Abstract
Rice is a staple and widely grown crop endowed with rich genetic diversity. As it is difficult to differentiate seeds of various rice varieties based on visual observation accurately, the harvested seeds and subsequent processed products are highly prone to adulteration with look-alike and low quality seeds by the dishonest traders. To protect the interests of importing countries and consumers, several methods have been employed over the last few decades for unambiguous discrimination of cultivars, accurate quantification of the adulterants, and for determination of cultivated geographical area. With recent advances in biotechnology, DNA based techniques evolved rapidly and proved successful over conventional non-DNA based methods to purge the problem of adulteration at commercial level. In the current review, we made an attempt to summarize the existing methods of adulteration detection and quantification in a comprehensive manner by providing Basmati as a case study to enable the traders to arrive at a quick resolution in choosing the apt method to eliminate the adulteration practice in the global rice industry.
Keywords: Adulteration, Basmati rice, Microsatellite, SSR markers
Introduction
Rice, being a staple food crop for over one third of the world’s population, has become a potential target for many unscrupulous traders for mixing with low grade, low cost grains/products and low nutritious adulterants to fetch profits with least efforts. In rice, thousands of varieties have been released worldwide since last century predominantly after the Green revolution in the 1960s (Siddiq et al. 2012). These varieties are not only adapted to diverse ecologies but also suits the tastes of the gamut of consumers. Attempts to develop superior quality varieties/products always ended with some of the poor quality ones. This led to existence of different quality standards with obvious price difference in the market (Nagaraju et al. 2002). This situation tempted the dishonest traders to go for adulteration of the genuine products to fetch additional profits.
Authenticity of rice products become a key issue in the food industry addressed to protect the interests of quality conscious consumers, stakeholders, and importing countries (Vlachos and Arvanitoyannis 2008). Adulteration in rice either adventitiously or deliberately is feasible right from crop harvest to till the grain reaches to the hands of the consumers. The common forms of rice prone to adulteration are brown rice, polished rice, rice flour, rice cake and rice bran oil. Many methods based on criteria such as morphological parameters, physico-chemical properties, DNA, protein, and metabolites have been developed to detect the genuineness of the agricultural food products (Arvanitoyannis 2008; Findlay et al. 1997; Li and Rutger 2000; Thind and Sogi 2005; Vaingankar and Kulkarni 1989; Zhang et al. 1992), yet there is an ambiguity in choosing one of them for commercial scale detection and quantification of adulteration. However, a few methods have proven fit to unravel the menace of adulteration (Vemireddy et al. 2007). With the availability of whole genome sequence of rice including indica (9311) and japonica (Nipponbare) cultivars, thereby resulted discovery of molecular markers have provided unprecedented avenues in the quality control of food products. Currently, DNA-based methods proved handy and robust enough for unambiguous detection and quantification of adulteration (Primrose et al. 2010; Woolfe and Primrose 2004). However, these methods are unsuitable to detect the geographical area of the product from where the product has been produced/cultivated. Geographical area of cultivation can be determined by isotopic and multi element analyses of rice varietal samples (Kelly et al. 2002).
The aim of this review was to provide an overview of the currently employed protocols for adulteration detection and quantification emphasizing the importance of DNA based protocols for eradicating this vital problem from the global rice industry with special reference to Basmati.
Methods for detection of adulteration
Adulteration is rife in almost all agricultural food products where distinguishing the adulterant with look-alike food products is difficult with naked eye/visual observation. Many of the food products targeted for adulteration are of high commercial value products and/or produced in high tonnage around the world. In any food product, adulteration may be due to 1) substitution with look-alike material of low cost, 2) substitution with low quality material, 3) dilution of the original product, and, 4) mislabelling of age and origin of the material. These are the four major criteria being followed by most of the unscrupulous traders for illegal adulteration to obtain profits out of their products. However, rice has largely been sold either as a brown rice form (exporting purpose) or polished rice form (available in domestic markets). Hence, high quality rices are being adulterated with low quality and low price rices by the traders. For instance, the good quality Basmati rices are being blended with look alike low cost non-Basmati rices.
