Abstract
Cue-induced methamphetamine seeking progressively increases after withdrawal (incubation of methamphetamine craving), but the underlying mechanisms are largely unknown. We determined whether this incubation is associated with alterations in candidate genes in dorsal striatum (DS), a brain area implicated in cue- and context-induced drug relapse. We first measured mRNA expression of 24 candidate genes in whole DS extracts after short (2 d) or prolonged (1 month) withdrawal in rats following extended-access methamphetamine or saline (control condition) self-administration (9 h/d, 10 d). We found minimal changes. Next, using fluorescence-activated cell sorting, we compared gene expression in Fos-positive dorsal striatal neurons, which were activated during “incubated” cue-induced drug-seeking tests after prolonged withdrawal, with nonactivated Fos-negative neurons. We found significant increases in mRNA expression of immediate early genes (Arc, Egr1), Bdnf and its receptor (Trkb), glutamate receptor subunits (Gria1, Gria3, Grm1), and epigenetic enzymes (Hdac3, Hdac4, Hdac5, GLP, Dnmt3a, Kdm1a) in the Fos-positive neurons only. Using RNAscope to determine striatal subregion and cell-type specificity of the activated neurons, we measured colabeling of Fos with Drd1 and Drd2 in three DS subregions. Fos expression was neither subregion nor cell-type specific (52.5 and 39.2% of Fos expression colabeled with Drd1 and Drd2, respectively). Finally, we found that DS injections of SCH23390 (C17H18ClNO), a D1-family receptor antagonist known to block cue-induced Fos induction, decreased incubated cue-induced methamphetamine seeking after prolonged withdrawal. Results demonstrate a critical role of DS in incubation of methamphetamine craving and that this incubation is associated with selective gene-expression alterations in cue-activated D1- and D2-expressing DS neurons.
Keywords: dorsal striatum, epigenetics, FACS, glutamate, methamphetamine, relapse
Introduction
Cue-induced drug seeking progressively increases or incubates after withdrawal (Grimm et al., 2001). This incubation phenomenon was first observed in rats trained to self-administer cocaine or heroin (Neisewander et al., 2000; Shalev et al., 2001), and subsequently observed in rats trained to self-administer methamphetamine (Shepard et al., 2004). A previous clinical study reported incubation of cue-induced methamphetamine craving in methamphetamine-dependent patients (Wang et al., 2013). Over the last decade, mechanistic studies have primarily focused on incubation of cocaine craving (Pickens et al., 2011; Loweth et al., 2014a). In contrast, only two previous studies began to explore mechanisms of incubation of methamphetamine craving (Caprioli et al., 2015; Li et al., 2015). In these studies, we determined cue-induced methamphetamine seeking in extinction tests performed after short (1–2 d) or prolonged (3–5 week) withdrawal periods. We found that reversible inactivation of central amygdala (but not basolateral amygdala, mPFC, or orbitofrontal cortex) selectively decreases “incubated” cue-induced methamphetamine seeking after prolonged withdrawal (Li et al., 2015). We also found that systemic injections of a positive modulator of mGluR2 (AZD8529) decreases incubated cue-induced methamphetamine seeking after prolonged forced (home-cage confinement) or voluntary (achieved via a discrete choice procedure between methamphetamine and palatable food) abstinence (Caprioli et al., 2015).
In the current study, we determined the role of dorsal striatum (DS) in the incubation of methamphetamine craving, because subregions of DS (dorsomedial, dorsolateral) contribute to context- and cue-induced heroin, cocaine, and alcohol seeking (Fuchs et al., 2005; Vanderschuren et al., 2005; Bossert et al., 2009; Wang et al., 2010a; Pacchioni et al., 2011; Corbit et al., 2012; Murray et al., 2012). Additionally, we previously used immunohistochemistry for the neuronal activity marker Fos (Cruz et al., 2014a) and reversible inactivation of DS to demonstrate a role for this brain region in context-induced reinstatement of methamphetamine seeking (Rubio et al., 2015). We also used fluorescence-activated cell sorting (FACS; Guez-Barber et al., 2011; Liu et al., 2014) and RNAscope in situ hybridization (ISH; Wang et al., 2012) methods and found that context-induced reinstatement was associated with selective increases in mRNA expression of NMDA receptor subunits Grin2a and Grin2b in Fos-positive but not Fos-negative neurons.
In the current study, we first determined whether time-dependent increases in cue-induced methamphetamine seeking are associated with time-dependent changes in mRNA expression of 24 candidate genes in whole DS extracts. We assessed three classes of candidate genes. The first class included BDNF and its receptor TrkB, previously implicated in incubation of cocaine craving and reinstatement of cocaine seeking (McGinty et al., 2010; Li and Wolf, 2015). The second class included glutamate receptors: AMPARs (Gria1, Gria2, Gria3), NMDARs (Grin1, Grin2a, Grin2b), mGluRs (Grm1, Grm5) implicated previously in incubation of cocaine craving (Ben-Shahar et al., 2013; Lee et al., 2013; Halbout et al., 2014; Loweth et al., 2014b) and reinstatement of cocaine seeking (Kalivas et al., 2009). The third class included genes encoding epigenetic enzymes involved in chromatin remodeling or DNA methylation (Kouzarides, 2007). These genes were chosen because of their roles in cocaine's physiological and behavioral effects (Robison and Nestler, 2011; Rogge and Wood, 2013; Schmidt et al., 2013; Sadri-Vakili, 2014).
We found minimal gene expression changes in whole DS extracts after short or prolonged withdrawal from methamphetamine. Therefore, we investigated whether incubated cue-induced methamphetamine seeking after prolonged withdrawal is associated with selective gene-expression changes in a subpopulation of DS neurons that were activated (Fos-positive) during the cue-induced drug-seeking test. We used FACS to isolate cue-activated Fos-positive DS neurons from the majority of Fos-negative neurons and RNAscope in situ hybridization to identify striatal subregion and cell type (Drd1- vs Drd2-expressing neurons) specificity of the Fos-positive neurons. To determine a causal role of DS in incubation of methamphetamine craving, we examined the effect of DS injections of the D1-family receptor antagonist SCH23390 (C17H18ClNO), because SCH23390 and a related drug (SCH39166, C19H20ClNO) block drug- and cue-induced Fos expression (Valjent et al., 2000; Ciccocioppo et al., 2001; Nair et al., 2011).
Materials and Methods
Subjects.
We used male Sprague Dawley rats (Charles River Laboratories; total n = 138), weighing 300–350 g before surgery and 325–375 g at the start of the drug self-administration procedure. We maintained the rats under a reverse 12 h light/dark cycle with food and water available ad libitum. We housed two rats per cage before surgery and then individually after surgery. We performed the experiments in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (eighth edition), under the protocols approved by the Animal Care and Use Committee of National Institute on Drug Abuse. We excluded 25 rats due to failure of catheter patency, incorrect cannula placements, failure to acquire reliable methamphetamine self-administration, or health-related issues.
Intravenous surgery.
We anesthetized the rats with ketamine and xylazine (80 and 10 mg/kg, i.p., respectively) or isoflurane gas (5% induction; 2–3% maintenance) and inserted Silastic catheters into each rat's jugular vein, as described previously (Lu et al., 2007; Bossert et al., 2009; Theberge et al., 2013). We injected the rats with buprenorphine (0.1 mg/kg, s.c.) or ketoprofen (2.5 mg/kg, s.c.) after surgery to relieve pain and inflammation, and we allowed the rats to recover 5–7 d before methamphetamine self-administration training. During the recovery and training phases, we flushed the catheters every 24–48 h with gentamicin (Butler Schein; 5 mg/ml) dissolved in sterile saline.
Intracranial surgery.
We implanted guide cannulas (23 gauge; Plastics One) 1 mm above the target sites. We set the nose bar at −3.3 mm and used the following DS coordinates from Bregma: dorsomedial striatum (DMS), anteroposterior (AP) +1.2 mm, mediolateral (ML) ±2.4 mm (8° angle), dorsoventral (DV) −4.3 mm; dorsal central striatum (DCS), AP +1.2 mm, ML ±3.2 mm (6° angle), DV −4.3 mm; and dorsolateral striatum (DLS), AP +1.2 mm, ML ±3.3 mm (2° angle), DV −4.3 mm. We anchored the cannulas to the skull with jeweler's screws and dental cement. We used the above coordinates based on our previous study (Bossert et al., 2009).
Intracranial injections.
We dissolved SCH23390 hydrochloride (Tocris Bioscience) in sterile saline and injected the drug bilaterally 15 min before starting the extinction test sessions. The dose of SCH23390 hydrochloride (concentration, 0.75 μg/1 μl/side for DCS; 0.75 μg/0.5 μl/side for DLS and DMS) is based on our previous study (Bossert et al., 2009). The injectors extended 1 mm below the tips of the guide cannulas. We injected vehicle (saline) or SCH23390 hydrochloride at a rate of 0.5 μl/min and left the injectors in place for an additional minute to allow diffusion. We connected the syringe pump (Harvard Apparatus) to 10 μl Hamilton syringes and attached the Hamilton syringes to the 30 gauge injectors via polyethylene-50 tubing. After testing, we extracted the rat brains and stored them in 10% formalin. We sectioned the rat brains (50 μm) using a Leica cryostat, stained the sections with cresyl violet, and verified cannula placements under a light microscope.
Apparatus.
We trained the rats in self-administration chambers located inside sound-attenuating cabinets and controlled by a Med Associates system. Each chamber had two levers located 8–9 cm above the floor. During self-administration training, presses on the retractable (active) lever activated the infusion pump (which delivered a methamphetamine or saline infusion); presses on the stationary (inactive) lever were not reinforced. For intravenous infusions, we connected each rat's catheter to a liquid swivel (Instech) via polyethylene-50 tubing protected by a metal spring. We then attached the liquid swivel to a 20 ml syringe via polyethylene-50 tubing and to a 22 gauge modified cannula (Plastics One).
Methamphetamine self-administration training.
We used an extended-access training procedure described previously by Theberge et al. (2013), Krasnova et al. (2014), and Li et al. (2015). We brought the rats to the self-administration room on their first day of training and housed them chronically in the self-administration chambers. We trained the rats to self-administer methamphetamine 9 h per day (three 3 h sessions, separated by 1 h in between sessions) under a fixed-ratio 1 (FR1) with 20 s time-out reinforcement schedule. We dissolved methamphetamine in saline, and the rats self-administered methamphetamine at a dose of 0.1 mg/kg/infusion over 3.5 s (0.10 ml/infusion). We trained the rats for 10 sessions over a 14 d period (off day every third or fourth day) to prevent loss of body weight during the training phase. [Note that methamphetamine-trained rats lose about 4–8 g after each day of training and regain the lost weight during the off day (Theberge et al., 2013; Krasnova et al., 2014; Li et al., 2015).]
