Abstract
Many microbial species form biofilms/mats under nutrient-limiting conditions, and fungal pathogens rely on this social behavior for virulence. In budding yeast, mat formation is dependent on the mucinlike flocculin Flo11, which promotes cell-to-cell and cell-to-substrate adhesion in mats. The biofilm/ mat assays described here allow the evaluation of the role of Flo11 in the formation of mats. Cells are grown on surfaces with different degrees of rigidity to assess their expansion and three-dimensional architecture, and the cells are also exposed to plastic surfaces to quantify their adherence. These assays are broadly applicable to studying biofilm/mat formation in microbial species.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
RECIPES: Please see the end of this protocol for recipes indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
Reagents
Crystal violet (1% w/v in H2O)
Distilled water, sterile
- Yeast strains of interest
- The Σ1278b background undergoes biofilm/mat formation (Gimeno et al. 1992). Commonly used laboratory strains have lost the ability to undergo biofilm/mat formation (Liu et al. 1996). A mutant defective for biofilm/mat formation (e.g., flo11Δ) should be included in the assay as a negative control.
- YEPD agar plates with varying agar concentrations (e.g., 0.3%, 2%, and 4%) <R>
- Do not invert 0.3% agar plates.
Equipment
Digital camera
Flat-end toothpicks, sterile
ImageJ software (http://imagej.nih.gov; Schneider et al. 2012)
Incubator set at 30°C
Light microscope with 100× objective
Nitrocellulose filters (circular)
Plastic wrap (e.g., Saran Wrap)
Polystyrene or polypropylene plate (96-well)
Spectrophotometer
METHOD
- Using a sterile toothpick, transfer cells of the yeast strain of interest to each of the plates listed below. Gently touch the toothpick containing cells to the center of each plate. On a separate set of plates, transfer cells of mutant strain defective for mat formation (a negative control; e.g., flo11Δ).
- YEPD plate with 0.3% agar
- 0.3% agar is optimal to observe colony spreading. On these plates, colonies can show a radial spoke pattern.
- YEPD plate with 2% agar
- 2% agar is optimal to observe mat architecture. It also allows assessment of invasive growth by the platewashing assay (see The Plate-Washing Assay: A Simple Test for Filamentous Growth in Budding Yeast [Cullen 2015]).
- YEPD plate with 4% agar
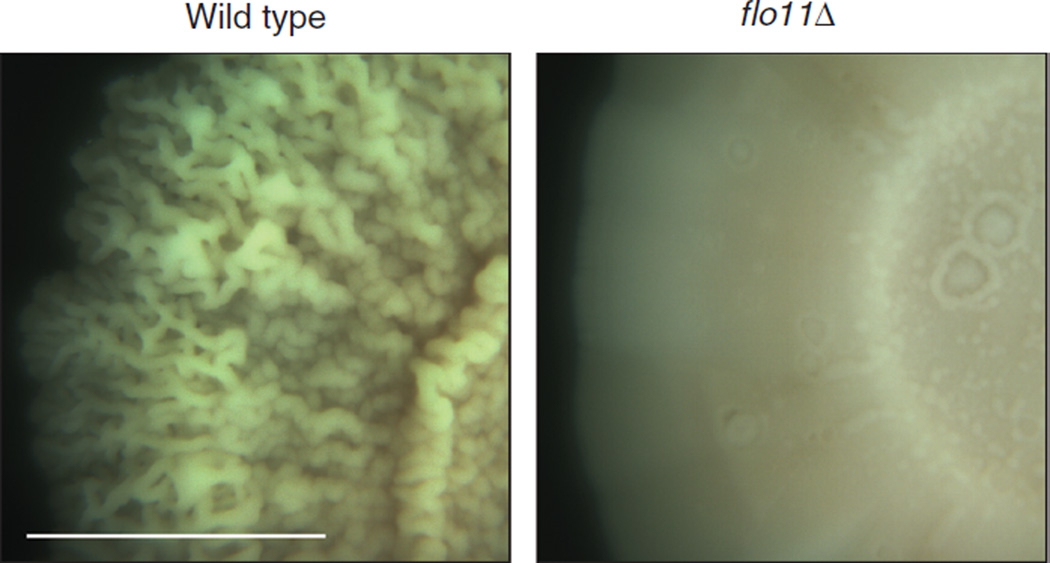
- 4% agar is optimal to observe colony ruffling and z-axis growth (see Fig. 1). The high surface rigidity reduces expansion in the plane of the xy-axis and promotes formation of dense architecturally complex mats that grow upward in the plane of the z-axis.