There are many sophisticated methods or protocols being employed over the past decades for unambiguous detection and precise quantification of the adulterant using DNA or protein or metabolites or chemical composition of the material being marketed. However, no one method detects the adulterants in all agricultural products. Different combination of methods/protocols are necessary to curb this menace effectively. Broadly, the methods employed for detection and quantification of adulteration are classified as non-DNA based and DNA based methods depending on the source material used.
Non-DNA based methods
Morphology and physico-chemical based methods
In the past, morphology based methods were the criterion for differentiating various rice groups (Thind and Sogi 2005; Vaingankar and Kulkarni 1989). Significant variation in price within similar kind of kernel morphology, made this criterion unsuccessful for differentiating the cultivars. For instance, premium Basmati rice has long grain and soft texture. There are many long grain rice varieties of low cost with kernel morphology of Basmati but with quality unmatched to it (Fig. 1). To determine the morphology of kernel traits, techniques such as electron microscopy (Beerh and Srinivas 1991) and image analysis (Carter et al. 2006; Kim et al. 1997) were used (Table 1). In addition, histological variation was also used to distinguish aromatic and non-aromatic rice varieties (Nadaf et al. 2006). Large to medium size metaxylem and phloem was reported in medium and non-scented varieties, while in strongly scented rice, metaxylem was much smaller in size. In aromatic rice varieties, normal phloem and xylem elements were found to be replaced by a largely undifferentiated paranchymatous cells. However, the morphology or histological based techniques were expensive, cumbersome and unfit for commercial scale detection of the adulterants.
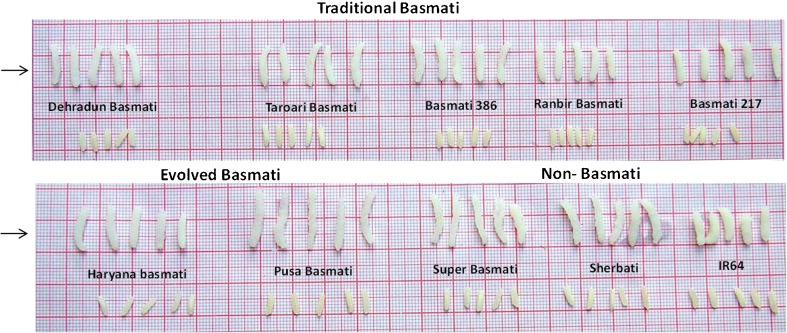
Fig. 1.
Comparison of grain shape (before and after cooking) among traditional and evolved Basmati and non-basmati varieties. Traditional Basmati varieties- Dehradun basmati, Taraori basmati, Basmati 386, Ranbir basmati and Basmati 217; Evolved Basmati varieties- Haryana basmati, Pusa basmati-1 and Superbasmati; Non-Basmati varieties- Sherbati and IR64. The length of the each thick sqare box is 10 mm
Table 1.