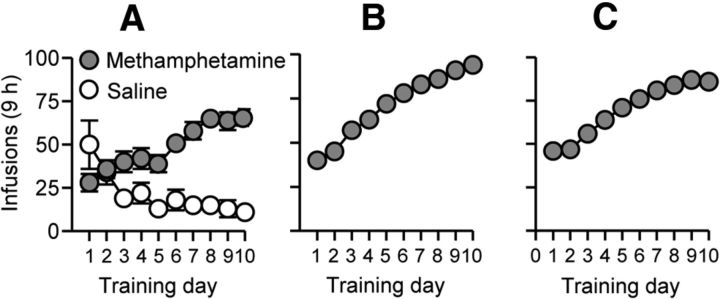
The sessions began with the extension of the active lever and the illumination of the red house light, which remained on for the duration of the each 3 h session. During training, active lever presses led to the delivery of a methamphetamine infusion and a compound 5 s tone/light cue [the tone and light modules (Med Associates) were located above the active lever]. During the 20 s time-out, we recorded nonreinforced lever presses. The daily training sessions started at the onset of the dark cycle and began with the extension of the active lever and the illumination of a red house light that remained on for the duration of the session. We set 35 infusions as the maximum for each 3 h session to prevent overdose. The red house light was turned off and the active lever retracted after the rats received the maximum infusions or at the end of each 3 h session. The training data from Experiments 1–3 are described in Figure 1.
Figure 1.
Methamphetamine and saline self-administration training. Data are the mean ± SEM number of methamphetamine (0.1 mg/kg/infusion) or saline infusions during the 10 9 h daily self-administration sessions for Experiment 1 (A: total, n = 23), Experiment 2 (B: total, n = 25), and Experiment 3 (C: total, n = 65). During training, active lever presses were reinforced on an FR1 20 s time-out reinforcement schedule, and methamphetamine or saline infusions were paired with a 5 s tone/light cue.
Withdrawal phase.
During this phase, we housed the rats individually in the animal facility and handled them three to four times per week.
Extinction tests.
We conducted all extinction tests immediately after the onset of the dark cycle. The sessions began with the extension of the active lever and the illumination of the red house light, which remained on for the duration of the session. Active lever presses during testing (the operational measure of cue-induced drug seeking in incubation of craving studies (Lu et al., 2004a; Pickens et al., 2011) resulted in contingent presentations of the tone/light cue, previously paired with methamphetamine infusions, but not the drug.
Food self-administration.
We trained the rats to self-administer food pellets (TestDiet, catalog #1811155) for 1 h per day during the middle of their dark cycle, under an FR1 20 s time-out reinforcement schedule; pellet delivery was paired with a 5 s light cue. To increase the rats' motivation to lever-press for the food pellets, we restricted their home cage's diet to 20 g/d and fed them after they finished the food self-administration session. Additionally, to facilitate acquisition of food self-administration, we gave the rats 1 h magazine training before the operant training during the first three training days. During magazine training, we presented the food pellets to the rats every 5 min; pellet delivery was paired with the 5 s light cue.
RNA extraction, cDNA synthesis, and qPCR for whole DS extracts.
We took rats directly from the home cage and performed live decapitation. We obtained a 2 mm coronal section containing the striatum using a brain matrix (ASI Instruments) and punched the DS with a 12 gauge metal needle. We froze tissue punches in Eppendorf tubes on dry ice and stored tissue at −80°C. For RNA extraction, we homogenized tissue punches in Trizol (Invitrogen) according to the protocol from the manufacturer. We purified RNA with RNeasy Micro columns (Qiagen). We confirmed the RNA purity by spectroscopy at 260/280 and 260/230 > 1.8. We reverse transcribed RNA into cDNA using iScript cDNA synthesis (Bio-Rad) and performed quantitative PCR (qPCR) using SYBR green and Applied Biosystems 7500 systems. We ran triplicates for each reaction and analyzed them using the ΔΔCt method with glyceraldehyde-3-phosphate dehydrogenase (Gapdh) as the housekeeping gene. See Table 1 for mRNA primer sequences.
Table 1.
Primer sequences for qPCR in whole DS extracts
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Gapdh | ACAGCAACAGGGTGGTGGAC | TTTGAGGGTGCAGCGAACTT |
| Bdnf | GTGTGTGTCTCTGCGCCTCAGTGGA | GAAGTGTACAAGTCCGCGTCCTTA |
| Trkb | CTACCTGGCATCCCAACACT | CTCGGTGGTGAATTTCCTGT |
| Gria1 | GCCAATGTAAAAAGGAATA | AACAGAAACGGTAAGTCATC |
| Gria2 | CATATTTCTGTCCTCCTTTC | CGTAGACTCCTCTTGAAAAC |
| Gria3 | TTGCTGCTTTCCTGACTGTG | CAGATGGTTCTGCGGATTTC |
| Grin1 | TTCACAGAAGTGCGATCTGG | CCTGATACCGAACCCATGTC |
| Grin2a | TCGAATCTGCCCACCTACTC | TTCTGTGACCAGTCCTGCTG |
| Grin2b | GAGAAGCACGGAGTGGTAGG | TCCTCGCTGATGTCGTACAG |
| Grm1 | CACACTCGGACAAAATCTA | CGATGACTTCATCTCTGTCT |
| Grm5 | ACTCAGCCTAGTTTATCCAG | AATAGGTACAGCCAAATGAT |
| Hdac1 | ACGGCATTGATGATGAGTCC | CTGAGCCGCACTGTAGGACC |
| Hdac2 | GGGCTGCTTCAACCTAACTG | TTCACAATCAAGGGCAACTG |
| Hdac3 | CCTGGCATTGACTCATAG | ATTAAGGCTCTTGGTGAAA |
| Hdac4 | TGCTGGGAAATGAGCTTGAG | AGCAGGTTTGACGCCTACAG |
| Hdac5 | ATCGCTACGACAATGGGAAC | CCAGCGGAGACTAGAACGAC |
| Sirt1 | CACCGATCCTCGAACAATTC | TCCCACAGGAAACAGAAACC |
| Sirt2 | ATCAGCAAGGCACCACTAGC | AAGGTCTTCCAGCTCCTTCC |
| Crebbp | TCAGGGTGACAGCAAGAATG | CATGCAGGTGGATCACAAAG |
| Suv39h1 | TGGAGAGTACCCGAATGGAC | GAGGACAAGTGGAGGCAGAC |
| Ehmt1(GLP) | CATTCCCTGTGTCAATGCTG | TCGAAGATCAAGGGTGGTTC |
| Ehmt2 (G9a) | ACGTGTGTTGACGACTGCTC | GGTTCTTGCAGCTTCTCCAG |
| Kdm1a | GCTACTCGTGTGTGCCTGT | TTCAGCACTCCCAAAGGAAG |
| Kmt2a(Mll1) | ATCCAGGCAACACAGAATCC | AGCCGTCATCACTCGAAATC |
| Dnmt3a | AACCTTCCTGGCATGAACAG | TTTCAGTGCACCAGAGGATG |
FACS.
We dissected DS from 2-mm-thick coronal sections between approximately Bregma anteroposterior +2.28 and +0.36 mm (Paxinos and Watson, 2005). We finely minced the tissue with razor blades on ice and transferred the tissue into 1 ml of ice-cold Hibernate A (HA-if; Brain Bits). After centrifuging the tissue for 2 min at 110 × g (4°C), we added 1 ml of Accutase (SCR005; Millipore) and mixed up and down four times before digesting the tissue for 30 min at 4°C with end-over-end mixing. We then centrifuged the tissue for 2 min at 960 × g (4°C) and resuspended the pellet in 0.6 ml of ice-cold Hibernate A. We first triturated each tissue sample three times in series using fire-polished glass pipettes with successively smaller diameters (1.3, 0.8, and 0.4 mm) and then conducted 3 additional trituration steps with 0.4-mm-diameter glass pipettes. Each trituration step consisted of triturating up and down 10 times followed by 2 min on ice to sediment the larger debris and undissociated cells. We combined supernatant from each trituration step to obtain dissociated cells in a total volume of 3.6 ml. We then fixed and permeabilized cells by adding the same volume of 100% of cold ethanol (−20°C) for 15 min on ice. After collecting the cells by centrifugation (1,700 × g, 4 min, 4°C), we resuspended the cells with 0.7 ml of cold PBS and then filtered the cells with 100 and 40 μm cell strainers (Falcon brand; BD Biosciences).
We incubated cells with PE-labeled anti-NeuN antibody (1:500 in PBS; FCMAB317PE, Millipore) and Alexa 647-labeled anti-c-Fos antibodies (1:500 in PBS; sc-253 AF647, Santa Cruz Biotechnology) for 30 min at 4°C and then washed the cells with 0.8 ml cold PBS. After collecting the cells by centrifugation (1300 × g, 3 min, 4°C), we washed the cells again with 1 ml cold PBS, followed by centrifugation (1300 × g, 3 min, 4°C), and resuspended the cells in 0.5 ml cold PBS for sorting in a FACSAria I cell sorting (BD Biosciences).
As we reported previously (Liu et al., 2014; Rubio et al., 2015), neurons can be identified based on the distinct forward (FSC) and side (SSC) scatter properties. DAPI (1 μg/ml, DNA staining) staining showed that ∼98% of the events in the neuron gate are DAPI-positive events (nucleated cells). After defining the cell population, we gated single cells by FCS width and height (∼98% of single cell populations were DAPI positive) and conducted subsequent sorting within this single-cell population. We sorted neurons according to PE (NeuN-immunopositive) and Alexa Fluor 647 (Fos-immunopositive) fluorescence signal. We set the threshold of Alexa Fluor 647 fluorescence signal based on background fluorescence signals of the No test group. We collected all NeuN-positive plus Fos-positive events (Fos-positive neurons) and a maximum of 5000 NeuN-positive plus Fos-negative events (Fos-negative neurons). We analyzed the data using FCS Express 4.
RNA extraction, cDNA synthesis, and qPCR from FACS-isolated neurons.
We collected sorted cells directly into 50 μl of the extraction buffer from the PicoPure RNA isolation kit (Arcturus Bioscience) and lysed the cells by pipetting up and down 10 times followed by incubation for 30 min at 42°C. After centrifuging the suspension at 3000 × g at 4°C for 2 min, we collected the supernatant for RNA extraction. We extracted RNA according to Picopure RNA isolation protocol and synthesized single-strand cDNA using the Superscript III first strand cDNA synthesis kit (Invitrogen) according to the manufacturer's protocol.
We used gene-targeted preamplification of cDNA as described previously (Liu et al., 2014; Rubio et al., 2015). Briefly, we used a pooled primer solution of 0.2× concentration of TaqMan ABI primer/probes (20× TaqMan gene expression assay as the stocking solution) and 80 nm of customized primer sets (Table 2). Each cDNA sample (7.5 μl) was mixed with the pooled primer solution (7.5 μl) and 15 μl of 2× TaqMan PreAmp Master Mix (Applied Biosystems). We preamplified cDNA in an ABI 9700 Thermal Cycler using the following program: 95°C hold for 10 min, denaturation at 90°C for 15 s, and annealing and extension at 60°C for 4 min (14 cycles). We diluted the preamplified cDNA product seven times before proceeding to qPCR. We performed qPCR in duplicates with a Fam-labeled probe for each target gene and a Vic-labeled probe for the endogenous control gene (NeuN). We used TaqMan Advanced Fast PCR Master Mix (Life Technologies) in 7500 Fast TaqMan instrument, using the following program: 95°C hold for 20 s, then 40 cycles with denaturation at 95°C for 3 s, and annealing and extension at 60°C for 30 s. We analyzed reactions using the ΔΔCt method with NeuN as the housekeeping gene.
