell Biol. 2009;21:4–13. doi: 10.1016/j.ceb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Leung KF, Dacks JB, Field MC. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 47.Kostelansky MS, et al. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–126. doi: 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 49.Nunoura T, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole AM, Gribaldo S. Eukaryotic origins: how and when was the mitochondrion acquired? Cold Spring Harb. Perspect. Biol. 2014;6:a015990. doi: 10.1101/cshperspect.a015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jorgensen SL, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl Acad. Sci. USA. 2012;109:E2846–E2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen SL, Thorseth IH, Pedersen RB, Baumberger T, Schleper C. Quantitative and phylogenetic study of the deep sea archaeal group in sediments of the Arctic Mid-Ocean spreading ridge. Front. Microbiol. 2013;4:299. doi: 10.3389/fmicb.2013.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 57.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durbin AM, Teske A. Archaea in organic-lean and organic-rich marine subsurface sediments: an environmental gradient reflected in distinct phylogenetic lineages. Front. Microbiol. 2012;3:168. doi: 10.3389/fmicb.2012.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pruesse E, Peplies J, Glockner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 67.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 68.Junier T, Zdobnov EM. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010;26:1669–1670. doi: 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugahara J, et al. SPLITS: a newprogram for predicting splitand intron-containing tRNA genes at the genome level. In Silico Biol. 2006;6:411–418. [PubMed] [Google Scholar]
- 74.Wolf YI, Makarova KS, Yutin N, Koonin EV. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol. Direct. 2012;7:46. doi: 10.1186/1745-6150-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kristensen DM, et al. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics. 2010;26:1481–1487. doi: 10.1093/bioinformatics/btq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- 77.Sukumaran J, Holder MT. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- 78.Viklund J, Ettema TJ, Andersson SG. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol. Biol. Evol. 2012;29:599–615. doi: 10.1093/molbev/msr203. [DOI] [PubMed] [Google Scholar]
- 79.Guy L, Saw JH, Ettema TJ. The Archaeal Legacy of Eukaryotes: A Phylogenomic Perspective. Cold Spring Harb. Perspect. Biol. 2014;6:a016022. doi: 10.1101/cshperspect.a016022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 81.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 82.Guy L, Spang A, Saw JH, Ettema TJ. ‘Geoarchaeote NAG1’ is a deeply rooting lineage of the archaeal order Thermoproteales rather than a new phylum. ISME J. 2014;8:1353–1357. doi: 10.1038/ismej.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.R Core Team R: A Language and Environment for Statistical Computing. 2014.
- 84.Venables WN, Ripley BD. Modern Applied Statistics with S. 4th edn Springer; 2002. [Google Scholar]
- 85.Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nature Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 87.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 89.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 91.Vallenet D, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 93.Yutin N, Puigbo P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS ONE. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Powell S, et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kawai M, et al. High frequency of phylogenetically diverse reductive dehalogenase-homologous genes in deep subseafloor sedimentary metagenomes. Front. Microbiol. 2014;5:80. doi: 10.3389/fmicb.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morono Y, Terada T, Hoshino T, Inagaki F. Hot-alkaline DNA extraction method for deep-subseafloor archaeal communities. Appl. Environ. Microbiol. 2014;80:1985–1994. doi: 10.1128/AEM.04150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nature Rev. Microbiol. 2010;8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dong JH, Wen JF, Tian HF. Homologs of eukaryotic Ras superfamily proteins in prokaryotes and their novel phylogenetic correlation with their eukaryotic analogs. Gene. 2007;396:116–124. doi: 10.1016/j.gene.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 100.Ettema TJ, Lindas AC, Bernander R. An actin-based cytoskeleton in archaea. Mol. Microbiol. 2011;80:1052–1061. doi: 10.1111/j.1365-2958.2011.07635.x. [DOI] [PubMed] [Google Scholar]
- 101.Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biol. Direct. 2009;4:9. doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goodson HV, Hawse WF. Molecular evolution of the actin family. J. Cell Sci. 2002;115:2619–2622. doi: 10.1242/jcs.115.13.2619. [DOI] [PubMed] [Google Scholar]
- 103.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guy L, Roat Kultima J, Andersson SGE. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.