List of non-DNA based methods for detection of adulteration in rice
| Method | Methodology | Component tested | Variety | Target component/Property studied | Reference |
|---|---|---|---|---|---|
| Morphology based methods | Length/Breadth ratio | Grain | Basmati and coarse, fine and super fine rice | Length and breadth ratio | Vaingankar and Kulkarni 1989; Thind and Sogi 2005 |
| Imaging system (using mirrors) | Kernels | Japonica rice | Area, diameter, perimeter of each kernel | Carter et al. 2006 | |
| Electron microscopy | Grain | Basmati rice | Histological factors | Beerh and Srinivas 1991 | |
| Histochemical analysis | Caryopsis | Basmati and other scented rice (Pusa and Ghansal) | Metaxylem and phloem elements and aroma | Nadaf et al. 2006 | |
| Cell Wall Material (CWMs) | Grains | Waxy rice and Non-waxy rice | Water solubility, CWMs, Sugars (ratio of arabinose to xylose) | Lai et al. 2007 | |
| Physico-chemical bsed methods | Kernel elongation | Grains | Basmati and NonBasamti rice | Kernel elongation | Siddiq 1982 |
| Cooking property | Grains | Orysa sativa | Hot water insoluble amylose | Bhattacharya 1982 | |
| Cooking Property | Grain | Basmati rice | Length increase upon cooking | Hirannaiah et al. 2001 | |
| Cooking property | Grains | Basmati and look-alike adulterants | Cooking behaviour, water uptake etc. | Vaingankar and Kulkarni 1989 | |
| Cooking property | Grains | Coarse IR8, Fine PR106 and Super fine Basmati 386 | Grain size, globulin, glutelin content | Thind and Sogi 2005 | |
| Cooking Quality with Wards Cluster Analysis | Grains | Basmati rice | Cooking and processing properties, amylose, proteins, flavour and texture | Bett-Garber et al. 2001 | |
| Aroma | Grain | Scented rice | Potassium hydroxide based test | Sood and Siddiq 1978 | |
| Aroma | Grain | Aromati, Basmati and Non Basmati rice | 2-acetyl-1-Pyrroline | Bergman et al. 2000 | |
| Aroma | Grain | Basmati rice and Aromatic rice | 2-acetyl-1-Pyrroline | Bhattacharjee et al. 2002 | |
| Solid Phase microextraction (SPME) | Grain | Jasmine rice | 2-acetyl-1-Pyrroline | Grimm et al. 2001 | |
| Alkaline Oxidation | Kernels | Basmati, Scented and Non-scented rice (American long grain rice) cultivar | Volatiles, Flavor | Agrawal and Sinha 1965 | |
| Alkaline oxidation | Stored grains | White and Brown fragrant rice (different storage periods) | Volatile compounds | Widjaja et al. 1996 | |
| Lipid Oxidation | Grain | Aromatic Brown rice | 2-acetyl-1-Pyrroline | Buttery et al. 1983 | |
| High Pressure Liquid Chromatography (HPLC) | Endosperm, Grains | Rice cultivars | Proteins | Huebner et al. 1990; Hamada 1996 | |
| Gas Chromatography Mass Spectrometry (GC MS) | Grains | Brown rice | Lipoxygenease based flavor | Suzuki et al. 1999 | |
| Gas Chromatography | Normal germinated rice | Rice | Antioxidant activity (ferulic and coumaric acids) | Rao and Muralikrishna 2004 | |
| Headspace Chromatography (HS GC) Nitrogen Phosphorus Detector (NPD) | Grain | Fragrant rice | Aroma | Sriseadka et al. 2006 | |
| Differential Scanning Calorimetry | Long grain rice | Basmati 370 | Starch gelatinization degree | Ahmed et al. 2008 | |
| Mid-infrared (MIR) Spectroscopy | Cell wall components | Oryza sativa | Suberin | Cai et al. 2011 | |
| Largo-Gosens et al. 2014 | |||||
| Near-infrared (NIR) Spectroscopy | Grain | Basmati and other long grain rice | Spectral data | Osborne et al. 1993 | |
| Near-infrared (NIR) Spectroscopy | Milled grain | Traditional and aromatic Italian rice | Textural changes of rice gels | Mariotti et al. 2009 | |
| Near-infrared (NIR) Spectroscopy | Milled grain | Rice cultivars (Koshihikari and Akitakomachi) | Protein histogram | Rittiron et al. 2005 | |
| Fourier Transform (FT) Near-infrared (NIR) Spectroscopy | Whole grain | Rough rice varieties | Cooling and processing quality of rice | Attaviroj et al. 2011 | |
| Flame Atomic Absorption Spectroscopy (FAAS) | Diets and Cereals | Brown and White rice | Content of minerals and trace elements | Srikumar 1993 | |
| Chemometric and Calibration of FT Raman (NIR-FT/Raman) Spectroscopy | Milled rice | Small, medium and long grain rice | Protein content | Himmelsbach et al. 2001 | |
