Abstract
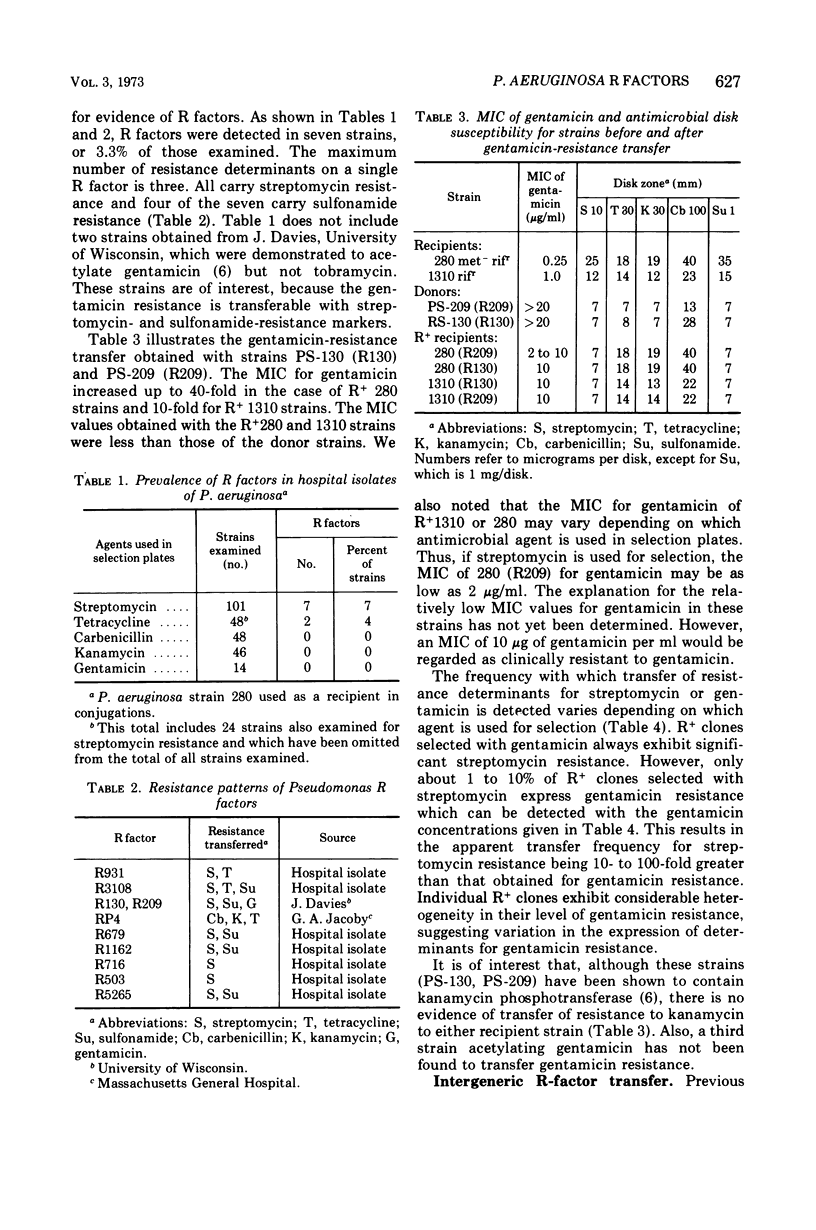
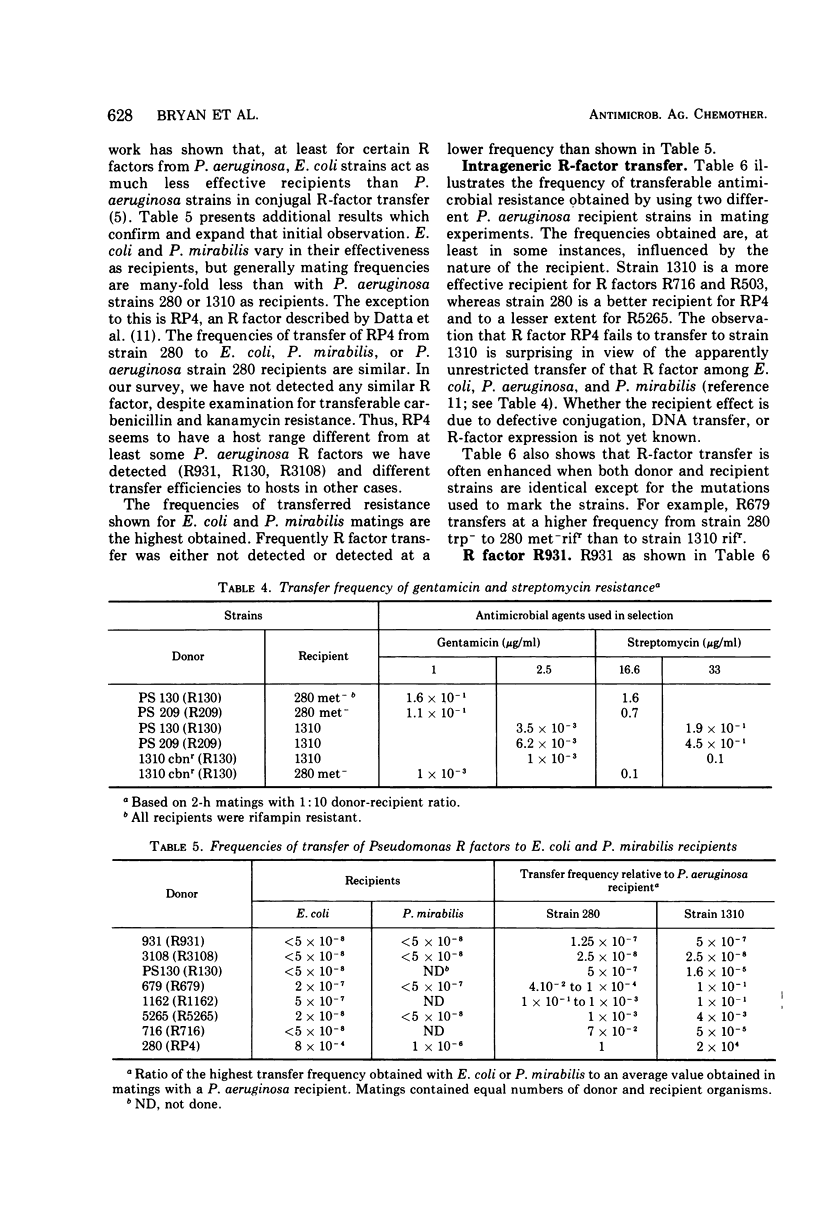
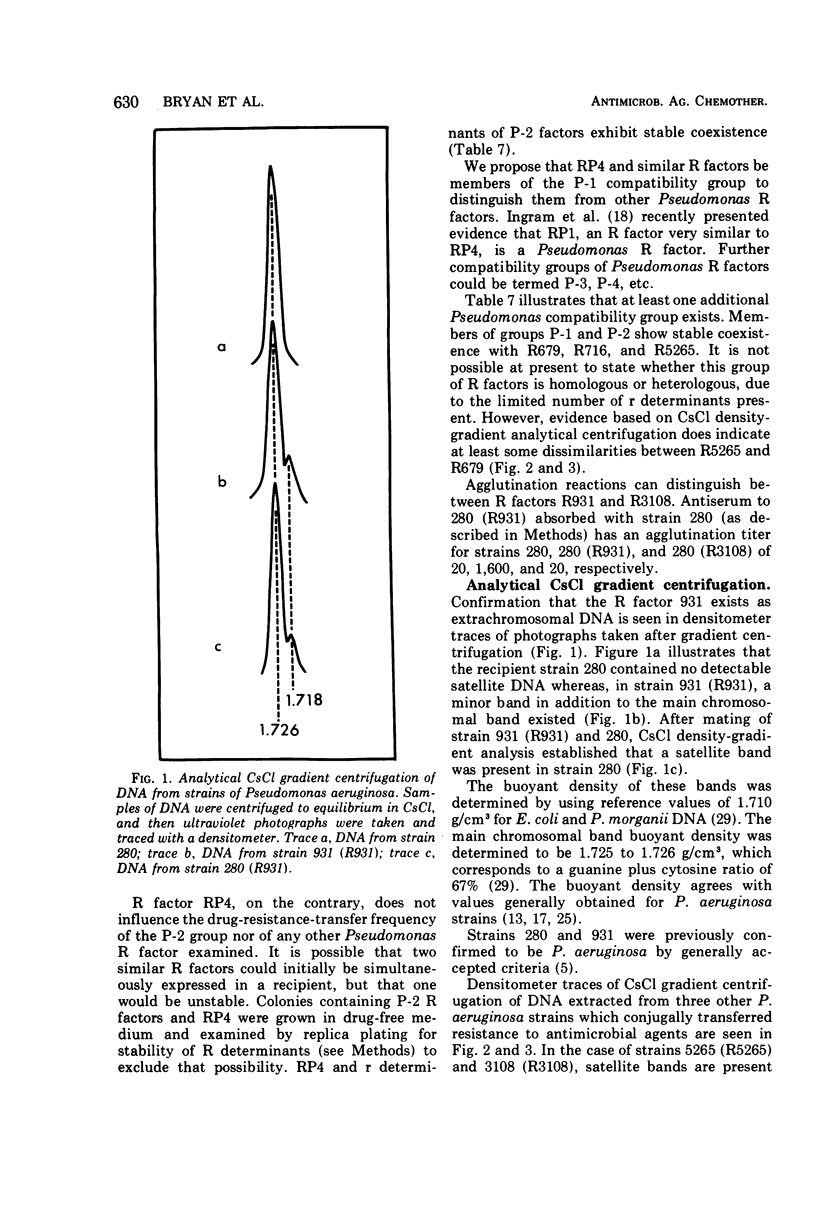
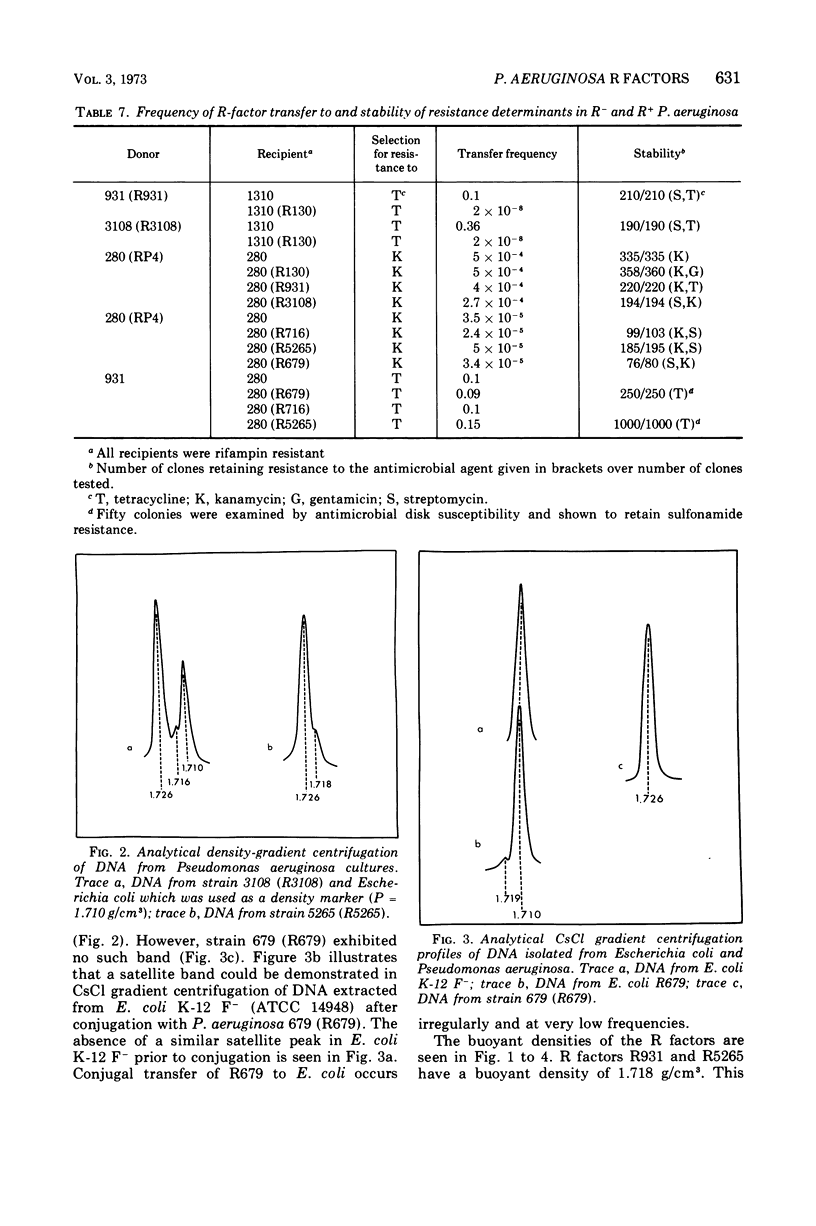
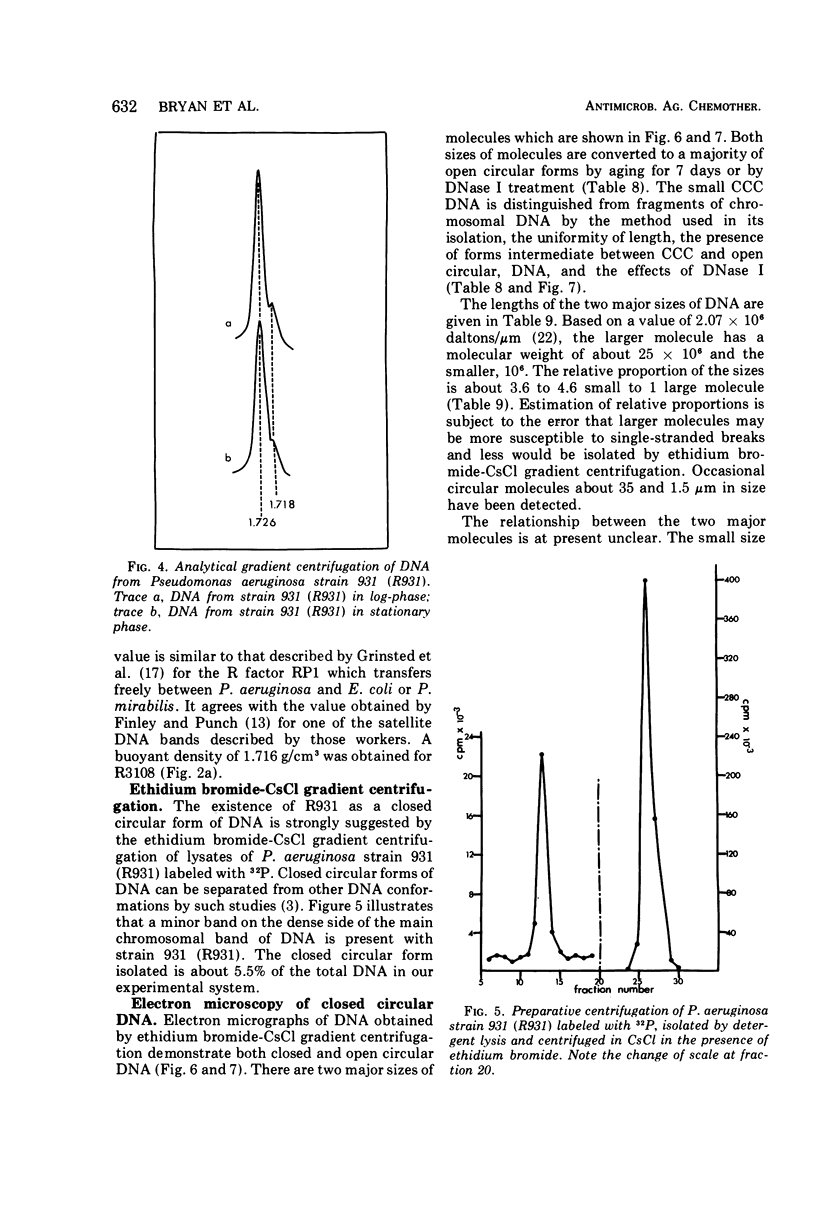
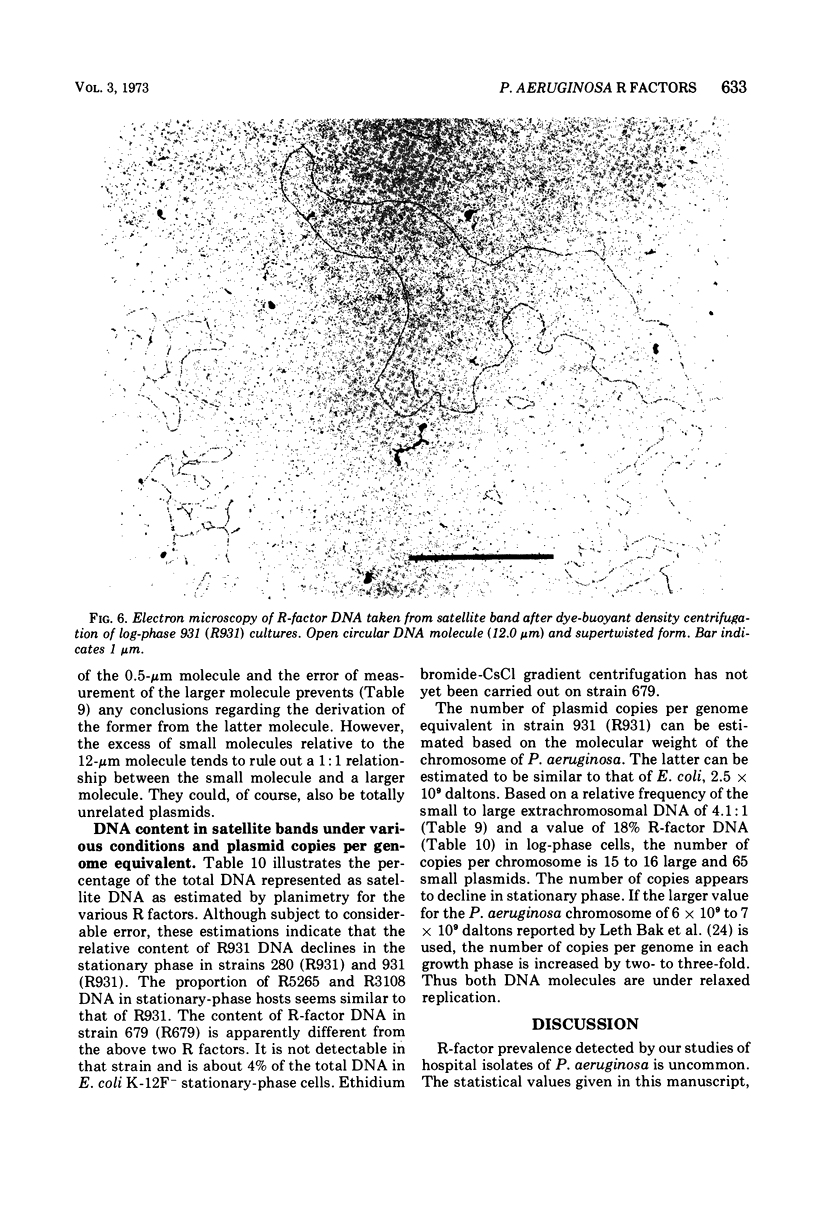
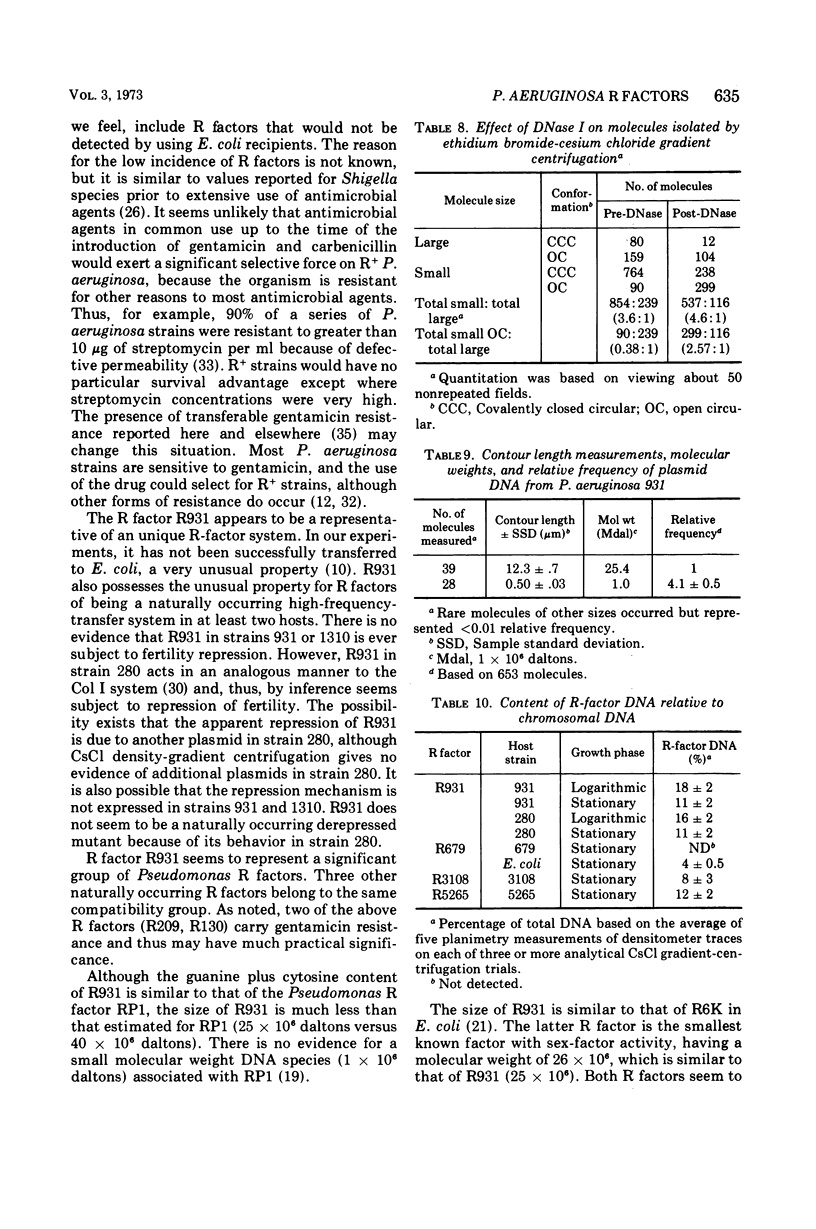
R factors were detected in 3.3% of 233 hospital isolates of Pseudomonas aeruginosa using P. aeruginosa recipients in conjugations. Transferred markers included streptomycin, tetracycline, and sulfonamide resistance. Gentamicin resistance was transferred from two strains previously shown to acetylate gentamicin. A group of R factors exemplified by R931 were characterized by failure to transfer to Escherichia coli recipients. Such R factors formed a single compatibility group when examined in a P. aeruginosa recipient. Other P. aeruginosa R factors, including RP4, showed stable coexistence with the R931 group. It is proposed that RP4 and similar R factors be members of the P-1 compatibility group and that R931, R3108, R209, and R130 be members of a group