Abstract
Better biomarkers that can discriminate between aggressive and indolent phenotypes of prostate cancer are urgently needed. In the first 20 years of the prostate-specific antigen (PSA) era, screening for prostate cancer has successfully reduced prostate cancer mortality, but has led to significant problems with overdiagnosis and overtreatment. As a result, many men are subjected to unnecessary prostate biopsies and overtreatment of indolent cancer in order to save one man from dying of prostate cancer. A novel blood test known as the 4Kscore® Test (OPKO Lab, Nashville, TN) incorporates a panel of four kallikrein protein biomarkers (total PSA, free PSA, intact PSA, and human kallikrein-related peptidase 2) and other clinical information in an algorithm that provides a percent risk for a high-grade (Gleason score ≥ 7) cancer on biopsy. In 10 peer-reviewed publications, the four kallikrein biomarkers and algorithm of the 4Kscore Test have been shown to improve the prediction not only of biopsy histopathology, but also surgical pathology and occurrence of aggressive, metastatic disease. Recently, a blinded prospective trial of the 4Kscore Test was conducted across the United States among 1012 men. The 4Kscore Test replicated previous European results showing accuracy in predicting biopsy outcome of Gleason score ≥ 7. In a recent case-control study nested within a population-based cohort from Västerbotten, Sweden, the four kallikrein biomarkers of the 4Kscore Test also predicted the risk for aggressive prostate cancer that metastasized within 20 years after the test was administered. These results indicate that men with an abnormal PSA or digital rectal examination result, and for whom an initial or repeat prostate biopsy is being considered, would benefit from a reflex 4Kscore Test to add important information to the clinical decision-making process. A high-risk 4Kscore Test result may be used to select men with a high probability of aggressive prostate cancer who would benefit from a biopsy of the prostate to prevent an adverse and potentially lethal outcome from prostate cancer. Men with a low 4Kscore Test result may safely defer biopsy.
Key words: Prostate cancer, Biomarker, High-grade prostate cancer, Screening
Prostate cancer is the most common cancer in men in the United States, accounting for an estimated 27% of all newly diagnosed cancers in 2014.1 Since the advent of screening for prostate cancer with serum prostate-specific antigen (PSA), we have seen a significant decline in prostate cancer mortality.1 Randomized clinical trials have reported a 20% to 40% reduction in death from prostate cancer in men undergoing routine screening compared with those who are not screened.2,3 However, these trials, and a trial showing little difference between opportunistic and systematic screening,4 have raised the concern for overdiagnosis and overtreatment of indolent prostate cancer. The fundamental concern is that an overwhelming number of men are subjected to interventions such as prostate biopsy in order to prevent one man’s death from prostate cancer.2,3
Prostate biopsy is an invasive procedure with significant complications, such as bleeding, urinary retention, and life-threatening infection. A recent population-based study from Ontario, Canada, revealed a fourfold increase to 4.1% for the rate of hospital admissions after prostate biopsy from 1996 to 2005, with 72% of admissions being due to infection.5 These risks, combined with the enormous anxiety involved in undergoing the procedure, present a significant burden to any man considering prostate cancer screening.
Today, most men diagnosed with prostate cancer have a tumor that is unlikely to pose a threat to their life expectancies. A recent systematic analysis suggested that up to 60% of prostate cancers diagnosed in contemporary studies can be safely observed without a need for immediate intervention.6 However, in the United States, because of the concern for possible undergrading of prostate cancer due to biopsy sampling error, 90% of men diagnosed with prostate cancer undergo treatment and approximately 66% will be confirmed to have indolent Gleason score 6 prostate cancer,7 suggesting a significant problem with overtreatment. Although treatment for localized prostate cancer provides excellent cancer control,8,9 it comes at a significant detriment to health-related quality of life (HRQoL). Previous studies have reported significant changes in HRQoL after primary treatment for prostate cancer, primarily in the domains of sexual and urinary function and bother.10–12 Given the physical and psychological burden of these secondary adverse events, many government agencies and patients are beginning to question the risks and benefits of prostate cancer screening and treatment.13
The United States Preventive Services Task Force recently advised against routine screening for prostate cancer, claiming that the risks of screening outweigh the benefits.13 However, 20% to 30% of men who are diagnosed with prostate cancer are found to have high-grade disease at presentation14; without screening, these men would lose their opportunity for cure. It is clear that new biomarkers or tests that promote the detection of both indolent and aggressive prostate cancer are unlikely to be helpful. We need tests that focus on the detection of aggressive tumors, not the indolent ones that are better left alone. Aggressive prostate cancer, for purposes of this review, is defined as cancer with a Gleason score ≥ 7 and tumors that are most likely to progress to metastatic disease and death. Targeted detection of aggressive prostate cancer would allow urologists to diagnose and treat those men most likely to benefit from aggressive intervention to avoid premature death. Conversely, those men harboring non-life-threatening disease would be able to avoid unnecessary interventions. The 4Kscore® Test (OPKO Lab, Nashville, TN) is a new blood test that accurately identifies the risk of aggressive prostate cancer. The 4Kscore Test plays an important clinical role as a reflex test prior to proceeding with initial prostate biopsy in men with an elevated PSA level or abnormal digital rectal examination (DRE) results, or after a prior negative biopsy and persistently abnormal PSA levels.
Biomarkers Available for the Early Detection of Aggressive Prostate Cancer
Unquestionably, the most commonly used marker for detection of prostate cancer is serum PSA. However, since its introduction in the early 1990s, the limitations of PSA in this regard have been widely recognized.4,13 Alternative methods of utilizing PSA, such as PSA velocity, have been reported to improve the specificity for prostate cancer detection.15,16 However, recent studies have also suggested that using PSA velocity can result in an increase of unnecessary biopsies, and adds very little to the predictive value of a single PSA measurement alone.17,18 Similarly, PSA density has been promoted as another method to improve prostate cancer detection.16 However, measurement of prostate volume is an invasive, time-consuming procedure. Although both PSA enhancements, at best, provide a slight improvement from PSA alone, these techniques are far from the magnitude of improvement needed to reliably discern between aggressive and indolent forms of prostate cancer.