Table 2.
Primer/probe sequences for FACS-isolated neurons
| Gene | TaqMan probe or primer/probe | Forward primer | Reverse primer |
|---|---|---|---|
| Pde10a | TCCCATCGAGACCGC | CGCTGAACCTCCACAACCA | CGCAGGCAGTCATCATCAAG |
| NeuN | CACTCCAACAGCGTGAC | GGCCCCTGGCAGAAAGTAG | TTCCCCCTGGTCCTTCTGA |
| Gapdh | CTCATGACCACAGTCCA | GACAACTTTGGCATCGTGGAA | CACAGTCTTCTGAGTGGCAGTGA |
| Actb | ATGAAGATCAAGATCATTGCT | AGAAGGAGATTACTGCCCTG | CCACCAATCCACACAGAGTACTT |
| Fos | Rn00487426_g1a | ||
| Erg1 | Rn00561138_m1a | ||
| Arc | Rn00571208_g1a | ||
| Bdnf | TGGTCAGTGGCTGGC | GGAGACCCTCCGCAACTGT | GAGCTATGATGTATCTTAGTGGGTATGAG |
| Trkb1 | CATGAAAGGCCCAGCTT | TGGCGAGACATTCCAAGTTTG | AGAGTCATCGTCGTTGCTGATG |
| Gria1 | Rn00709588_m1a | ||
| Gria2 | Rn00568514_m1a | ||
| Gria3 | Rn00583547_m1a | ||
| Grin1 | Rn01436038_m1a | ||
| Grin2a | Rn00561341_m1a | ||
| Grin2b | Rn00680474_m1a | ||
| Grm1 | Rn01440619_m1a | ||
| Grm5 | Rn00690337_m1a | ||
| Hdac1 | Rn01519308_g1a | ||
| Hdac2 | Rn01193634_g1a | ||
| Hdac3 | Rn00584926_m1a | ||
| Hdac4 | Rn01427040_m1a | ||
| Hdac5 | Rn01464245_m1a | ||
| Sirt1 | CAGTGTCATGGTTCCTT | TTGCAGGAATCCAAAGGATCA | CAAATCAGGCAAGATGCTGTTG |
| Sirt2 | Rn01457502_m1a | ||
| Crebbp | Rn01427040_m1a | ||
| Suv39h1 | Rn01528294_g1a | ||
| Ehmt1 (GLP) | Rn01435512_m1a | ||
| Ehmt2 (G9a) | Rn0152918_m1a | ||
| Kdm1a | Rn01181029_m1a | ||
| Kmt2a (Mll1) | CAAACAGACTGACCAGCC | GCCCAGCTCTGCAAGATAGAGA | ATCACTTTCTTGACCCTGTGCTTT |
| Dnmt3a | Rn01027162_g1a |
aTaqMan catalog number.
To confirm that NeuN is the appropriate housekeeping gene under our experimental conditions, we analyzed NeuN Ct value by using the geometric means of the Ct values among Gapdh, Pde10a (phosphodiesterase 10a), and β-actin and found that there were no differences of NeuN expression across different experimental conditions (data not shown). We also verified the uniformity of the preamplification step by comparing cDNA templates from the unamplified and preamplified samples. All ΔΔCt values of the tested genes between preamplified and unamplified cDNA samples were within the range of ±1.5 for all target genes between the preamplified and unamplified samples (data not shown).
RNAscope in situ hybridization assay.
We performed RNA in situ hybridization for Fos, Drd1, and Drd2 mRNAs as described previously (Rubio et al., 2015). Immediately after dissecting DS tissue for FACS (see FACS section, above), we froze the rest of the rat brain directly in isopentane (kept on dry ice) for 20 s before we transferred the brain in a sealed bag to −80°C for long-term storage. After equilibrating brains in a cryostat (CM 3050S) at −20°C for 2 h, we collected 16 μm brain slices at approximately Bregma +0.24 mm (Paxinos and Watson, 2005) and mounted slices directly onto Super Frost Plus slides (Fisher Scientific). We left slides at −20°C for 10 min and then stored them at −80°C until ISH assay.
We used RNAscope Multiplex Fluorescent Reagent Kit (Advanced Cell Diagnostics) and performed ISH assay according to the user manual for fresh frozen tissue. On the first day, we fixed brain slices in 10% neutral buffered formalin (Fisher Scientific) for 20 min at 4°C. We rinsed the slices three times in PBS and dehydrated the slices in 50, 70, 100, and 100% ethanol. We stored slices in fresh 100% ethanol overnight at −20°C. On the second day, we first dried the slides at room temperature for 10 min. To limit the spreading of solutions, we drew a hydrophobic barrier on slides around brain slices. We then treated the slides with protease solution (pretreatment 4) at room temperature for 20 min and then washed it off. We applied 1× target probes for Fos, Drd1, and Drd2 to the slides and incubated them at 40°C for 2 h in the HybEZ oven.
Each RNAscope target probe contains a mixture of 20 ZZ oligonucleotide probes that are bound to the target RNA: Fos-C3 probe (GenBank accession number NM_022197.2; target nt region, 473–1497); Drd1-C1 probe (GenBank accession number NM_012546.2; target nt region, 104–1053), and Drd2-C2 probe (GenBank accession number NM_012547.1; target nt region, 445–1531). Next, we incubated the slides with preamplifier and amplifier probes (AMP1, 40°C for 30 min; AMP2, 40°C for 15 min; AMP3, 40°C for 30 min). We then incubated the slides with fluorescently labeled probes by selecting a specific combination of colors associated with each channel: green (Alexa 488 nm), orange (Alexa 550 nm), and far red (Alexa 647 nm). We used AMP4 Alt4 to detect triplex Fos, Drd1, and Drd2 in far red, green, and orange. Last, we incubated sections for 20 s with DAPI. We washed the slides with 1× washing buffer two times in between incubation. After air drying the slides we coverslipped them with a Vectashield fluorescent mounting medium (H-1400; Vector Laboratories).
We captured fluorescent images of DS with a Rolera EM-C2 (QImaging) on a Nikon Eclipse E800 microscope and analyzed them with Adobe Photoshop CS5. We set the criteria for Fos-positive cells as 20 total positive pixels after adjusting threshold with ImageJ software. Based on this criterion, we excluded data from one of the six rats that had very low numbers of Fos-positive cells in the different striatal subregions.
Experiment 1: mRNA expression of candidate genes in whole DS extracts after short and prolonged withdrawal.
Our original goal in Experiment 1 was to identify candidate gene(s) that show time-dependent changes that parallel with the time course of cue-induced methamphetamine seeking after withdrawal, and subsequently to use viral-vector genetic approaches to manipulate the expression of the identified gene(s) during the withdrawal period and determine the causal role of the identified genes in the incubation of methamphetamine seeking. In Experiment 1, we used a 2 × 2 factorial design that included the between-subjects factor of Training Condition (saline, methamphetamine) and Withdrawal Day (Day 2, Day 35). We did not perform the cue-induced drug-seeking extinction test in Experiment 1 because we were interested in determining time-dependent gene expression changes without the potential confound of rapid gene expression changes that can occur during the extinction tests.
We performed intravenous surgery on four groups of rats (total n = 23) and trained them to self-administer either saline (n = 8) or methamphetamine (n = 15) in two independent cohorts of rats. We performed live decapitation after short (2 d) or prolonged (35 d) withdrawal periods and collected DS tissue for measuring mRNA expression of candidate genes, including Bdnf and Trkb, glutamate receptors, and epigenetic enzymes (Fig. 2B). The number of rats was four per group for the two saline groups and seven or eight for the two methamphetamine groups (the data of the saline groups was combined for the qPCR analyses). [Note that we chose to examine gene expression changes in the entire DS instead of its different subregions to keep the anatomical area consistent between Experiment 1 and Experiment 2 (FACS). When we conducted the FACS part of Experiment 2, our single-subject FACS procedure was not sensitive enough to reliably separate Fos-positive from Fos-negative neurons in brain areas smaller than the entire DS (e.g., dorsomedial and dorsolateral striatum; Liu et al., 2014).]
Figure 2.
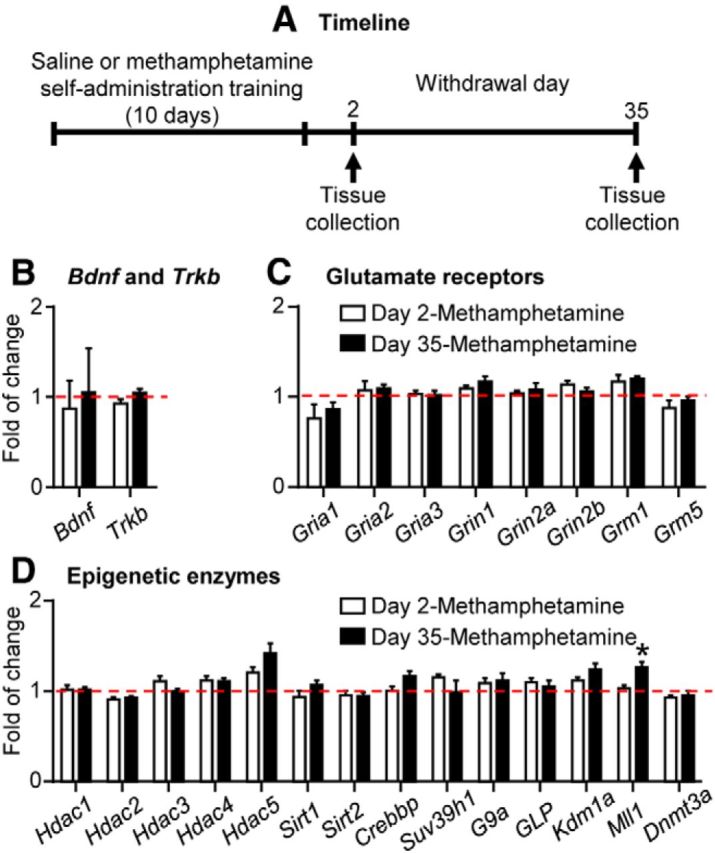
Minimal changes in mRNA expression of candidate genes in whole DS extracts after short and prolonged withdrawal (Experiment 1). A, Timeline of the experiment. B–D, Bdnf and Trkb, glutamate receptors, and epigenetic enzyme mRNA expression in whole DS extracts. Data are presented as folds of mean values in the saline group (combined data of saline Day 2 and Day 35; red dashed lines). *p < 0.005 (different from saline); n = 7–8 per group. Error bars indicate SEM.
Experiment 2: mRNA expression of candidate genes in DS Fos-positive neurons after prolonged withdrawal.