termed P-2. The buoyant densities of all R factors examined were similar, about 1.716 to 1.719 g/cm3. The content of R-factor deoxyribonucleic acid (DNA) relative to the total DNA varied among the different R factors, ranging from about 18 ± 2% in log-phase cells of 931 (R931) to undetectable for 679 (R679). However, R679, which transferred from strain 679 at extremely low and irregular frequencies to an E. coli host, was shown to represent about 4% R-factor DNA in that host. The relative DNA content of R931 appeared to decline in the stationary growth phase of 931 (R931) or 280 (R931). R931 covalently closed circular DNA was isolated by ethidium bromide-CsCl gradient centrifugation and examined by electron microscopy. Two major molecular distributions existed, having contour lengths of 0.5 and 12.4 μm. The molecular weights were estimated to be 106 and 25 × 106. Both molecules were under relaxed replication control. R factor R931 exists as a naturally occurring high-frequency transfer system in P. aeruginosa strains 931 and 1310. However, in strain 280 it acts as if subject to fertility repression. Other members of the P-2 compatibility group also are high-frequency transfer systems in the natural host and in strain 1310. RP4 is restricted from recipient strain 1310. Some additional recipient effects were noted in that strains 1310 or 280 sometimes differed in recipient effectiveness with a given donor. Agglutination reactions with absorbed antiserum were able to distinguish between two members of the same R-factor compatibility group, R931 and R3108.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson E. S., Lewis M. J. Characterization of a transfer factor associated with drug resistance in Salmonella typhimurium. Nature. 1965 Nov 27;208(5013):843–849. doi: 10.1038/208843a0. [DOI] [PubMed] [Google Scholar]
- Ansz H. S., Zandberg J., van de Pol J. H., van Bruggen E. F. Circular DNA from Shigella paradysenteriae. Eur J Biochem. 1969 Jun;9(2):156–159. doi: 10.1111/j.1432-1033.1969.tb00589.x. [DOI] [PubMed] [Google Scholar]