PCA3 and TMPRSS2:ERG fusion are biomarkers measured in the urine immediately following vigorous prostatic massage. Both biomarkers have been actively studied for their ability to stratify prostate cancer risk.16 PCA3 expresses noncoding RNA that is highly overexpressed in prostate tumor tissue compared with normal prostate tissue, whereas TMPRSS2:ERG fusion represents a chromosomal rearrangement of the TMPRSS2 androgen-related gene with ERG or ETV1, two transcription factors from the ETS family. Although both of these markers have been shown to be associated with Gleason grade,19 they have serious limitations. For PCA3, the results can be difficult to interpret, partly due to lack of an optimal cutoff for a positive test result. Furthermore, although PCA3 is present at elevated levels in prostate tissue, it has not been consistently linked to the detection of more advanced or aggressive disease.16 The TMPRSS2:ERG fusion has been studied with equivocal results on its specificity for aggressive prostate cancer.16
The Prostate Health Index (PHI) is a blood-based, multibiomarker test that combines free and total PSA with [-2]proPSA, a precursor form of free PSA, in the following algorithm: [(p2PSA/fPSA) × √tPSA].16,20 In a prospective study of 892 men, PHI displayed an area under the receiver operator curve (AUC) of 0.72 for high-grade disease defined as Gleason score ≥ 4 + 3 = 7. A recent meta-analysis of 16 published studies assessing PHI for the detection of high-grade (Gleason score ≥ 7) prostate cancer reported a pooled sensitivity of 0.90 (95% confidence interval [CI], 0.87–0.92), and pooled specificity of only 0.17 (95% CI, 0.14–0.19).21 The pooled AUC of PHI for high-grade disease was only 0.67 (95% CI, 0.57–0.77). In a recent multicenter European prospective trial of 489 men undergoing radical prostatectomy, the PHI test was found to be a significant predictor of unfavorable histopathology at surgery.22 However, the authors found that using PHI added very little net benefit for clinical decision making, and questioned its clinical utility. The PHI clinical data have been used to report risk of aggressive cancer as a four-tiered probability based on various PHI cutoffs.20 However, the risk score has not been calibrated or validated prospectively. There are also no published long-term outcomes studies associated with the clinical use of the PHI test in either predicting development of metastatic disease or demonstrating the safety of deferring biopsy in men with a low PHI result.
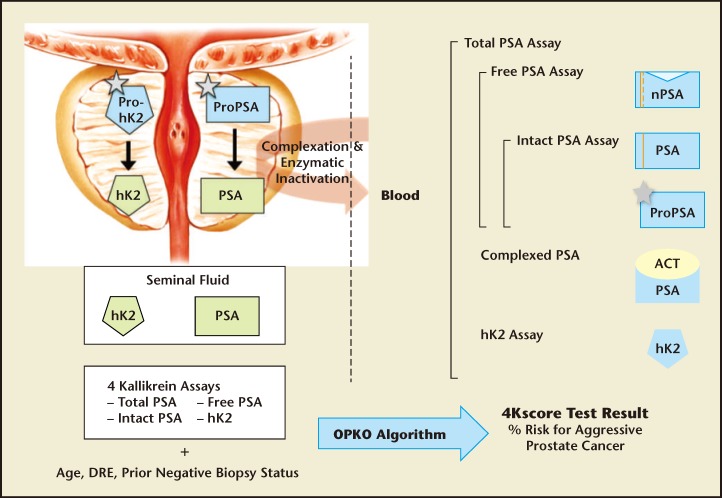
The Four-Kallikrein Panel Tissue kallikrein and kallikrein-related enzymes are a family of 15 closely related serine proteases with high homology.23 Although messenger RNA expression of all 15 kallikreins can be detected in prostate tissue, by far the dominant kallikreins are KLK2 (the gene that produces the enzyme human kallikrein-2 [hK2]) and KLK3 (the gene that produces the enzyme human kallikrein-3, or PSA).24 These kallikreins, and various molecular forms of each, have been extensively investigated for their utility in detecting aggressive prostate cancer. Monoclonal antibodies and immunoassays specific to total PSA, free PSA, intact PSA, and hK2 (Figure 1) have been developed; the measurement of all of these prostate-specific kallikreins has proven useful for the detection of aggressive prostate cancer.24
Figure 1.
Active forms of prostate-specific antigen (PSA; human kallikrein-3) and human kallikrein-2 (hK2) are shown in green, inactive forms are shown in blue. In the prostate, propeptides (represented as grey stars) are removed from proPSA and prohK2, leaving the mature, catalytically active forms. PSA and hK2 are released at high concentrations (mg/mL level) into prostatic fluid to liquefy semen delivered by the vas deferens. The prostatic kallikreins are found in blood at the ng/mL level (total PSA and free PSA) or pg/mL level (intact PSA and hK2), approximately 1 million-fold lower concentration than levels in seminal fluid. Blood contains a variety of forms of PSA. The most abundant form in blood is PSA complexed with a-1-antichymotrypsin (ACT). Free PSA not complexed with ACT has several molecular forms (nicked, intact, and proPSA). Commercial total PSA assays measure all forms of PSA (complexed and free) and the highly homologous kallikrein, hK2. The 4Kscore Test® (OPKO Lab, Nashville, TN) utilizes four specific kallikrein immunoassays: total PSA, free PSA, intact PSA, and hK2. The patient’s age, digital rectal examination (DRE) results (nodules/no nodules), and prior negative biopsy status (yes/no) are combined in an algorithm to produce the 4Kscore Test result: an individual patient’s risk of having aggressive cancer. The result is reported on a continuous scale from < 1% to > 95%. Adapted from Lilja H et al.24
In prostate cancer, there is dysregulation and overexpression of both PSA and hK2, and levels of both intact PSA and hK2 increase as prostate cancer becomes more undifferentiated.25 These kallikreins directly and indirectly alter cell growth regulation, increase extracellular matrix degradation and remodeling, and promote cell invasion and angiogenesis—all of which contribute to prostate cancer progression and metastasis.26 As a result, these kallikreins show tremendous merit in differentiating between the aggressive versus indolent forms of prostate cancer. Furthermore, their levels show little variability for up to 12 days in men undergoing evaluation for prostate cancer, making them diagnostically useful.27
Evidence Supporting the Four-Kallikrein Panel in the Detection of Aggressive Prostate Cancer
The clinical utility of the four-kallikrein panel in the 4Kscore Test is based on testing over 15,000 patients reported in 10 peer-reviewed publications. The patients were part of the European Randomized Study of Prostate Cancer Screening (ERSPC), the Malmö Diet and Cancer Study, the Västerbotten Intervention Project, and the validation study recently conducted in the United States by OPKO Health (Table 1). The first clinical report of the four-kallikrein panel was among 740 previously unscreened men who underwent biopsy for a PSA ≥ 3.0 ng/mL during the first round of screening in the Göteborg arm of the ERSPC.28 Among the 740 men in the cohort, 152 (21%) were diagnosed with a Gleason score 6 cancer, whereas 40 (5%) were diagnosed with a Gleason score ≥ 7 cancer. The AUC for detecting Gleason score ≥ 7 prostate cancer for a base model consisting of age, total PSA, and DRE was compared with a similar model with the addition of the four-kallikrein panel. The authors reported that the AUC increased from 0.87 in the base model to 0.90 in the full clinical model that incorporates the four-kallikrein panel results, age, and DRE. Using this full clinical model, which closely approximates the current 4Kscore Test algorithm, only 1 out of 40 Gleason score ≥ 7 tumors would have had a delayed diagnosis of their disease and 60% of prostate biopsies would have been avoided.