| Fluorescence and UV Spectroscopy | Grains | Long and medium rice | Surface lipid content | Gangidi et al. 2002 | |
| Near-infrared spectroscopy (NIR) and Modified Partial Least Square (MPLS) regression | Grains | Korean domestic rice and Foreign rice | Images | Kim et al. 2003 | |
| Near-infrared (NIR) Reflectance Spectroscopy | Stored seed | Rice | Spectral data from seed of different storage years | Dengsheng and Xiaoli 2007 | |
| 1H Nuclear Magentic Resonance (NMR) Spectroscopy and chemometric tools | Grains | Basmati, Non Basmati long grain and round grain rice | Geographical origin based on trace elements | Monakhova et al. 2013 | |
| Spectrophotometry | Seed | Mixture of rice bran and mustard oil | Visible spectra | Jha 1980 | |
| Stable Isotope carbon Analysis | Rice vinegar | Rice vinegar and Cane sugar | 13C value of acetic acid content | Kanno et al. 1989 | |
| Near-infrared (NIR) spectrometry | Rice wine | Shaoxing and non Shaoxing rice wines | Spectral data | Shen et al. 2010 | |
| Fourier Transform Near-infrared (FT NIR) NIR spectroscopy | Rice wine | Chinese rice wine | Tartartic acid | Yu et al. 2007 | |
| Marker based | Vegetable oil | Vegetable oil | 2-acetylfuran −3 glucopyranoside | Xue et al. 2013 | |
| Protein based methods | Acrylamide gel electrophoresis | Grains | Subspecies of Oryza sativa, namely, indica, japonica and javanica, and a group of intermediate types from North East India, | soluble proteins | Siddiq et al. (1972) |
| Isozymes based gel electrphoresis | Grains | 1,688 traditional rices from Asia | Enzymes | Glaszman 1987 | |
| Sodium Dodecyl Sulfate Polyacrylamide (SDS PAGE) | Seeds | Basmati 370, Brazilian and Japanese Upland rice | Protein separation | Montalvan et al. 1998 | |
| Western blotting | Grain | Basmati rice and look-alike adulterants | Aleurone specific profile | Singh et al. 2004 | |
| Western blotting | Rice tissues | Oryza sativa | Heat Shock Proteins and Elongation factor | Li et al. 2011 | |
| Kjeldahl Method and Soxhlet Apparatus | Grains | Rice cultivars of Brazil | Ash, Crude Fat, Crude Protein, Nitrogen | Storck et al. 2005 |
Physico-chemical properties of starch such as water uptake, loss of solids in cooking water (Vaingankar and Kulkarni 1989), protein characteristics (Thind and Sogi 2005), and grain length after cooking (Siddiq 1982) were also used for cultivar identification and quantification of adulteration. Aside, scent determination of Basmati rices by treating with 1.7 % potassium hydroxide (KOH) solution (Sood and Siddiq 1978) and quantification of aromatic compounds by chromatography (Lorieux et al. 1996; Widjaja et al. 1996) were also used. Water solubility and sugar compositions of the rice cell wall materials (CWMs) to also reposted to be differentiated the rice cultivars (Lai et al. 2007). In another study, Principal Component Analysis (PCA) and multiple linear regression analysis has been applied to identify the level of Jasmine rice adulteration using textural and pasting measurements and suggested to use physico-chemical properties in combination with multivariate analysis for adulteration detection (Pitiphunpong et al. 2011). Also, Thind and Sogi (2005) used physical, cooking and protein characters as the authenticity criterion for distinguishing coarse (IR8), fine (PR106) and super fine (Basmati386) rice cultivars and inferred that length-breadth (LB) ratio and globulin and glutelin contents are differentiating these three cultivars.
The chemical causing the popcorn like smell in all aromatic rice cultivars identified as 2-acetyl-1-pyrroline three decades ago by Buttery et al. (1983), hence, also used to distinguish popular aromatic varieties from non-aromatic look alike long grain as obvious price difference exists between them. However, the aroma alone may not be sufficient to differentiate popular Basmati rices from another look alike non-aromatic varieties like Sharbati, a common adulterant in Basmati export as the former has other unique cooking and textural properties which no other rice group has.