As described in Results, the main finding in Experiment 1 was that extended-access methamphetamine self-administration and short or prolonged withdrawal periods had minimal effects on the expression levels of our target genes in whole DS extracts. Therefore, in Experiment 2 we determined whether expression changes in the target genes selectively occur in a small group of DS neurons that were activated (Fos positive) during the prolonged withdrawal cue-induced methamphetamine seeking test. We used both within-subjects and between-subjects control conditions. The within-subject control condition was the Fos-negative DS neurons of the group of rats that underwent the prolonged withdrawal extinction test. The between-subjects control condition was the Fos-negative DS neurons of the rats from the No test group whose experimental conditions were identical to those of the prolonged withdrawal methamphetamine group in Experiment 1. We could not use expression levels in Fos-positive neurons in the No test group to compare with Fos-positive neurons in the Extinction test group, because this would be comparing different sets of neurons. Based on our previous studies showing cue selectivity of Fos-expressing neuronal ensembles (Koya et al., 2009; Bossert et al., 2011; Fanous et al., 2012; Cruz et al., 2014b), the Fos-positive neurons in the No test group were unlikely to be the same neurons that were selectively activated by cues in the Extinction test group. In other words, the cue-activated Fos-positive neurons (as well as Fos-negative neurons) in the Extinction test group were most likely Fos-negative before extinction tests. In addition, DS from rats in the No Extinction test group contained, as would be expected, very low numbers of Fos-positive neurons, preventing reliable measurement. For these reasons, expression levels in Fos-negative neurons (and not Fos-positive neurons) in the No test group were the most valid indicators of basal levels of mRNA expression before cue activation in the extinction tests. Thus, to assess alterations of mRNA expression that were due to cue-induced extinction in both Fos-positive and Fos-negative neurons, we compared expression levels in these neurons to the basal levels most likely found in Fos-negative neurons in the no test group. For both groups, we separated the Fos-positive and Fos-negative neurons using FACS and then performed qPCR to quantify gene expression in these selected neuronal populations.
We performed intravenous surgery on two groups of rats (total, n = 25) and trained them to self-administer methamphetamine as described above. We then either tested (n = 15) or did not test (n = 10) the rats for cue-induced methamphetamine seeking (2 h test) on 6 individual days (Extinction test, n = 2–3 rats per day; No test, n = 1–2 rats per day) after 30 to 50 withdrawal days. At the end of the extinction tests, we collected DS tissues from both Extinction test and No test rats and processed the tissue for FACS as described above. Next we performed RNA extraction, cDNA synthesis, gene-targeted preamplification, and qPCR on FACS-isolated neurons (see FACS section, above). In FACS-isolated Fos-positive and Fos-negative neurons, we measured mRNA expression of the same genes we assessed in whole DS extracts in Experiment 1. Note that we performed the extinction tests over 20 d, because of technical and scheduling issues related to the FACS assay and because the Johns Hopkins FACS facility did not allow us to test more than two or three rats per day during weekdays.
To identify the striatal subregion and cell-type specificity of Fos-positive neurons, we also processed a subset of rats for the RNAscope ISH assay as described above. We first compared brain tissue from three rats in the No test group and five rats from the Extinction test group for Fos mRNA expression in dorsolateral, central, and dorsomedial striatum. Next, we performed triple labeling for Fos mRNA and for mRNA of D1 and D2 dopamine receptors (Drd1 and Drd2) in these three striatal subregions in the five rats from the Test group. Finally, in Experiment 2, we did not include a saline self-administration control group, because in Experiment 1 we found minimal differences in the expression of the candidate genes in whole DS extracts from rats that previously self-administered saline versus methamphetamine.
Experiment 3: Effect of dorsal striatum injections of the D1-like receptor antagonist SCH23390 on extinction responding after short and prolonged withdrawal.
The goal of Experiment 3 was to determine the causal role of DS in incubation of methamphetamine craving. For this purpose, we injected the D1-family receptor antagonist SCH23390 into the DS and determined its effect on both nonincubated cue-induced methamphetamine seeking in the extinction test performed after short (2 d) withdrawal and incubated cue-induced methamphetamine seeking after prolonged (1 month) withdrawal. We first injected SCH23390 into the central subregion of the DS, because our RNAscope in situ hybridization data from Experiment 2 showed that the Fos-positive neurons are similarly distributed in the dorsomedial, dorsolateral, and the central areas between these two subregions (see Results). We used a 2 × 2 between-subjects experimental design that included the factors Withdrawal Period (2 d, 1 month) and SCH23390 Dose (vehicle, 0.75 μg/1 μl/side).
We performed intravenous surgery on four groups of rats (total, n = 36; n = 8–10 per group) and implanted them with bilateral guide cannulas 1 mm above the DCS. We then trained them for methamphetamine self-administration in two independent cohorts of rats. We determined the effect of DS injections of saline or SCH23390 (0.75 μg/1 μl/side, 15 min pretreatment time) on cue-induced methamphetamine seeking in the extinction tests (3 h sessions) after short (2 d) or prolonged (1 month) withdrawal periods. We used a larger infusion volume (1 μl) and aimed at the central DS, because we did not observe regional specificity of the cue-activated Fos-positive cells in the different DS subregions (see Results).
To further examine whether the effect of SCH23390 is selective for dorsolateral versus dorsomedial striatum, we also injected SCH23390 into DLS or DMS and determined its effect on incubated cue-induced methamphetamine seeking after prolonged (1 month) withdrawal. We performed intravenous surgery on four groups of rats (total, n = 29; n = 6–8 per group) and implanted them with bilateral guide cannulas 1 mm above the DMS or DLS. After training them for methamphetamine self-administration, we determined the effect of DLS or DMS injections of saline or SCH23390 (0.75 μg/0.5 μl/side, 15 min pretreatment time) on cue-induced methamphetamine seeking in the extinction tests (3 h sessions) after prolonged (1 month) withdrawal. [Note that we did not examine the effect of SCH23390 in DLS or DMS after 2 d withdrawal based on our result showing no effect of SCH23390 in DCS on withdrawal day 2 (see Results).]
Finally, to confirm that the inhibitory effect of DCS, DMS, and DLS SCH23390 injections on extinction responding after prolonged withdrawal (see Results) is not due to motor deficits, we trained rats from the 1 month withdrawal vehicle group (DCS, n = 8; DLS, n = 8; DMS, n = 8) to lever press for food pellets for 7 d (1 h/d). We then injected them with vehicle or SCH23390 into the DCS (0.75 μg/1 μl/side) or DMS or DLS (0.75 μg/0.5 μl/side), 15 min before the 1 h food self-administration session on the eighth and ninth days; we counterbalanced the order of the vehicle and SCH23390 injections.
Statistical analysis.
We analyzed the behavioral and molecular data with SPSS (version 20) or Prism GraphPad (version 5) using mixed ANOVAs, ANCOVAs, one-way ANOVA, or t test, as appropriate. We followed significant interactions or main effects with Fisher PLSD post hoc tests. We indicate the between- and within-subjects factors of the different analyses in the Results section. To account for the multiple statistical comparisons of the mRNA expression data of the 24 genes in Experiment 1 (whole DS extracts) and the 27 genes in Experiment 2 (FACS), we set a conservative and more stringent significance criterion for these data (p < 0.005); these statistical comparisons are listed in Table 3. We also excluded outliner values (3 SDs outside the group mean) and samples with undetectable Ct values from the data presentation and statistical analysis of the molecular data.
Table 3.
Statistics for mRNA expression of genes in both homogenate dorsal striatum and FACS sorted dorsal striatal neurons
| Genes | Whole DS extracts (one-way ANOVA) | FACS |
|
|---|---|---|---|
| Paired t test (extinction test; Fos positive vs Fos negative) | Unpaired t test (Fos negative; no test vs extinction test) | ||
| Immediate early genes | |||
| Fos | t(14) = 5.5, p = 0.00008* | t(22) = 3.0, p = 0.006 | |
| Egr1 | t(13) = 4.2, p = 0.001* | t(21) = 0.6, p = 0.5 | |
| Arc | t(14) = 5.3, p = 0.0001* | t(22) = 0.2, p = 0.2 | |
| Epigenetic enzyme genes | |||
| Hdac1 | F(2,20) = 0.1, p = 0.9 | t(13) = 1.3, p = 0.2 | t(22) = 2.2, p = 0.04 |
| Hdac2 | F(2,19) = 1.6, p = 0.2 | t(14) = 2.5, p = 0.03 | t(23) = 1.8, p = 0.1 |
| Hdac3 | F(2,20) = 1.0, p = 0.4 | t(13) = 3.7, p = 0.003* | t(22) = 0.4, p = 0.7 |
| Hdac4 | F(2,18) = 1.1, p = 0.5 | t(13) = 5.6, p = 0.00008* | t(21) = 1.7, p = 0.1 |
| Hdac5 | F(2,20) = 4.5 p = 0.02 | t(14) = 4.4, p = 0.0007* | t(22) = 0.6, p = 0.5 |
| Sirt1 | F(2,20) = 1.1, p = 0.3 | t(13) = 1.4, p = 0.2 | t(22) = 1.1, p = 0.3 |
| Sirt2 | F(2,20) = 0.2, p = 0.8 | t(13) = 2.4, p = 0.03 | t(22) = 1.8, p = 0.1 |
| Crebbp | F(2,20) = 1.9, p = 0.2 | t(13) = 2.8, p = 0.01 | t(22) = 1.4, p = 0.2 |
| Suv 39h1 | F(2,19) = 1.2, p = 0.3 | t(11) = 1.9, p = 0.09 | t(20) = 1.8, p = 0.1 |
| G9a | F(2,20) = 0.8, p = 0.5 | t(14) = 0.1, p = 0.9 | t(22) = 1.4, p = 0.2 |
| GLP | F(2,20) = 0.9, p = 0.4 | t(13) = 4.0, p = 0.002* | t(22) = 2.5, p = 0.02 |
| Kdm1a | F(2,20) = 2.7, p = 0.1 | t(13) = 3.9, p = 0.002* | t(22) = 2.4, p = 0.02 |
| Mll1 | F(2,20) = 7.1, p = 0.0048* | t(14) = 1.6, p = 0.1 | t(22) = 0.4, p = 0.7 |
| Dnmt3a | F(2,19) = 0.9 p = 0.4 | t(13) = 8.3, p = 0.0001* | t(22) = 2.6, p = 0.01 |
| Glutamate receptor genes | |||
| Gria1 | F(2,20) = 1.1, p = 0.4 | t(13) = 3.9, p = 0.002* | t(22) = 1.3, p = 0.2 |
| Gria2 | F(2,20) = 0.4, p = 0.6 | t(14) = 3.9, p = 0.002* | t(23) = 1.6, p = 0.1 |
| Gria3 | F(2,18) = 0.1, p = 0.9 | t(13) = 3.4, p = 0.005* | t(22) = 1.5, p = 0.1 |
| Grin1 | F(2,18) = 1.4, p = 0.3 | t(14) = 0.1, p = 1.0 | t(22) = 0.6, p = 0.6 |
| Grin2a | F(2,17) = 0.9, p = 0.4 | t(14) = 4.3, p = 0.0007* | t(23) = 0.4, p = 0.7 |
| Grin2b | F(2,19) = 1.4, p = 0.3 | t(14) = 2.5, p = 0.03 | t(22) = 0.1, p = 0.9 |
| Grm1 | F(2,17) = 4.9, p = 0.02 | t(12) = 5.4, p = 0.0002* | t(21) = 0.4, p = 0.7 |
| Grm5 | F(2,20) = 0.8, p = 0.4 | t(14) = 0.2, p = 0.9 | t(23) = 0.6, p = 0.5 |
| Growth factor genes | |||
| Bdnf | F(2,18) = 0.1, p = 0.9 | t(13) = 6.1, p = 0.00004* | t(22) = 2.1, p = 0.04 |
| Trkb | F(2,19) = 3.1, p = 0.07 | t(11) = 3.6, p = 0.004* | t(19) = 0.2, p = 0.8 |
We analyzed the data of the whole DS extracts using a between-subjects factor one-way ANOVA including three groups: saline (combined data of saline day 2 and day 35), methamphetamine day 2, and methamphetamine day 35. We analyzed the FACS data using a within-subjects paired t test comparing Fos-positive to Fos-negative neurons in the extinction test group (n = 15). We also analyzed the FACS data using a between-subjects unpaired t test comparing Fos-negative neurons between the no test (n = 10) and extinction test groups.