- YEPD plate with 4% agar plate with a nitrocellulose filter placed on top
- This plate maximizes complex colony morphology.
- Maintain the plates in a 30°C incubator in a location where vibrations are minimized. Examine mat expansion visually starting at 24 h and continuing over the course of several weeks.
- Mats can be defined by their FLO11-dependent colony architecture and degree of expansion in the x-, y-, and z-axes by visual inspection (e.g., see Fig. 1).
- Photograph the biofilms.
- Measure the mat areas by photographing the plates and analyzing the photos with ImageJ software (http://imagej.nih.gov; Schneider et al. 2012).
- Examine the ruffled morphology characteristic of mats by photography with a digital camera at 1× to 5× magnification.
- To separate adherent from nonadherent cells, use an overlay adhesion assay (Reynolds et al. 2008).
- Place plastic wrap over the agar plate, and gently pull the plastic wrap off the plate.
- When performing this action, the mat may or may not be removed from the agar. This distinguishes cells that adhere to the plastic from cells that adhere to the agar.
- Photograph both the plate and the wrap.
- (Optional) Remove cells for additional analysis.
- Quantify the degree of cell adhesion by measuring the adhesion of yeast cells to a plastic surface.
- At various times after mat formation has occurred, remove cells using a toothpick. Resuspend in 500 µL of H2O.
- Always compare the cells of interest with flo11Δ cells (a negative control).
- Adjust the optical density of the cells to A600 = 2.0.
- Add 100 µL of the cell suspension to a 96-well polystyrene or polypropylene plate.
- Incubate the cells for 4 h at 25°C to allow the cells to settle to the bottom of the wells.
- Add 100 µL of 1% crystal violet to each well. Incubate for 20 min at 25°C.
- Wash wells five times with water and photograph adherent cells by with a digital camera or by microscopy at 10×.
- Wild-type cells will adhere to the plastic surface and will be violet in color. In contrast, flo11Δ cells will not adhere, leaving a transparent plastic surface that is relatively free of cells.
FIGURE 1.
Sample results from a biofilm/mat assay. When grown on medium containing a high agar concentration (YEPD medium containing 4% agar), wild-type cells (left) produce a ruffled pattern, whereas flo11Δ mutant cells (right) do not. Bar, 1 cm. Magnification, 3×.
RELATED INFORMATION
For more discussion on the biology of biofilm/mat formation and applications of the assays described here, see Reynolds and Fink (2001), Blankenship and Mitchell (2006), and Karunanithi et al. (2012).
RECIPES
YEPD Agar Plates
| Reagent | Quantity |
|---|---|
| Bacto-agar (2%) | 20 g |
| YEPD liquid medium | 1 L |
Add Bacto-agar to YEPD liquid medium in a 2-L flask and autoclave. Fill sterile Petri dishes with 30–40 mL of autoclaved medium.
Yeast Extract-Peptone-Dextrose Growth Medium (YEPD)
| Reagent | Quantity (for 1 L) | Final concentration (w/v) |
|---|---|---|
| Bacto peptone | 20 g | 2% |
| Yeast extract | 10 g | 1% |
| Dextrose | 20 g | 2% |
| H2O | to 1 L |
Sterilize by autoclaving.
ACKNOWLEDGMENTS
The author thank T. Reynolds for engaging discussions about biofilm/mat form growth. P.J.C. is supported from a U.S. Public Health Service grant (GM098629).
REFERENCES
- Blankenship JR, Mitchell AP. How to build a biofilm: A fungal perspective. Curr Opin Microbiol. 2006;9:588–594. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Cullen PJ. The plate-washing assay: A simple test for filamentous growth in budding yeast. Cold Spring Harb Protoc. 2015 doi: 10.1101/pdb.prot085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeastS. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Karunanithi S, Joshi J, Chavel C, Birkaya B, Grell L, Cullen PJ. Regulation of mat responses by a differentiation MAPK pathway in Saccharomyces cerevisiae. PLoS ONE. 2012;7:e32294. doi: 10.1371/journal.pone.0032294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- Reynolds TB, Jansen A, Peng X, Fink GR. Mat formation in Saccharomyces cerevisiae requires nutrient and pH gradients. Eukaryot Cell. 2008;7:122–130. doi: 10.1128/EC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]