Osborne et al. (1997) reported that it was feasible to use near infrared transmission (NIR) spectroscopy to classify 9 Basmati or other rice samples on 200 g bulk samples. However, NIR spectra of individual grains misclassified 8 % of basmati and 14 % of the other rices. Subsequently, a number of analytical techniques such as Gas Chromatography (Rao and Muralikrishna 2004), Gas chromatography in conjunction with Mass Spectrometry (Suzuki et al. 1999), High Pressure Liquid Chromatography (Huebner et al. 1990; Hamada 1996), Calorimetry (Ahmed et al. 2008), Mid or Near-Infrared (NIR) Spectroscopy (Largo-Gosens et al. 2014; Osborne et al. 1993), Fourier Transform NIR Spectroscopy (Attaviroj et al. 2011), Fluorescence and UV spectroscopy (Gangidi et al. 2002), Flame Atomic Absorption Spectroscopy (Srikumar 1993), have been used for cultivar discrimination either alone or in conjunction with the application of chemometric or multivariate analysis methods like Principal Component Analysis, Discriminant Analysis, and Cluster Analysis, and Partial Least Squares (Singhal et al. 1997; Vlachos and Arvanitoyannis 2008) (Table 1).
Though the physico-chemical methods are simple and cost effective, none of them are found suitable for reliable detection of adulteration at commercial level, as these traits are highly influenced by environment as well as the genotype of the plant. This situation prompted researchers to develop approaches which confer more stable results.
Protein based methods
Among non-DNA based methods, protein-based methods also played important role in detection of the adulterants. Important protein-based methods includes isozymes based gel electrphoresis, immunoassays (Vlachos and Arvanitoyannis 2008), Sodium dodecyl sulfate polyacrylamide gels (SDS-PAGE) (Montalvan et al. 1998), Western blotting (Li et al. 20011; Singh et al. 2004) and Kjeldahl method and Soxhlet apparatus (Storck et al. 2005) (Table 1).
The protein-based methods especially of practical value in grouping of the cultivars rather than identification of the specific cultivars. Initially, Siddiq et al. (1972) reported variation in soluble proteins of three subspecies of Oryza sativa, namely, indica, japonica and javanica, and a group of intermediate types collected from North East India, using acrylamide gel electrophoresis. Subsequently, six rice varietal groups which includes two major groups viz., Group I (indica) and Group VI (japonica), two minor groups Group II (aus) and V (Basmati) and two satellite ones Group III and IV (deepwater rices) have been reported using eight isozyme markers detected by starch gel electrophoresis (Glaszman 1987).
Montalvan et al. (1998) by means of SDS-PAGE separated the rice grain proteins from Brazilian and Japanese upland rice cultivars electrophoretically. Densitometric scanning of the electrophoretic profiles allowed the estimation of the relative concentration of protein fractions, which were used as variables between the rice cultivars. carried out. Western blotting also used to detect the expression of target proteins in rice samples representing different rice tissues at different developmental stages (Singh et al. 2004; Li et al. 2011).
DNA-based methods
Of all the methods of adulteration detection, DNA-based techniques have wider acceptability due to their simplicity, accessibility, repeatability and rapidness. Advances in the development of DNA markers and cost effective sequencing techniques, renders DNA based methods more popular.