Table 1.
Summary of 10 Peer-reviewed Studies Investigating the Four-kallikrein Panel of the 4Kscore Test in Multiple Cohorts Totaling 15,139 Subjects
| 4Kscore | % Reduction | |||||
|---|---|---|---|---|---|---|
| Study | Trial | Cohort | Endpoint | Subjects | AUC | (Intervention) |
| Vickers AJ et al28 | ERSPC | Not screened | Bx histology | 740 | 0.90 | 57 (Bx) |
| Benchikh A et al29 | ERSPC | Not screened | Bx histology | 262 | 0.87 | 49 (Bx) |
| Vickers A et al30 | ERSPC | Not screened | Bx histology | 2914 | 0.84 | 51 (Bx) |
| Vickers AJ et al31 | ERSPC | Screened | Bx histology | 1501 | 0.80 | 36 (Bx) |
| Vickers AJ et al32 | ERSPC | Screened | Bx histology | 1241 | 0.83 | 41 (Bx) |
| Gupta A et al33 | ERSPC | Prior biopsy | Bx histology | 925 | 0.87 | 82 (Bx) |
| Parekh DJ et al36 | US Validation | Screened | Bx histology | 1012 | 0.82 | 43 (Bx) |
| Study | ||||||
| Carlsson S et al34 | ERSPC | Post RP | Radical prostatectomy | 392 | 0.84 | 14 (RP) |
| histology | ||||||
| Vickers AJ et al42 | MDC | Not screened | Prostate cancer: | 792 | 0.82 | 42 (Bx) |
| any, palpable, | ||||||
| metastatic | ||||||
| Stattin P et al35 | VIP | Not screened | Distant prostate | 5360 | – | – |
| cancer metastasis | ||||||
| Total subjects studied | 15,139 |
AUC, area under the operating curve; Bx, biopsy; ERSPC, European Randomized Study of Prostate Cancer Screening; MDC, Malmö Diet and Cancer Study; RP,radical prostatectomy; VIP, Västerbotten Intervention Project.
4Kscore® Test; OPKO Lab, Nashville, TN.
The clinical utility was also studied in a different cohort of 262 men undergoing biopsy for a PSA ≥ 3.0 ng/mL and further clinical work-up in the French arm of the ERSPC.29 Among these men, the AUC for detecting a Gleason score ≥ 7 cancer increased from 0.77 for a base model (age, total PSA, and DRE) to 0.87 for a model including the four-kallikrein panel. In yet another validation, the four-kallikrein panel was studied in an independent cohort of 2914 unscreened men participating in the first round of the Rotterdam arm of the ERSPC.30 The study design provided 728 participants in the training set and 2186 participants in the validation set. Among men in the validation set, 219 (10%) were found to have Gleason score ≥ 7 prostate cancer, with a similar percentage noted in the training set. As in the previous studies, the authors found the AUC increased, in this case from 0.81 for the clinical base model (age, total PSA, and DRE) to 0.84 for a model incorporating the four-kallikrein panel for the detection of Gleason score ≥ 7 prostate cancer. Application of the four-kallikrein panel to 1000 men with elevated PSA would result in 513 men avoiding a biopsy while missing only 12 of 100 Gleason score ≥ 7 cancers. Results from these studies consistently show the four-kallikrein panel can effectively identify high-grade disease while reducing the number of unnecessary biopsies 49% to 57% among men being screened for the first time.
The test has also been validated in men who have undergone prior screening. Among 1501 previously screened men with elevated PSA who underwent their first biopsy in the second and third round of the ERSPC Rotterdam arm, 91 patients (23% of all cancers and 6% of all patients) had Gleason score ≥ 7 cancers.31 The AUC for detecting significant cancer increased from 0.71 in a base clinical model (age, DRE, PSA) to 0.80 by adding the four-kallikrein panel to the model. Using the four-kallikrein panel to decide on the need for biopsy would have avoided 362 biopsies among 1000 men, and missed only 4 high-grade cancers. These findings were confirmed in a second study of 1241 men from Sweden who underwent biopsy for an elevated PSA in the second or later round of screening in the ERSPC Gothenburg arm.32 Among these men, 53 (16% of all cancers and 4% of all patients) had Gleason score ≥ 7 cancer. Incorporating the four-kallikrein panel into a clinical model of age, PSA, and DRE increased the AUC from 0.72 to 0.83. Again, the four-kallikrein panel allowed a 41% reduction in biopsies and missed only one high-grade cancer. Even among men with a previous negative biopsy result, the four-kallikrein panel reduces unnecessary biopsies with little compromise in detecting high-grade tumors.
