Abstract
Necrotizing enterocolitis (NEC) is a severe form of bowel disease that develops in premature infants. Although animal data and human studies suggest that aberrant activation of the intestinal immune system contributes to NEC, the pathogenesis remains unclear. We hypothesized that inherited defects in the regulation of Toll-like receptor signaling can contribute to NEC susceptibility in premature infants. A forward genetic screen done in an infant with lethal NEC using exome sequencing identified a novel stop mutation (p.Y168X) and a rare missense variant (p.S80Y) in SIGIRR, a gene that inhibits intestinal Toll-like receptor signaling. Functional studies carried out in human embryonic kidney cells and intestinal epithelial cells demonstrated that SIGIRR inhibited inflammation induced by lipopolysaccharide, a cell wall component of Gram-negative bacteria implicated in NEC. The genetic variants identified in the infant with NEC resulted in loss of SIGIRR function and exaggerated inflammation in response to lipopolysaccharide. Additionally, Sanger sequencing identified missense, stop, or splice region SIGIRR variants in 10 of 17 premature infants with stage II+ NEC. To the best of our knowledge, this is one of the first reports of a phenotype associated with SIGIRR in humans. Our data provide novel mechanistic insight into the probable causation of NEC and support additional investigation of the hypothesis that inherited defects in the regulation of innate immune signaling can contribute to NEC susceptibility in premature infants.
Necrotizing enterocolitis (NEC), a severe form of bowel disease that develops in 5% to 14% of premature infants, has a mortality rate of 25% to 40%.1 Although risk factors that contribute to mucosal injury (patent ductus arteriosus, hypoxia) or aberrant intestinal colonization (formula feeds) are implicated in NEC, its pathogenesis remains unclear.1–4 Animal data and human studies suggest that aberrant activation of intestinal immune responses by gut bacteria can trigger inflammation and mucosal injury in NEC.3–5 In the immature intestine, the Toll-like receptor (TLR) family of pathogen recognition receptors maintains the critical balance between bacterial tolerance and intolerance.3,6 We hypothesized that loss of function variants in genes that inhibit intestinal TLR signaling will contribute to NEC pathogenesis. In this study, we identified a potential candidate TLR pathway gene that can contribute to NEC susceptibility.
Methods
Infant Recruitment
Infants were recruited under the auspices of an institutional review board–approved study to investigate the impact of genetic variation on diseases of prematurity. After informed consent was obtained, a blood sample was collected for DNA extraction. Deidentified clinical data were stored in a password-protected database.
Exome Capture and Analysis
We extracted DNA from blood samples by using the FlexiGene DNA kit (Qiagen, Inc, Alameda, CA). We performed exome capture by using 5 μg of genomic DNA with the Agilent v4 Exome capture kit (Agilent, Inc, Santa Clara, CA). The captured fragments were sequenced on the HiSeq 2000 with Illumina’s TruSeq technology (Illumina, Inc, San Diego, CA) to an average depth of >40 reads per target base. Image files were processed into binary read files, transferred to an analysis server, and aligned to the human reference sequence hg19/GRCh37 via the Illumina CASAVA pipeline. Variants with respect to GRCh37 were identified and imported into our clinical laboratory’s variant annotation tool “Carpe Novo” and then annotated for functional impact.7
Statistical Analysis
Data are represented as mean ± SD. Changes in nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) activation with lipopolysaccharide (LPS) were expressed relative to controls and compared between groups through analysis of variance (ANOVA). For interleukin-8 (IL-8) protein levels, absolute values were compared between groups through ANOVA. Interleukin-6 (IL-6) and inducible nitric oxide synthase (iNOS) RNA levels were expressed as a fold-change relative to controls and analyzed with ANOVA. The post hoc Tukey test was used in conjunction with ANOVA to correct for multiple comparisons. A P < .05 was considered significant.