*Significant group differences for whole DS extracts, and significant difference between Fos-positive and Fos-negative cells for FACS sorting, p < 0.005.
Results
Methamphetamine self-administration training (Experiments 1–3)
The methamphetamine-trained rats in Experiments 1–3 increased their drug intake over time, whereas the self-administration behavior of the saline-trained rats in Experiment 1 decreased over time (Fig. 1). The statistical analysis of Experiment 1, which included the between-subjects factor Drug (saline, methamphetamine) and the within-session factor Session (1–10), showed a significant interaction between the two factors (F(9,189) = 17.8, p < 0.01). The statistical analysis of Experiments 2 and 3, which included the within-session factor Session (1–10), showed a significant effect of this factor (Experiment 2, F(9,216) = 24.1, p < 0.01; Experiment 3, F(9,576) = 122.3, p < 0.01). In Experiment 3, we lost the ninth day training data for five rats due to equipment failure; for data analysis and presentation, we estimated these data with the mean of the 8th and 10th days of training data of each rat.
Minimal changes in mRNA expression of candidate genes in whole DS extracts after short and prolonged abstinence (Experiment 1)
We pooled the data from the saline-trained rats in the Day 2 and Day 35 groups (n = 8) because there were no differences in the mRNA expression of the various genes between these groups. We analyzed the data with one-way ANOVA that included the between-subjects factor Group (saline, methamphetamine Day 2, methamphetamine Day 35). This analysis showed significant group differences in the mRNA expression of Mll1, an H3K4 methyltransferase, due to increased expression levels in the methamphetamine withdrawal Day 35 group (F(2,20) = 7.1, p < 0.005). In contrast, no group differences were observed in DS mRNA levels of the other 23 candidate genes (Table 3). Together, these results indicate that methamphetamine self-administration and subsequent short or prolonged withdrawal periods produced minimal alterations in expression levels of the candidate genes in whole DS extracts.
Robust changes in mRNA expression of candidate genes in DS cue-activated Fos-positive neurons after prolonged withdrawal (Experiment 2)
FACS-sorting of Fos-positive neurons
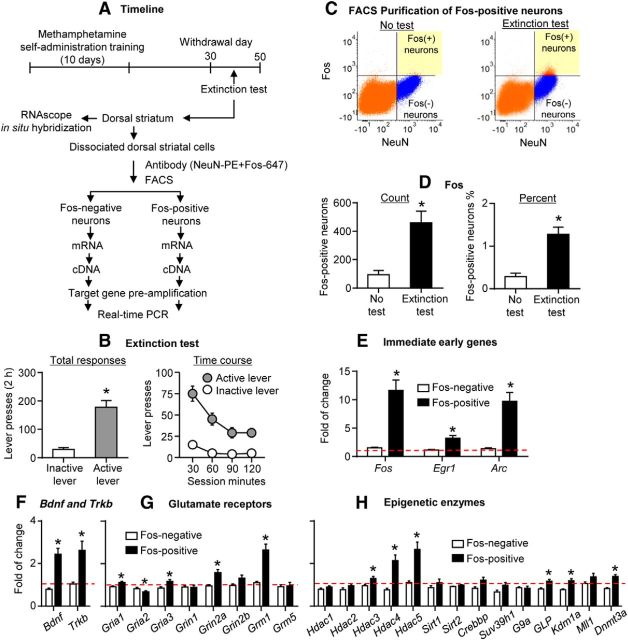
We used FACS as described previously (Liu et al., 2014; Rubio et al., 2015) and obtained four-fold more Fos-positive neurons in DS of rats that underwent extinction tests than in rats in the no test group (t(23) = 3.5, p < 0.01; Fig. 3C,D). Additionally, the percentage of Fos-positive neurons was higher in the Extinction test group than in the No test group (t(23) = 4.5, p < 0.01; Fig. 3D). The total number of DS neurons obtained by FACS was not different between the Extinction test (51,848 ± 6296) and No test (46,253 ± 3977) groups.
Figure 3.
Robust changes in mRNA expression of candidate genes in DS cue-activated Fos-positive neurons after prolonged withdrawal (Experiment 2, FACS). A, Timeline of the experiment. B, Extinction tests performed between withdrawal days 30 and 50. Data are the mean ± SEM of responses on the previously active lever and on the inactive lever during the 2 h extinction test. During testing, lever presses led to contingent presentations of the tone/light cue paired previously with methamphetamine infusions during training. *p < 0.001 (different from inactive lever); n = 15. C, An example of FACS sorting of NeuN-positive (x-axis) and Fos-positive or Fos-negative (y-axis) cells (Fos-positive and Fos-negative neurons) in DS. Left and right, Sorting of Fos-positive and Fos-negative neurons, respectively. Fos-positive and Fos-negative neurons are located in the top right and bottom right quadrants, respectively. D, FACS-sorted Fos-positive neurons. Left, Total number. Right, Percentage of total NeuN-positive neurons. *p < 0.01 (different from the No test group); n = 10–15 per group. E–H, Gene expression in FACS-sorted Fos-positive neurons in the Extinction test group. The immediate early genes (Fos, Egr1, Arc), Bdnf and Trkb, Gria 1, Gria3, Grin2a, Grm1, Hdac3, Hadc4, Hdac5, GLP, Kdm1a, and Dnmt3a increased. Gria2 decreased. Data are presented as folds of mean values in Fos-negative neurons from the No test group (red dashed line). *p < 0.005 (different from Fos-negative neurons). Error bars indicate SEM.
mRNA expression of immediate early genes and candidate genes after FACS sorting
Analysis of gene expressions in Fos-negative neurons showed no difference between the Extinction test and No test groups (unpaired t tests; Table 3). Thus, we performed the analysis in Fos-positive and Fos-negative neurons within the Extinction test group (paired t test). As expected, we found that Fos mRNA increased in the Fos-positive neurons (Fig. 3E; t(14) = 5.5, p < 0.005). In addition, other immediate early genes (IEGs) were increased in the Fos-positive neurons after the extinction tests (Fig. 3E), including Egr1 (t(13) = 4.2, p < 0.005) and Arc (t(14) = 5.3, p < 0.005). These results indicate that the FACS-sorted Fos-positive neurons represent activated neuronal populations in DS.
The mRNA expression of many of candidate genes was significantly higher in cue-activated DS Fos-positive neurons than in Fos-negative neurons (Fig. 3F–H, Table 3). These include Bdnf (brain long 3′ UTR form) and it receptor Trkb (Fig. 3F), genes encoding glutamate receptors (Gria1, Gria3, Grin2a and Grm1; Fig. 3G), and genes encoding epigenetic enzymes (Hdac3, Hdac4, Hdac5, GLP, Kdm1a, Dnmt3a; Fig. 3H). We also observed a significant decrease in the expression of Gria2 mRNA in the Fos-positive neurons (Fig. 3G). These results demonstrate that the gene expression profile of DS neurons that was activated during the test for incubated cue-induced methamphetamine seeking is significantly different from that of the nonactivated or lesser activated Fos-negative neurons.
Dorsal striatum subregion and cell-type specificity of Fos-positive neurons using RNAscope
We found no evidence for DS subregion or cell-type specificity of the cue-activated (Fos-positive) DS neurons (Fig. 4B,C). Regarding subregion specificity, the analysis of Fos-positive neurons in the three DS subregions (DLS, DCS, and DMS) included the between-subjects factor Test Condition (No test, Extinction test) and the within-subjects factor DS Subregion (DLS, DCS, DMS). This analysis showed a significant main effect of Test Condition (F(1,6) = 45.5, p < 0.001), but no effects of DS Subregion or an interaction between the two factors. Regarding cell-type specificity, we found that 52.5 and 39.2% of the Fos-positive cells coexpressed with Drd1 or Drd2, respectively (2.0% of the Fos-positive cells did not coexpress Drd1 or Drd2, and 6.3% of the Fos-positive cells could not be clearly identified as either Drd1- or Drd2-expressing cells). Representative pictures of Fos/Drd1/Drd2 triple labeling by RNAscope in situ hybridization are shown in Figure 4, D and E.
Figure 4.
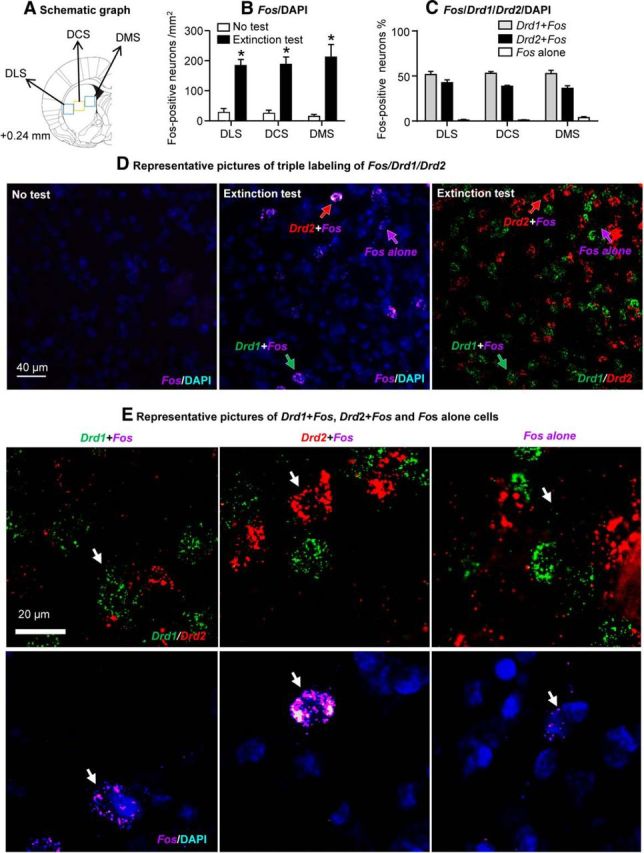
Lack of subregional and cell-type specificity of cue-activated (Fos-positive) DS neurons. A, Schematic illustration of DS subregions sampled for RNAscope in situ hybridization (Paxinos and Watson, 2005). B, Fos-positive cells in the three DS subregions in rats that were not tested (No test, n = 3) or tested for cue-induced methamphetamine seeking in an extinction test (Extinction test, n = 5) after prolonged (30–50 d) withdrawal. p < 0.001 (different from No test). C, Percentages of Fos-positive cells (Extinction test rats, n = 5) that coexpress Drd1 (Drd1+Fos), Drd2 mRNA (Drd2+Fos), or neither (Fos alone) in DS. D, Representative pictures of Fos labeling in the both No test (left) and Extinction test groups (middle), and Drd1 or Drd2 labeling in the Extinction test group (right). E, Representative pictures (higher magnification of the middle and right panels of Fig. 4C) of Drd1+Fos (left), Drd2+Fos (middle), and Fos alone (right) cells (Fos, violet; Drd1, green; Drdr2, red; DAPI, blue). Arrows indicate representative cells. The RNAscope in situ hybridization data are from the posterior DS of eight rats from Experiment 2 (FACS). Error bars indicate SEM.