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Bauer W., Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968 Apr 14;33(1):141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Bryan L. E., Van Den Elzen H. M., Tseng J. T. Transferable drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1972 Jan;1(1):22–29. doi: 10.1128/aac.1.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinska M., Benveniste R., Davies J., Daniels P. J., Weinstein J. Gentamicin resistance in strains of Pseudomonas aeruginosa mediated by enzymatic N-acetylation of the deoxystreptamine moiety. Biochemistry. 1972 Feb 29;11(5):761–765. doi: 10.1021/bi00755a013. [DOI] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Miller C. A. Non-chromosomal antibiotic resistance in bacteria. II. Molecular nature of R-factors isolated from Proteus mirabilis and Escherichia coli. J Mol Biol. 1970 Jun 28;50(3):671–687. doi: 10.1016/0022-2836(70)90092-6. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Compatibility groups among fi - R factors. Nature. 1971 Nov 26;234(5326):222–223. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Host ranges of R factors. J Gen Microbiol. 1972 May;70(3):453–460. doi: 10.1099/00221287-70-3-453. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. E., Rownd R. Transmissible multiple drug resistance in Enterobacteriaceae. Science. 1972 May 19;176(4036):758–768. doi: 10.1126/science.176.4036.758. [DOI] [PubMed] [Google Scholar]
- Finley R. B., Punch J. D. Satellite deoxyribonucleic acid in Pseudomonas aeruginosa. Can J Microbiol. 1972 Jul;18(7):1003–1006. doi: 10.1139/m72-156. [DOI] [PubMed] [Google Scholar]
- Fullbrook P. D., Elson S. W., Slocombe B. R-factor mediated beta-lactamase in Pseudomonas aeruginosa. Nature. 1970 Jun 13;226(5250):1054–1056. doi: 10.1038/2261054a0. [DOI] [PubMed] [Google Scholar]
- Goebel W., Schrempf H. Isolation of minicircular deoxyribonucleic acids from wild strains of Escherichia coli and their relationship to other bacterial plasmids. J Bacteriol. 1972 Sep;111(3):696–704. doi: 10.1128/jb.111.3.696-704.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan J. R., Gillies R. R. Further studies in the pyocine typing of Pseudomonas pyocyanea. J Med Microbiol. 1969 Feb;2(1):17–25. doi: 10.1099/00222615-2-1-17. [DOI] [PubMed] [Google Scholar]