Another study within the ERSPC involved 925 men who underwent repeat biopsy for an elevated PSA after a previous negative biopsy. In this cohort, 110 prostate cancers were detected (12% of all patients), of which 18 (2%) were Gleason score ≥ 7.33 The four-kallikrein panel increased the AUC for aggressive cancer detection from 0.76 to 0.87 when compared with a base clinical model with age, total PSA, and DRE. Based on this study, if 1000 men with a prior negative prostate biopsy result were to be tested, 817 of 1000 patients (82%) would have been able to avoid a biopsy whereas only 3 Gleason score ≥ 7 cancers would be missed. These studies combined have demonstrated the clinical utility of the four-kallikrein panel in selecting men for biopsy in both screened and nonscreened populations, regardless of whether it was performed prior to initial biopsy or a repeat biopsy after a previous negative biopsy result.
The four-kallikrein panel has also shown merit for treatment decision making. In a cohort of 392 men who were diagnosed with prostate cancer and underwent radical prostatectomy in the Rotterdam arm of the ERSPC trial, the addition of the four-kallikrein panel improved the prediction of aggressive histopathology from 0.81 to 0.84 compared with a clinical model of age, stage, PSA, and biopsy Gleason and cores data.34 In this study, aggressive disease was defined as extracapsular extension, tumor volume > 0.5 cm3, or any Gleason grade ≥ 4 tumor in the radical prostatectomy specimen. The authors reported that adding the use of the four-kallikrein panel in a clinical prediction model would allow a reduction of 135 (14%) unnecessary surgeries per 1000 men.
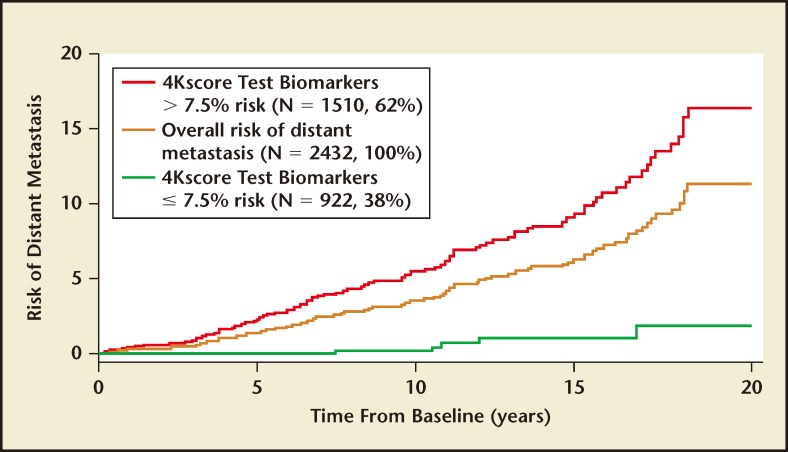
The typical endpoint for most prostate cancer biomarker studies is Gleason score on biopsy. A more clinically relevant outcome is progression to metastases and death, as most men will die with prostate cancer, not from it. In a recently published, nested, case-control study on a population-based cohort from Västerbotten, Sweden, 40,379 men provided blood samples at ages 40, 50, and 60 years from 1986 to 2009.35 Among the 12,542 men who had > 15 years of follow-up, the Swedish Cancer Registry identified 1423 incident cases of prostate cancer and 235 cases with distant metastasis. Among men with an elevated PSA, the concordance index for long-term risk of metastasis was 0.850 to 0.875 depending on the age group and PSA threshold. Among 2432 men with a PSA of 3 ng/mL or higher at age 60, using a 7.5% risk of Gleason 7 or higher cancer as a cutoff, the 4-kallikrein panel was able to identify 38% of the population who were low risk for prostate cancer progression. Over 15 years, the risk of prostate cancer metastasis in these men was only 1% (Figure 2). The four-kallikrein panel was able to discriminate between those who need further testing due to a high risk for prostate cancer progression to metastases and death versus those who can safely forego prostate biopsy for a low likelihood of ever having prostate cancer progression.
Figure 2.
All 60-year-old men with PSA ≥3 ng/mL (N = 2432) in the Västerbotten Intervention Project (VIP) were stratified into two groups: men with ≥ 7.5% 4Kscore Test® (OPKO Lab, Nashville, TN) value (high risk) versus men with < 7.5% 4Kscore Test value (low risk). The absolute risk for metastatic prostate cancer at 5, 10, 15, and 20 years for the low-risk group (green line) was 0.0%, 0.2%, 1.0%, and 1.8% risk. For the high-risk group (red line) the risk for metastatic prostate cancer was 2.4%, 5.6%, 9.9%, and 16.4%. Adapted from Stattin P et al.35
Prospective US Clinical Trial: The US Validation Study
The European studies of four-kallikrein panel, or 4Kscore Test, demonstrated that a combination of four blood kallikrein levels in a calibrated statistical model could provide an accurate estimate of an individual patient’s probability of having a Gleason score ≥ 7 cancer if a biopsy of the prostate were to be performed. However, the studies prior to 2013 were retrospective, included predominantly white populations, largely involved sextant biopsy, and the Gleason grading system used did not follow contemporary guidelines. The 4Kscore Test had never been validated prospectively with a prespecified endpoint for detection of high-grade (Gleason score ≥ 7) prostate cancer, or evaluated in a population of men from the United States. Beginning in October 2013, and concluding in April 2014, subjects were enrolled in an OPKO Health-sponsored clinical study involving 26 independent urology clinics across the nation in a prospective, blinded validation of the 4Kscore Test. The US Validation Study enrolled men who were scheduled to undergo prostate biopsy by a urologist for clinical suspicion of prostate cancer due to abnormal PSA value or DRE result (or other reason) and were continuously enrolled at each of the centers.36 The inclusion criteria were purposely broad to allow an enrollment of a wide range of ages and PSA values representative of current US clinical practice. All men provided a sample of blood prior to undergoing a minimum 10-core prostate biopsy.