Modeling Approach to Demonstrate Increased Prevalence of SIGIRR Variants in Infants With NEC
To determine whether there is a greater prevalence of potentially deleterious SIGIRR variants in infants with NEC when compared with the general population, we used the strategy recommended by MacArthur et al.8 We constructed a variation site frequency spectrum (SFS) of SIGIRR variants from the exome variant server control cohort. The null model of the SFS considers identifying the pathogenic classes of variant present in that gene in the control cohort and calculating the probability of sampling variants of the same class of pathogenicity in the study population. Specifically, we evaluated the rate of predicted “null mutations,” splice region mutations (ie, a variant occurring within 10 bases of the canonical splice site), and missense variants predicted by PolyPhen-2 to be damaging to the SIGGR function. Among the 18 cases of NEC we identified 5 potentially deleterious variants (Table 1). Performing the same estimates on the exome variant server cohort, we identified 380 variants (Table 2). In the exome variant server cohort, not all loci had the same allele counts. Therefore, for statistical analysis we took the site with the lowest number of cases as the denominator. Performing a χ2 test (P < .001, Pearson’s χ2) we obtained a significance result.
TABLE 1.
Distribution of SIGIRR Variants in Infants With Stage II+ NEC
| Infant | Variant rs Number | Protein Change | MAF | GA, wk | Bwt, g | Gender | Race | Feed Type | PDA | NEC Stage | DOL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | Novel and rs117739035 | p.Y168X, p.S80Y | Novel, 0.02 | 23 | 560 | M | AA | BOTH | Yes | 3 | 51 |
| 2 | 29 | 1240 | F | AA | FM | No | 3 | 11 | |||
| 3 | rs111819059 | p.P115R | 0.006 | 26 | 680 | F | Cau | BM | Yes | 2 | 64 |
| 4 | 24 | 800 | M | Cau | BOTH | Yes | 2 | 30 | |||
| 5b | rs3210908 | p.Q312R | 0.18 | 30 | 1310 | M | Cau | BM | No | 2 | 18 |
| 6b | rs3210908 | p.Q312R | 0.18 | 30 | 1340 | M | Cau | BM | No | 2 | 14 |
| 7 | 24 | 660 | M | Cau | BOTH | Yes | 2 | 64 | |||
| 8 | rs3210908 | p.Q312R | 0.18 | 29 | 1475 | M | Cau | BM | No | 2 | 12 |
| 9 | 26 | 850 | M | AA | BOTH | No | 3 | 26 | |||
| 10c | Novel and rs117739035 | p.Y168×, p.S80Y | Novel, 0.02 | 23 | 630 | M | AA | BOTH | Yes | 2 | 61 |
| 11 | 28 | 1140 | F | AA | BOTH | No | 3 | 10 | |||
| 12 | rs111819059 | p.P115R | 0.006 | 24 | 580 | M | Cau | BM | No | 3 | 16 |
| 13 | 25 | 940 | M | Cau | FM | Yes | 3 | 14 | |||
| 14 | rs3210908 | p.Q312R | 0.18 | 24 | 780 | F | AA | BM | No | 3 | 13 |
| 15 | rs201897529d | Splice region variant | 0.002 | 25 | 780 | F | Cau | FM | Yes | 3 | 40 |
| 16 | rs111819059 | p.P115R | 0.006 | 24 | 300 | F | Cau | BM | Yes | 2 | 65 |
| 17 | 27 | 1025 | F | AA | FM | No | 3 | 13 | |||
| 18 | rs3210908 | p.Q312R | 0.18 | 29 | 1320 | F | Cau | BM | No | 2 | 14 |
AA, African American; BM, breast milk; BOTH, breast milk and formula feeds; Bwt, birth wt; Cau, Caucasian; DOL, day of life for NEC onset; FM, formula milk; GA, gestational age; PDA, patent ductus arteriosus; rs number, reference single nucleotide polymorphism number.
Infant 1 was the proband.
Another pair of twins.
Twin of infant 1.
Variant predicted by SpliceAid (http://www.introni.it/splicing.html) to alter exon splicing.16
TABLE 2.
Modeling Approach to Demonstrate Greater Prevalence of SIGIRR Variants in Infants With NEC
| Cohort | Total Tested Alleles | Missense Variants | Splice Region Variants | Stop Variants | Total Deleterious Alleles | Normal Alleles |
|---|---|---|---|---|---|---|
| NEC | 36 | 2 | 1 | 2 | 5 | 31 |
| Exome variant server | 12 317 | 352 | 27 | 1 | 380 | 11 937 |
SFS analysis was performed to compare frequency of deleterious SIGIRR alleles in our cohort with the exome variant cohort (see the Methods section for description). This revealed an increase in SIGIRR variants in the NEC cohort compared with the general population (P < .001).
Other Methods
These methods are described in the Supplemental Material.