- Grinsted J., Saunders J. R., Ingram L. C., Sykes R. B., Richmond M. H. Properties of a R factor which originated in Pseudomonas aeruginosa 1822. J Bacteriol. 1972 May;110(2):529–537. doi: 10.1128/jb.110.2.529-537.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L., Sykes R. B., Grinsted J., Saunders J. R., Richmond M. H. A transmissible resistance element from a strain of Pseudomonas aeruginosa containing no detectable extrachromosomal DNA. J Gen Microbiol. 1972 Sep;72(2):269–279. doi: 10.1099/00221287-72-2-269. [DOI] [PubMed] [Google Scholar]
- Kontomichalou P., Mitani M., Clowes R. C. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under either relaxed or stringent control. J Bacteriol. 1970 Oct;104(1):34–44. doi: 10.1128/jb.104.1.34-44.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEDERBERG J., LEDERBERG E. M. Replica plating and indirect selection of bacterial mutants. J Bacteriol. 1952 Mar;63(3):399–406. doi: 10.1128/jb.63.3.399-406.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D. Molecular weights of coliphages and coliphage DNA. 3. Contour length and molecular weight of DNA from bacteriophages T4, T5 and T7, and from bovine papilloma virus. J Mol Biol. 1970 Dec 28;54(3):557–565. doi: 10.1016/0022-2836(70)90126-9. [DOI] [PubMed] [Google Scholar]
- Mandel M. Deoxyribonucleic acid base composition in the genus Pseudomonas. J Gen Microbiol. 1966 May;43(2):273–292. doi: 10.1099/00221287-43-2-273. [DOI] [PubMed] [Google Scholar]
- Roe E., Jones R. J., Lowbury E. J. Transfer of anibioic resistanceetween Pseudomonas aeruginosa, Escherichia coli, and other gram-negative bacilli in rns. Lancet. 1971 Jan 23;1(7691):149–152. doi: 10.1016/s0140-6736(71)91930-1. [DOI] [PubMed] [Google Scholar]
- Rush M. G., Gordon C. N., Warner R. C. Circular deoxyribonucleic acid from Shigella dysenteriae Y6R. J Bacteriol. 1969 Nov;100(2):803–808. doi: 10.1128/jb.100.2.803-808.