The 4Kscore Test is identical to the four-kallikrein panel previously studied in the ERSPC cohorts, incorporating the four-kallikrein panel of total PSA, free PSA, intact PSA, and hK2 plus age and DRE, but needed to address certain modifications for the US Validation Study: (1) the 4Kscore Test uses a fresh plasma specimen not a frozen serum specimen as described in the previous European studies; (2) the 4Kscore Test incorporates risk adjustment for a prior negative biopsy, as this can significantly lower overall risk for aggressive prostate cancer; (3) the PSA and free PSA assays used in the European studies were ProStatus™ assays (PerkinElmer, Waltham, MA). These assays are not US Food and Drug Administration (FDA) approved, so the 4Kscore Test was adapted to use the FDA-approved Elecsys Total PSA (Roche Diagnostics, Indianapolis, IN) and the Elecsys Free PSA (Roche Diagnostics) as part of the algorithm.
Because of the modifications just described, the first 300 consecutively enrolled subjects who met all eligibility requirements were used to check the calibration of the 4Kscore Test algorithm for high-grade disease prediction against the prostate biopsy results. The 4Kscore Test calibration was found to be near perfect and no adjustments were needed prior to proceeding to the validation phase of the trial.
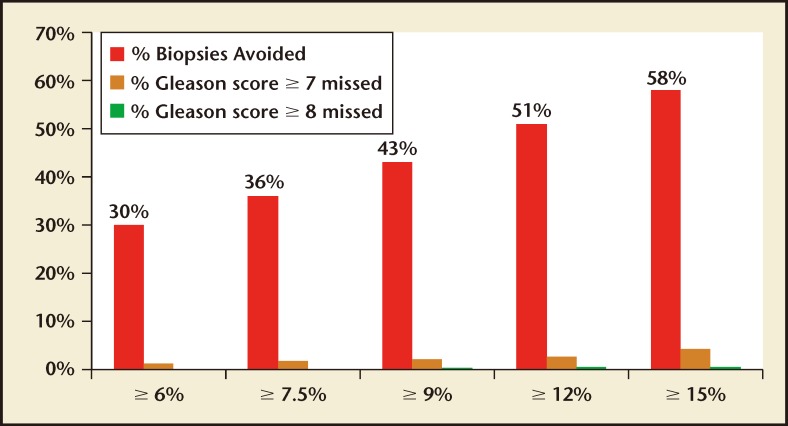
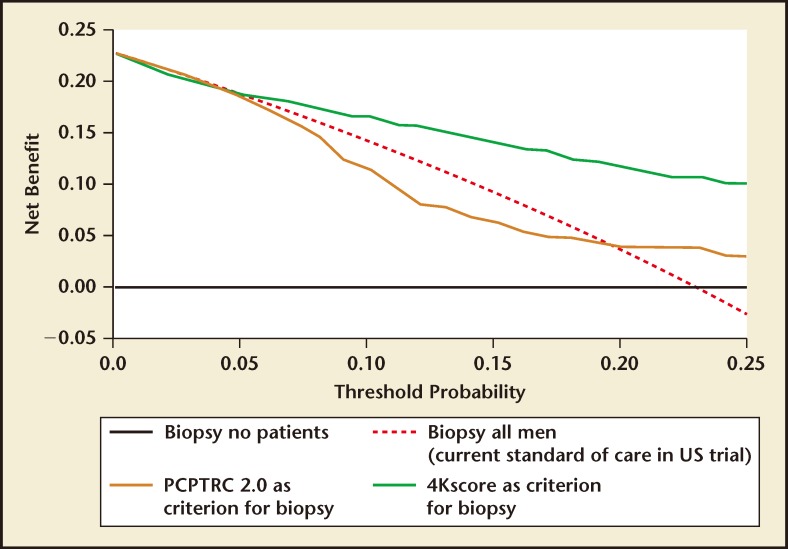
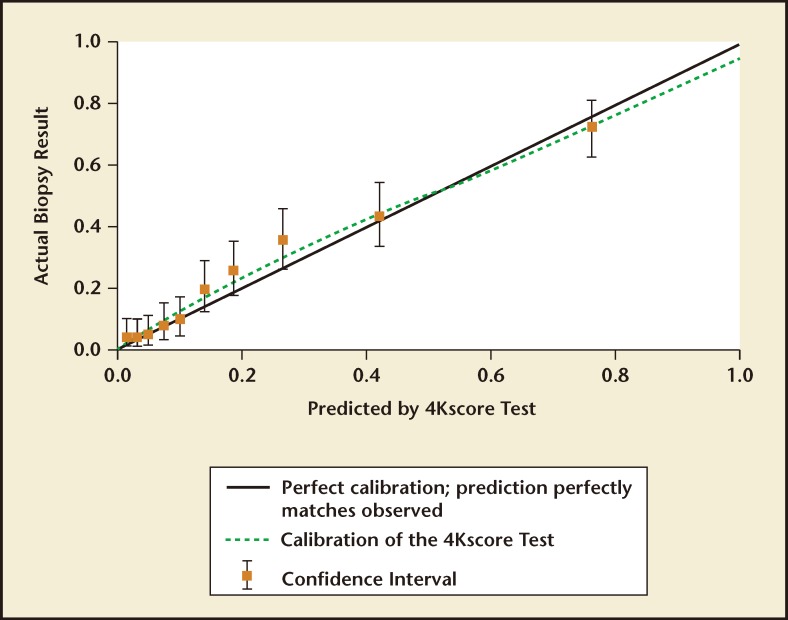
The validation phase of the US Validation Study consisted of 1012 men, with 239 (24%) found to have a low-grade Gleason score 6 cancer and 231 (23%) found to have a Gleason score ≥ 7 cancer of the prostate. The 4Kscore Test showed excellent calibration in the validation phase of the US Validation Study, with predictions of high-grade disease risk closely matching the actual percent of Gleason score ≥ 7 found at prostate biopsy (Figure 3). The AUC for predicting a Gleason score ≥ 7 cancer for the 4Kscore Test was 0.82, consistent with the findings in all the previous European cohorts studied. The authors used various thresholds to illustrate the clinical implications of using the test (Figure 4), and reported that using a 7.5% risk of Gleason score ≥ 7 cancer as a cutoff for biopsy would result in a 36% reduction in the number of unnecessary biopsies, while delaying diagnosis of only 17 (1.7%) significant tumors. The majority of these delayed diagnoses were Gleason score 314 tumors and no men whose biopsy would have been avoided had true high-risk (Gleason score ≥ 8) disease. A decision curve analysis,37 used to determine the net benefit of providing benefit through true positives versus doing harm through false positives, was also investigated. The 4Kscore Test was compared using decision curve analysis to the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) 2.0, a commonly used individual risk calculator derived from the placebo arm of the Prostate Cancer Prevention Trial. Unlike the 4Kscore Test, the PCPTRC 2.0 provides an individual risk prediction for both indolent and aggressive cancer.38 The 4Kscore Test was found to have a higher net benefit compared with the PCPTRC 2.0 as a decision tool for aggressive prostate biopsy across all clinically relevant risk thresholds (Figure 5). The AUC for the 4Kscore Test (0.82) was significantly higher (P < .0001) than the AUC of the PCPTRC 2.0 (0.74) for aggressive cancer. Moreover, when comparing AUC of the 4Kscore Test among white and black men, the authors found that the CIs for the difference in AUC crossed zero (−0.064 to 0.119), implying no significant difference for the 4Kscore Test performance between white and black men.