Results and Discussion
SIGIRR Mutations in Proband With NEC
The proband was born at 23 weeks and 4 days’ gestation with a birth weight of 560 g. On day of life (DOL) 50, while on full enteral feeds, the infant developed apneas of increasing severity, with 1 large gastric residual warranting increase in respiratory support. On DOL 51, he was noted to have abdominal distension, and radiography showed diffuse pneumatosis intestinalis. Despite intensive medical care there was clinical deterioration warranting surgery, which revealed NEC totalis with pan-pneumatosis and necrosis. We performed whole exome sequencing followed by variant identification by using the Illumina Hi-Seq 2000 and a custom bioinformatics tool developed in our institution.7 We screened >19 000 exonic variants to identify complete loss-of-function variants. Variant prioritization based on gene function, relevance to immune signaling, and gastrointestinal disease identified 2 potentially deleterious variants in SIGIRR (NM_021805.2), a gene that inhibits TLR signaling.9,10 The stop variant (p.Y168X, c.504C>G) is novel because it is not found in >6200 individuals belonging to the National Heart, Lung, and Blood Institute exome cohort (snp.gs.washington.edu/EVS). The other variant (p.S80Y, c.239C>A, rs117739035) has a mean allele frequency (MAF) of ∼0.02. We performed Sanger sequencing to confirm the presence of both SIGIRR variants in our proband.
SIGIRR Variants Result in Unregulated TLR4-Mediated Inflammation
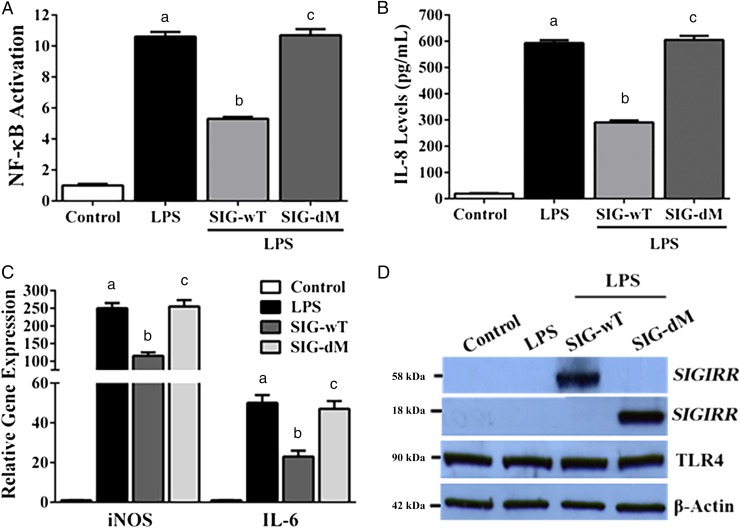
Activation of TLR4, which senses LPS from Gram-negative bacteria, is posited to be a central event in NEC pathogenesis.3,5,11 Therefore, we investigated whether SIGIRR variants dysregulated TLR4-mediated inflammation. We carried out functional analyses in human embryonic kidney cell line (HEK293) by using LPS. LPS-induced NF-κB activation (a sign of inflammation) and IL-8 protein expression at 20 hours was strongly inhibited in cells transfected with wild-type SIGIRR (SIG-wT) (Fig 1). Similarly, SIGIRR attenuated LPS-induced IL-6 and iNOS RNA expression at 8 hours (Fig 1). Transfection with mutant SIGIRR (SIG-dM) encoding both variants (p.Y168× and p.S80Y) identified in the infant with NEC abolished SIGIRR function and restored TLR4-mediated NF-κB activation, IL-8, IL-6, and iNOS expression (Fig 1). These data show that although SIGIRR inhibits inflammation mediated by bacterial ligands, the variants identified in our proband result in loss of regulation of LPS-mediated inflammation.
FIGURE 1.