Causal role of DS in incubation of methamphetamine craving (Experiment 3)
In this final experiment, we used the D1-family receptor antagonist, which is known to block cue- and drug-induced Fos induction in the striatum and other brain areas (Ciccocioppo et al., 2001; Nair et al., 2011), to demonstrate a causal role of DS in incubation of methamphetamine craving.
SCH23390 injections into the DCS decreased cue-induced methamphetamine seeking after prolonged (1 month) but not short (2 d) withdrawal (Fig. 5). Analysis of active lever presses (inactive lever presses as a covariate) showed a significant interaction of Withdrawal Period (2 d, 1 month) by SCH23390 Dose (0, 0.75 μg/side) (F(1,32) = 15.3, p < 0.01). Analysis of time courses of active lever presses showed a significant triple interaction of Withdrawal Period by SCH23390 Dose by Session Hour (F(2,64) = 4.3, p < 0.05).
Figure 5.
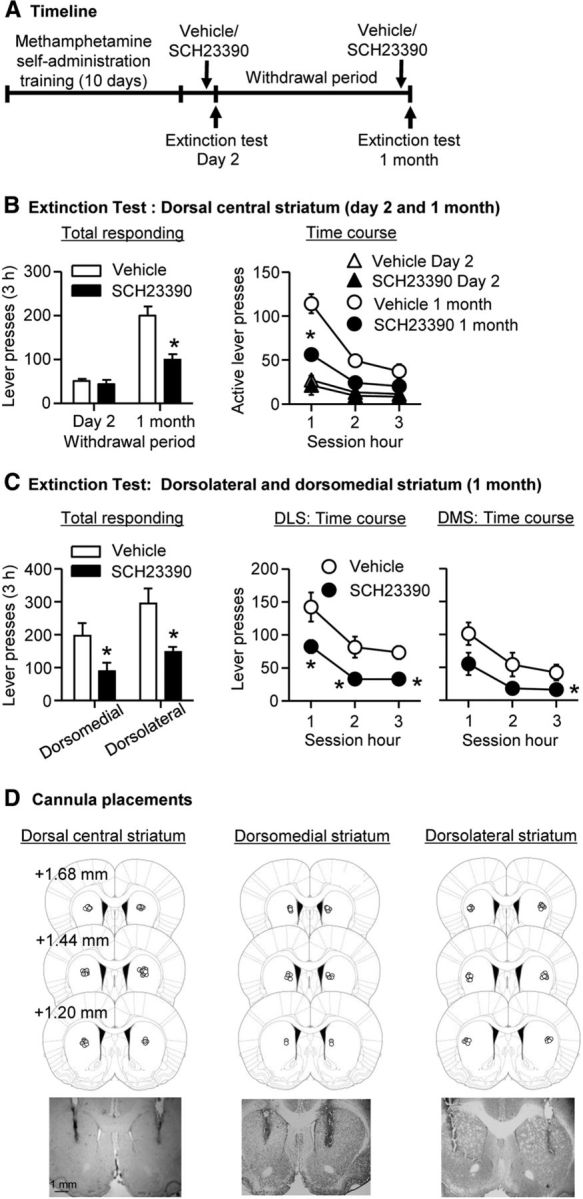
SCH23390 injection into DS decreased cue-induced methamphetamine seeking after prolonged but not early withdrawal. A, Timeline of the experiment. B, Extinction tests after 2 (n = 8–10) and 30 (n = 8–10) withdrawal days after bilateral injections of vehicle or SCH23390 (0.75 μg/1 μl/side) into dorsal central striatum. During testing, lever presses led to contingent presentations of the tone/light cue previously paired with methamphetamine infusions during training, but did not result in methamphetamine infusions. Data are the mean ± SEM of responses on the previously active lever during the extinction tests. C, Extinction tests after 30 (n = 6–8) withdrawal days after bilateral injections of vehicle or SCH23390 (0.75 μg/0.5 μl/side) into dorsomedial or dorsolateral striatum. D, Approximate placement (millimeters from Bregma; Paxinos and Watson, 2005) of injection tips (vehicle, open circles; SCH23390, closed circles) and representative cannula placements. *p < 0.05 (different from vehicle).
SCH23390 injections into DS had no effect on inactive lever presses, which were significantly lower than active lever presses; inactive lever presses were somewhat higher after 1 month of withdrawal (range, 0–71 lever presses per 3 h in the different groups) than after 2 d (range, 1 to 27 lever presses). The latter effect, which is commonly observed in our incubation of craving studies (Lu et al., 2004a; Airavaara et al., 2011; Theberge et al., 2013), likely reflects response generalization after prolonged withdrawal (Shalev et al., 2002).
Additionally, SCH23390 injections into either DLS or DMS decreased incubated cue-induced methamphetamine seeking after prolonged (1 month) withdrawal. Analysis of active lever presses (inactive lever presses as a covariate), which included the between-subjects factors DS Area (DMS, DLS) and SCH23390 Dose (0, 0.75 μg/side) and the within-subjects factor Session Hour, showed significant effects of SCH23390 Dose (F(1,24) = 10.5, p < 0.01) and Session Hour (F(1,24) = 16.5, p < 0.01); the main effect of DS area or the interaction among the three factors were not significant. Finally, SCH23390 injections into DLS and DMS had no effect on inactive lever presses, which were significantly lower than active lever presses.
Finally, to rule out that the effect of SCH23390 is due to a motor deficit, we trained rats (DCS, n = 8; DLS, n = 8; DMS, n = 8) to self-administer palatable or preferred food pellets (Calu et al., 2014) and determined the effect of vehicle or SCH23390 injections on ongoing food-reinforced responding. We found that DS SCH23390 injections had no effect on food self-administration. The numbers of food pellets and active lever presses (mean ± SEM), respectively, during the 1 h test sessions were 120 ± 9 and 414 ± 70 (DCS), 72 ± 11 and 169 ± 39 (DLS), 115 ± 13 and 619 ± 278 (DMS) after vehicle injections, and 115 ± 13 and 343 ± 64 (DCS), 70 ± 11 and 137 ± 28 (DLS), 112 ± 13 and 458 ± 176 (DMS) after SCH23390 injections.
Discussion
We report four main findings. First, extended-access methamphetamine self-administration and subsequent short or prolonged withdrawal produced only one persistent alteration of mRNA out of the 24 candidate genes in whole DS extracts. Second, after prolonged withdrawal, we observed significant differences in mRNA expression for 13 of the 24 candidate genes between cue-activated Fos-positive and Fos-negative FACS-sorted DS neurons. These cue-activated Fos-positive DS neurons not only exhibited the expected increased expression of other IEGs (Egr1, Arc), but also demonstrated unique gene alterations, including increased expression of Bdnf and Trkb, glutamate receptor subunits, and epigenetic enzymes. Third, the cue-activated Fos-positive neurons did not show anatomical specificity within the DS subregions (dorsolateral, center, dorsomedial) and were colocalized with both Drd1- and Drd2-expressing neurons. Finally, blockade of DS D1-family receptors selectively decreased incubated cue-induced methamphetamine seeking after prolonged withdrawal. Our results demonstrate a critical role of DS in incubation of methamphetamine craving and that this incubation is associated with selective gene-expression alterations in the cue-activated Fos-positive neurons.
The use of FACS to study molecular characteristics of cue-activated Fos-positive neurons
The first application of FACS in adult brain cells in vivo was to purify dopamine D1- and D2-expressing striatal neurons in transgenic mice (Lobo et al., 2006; Lobo, 2009). We subsequently developed antibody-based FACS methods to purify activated (Fos-positive) neurons from adult wild-type rats (Guez-Barber et al., 2011; Liu et al., 2014). We used these methods to determine gene-expression changes in Fos-positive neurons in striatum and cortex in studies on cocaine-induced locomotor sensitization (Guez-Barber et al., 2011), incubation of heroin craving (Fanous et al., 2013), and context-induced reinstatement of methamphetamine seeking (Rubio et al., 2015). The main finding from the first two studies was that the Fos-positive neurons primarily showed higher expression of other IEGs (Guez-Barber et al., 2011; Fanous et al., 2013). In the third study, we found higher expression of IEGs in Fos-positive neurons, as well as increased expression of Grin2a and Grin2b mRNA in these neurons; the expression of other glutamate receptors and gene targets was similar in Fos-positive and Fos-negative neurons (Rubio et al., 2015).
Here, we have identified more extensive alterations in gene expression (>50% of the target genes) in Fos-positive neurons. Our results also confirm previous results of higher expression of mRNA for other IEGs (Egr1, Arc) in the Fos-positive neurons. These findings support the notion that Fos-positive neurons represent physiologically activated neurons (Cruz et al., 2014a) and demonstrate the utility of using FACS to study molecular alterations in neurons selectively activated by drug cues or contexts (Cruz et al., 2013). In contrast to the minimal changes in whole DS extracts after prolonged withdrawal from methamphetamine, cue-activated Fos-positive neurons exhibited a molecular profile that was substantially different from the Fos-negative neurons. It is important to note that DS from rats in the No test group also contained low numbers of Fos-positive neurons. However, based on our previous studies showing cue selectivity of Fos-expressing neuronal ensembles (Koya et al., 2009; Bossert et al., 2011; Fanous et al., 2012; Cruz et al., 2014b), the Fos-positive neurons in the No test group were unlikely to be the same neurons that were selectively activated by cues in the Extinction test group. Indeed, most of the cue-activated neurons were likely Fos-negative before extinction tests. Thus, comparisons of gene expression between the different sets of Fos-positive neurons in the No test and Extinction test groups would not be an informative comparison.