Figure 4.
Graph displaying the percentage of biopsies avoided and the proportion of Gleason score ≥ 7, and Gleason score ≥ 8 cancers missed at various 4Kscore Test® (OPKO Lab, Nashville, TN) thresholds for performing a biopsy based on the US Validation Study. Adapted from Parekh D et al.36
Figure 5.
Decision curve analysis demonstrating the higher net benefit of the 4Kscore Test® (OPKO Lab, Nashville, TN) (green line) compared with biopsy all (dashed line) and the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) 2.0 (yellow line) across all relevant risk threshold values for clinical decision making from the US Validation Study. Adapted from Parekh D, et al.36
These analyses in the US Validation Study show that the 4Kscore Test accurately identifies risk for high-grade prostate cancer at prostate biopsy and provides new information to facilitate informed decision making concerning whether to undergo a prostate biopsy.
Discussion
A significant strength of the 4Kscore Test is that it is a well-calibrated algorithm for predicting the risk of high-grade prostate cancer prior to biopsy of the prostate and histopathology after radical prostatectomy. Long-term outcomes data have displayed that the biomarkers in the 4Kscore Test can predict the risk of prostate cancer metastases up to 20 years in advance. Two of the four biomarkers of the 4Kscore Test, total PSA and free PSA, are well-established tests and have been used previously for prostate cancer detection. However, neither total PSA nor the free:total PSA ratio has provided sufficient specificity to address the overdiagnosis and overtreatment dilemma facing early detection programs. In the 4Kscore Test, intact PSA and hK2 have been added to total PSA and free PSA to enhance the accuracy for aggressive disease prediction,25,26 and combined with clinical information (age, DRE, and whether there has been a prior negative biopsy result), the test reports a risk score predicting the probability of a Gleason score ≥ 7 cancer on a continuous scale from < 1% to > 95%. The 4Kscore Test, referenced in the scientific literature as the four-kallikrein panel, has been extensively studied in multiple cohorts, where it enhances the prediction of aggressive prostate cancer beyond available clinical information or other available biomarkers. The recent prospective US Validation Study confirms that these findings apply in contemporary clinical practice in the United States. The 4Kscore Test has consistently outperformed other available biomarkers in the detection of aggressive cancer on biopsy of the prostate. The accuracy of the 4Kscore Test was demonstrated in the consistently high AUC observed in multiple cohorts (0.80–0.90) and proves it is a robust and valuable tool for clinical decision making.
Another strength of the 4Kscore Test is its focus on aggressive, or lethal, cancer. Recent data from randomized trials have suggested there is little or no difference in prostate cancer mortality between men with low-risk cancer who underwent radical therapy versus those who were observed.39,40 However, several studies have reported significant detriments in HRQoL after treatment,10–12 suggesting that the benefits of treating low-risk prostate cancer do not outweigh the risks. Therefore, more efforts need to be concentrated on the identification of aggressive tumors, for which treatment can be helpful and a favorable risk-to-benefit ratio can be attained.41 The four-kallikrein biomarkers that serve as the backbone of the 4Kscore Test have demonstrated excellent accuracy in detecting aggressive cancer, both in the biopsy and the radical prostatectomy specimens.34,36 Furthermore, it is the only test, other than PSA, that has been linked to long-term endpoints, such as prostate cancer metastasis.35 By demonstrating an association to more meaningful endpoints and showing an ability to discriminate between lethal and nonlethal prostate cancer, the 4Kscore Test allows us to avoid prostate biopsy in men whose cancers are better left undetected, and focus intervention on men who are most likely to benefit from it.
A final attribute of the 4Kscore Test that is unique is the accuracy of its individualized probability prediction. The test result is a personalized positive predictive value of finding Gleason score ≥ 7 cancer on biopsy of the prostate. This provides the patient and clinician with quantifiable information to aid in clinical decision making. For instance, a healthy, risk-averse man may favor a low threshold probability of aggressive cancer (eg, 6%) before proceeding to a biopsy, whereas an older man with shorter life expectancy due to comorbidities may opt for a much higher 4Kscore Test threshold (eg, 15%) before considering a biopsy. Such individualized risk prediction allows urologists and patients to better address the individual’s personal risk prior to initiating invasive procedures and results in more informed shared decision making.
Future Studies
Although these studies have informed us about the value of the 4Kscore Test in detecting aggressive prostate cancer, future studies have been planned to further investigate and better characterize the impact of the 4Kscore Test in other areas of clinical practice. For instance, a recent prospective trial enrolled a wide variety of men of all ages and PSA values and provides an opportunity to investigate the performance of the 4Kscore Test in various subpopulations at both the low and the high end of the PSA spectrum. There is also a large prostate biopsy for men on active surveillance, and we plan to study how the 4Kscore Test may address this need.40
Conclusions
The 4Kscore Test is a reflex blood test for men who have an abnormal PSA or DRE result and are being considered for an initial or repeat prostate biopsy after a prior negative biopsy result. The 4Kscore Test can accurately identify men with aggressive prostate cancer and the risk of developing prostate cancer metastases within 20 years. Therefore, using the test to select out men with a significant probability of aggressive prostate cancer should allow us to avoid premature death from prostate cancer, while safely avoiding harmful interventions in men who do not require them. Men determined to be at low risk by the 4Kscore Test have a near zero chance of metastasis within 10 years, even if not followed further; therefore, they can be carefully monitored with serial PSA level and 4Kscore testing in place of biopsy.