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Sykes R. B., Richmond M. H. Intergeneric transfer of a beta-lactamase gene between Ps. aeruginosa and E. coli. Nature. 1970 Jun 6;226(5249):952–954. doi: 10.1038/226952a0. [DOI] [PubMed] [Google Scholar]
- Tanaka N. Biochemical studies on gentamicin resistance. J Antibiot (Tokyo) 1970 Sep;23(9):469–471. doi: 10.7164/antibiotics.23.469. [DOI] [PubMed] [Google Scholar]
- Tseng J. T., Bryan L. E., Van den Elzen H. M. Mechanisms and spectrum of streptomycin resistance in a natural population of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1972 Sep;2(3):136–141. doi: 10.1128/aac.2.3.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE T. EPISOME-MEDIATED TRANSFER OF DRUG RESISTANCE IN ENTEROBACTERIACEAE. VI. HIGH-FREQUENCY RESISTANCE TRANSFER SYSTEM IN ESCHERICHIA COLI. J Bacteriol. 1963 Apr;85:788–794. doi: 10.1128/jb.85.4.788-794.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchitz J. L., Gerbaud G. R. Classification de plasmides conférant la résistance a la gentamicine. Ann Inst Pasteur (Paris) 1972 Sep;123(3):333–339. [PubMed] [Google Scholar]