Effect of SIGIRR variants on LPS-induced inflammation; HEK293 cells transfected overnight with plasmids encoding empty plasmid, the reference allele (SIG-wT), and both SIGIRR variants (SIG-dM) were treated with LPS (100 ng/mL). Culture supernatants or cell lysate protein were used for experiments. A, NF-κB activation was quantified in culture supernatants 20 hours after LPS, as described in the Methods section. Fold-increase in NF-κB activation relative to control is shown (n = 4). P < .05 for all comparisons. B, IL-8 protein was quantified 20 hours after LPS treatment in cell culture supernatants by enzyme-linked immunosorbent assay (n = 4). P < .05 for all comparisons. C, IL-6 and iNOS RNA expression were quantified in cell lysates by quantitative reverse transcription PCR 8 hours after LPS treatment. Fold-increase in expression relative to control is shown (n = 3). P < .05 for all comparisons. D, Whole cell-lysate protein (20 μg) obtained 20 hours after LPS treatment was immunoblotted for SIGIRR, TLR4, and β-actin (n = 4). Note: The stop variant in mutant SIGIRR plasmid (SIG-dM) encodes a truncated SIGIRR protein (18 kDa). aControl versus LPS. bLPS versus LPS + SIG-wT. cLPS + SIG-wT versus LPS + SIG-dM.
To investigate whether SIGIRR variants alter intestinal signaling, we performed studies in IEC-18, a nontransformed small intestinal epithelial cell line.12 Although SIGIRR (SIG-wT) inhibited LPS-induced NF-κB activation, IL-8, IL-6, and iNOS RNA expression, mutant SIGIRR (SIG-dM) abolished SIGIRR function and restored LPS responsiveness (Supplemental Fig 2). Furthermore, SIGIRR mediated inhibition of LPS-induced cleaved caspase-3 expression was abolished with mutant SIGIRR (Supplemental Fig 2). These data show that SIGIRR inhibits TLR4-mediated inflammation and apoptotic signaling in intestinal epithelial cells.
SIGIRR Variants in Other Infants With NEC
We sequenced the coding region of SIGIRR in 17 premature infants with stage II+ NEC. The distribution of clinical and epidemiologic variables in infants with NEC is shown in Table 1. The twin of our proband who survived stage II NEC had both the p.Y168× and p.S80Y variants. Five infants had the missense p.Q312R variant (rs3210908, MAF = 0.18), 3 had the rare missense p.P115R variant (rs111819059, MAF = 0.006), 1 had the rare splice region variant (rs201897529, MAF = 0.002), and the other 7 did not have missense, splice, stop, or frameshift exonic SIGIRR variants. We modeled the SFS approach to compare the prevalence of potentially deleterious SIGIRR variants among NEC infants (n = 18) with the exome variant server cohort.8 This approach revealed a significant excess of deleterious SIGIRR variants (P < .001) in our cohort compared with the general population (Table 2). Additionally, we sequenced DNA from 20 preterm infants without NEC. We did not find the novel (p.Y168×), rare exonic (p.S80Y, p.P115R), or rare splice region variant (rs201897529). Only the benign, common missense variant (p.Q312R) was found in 4 infants. These data show that functional or rare SIGIRR variants are more common in infants with NEC.
SIGIRR and NEC
SIGIRR is a major negative regulator of intestinal inflammation mediated by TLRs.9,10,13 However, to the best of our knowledge this is one of the first reports of a phenotype associated with SIGIRR in humans. Of the 2 variants identified in our proband and his twin, the stop variant (p.Y168×) has not been found among the genome of >6000 adults. Although the twin of our proband also had both the SIGIRR variants, he survived stage II NEC. Whether differences in their outcomes were caused by presence of other clinical risk factors or the effect of modifier genes remains unclear. Additionally, we found both rare and common missense or splice region variants in 9 of 16 infants with NEC. By comparing the frequency of SIGIRR variants in our cohort with the general population, we found more SIGIRR variants in infants with NEC. Furthermore, among 20 premature infants without NEC, we did not find functional or rare SIGIRR variants. NEC is a complex disease involving interactions between clinical variables, developmental immaturity, gut microbiota, and genetic factors.1 Although our data support a role for SIGIRR variants in NEC, additional studies are needed to evaluate how SIGIRR variants interact with known risk factors to cause disease.
Our functional data suggest that SIGIRR variants may contribute to NEC through loss of inhibition of TLR4-mediated inflammation. Activation of TLR4-mediated inflammation has been implicated in NEC pathogenesis in rodent models and humans.5,14 Though consistent with these studies, our data suggest that genetic factors can contribute to selective activation of innate immune pathways in infants with NEC. Similarly, a role for reduced SIGIRR function in NEC has been suggested by Nanthakumar et al,4 who showed less SIGIRR expression in ileal tissue resected from infants with NEC. We speculate that mucosal injury and gut bacteria elicit unregulated TLR-mediated inflammation in infants with SIGIRR variants causing NEC.15
Conclusions
Our data provide novel insight into the probable causation of NEC and support the hypothesis that inherited defects in genes that inhibit intestinal innate immune signaling can contribute to NEC. Adequately powered studies are needed to study the impact of SIGIRR variants and variant–clinical variable interactions in the causation of NEC.