Main Points.
Prostate-specific antigen (PSA) screening for prostate cancer has successfully reduced prostate cancer mortality, but has led to significant problems of overdiagnosis and overtreatment. The major concern is that an overwhelming number of men are subjected to interventions such as prostate biopsy in order to prevent one man’s death from prostate cancer.
The most commonly used marker for detection of prostate cancer is serum PSA. However, the limitations of PSA are widely recognized. Alternative methods of utilizing PSA, such as PSA velocity and PSA density, have been reported to improve the specificity for prostate cancer detection. Although both PSA enhancements, at best, provide a slight improvement from PSA alone, these techniques are far from the magnitude of improvement needed to reliably discern between aggressive and indolent forms of prostate cancer.
The 4Kscore® Test (OPKO Lab, Nashville, TN) is a new blood test that accurately identifies the risk of aggressive prostate cancer; it plays an important clinical role as a reflex test prior to proceeding with initial prostate biopsy in men with an elevated PSA level or abnormal digital rectal examination results.
The four-kallikrein biomarkers that serve as the backbone of the 4Kscore Test have demonstrated excellent accuracy in detecting aggressive cancer, both in the biopsy and the radical prostatectomy specimens. By demonstrating an association to more meaningful endpoints and showing an ability to discriminate between lethal and nonlethal prostate cancer, the 4Kscore Test allows us to safely avoid prostate biopsy in men whose cancers are better left alone, and focus intervention on men who are most likely to benefit from it.
The test result itself is a personalized positive predictive value of finding Gleason score ≥ 7 cancer on biopsy of the prostate. This provides the patient and clinician with quantifiable information to aid in clinical decision making.
Figure 3.
A calibration plot showing the risk for Gleason score ≥ 7 prostate cancer predicted by the 4Kscore Test® (OPKO Lab, Nashville, TN) compared with the actual prostate biopsy results. Data are from the US Validation Study. Perfect calibration is noted by the solid black line; the green line displays the calibration of the 4Kscore Test. The results show near perfect correlation between the risk predicted by the 4Kscore Test versus actual risk for high-grade prostate cancer determined by biopsy. Adapted from Parekh D et al.36
Footnotes
The authors report no real or apparent conflict of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Schröder FH, Hugosson J, Roobol MJ, et al. ERSPC Investigators. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 3.Hugosson J, Carlsson S, Aus G, et al. Mortality results from the Göteborg randomised population-based prostate-cancer screening trial. Lancet Oncol. 2010;11:725–732. doi: 10.1016/S1470-2045(10)70146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andriole GL, Crawford ED, Grubb RL, 3rd, et al. PLCO Project Team. Prostate cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: mortality results after 13 years of follow-up. J Natl Cancer Inst. 2012;104:125–132. doi: 10.1093/jnci/djr500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nam RK, Saskin R, Lee Y, et al. Increasing hospital admission rates for urological complications after transrectal ultrasound guided prostate biopsy. J Urol. 2013;189(1 suppl):S12–S18. doi: 10.1016/j.juro.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Dall’Era MA, Albertsen PC, Bangma C, et al. Active surveillance for prostate cancer: a systematic review of the literature. Eur Urol. 2012;62:976–983. doi: 10.1016/j.eururo.2012.05.072. [DOI] [PubMed] [Google Scholar]
- 7.Jalloh M, Myers F, Cowan JE, et al. Racial variation in prostate cancer upgrading and upstaging among men with low-risk clinical characteristics. Eur Urol. 2015;67:451–457. doi: 10.1016/j.eururo.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 8.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 9.Cooperberg MR, Vickers AJ, Broering JM, Carroll PR. Comparative risk-adjusted mortality outcomes after primary surgery, radiotherapy, or androgendeprivation therapy for localized prostate cancer. Cancer. 2010;116:5226–5234. doi: 10.1002/cncr.25456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Resnick MJ, Koyama T, Fan KH, et al. Long-term functional outcomes after treatment for localized prostate cancer. N Engl J Med. 2013;368:436–445. doi: 10.1056/NEJMoa1209978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Punnen S, Cowan JE, Chan JM, et al. Long-term health-related quality of life after primary treatment for localized prostate cancer: results from the CaPSURE Registry [published online ahead of print Sept. 18, 2014] Eur Urol. doi: 10.1016/j.eururo.2014.08.074. doi: 10.1016/j.eururo.2014.08.074. [DOI] [PubMed] [Google Scholar]
- 12.Barry MJ, Gallagher PM, Skinner JS, Fowler FJ., Jr Adverse effects of robotic-assisted laparoscopic versus open retropubic radical prostatectomy among a nationwide random sample of Medicare-age men. J Clin Oncol. 2012;30:513–518. doi: 10.1200/JCO.2011.36.8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moyer VA US Preventive Services Task Force, authors. Screening for prostate cancer: US Preventive Services Task Force recommendation statement. Ann Intern Med. 2012;157:120–134. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 14.Punnen S, Cooperberg MR. The epidemiology of high-risk prostate cancer. Curr Opin Urol. 2013;23:331–336. doi: 10.1097/MOU.0b013e328361d48e. [DOI] [PubMed] [Google Scholar]