We found that mRNAs for Bdnf and its receptor Trkb were higher in Fos-positive than in Fos-negative neurons. These results extend those from previous studies on the role of accumbens BDNF in the incubation of cocaine craving and cue-induced cocaine seeking (Grimm et al., 2003; Lu et al., 2004b; Graham et al., 2007; Li et al., 2013) and DS BDNF in the escalation of cocaine self-administration (Im et al., 2010). We also found increased expression of Grin2a mRNA in DS Fos-positive neurons, extending our findings using the context-induced reinstatement model (Rubio et al., 2015). Additionally, Fos-positive DS neurons showed increased Gria1, Gria3, and Grm1 mRNA and decreased Gria2 mRNA. These results extend previous studies of accumbens glutamatergic transmission in incubation of cocaine craving (Conrad et al., 2008; Lee et al., 2013; Loweth et al., 2014a,b; Ma et al., 2014).
A main finding from our study was the extensive alterations in mRNA expression of epigenetic enzymes (Hdac3, Hdac4, Hdac5, GLP, Kdm1a, Dnmt3a) in the cue-activated Fos-positive neurons. These results extend previous studies on the changes of these epigenetic enzymes in ventral striatum (accumbens) after cocaine exposure (Renthal et al., 2007; LaPlant et al., 2010; Maze et al., 2010; Robison and Nestler, 2011; Taniguchi et al., 2012; Rogge and Wood, 2013). The findings of a selective increase of Dnmt3a mRNA in Fos-positive DS neurons extend previous finding showing increased Dnmt3a mRNA in accumbens 28 d after chronic cocaine exposure (LaPlant et al., 2010). In contrast, our data on Hdac3, Hdac4, and Hdac5 are surprising based on several studies showing that HDAC3, HDAC4, and HDAC5 in accumbens negatively regulate cocaine reward (Renthal et al., 2007; Wang et al., 2010b; Taniguchi et al., 2012; Rogge et al., 2013). However, direct comparison should be made with caution because of differences in the behavioral procedures, the psychostimulant drug, and the striatal subregion. Finally, our data extended previous results by demonstrating that the changes in epigenetic enzymes expression are more pronounced in a small minority of cue-activated neurons that are more likely to encode learned behaviors in addiction models.
A challenge for future studies is to map the selective histone and DNA modifications at specific gene targets that are mediated by alterations in epigenetic enzyme expression in the Fos-positive DS neurons and to understand how altered expression of the targeted genes (which might include Bdnf, Trkb, and those encoding glutamate receptor subunits) contributes to incubation of methamphetamine craving.
Role of dorsal striatum in incubation of methamphetamine craving
We found that SCH23390 injections into the central DS selectively decreased incubated cue-induced methamphetamine seeking. We chose SCH23390 for DS injections because D1-family receptor antagonists are known to block drug- and cue-induced Fos induction in striatum and other brain areas (Valjent et al., 2000; Ciccocioppo et al., 2001; Nair et al., 2011). We used an infusion volume (1 μl) that is twice the volume we (Bossert et al., 2009) and others (Vanderschuren et al., 2005) used previously in studies on the role of DS in cue- and context-induced drug seeking because we wanted to inhibit activity in the whole DS. Based on the study by Vanderschuren et al. (2005) on the role of DLS in cue-induced cocaine seeking and several subsequent studies showing that DLS and DMS play different roles in drug seeking and taking (Bossert et al., 2009; Jeanblanc et al., 2009; Wang et al., 2010a; Corbit et al., 2012; Murray et al., 2012; Rubio et al., 2015), we also examined the effect of SCH23390 in these subregions on cue-induced of methamphetamine seeking after prolonged withdrawal. We found that SCH23390 injection into either DS subregion decreased incubated cue-induced methamphetamine seeking. These data are consistent with RNAscope in situ hybridization results, which showed similar cue-induced Fos induction in DLS, DMS, and DCS.
Finally, an important finding in our study was that Fos was induced in both Drd1- and Drd2-expressing neurons, indicating that both striatal cell types are activated by cues previously associated with methamphetamine self-administration during the prolonged withdrawal extinction (incubation) tests. This finding is consistent with our previous finding using the context-induced reinstatement model (Rubio et al., 2015) and the previous study by Badiani et al. (1999) on amphetamine's effect in a novel context on striatal Fos induction. These findings are not surprising, because when rats are active in a non-home cage environment, the main signal transduction pathway involves glutamatergic excitation of calcium influx, which initiates MAP/ERK-mediated CREB phosphorylation and Fos expression in both D1- and D2-expressing DS neurons (Cruz et al., 2014a).
Concluding remarks
The main novel finding of our study is that incubation of methamphetamine craving is associated with unique persistent gene-expression changes, including Bdnf and Trkb, glutamate receptors, and epigenetic enzymes that are observed selectively in a small number of cue-activated Fos-positive DS neurons. These changes were not detected in whole DS extracts in the classical comparison between drug-naive and drug-experienced rats after short and prolonged withdrawal periods before extinction tests. This pattern of results demonstrate the unique ability of the FACS method to identify molecular alterations in neurons selectively activated by drug-associated cues and contexts during relapse tests in animal models (Cruz et al., 2013). Another important finding in our study, derived from the RNAscope method, is that the cue-activated Fos-positive neurons are similarly distributed in different subregions of the DS and comprised of both D1- and D2-type DS neurons. Finally, we used classical pharmacology to demonstrate a causal role of DS in incubation of methamphetamine craving, extending results from previous studies on the role of this brain region in cue- and context-induced drug seeking and relapse (Belin et al., 2009; Badiani et al., 2011; Bossert et al., 2013).
Footnotes
This work was supported by the intramural (Y.S., B.T.H.) and extramural (P01DA008227; E.J.N.) divisions of the NIDA, and an intramural–extramural award from the NIDA Intramural Research Program (Y.S., X.L., E.J.N.). F.J.R. was supported by an appointment to the NIDA Research Participation Program sponsored by the National Institutes of Health and administered by the Oak Ridge Institute for Science and Education, and received additional financial support from a Becas-Chile scholarship managed by CONICYT and the Universidad de los Andes, Santiago, Chile. The Johns Hopkins FACS Core facility was supported by Award P30AR053503 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institute of Health.
The authors declare no competing financial interests.
References
- Airavaara M, Pickens CL, Stern AL, Wihbey KA, Harvey BK, Bossert JM, Liu QR, Hoffer BJ, Shaham Y. Endogenous GDNF in ventral tegmental area and nucleus accumbens does not play a role in the incubation of heroin craving. Addict Biol. 2011;16:261–272. doi: 10.1111/j.1369-1600.2010.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani A, Oates MM, Day HE, Watson SJ, Akil H, Robinson TE. Environmental modulation of amphetamine-induced c-fos expression in D1 versus D2 striatal neurons. Behav Brain Res. 1999;103:203–209. doi: 10.1016/S0166-4328(99)00041-8. [DOI] [PubMed] [Google Scholar]
- Badiani A, Belin D, Epstein D, Calu D, Shaham Y. Opiate versus psychostimulant addiction: the differences do matter. Nat Rev Neurosci. 2011;12:685–700. doi: 10.1038/nrn3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin D, Jonkman S, Dickinson A, Robbins TW, Everitt BJ. Parallel and interactive learning processes within the basal ganglia: relevance for the understanding of addiction. Behav Brain Res. 2009;199:89–102. doi: 10.1016/j.bbr.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar O, Sacramento AD, Miller BW, Webb SM, Wroten MG, Silva HE, Caruana AL, Gordon EJ, Ploense KL, Ditzhazy J, Kippin TE, Szumlinski KK. Deficits in ventromedial prefrontal cortex group 1 metabotropic glutamate receptor function mediate resistance to extinction during protracted withdrawal from an extensive history of cocaine self-administration. J Neurosci. 2013;33:495–506. doi: 10.1523/JNEUROSCI.3710-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Wihbey KA, Pickens CL, Nair SG, Shaham Y. Role of dopamine D(1)-family receptors in dorsolateral striatum in context-induced reinstatement of heroin seeking in rats. Psychopharmacology. 2009;206:51–60. doi: 10.1007/s00213-009-1580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Stern AL, Theberge FR, Cifani C, Koya E, Hope BT, Shaham Y. Ventral medial prefrontal cortex neuronal ensembles mediate context-induced relapse to heroin. Nat Neurosci. 2011;14:420–422. doi: 10.1038/nn.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Marchant NJ, Calu DJ, Shaham Y. The reinstatement model of drug relapse: recent neurobiological findings, emerging research topics, and translational research. Psychopharmacology. 2013;229:453–476. doi: 10.1007/s00213-013-3120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calu DJ, Chen YW, Kawa AB, Nair SG, Shaham Y. The use of the reinstatement model to study relapse to palatable food seeking during dieting. Neuropharmacology. 2014;76:395–406. doi: 10.1016/j.neuropharm.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli D, Venniro M, Zeric T, Li X, Adhikary S, Madangopal R, Marchant NJ, Lucantonio F, Schoenbaum G, Bossert JM, Shaham Y. Effect of the novel positive allosteric modulator of mGluR2 AZD8529 on incubation of methamphetamine craving after prolonged voluntary abstinence in a rat model. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.02.018. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo R, Sanna PP, Weiss F. Cocaine-predictive stimulus induces drug-seeking behavior and neural activation in limbic brain regions after multiple months of abstinence: reversal by D(1) antagonists. Proc Natl Acad Sci U S A. 2001;98:1976–1981. doi: 10.1073/pnas.98.4.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad KL, Tseng KY, Uejima JL, Reimers JM, Heng LJ, Shaham Y, Marinelli M, Wolf ME. Formation of accumbens GluR2-lacking AMPA receptors mediates incubation of cocaine craving. Nature. 2008;454:118–121. doi: 10.1038/nature06995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit LH, Nie H, Janak PH. Habitual alcohol seeking: time course and the contribution of subregions of the dorsal striatum. Biol Psychiatry. 2012;72:389–395. doi: 10.1016/j.biopsych.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Koya E, Guez-Barber DH, Bossert JM, Lupica CR, Shaham Y, Hope BT. New technologies for examining the role of neuronal ensembles in drug addiction and fear. Nat Rev Neurosci. 2013;14:743–754. doi: 10.1038/nrn3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Rubio FJ, Hope BT. Using c-fos to study neuronal ensembles in corticostriatal circuitry of addiction. Brain Res. 2014a doi: 10.1016/j.brainres.2014.11.005. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Babin KR, Leao RM, Goldart EM, Bossert JM, Shaham Y, Hope BT. Role of nucleus accumbens shell neuronal ensembles in context-induced reinstatement of cocaine-seeking. J Neurosci. 2014b;34:7437–7446. doi: 10.1523/JNEUROSCI.0238-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous S, Goldart EM, Theberge FR, Bossert JM, Shaham Y, Hope BT. Role of orbitofrontal cortex neuronal ensembles in the expression of incubation of heroin craving. J Neurosci. 2012;32:11600–11609. doi: 10.1523/JNEUROSCI.1914-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous S, Guez-Barber DH, Goldart EM, Schrama R, Theberge FR, Shaham Y, Hope BT. Unique gene alterations are induced in FACS-purified Fos-positive neurons activated during cue-induced relapse to heroin seeking. J Neurochem. 2013;124:100–108. doi: 10.1111/jnc.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RA, Evans KA, Ledford CC, Parker MP, Case JM, Mehta RH, See RE. The role of the dorsomedial prefrontal cortex, basolateral amygdala, and dorsal hippocampus in contextual reinstatement of cocaine seeking in rats. Neuropsychopharmacology. 2005;30:296–309. doi: 10.1038/sj.npp.1300579. [DOI] [PubMed] [Google Scholar]