Supplementary Material
Acknowledgments
We thank Dr Xiaoxia Li, PhD (Cleveland Clinic Foundation, Cleveland, OH), for the wild-type SIGIRR plasmid and Dr James Verbsky, MD, PhD (Medical College of Wisconsin, Milwaukee, WI), for the IL-8 antibody. We also thank Dr John M. Routes, MD (Medical College of Wisconsin, Milwaukee, WI), for general guidance. We thank Laura Lane, BSN, and Kathleen Meskin, BSN, for help with clinical data collection.
Footnotes
Dr Sampath provided the overall concept and design, supervised the experiments, and drafted the manuscript; Dr Dimmock provided the overall study concept and design and reviewed and revised the manuscript; Dr Ramchandran acquired data, reviewed and revised the manuscript, and supervised certain aspects of the study; Ms Menden acquired, analyzed, and interpreted the data and wrote the manuscript; and Mr Helbling, Dr Gastonguay, and Dr Li acquired, analyzed, and interpreted the data.
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: V.S. and H.M. are supported in part by grant 8KL2TR000056, Clinical and Translational Science Institute of Southeast Wisconsin, and V.S. is supported by Children’s Research Institute funds. A.G., K.L., and R.R. are partially supported by National Institutes of Health grants HL09712, HL102745, and HL112639 and by Children’s Research Institute startup funds. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med. 2011;364(3):255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caplan MS, Simon D, Jilling T. The role of PAF, TLR, and the inflammatory response in neonatal necrotizing enterocolitis. Semin Pediatr Surg. 2005;14(3):145–151 [DOI] [PubMed] [Google Scholar]
- 3.Afrazi A, Sodhi CP, Richardson W, et al. New insights into the pathogenesis and treatment of necrotizing enterocolitis: toll-like receptors and beyond. Pediatr Res. 2011;69(3):183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nanthakumar N, Meng D, Goldstein AM, et al. The mechanism of excessive intestinal inflammation in necrotizing enterocolitis: an immature innate immune response. PLoS ONE. 2011;6(3):e17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sodhi CP, Neal MD, Siggers R, et al. Intestinal epithelial toll-like receptor 4 regulates goblet cell development and is required for necrotizing enterocolitis in mice. Gastroenterology. 2012;143(3):708–718.e1–e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santaolalla R, Abreu MT. Innate immunity in the small intestine. Curr Opin Gastroenterol. 2012;28(2):124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–262 [DOI] [PubMed] [Google Scholar]
- 8.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wald D, Qin J, Zhao Z, et al. SIGIRR, a negative regulator of toll-like receptor–interleukin 1 receptor signaling. Nat Immunol. 2003;4(9):920–927 [DOI] [PubMed] [Google Scholar]
- 10.Garlanda C, Anders HJ, Mantovani A. TIR8/SIGIRR: an IL-1R/TLR family member with regulatory functions in inflammation and T cell polarization. Trends Immunol. 2009;30(9):439–446 [DOI] [PubMed] [Google Scholar]
- 11.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801 [DOI] [PubMed] [Google Scholar]
- 12.Ma TY, Hollander D, Bhalla D, Nguyen H, Krugliak P. IEC-18, a nontransformed small intestinal cell line for studying epithelial permeability. J Lab Clin Med. 1992;120(2):329–341 [PubMed] [Google Scholar]
- 13.Xiao H, Gulen MF, Qin J, et al. The toll-interleukin-1 receptor member SIGIRR regulates colonic epithelial homeostasis, inflammation, and tumorigenesis. Immunity. 2007;26(4):461–475 [DOI] [PubMed] [Google Scholar]
- 14.Jilling T, Simon D, Lu J, et al. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J Immunol. 2006;177(5):3273–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev. 2012;249(1):158–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piva F, Giulietti M, Burini AB, Principato G. SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Hum Mutat. 2012;33(1):81–85 [DOI] [PubMed] [Google Scholar]
- 17.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.