- 15.Greene KL, Albertsen PC, Babaian RJ, et al. American^Urological^Association. J Urol. 2013;189(1 suppl):S2–S11. doi: 10.1016/j.juro.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Cary KC, Cooperberg MR. Biomarkers in prostate cancer surveillance and screening: past, present, and future. Ther Adv Urol. 2013;5:318–329. doi: 10.1177/1756287213495915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulati R, Gore JL, Etzioni R. Comparative effectiveness of alternative prostate-specific antigenbased prostate cancer screening strategies: model estimates of potential benefits and harms. Ann Intern Med. 2013;158:145–153. doi: 10.7326/0003-4819-158-3-201302050-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers AJ, Till C, Tangen CM, et al. An empirical evaluation of guidelines on prostate-specific antigen velocity in prostate cancer detection. J Natl Cancer Inst. 2011;103:462–469. doi: 10.1093/jnci/djr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin DW, Newcomb LF, Brown E, et al. Canary Prostate Active Surveillance Study Investigators. Urinary TMPRSS2:ERG and PCA3 in an active surveillance cohort: results from a baseline analysis in the Canary Prostate Active Surveillance Study. Clin Cancer Res. 2013;19:2442–2450. doi: 10.1158/1078-0432.CCR-12-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catalona WJ, Partin AW, Sanda MG, et al. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J Urol. 2011;185:1650–1655. doi: 10.1016/j.juro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Wang M, Wang L, et al. Diagnostic ability of %p2PSA and prostate health index for aggressive prostate cancer: a meta-analysis. Sci Rep. 2014;4:5012. doi: 10.1038/srep05012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fossati N, Buffi NM, Haese A, et al. Preoperative prostate-specific antigen isoform p2PSA and its derivatives, %p2PSA and prostate health index, predict pathologic outcomes in patients undergoing radical prostatectomy for prostate cancer: results from a multicentric European prospective study [published online ahead of print Aug. 16, 2014] Eur Urol. doi: 10.1016/j.eururo.2014.07.034. 10.1016/j.eururo.2014.07.034. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Avgeris M, Mavridis K, Scorilas A. Kallikrein-related peptidases in prostate, breast, and ovarian cancers: from pathobiology to clinical relevance. Biol Chem. 2012;393:301–317. doi: 10.1515/hsz-2011-0260. [DOI] [PubMed] [Google Scholar]
- 24.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 25.Denmeade SR, Sokoll LJ, Dalrymple S, et al. Dissociation between androgen responsiveness for malignant growth vs. expression of prostate specific differentiation markers PSA, hK2, and PSMA in human prostate cancer models. Prostate. 2003;54:249–257. doi: 10.1002/pros.10199. [DOI] [PubMed] [Google Scholar]
- 26.Steuber T, Nurmikko P, Haese A, et al. Discrimination of benign from malignant prostatic disease by selective measurements of single chain, intact free prostate specific antigen. J Urol. 2002;168:1917–1922. doi: 10.1016/S0022-5347(05)64263-2. [DOI] [PubMed] [Google Scholar]
- 27.Christensson A, Bruun L, Björk T, et al. Intraindividual short-term variability of prostate-specific antigen and other kallikrein markers in a serial collection of blood from men under evaluation for prostate cancer. BJU Int. 2011;107:1769–1774. doi: 10.1111/j.1464-410X.2010.09761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vickers AJ, Cronin AM, Aus G, et al. A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Göteborg, Sweden. BMC Med. 2008;6:19. doi: 10.1186/1741-7015-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benchikh A, Savage C, Cronin A, et al. A panel of kallikrein markers can predict outcome of prostate biopsy following clinical work-up: an independent validation study from the European Randomized Study of Prostate Cancer screening, France. BMC Cancer. 2010;10:635. doi: 10.1186/1471-2407-10-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers A, Cronin A, Roobol M, et al. Reducing unnecessary biopsy during prostate cancer screening using a four-kallikrein panel: an independent replication. J Clin Oncol. 2010;28:2493–2498. doi: 10.1200/JCO.2009.24.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickers AJ, Cronin AM, Roobol MJ, et al. A four-kallikrein panel predicts prostate cancer in men with recent screening: data from the European Randomized Study of Screening for Prostate Cancer, Rotterdam. Clin Cancer Res. 2010;16:3232–3239. doi: 10.1158/1078-0432.CCR-10-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vickers AJ, Cronin AM, Aus G, et al. Impact of recent screening on predicting the outcome of prostate cancer biopsy in men with elevated prostate-specific antigen; data from the European Randomized Study of Prostate Cancer Screening in Gothenburg, Sweden. Cancer. 2010;116:2612–2620. doi: 10.1002/cncr.25010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta A, Roobol MJ, Savage CJ, et al. A four-kallikrein panel for the prediction of repeat prostate biopsy: data from the European Randomized Study of Prostate Cancer Screening in Rotterdam, Netherlands. Br J Cancer. 2010;103:708–714. doi: 10.1038/sj.bjc.6605815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlsson S, Maschino A, Schröder F, et al. Predictive value of four kallikrein markers for pathologically insignificant compared with aggressive prostate cancer in radical prostatectomy specimens: results from the European Randomized Study of Screening for Prostate Cancer Section Rotterdam. Eur Urol. 2013;64:693–699. doi: 10.1016/j.eururo.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stattin P, Vickers AJ, Sjoberg DD, et al. Improving the specificity of screening for lethal prostate cancer using prostate-specific antigen and a panel of kallikrein markers: a nested case-control study [published online ahead of print Feb. 11, 2015] Eur Urol. doi: 10.1016/j.eururo.2015.01.009. doi: 10.1016/j. eururo.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parekh DJ, Punnen S, Sjoberg DD, et al. A multiinstitutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer [published online ahead of print Oct. 27, 2014] Eur Urol. doi: 10.1016/j.eururo.2014.10.021. doi: 10.1016/j.eururo.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 37.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ankerst DP, Hoefler J, Bock S, et al. Prostate Cancer Prevention Trial risk calculator 2.0 for the prediction of low- vs high-grade prostate cancer. Urology. 2014;83:1362–1367. doi: 10.1016/j.urology.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilt TJ, Brawer MK, Jones KM, et al. Prostate Cancer Intervention versus Observation Trial (PIVOT) Study Group, authors. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367:203–213. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bill-Axelson A, Holmberg L, Garmo H, et al. Radical prostatectomy or watchful waiting in early prostate cancer. N Engl J Med. 2014;370:932–942. doi: 10.1056/NEJMoa1311593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu X, Albertsen PC, Andriole GL, et al. Risk-based prostate cancer screening. Eur Urol. 2012;61:652–661. doi: 10.1016/j.eururo.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vickers AJ, Gupta A, Savage CJ, et al. A panel of kallikrein marker predicts prostate cancer in a large, population-based cohort followed for 15 years without screening. Cancer Epidemiol Biomarkers Prev. 2011;20:255–261. doi: 10.1158/1055-9965.EPI-10-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]