- Graham DL, Edwards S, Bachtell RK, DiLeone RJ, Rios M, Self DW. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci. 2007;10:1029–1037. doi: 10.1038/nn1929. [DOI] [PubMed] [Google Scholar]
- Grimm JW, Hope BT, Wise RA, Shaham Y. Incubation of cocaine craving after withdrawal. Nature. 2001;412:141–142. doi: 10.1038/35084134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y. Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving. J Neurosci. 2003;23:742–747. doi: 10.1523/JNEUROSCI.23-03-00742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guez-Barber D, Fanous S, Golden SA, Schrama R, Koya E, Stern AL, Bossert JM, Harvey BK, Picciotto MR, Hope BT. FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. J Neurosci. 2011;31:4251–4259. doi: 10.1523/JNEUROSCI.6195-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbout B, Bernardi RE, Hansson AC, Spanagel R. Incubation of cocaine seeking following brief cocaine experience in mice is enhanced by mGluR1 blockade. J Neurosci. 2014;34:1781–1790. doi: 10.1523/JNEUROSCI.1076-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc J, He DY, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. J Neurosci. 2009;29:13494–13502. doi: 10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, LaLumiere RT, Knackstedt L, Shen HW. Glutamate transmission in addiction. Neuropharmacology. 2009;56:169–173. doi: 10.1016/j.neuropharm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Koya E, Golden SA, Harvey BK, Guez-Barber DH, Berkow A, Simmons DE, Bossert JM, Nair SG, Uejima JL, Marin MT, Mitchell TB, Farquhar D, Ghosh SC, Mattson BJ, Hope BT. Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nat Neurosci. 2009;12:1069–1073. doi: 10.1038/nn.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnova IN, Marchant NJ, Ladenheim B, McCoy MT, Panlilio LV, Bossert JM, Shaham Y, Cadet JL. Incubation of methamphetamine and palatable food craving after punishment-induced abstinence. Neuropsychopharmacology. 2014;39:2008–2016. doi: 10.1038/npp.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, Xiao G, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BR, Ma YY, Huang YH, Wang X, Otaka M, Ishikawa M, Neumann PA, Graziane NM, Brown TE, Suska A, Guo C, Lobo MK, Sesack SR, Wolf ME, Nestler EJ, Shaham Y, Schlüter OM, Dong Y. Maturation of silent synapses in amygdala-accumbens projection contributes to incubation of cocaine craving. Nat Neurosci. 2013;16:1644–1651. doi: 10.1038/nn.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wolf ME. Multiple faces of BDNF in cocaine addiction. Behav Brain Res. 2015;279:240–254. doi: 10.1016/j.bbr.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, DeJoseph MR, Urban JH, Bahi A, Dreyer JL, Meredith GE, Ford KA, Ferrario CR, Loweth JA, Wolf ME. Different roles of BDNF in nucleus accumbens core versus shell during the incubation of cue-induced cocaine craving and its long-term maintenance. J Neurosci. 2013;33:1130–1142. doi: 10.1523/JNEUROSCI.3082-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zeric T, Kambhampati S, Bossert JM, Shaham Y. The central amygdala nucleus is critical for incubation of methamphetamine craving. Neuropsychopharmacology. 2015;40:1297–1306. doi: 10.1038/npp.2014.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Rubio FJ, Bossert JM, Marchant NJ, Fanous S, Hou X, Shaham Y, Hope BT. Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS) J Neurochem. 2014;128:173–185. doi: 10.1111/jnc.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK. Molecular profiling of striatonigral and striatopallidal medium spiny neurons past, present, and future. Int Rev Neurobiol. 2009;89:1–35. doi: 10.1016/S0074-7742(09)89001-6. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Loweth JA, Tseng KY, Wolf ME. Adaptations in AMPA receptor transmission in the nucleus accumbens contributing to incubation of cocaine craving. Neuropharmacology. 2014a;76:287–300. doi: 10.1016/j.neuropharm.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loweth JA, Scheyer AF, Milovanovic M, LaCrosse AL, Flores-Barrera E, Werner CT, Li X, Ford KA, Le T, Olive MF, Szumlinski KK, Tseng KY, Wolf ME. Synaptic depression via mGluR1 positive allosteric modulation suppresses cue-induced cocaine craving. Nat Neurosci. 2014b;17:73–80. doi: 10.1038/nn.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Grimm JW, Hope BT, Shaham Y. Incubation of cocaine craving after withdrawal: a review of preclinical data. Neuropharmacology. 2004a;47(Suppl 1):214–226. doi: 10.1016/j.neuropharm.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Lu L, Dempsey J, Liu SY, Bossert JM, Shaham Y. A single infusion of brain-derived neurotrophic factor into the ventral tegmental area induces long-lasting potentiation of cocaine seeking after withdrawal. J Neurosci. 2004b;24:1604–1611. doi: 10.1523/JNEUROSCI.5124-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Uejima JL, Gray SM, Bossert JM, Shaham Y. Systemic and central amygdala injections of the mGluR(2/3) agonist LY379268 attenuate the expression of incubation of cocaine craving. Biol Psychiatry. 2007;61:591–598. doi: 10.1016/j.biopsych.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Ma YY, Lee BR, Wang X, Guo C, Liu L, Cui R, Lan Y, Balcita-Pedicino JJ, Wolf ME, Sesack SR, Shaham Y, Schlüter OM, Huang YH, Dong Y. Bidirectional modulation of incubation of cocaine craving by silent synapse-based remodeling of prefrontal cortex to accumbens projections. Neuron. 2014;83:1453–1467. doi: 10.1016/j.neuron.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty JF, Whitfield TW, Jr, Berglind WJ. Brain-derived neurotrophic factor and cocaine addiction. Brain Res. 2010;1314:183–193. doi: 10.1016/j.brainres.2009.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JE, Belin D, Everitt BJ. Double dissociation of the dorsomedial and dorsolateral striatal control over the acquisition and performance of cocaine seeking. Neuropsychopharmacology. 2012;37:2456–2466. doi: 10.1038/npp.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SG, Navarre BM, Cifani C, Pickens CL, Bossert JM, Shaham Y. Role of dorsal medial prefrontal cortex dopamine D1-family receptors in relapse to high-fat food seeking induced by the anxiogenic drug yohimbine. Neuropsychopharmacology. 2011;36:497–510. doi: 10.1038/npp.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisewander JL, Baker DA, Fuchs RA, Tran-Nguyen LT, Palmer A, Marshall JF. Fos protein expression and cocaine-seeking behavior in rats after exposure to a cocaine self-administration environment. J Neurosci. 2000;20:798–805. doi: 10.1523/JNEUROSCI.20-02-00798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacchioni AM, Gabriele A, See RE. Dorsal striatum mediation of cocaine-seeking after withdrawal from short or long daily access cocaine self-administration in rats. Behav Brain Res. 2011;218:296–300. doi: 10.1016/j.bbr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 5 Ed. Amsterdam: Elsevier Academic; 2005. [Google Scholar]
- Pickens CL, Airavaara M, Theberge F, Fanous S, Hope BT, Shaham Y. Neurobiology of incubation of cocaine craving. Trends Neurosci. 2011;34:411–420. doi: 10.1016/j.tins.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Wood MA. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology. 2013;38:94–110. doi: 10.1038/npp.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Singh H, Dang R, Wood MA. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci. 2013;33:6623–6632. doi: 10.1523/JNEUROSCI.4472-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio FJ, Liu QR, Li X, Cruz FC, Leão RM, Warren BL, Kambhampati S, Babin KR, McPherson KB, Cimbro R, Bossert JM, Shaham Y, Hope BT. Unique molecular alterations in Fos-expressing dorsal striatal neurons activated during context-induced reinstatement of methamphetamine seeking. J Neurosci. 2015;35:5625–5639. doi: 10.1523/JNEUROSCI.4997-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Vakili G. Cocaine triggers epigenetic alterations in the corticostriatal circuit. Brain Res. 2014 doi: 10.1016/j.brainres.2014.09.069. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HD, McGinty JF, West AE, Sadri-Vakili G. Epigenetics and psychostimulant addiction. Cold Spring Harb Perspect Med. 2013;3:a012047. doi: 10.1101/cshperspect.a012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev U, Morales M, Hope B, Yap J, Shaham Y. Time-dependent changes in extinction behavior and stress-induced reinstatement of drug seeking following withdrawal from heroin in rats. Psychopharmacology. 2001;156:98–107. doi: 10.1007/s002130100748. [DOI] [PubMed] [Google Scholar]
- Shalev U, Grimm JW, Shaham Y. Neurobiology of relapse to heroin and cocaine seeking: a review. Pharmacol Rev. 2002;54:1–42. doi: 10.1124/pr.54.1.1. [DOI] [PubMed] [Google Scholar]
- Shepard JD, Bossert JM, Liu SY, Shaham Y. The anxiogenic drug yohimbine reinstates methamphetamine seeking in a rat model of drug relapse. Biol Psychiatry. 2004;55:1082–1089. doi: 10.1016/j.biopsych.2004.02.032. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Carreira MB, Smith LN, Zirlin BC, Neve RL, Cowan CW. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron. 2012;73:108–120. doi: 10.1016/j.neuron.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theberge FR, Li X, Kambhampati S, Pickens CL, St Laurent R, Bossert JM, Baumann MH, Hutchinson MR, Rice KC, Watkins LR, Shaham Y. Effect of chronic delivery of the Toll-like receptor 4 antagonist (+)-naltrexone on incubation of heroin craving. Biol Psychiatry. 2013;73:729–737. doi: 10.1016/j.biopsych.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Corvol JC, Pages C, Besson MJ, Maldonado R, Caboche J. Involvement of the extracellular signal-regulated kinase cascade for cocaine-rewarding properties. J Neurosci. 2000;20:8701–8709. doi: 10.1523/JNEUROSCI.20-23-08701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderschuren LJ, Di Ciano P, Everitt BJ. Involvement of the dorsal striatum in cue-controlled cocaine seeking. J Neurosci. 2005;25:8665–8870. doi: 10.1523/JNEUROSCI.0925-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Shi J, Chen N, Xu L, Li J, Li P, Sun Y, Lu L. Effects of length of abstinence on decision-making and craving in methamphetamine abusers. PLoS One. 2013;8:e68791. doi: 10.1371/journal.pone.0068791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lanfranco MF, Gibb SL, Yowell QV, Carnicella S, Ron D. Long-lasting adaptations of the NR2B-containing NMDA receptors in the dorsomedial striatum play a crucial role in alcohol consumption and relapse. J Neurosci. 2010a;30:10187–10198. doi: 10.1523/JNEUROSCI.2268-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lv Z, Hu Z, Sheng J, Hui B, Sun J, Ma L. Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology. 2010b;35:913–928. doi: 10.1038/npp.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]