Molecular markers systems
The availability of thousands of DNA markers and cheaper sequencing methods offer unprecedented applications of DNA based methods to unravel the authentication crisis in food industry in general and rice market in particular (Arvanitoyannis 2008; Blair et al. 2002; Lockley and Bardsley 2008; Dhanya and Sasikumar 2010; Saini et al. 2004; Voorhuijzen et al. 2012) (Table 2). Wide array of DNA-based markers are available for cultivar identification which includes random amplified polymorphic DNA (RAPD) Choudhury et al. (2001), restriction fragment length polymorphism (RFLP) (Zhang et al. 1992), amplified fragment length polymorphisms (AFLP) (Mackill et al. 1996), fluorescent labelled inter simple sequence repeat (F-ISSR) Nagaraju et al. (2002), insertion and deletions (indels) (Steele et al. 2008), single nucleotide polymorphism (SNP) (Shirasawa et al. 2006) and microsatellites (Bligh 2000; Archak et al. 2007; Vemireddy et al. 2007) (Table 2). Each type of marker has its own pros and cons in their applicability. Of all the markers employed for varietal identification, microsatellite or simple sequence repeat (SSR) markers are the most informative in terms of reproducibility, genetic informativeness and applicability (McCouch et al. 2002; Vemireddy et al. 2007; Ashfaq and Khan 2012). Microsatellites, a short stretch of tandomly repeated region of less than 100 bp, have many advantages over other markers like abundance, co-dominant nature, PCR based markers and highly variable. As of now, more than 50,000 microsatellite markers are on hand in the rice. Microsatellite DNA markers have been extensively used in mapping and tagging genes, study of diversity, phylogeny and population genetics, disease diagnosis, and forensic investigations (Coburn et al. 2002; Srividhya et al. 2010; Yadav et al. 2013; Choudhury et al. 2013; Vemireddy et al. 2007). In addition, microsatellite markers have the potential to be used for unequivocal detection and quantification of adulteration in Basmati rice (Bligh 2000; Archak et al. 2007; Vemireddy et al. 2007). Recently, evenly-distributed hypervariable microsatellite markers were reported to be showed more polymorphism than non-hypervariable SSRs (Narshimulu et al. 2011) and also used for estimating the temporal trends of the genetics diversity over decadal periods (Choudhury et al. 2013).
Table 2.
Discrimination of rice cultivars by DNA markers
| Marker type | Fragment separation method | DNA Markers | Rice Cultivars | Reference |
|---|---|---|---|---|
| RFLP | Agarose | 43 RFLP probes | Indica and Japonica varieties | Zhang et al. (1992) |
| RAPD | Agarose | OPA 13, OPA 16, OPA 17, OPA 19, OPB 8, OPB11, OPB 13, OPB 18, OPN 1 and OPN 17 | Basmati varieties | Choudhury et al. (2001) |
| Microsatellites | Slab-gel | RM1,RM19,RM208,RM55,RM201,RM219,RM225,RM229,RM202,M2 and M16 | Basmati varieties | Bligh (2000) |
| Microsatellites | Slab-gel | RM163, RM171, RM72, RM238, RM102, RM16, RM302, RM330 and RM16 | Basmati varieties | Nagaraju et al. (2002) |
| Microsatellites | Agarose | RM1, RM21, RM38, RM170, RM210, RM226 and RM229 | Basmati varieties | Siwach et al. (2004) |
| Microsatellites | Slab-gel | RM1, RM5, RM103, RM135,RM171, RM174, RM222 and RM252 | Basmati varieties | Jain et al. (2004) |
| Microsatellites | Polyacrilamide gel | RM1,RM5,RM44,RM110,RM229, RM234,RM171,RM242 and RM255 | Basmati varieties | Pal et al. (2004) |
| Microsatellites | Capillary electrophoresis | RM1,RM171,RM202,RM44,RM55,RM348,RM72 and RM241 | Basmati varieties | Archak et al. (2007) |
| Microsatellites | Agarose, Slab-gel and Capillary electrophoresis | RM1,RM171,RM202,RM44,RM55,RM348,RM72 and RM241 | Basmati varieties | Vemireddy et al. (2007) |
| Microsatellites | Agarose | RM11/147 | BR-11 | Rahman et al. (2007) |
| RM151/289 | Badshabhog | |||
| RM153/178 | BR-19 | |||
| Microsatellites | Agarose | RM70, RM334, RM475, RM219, RM206, RM336, RM547, RM164, RM335, and RM276 | Parental lines and hybrids and some popular varieties | Sundaram et al. (2008) |
| Microsatellites | Agarose | RM 206, RM 216, RM 258 and RM263 | Hybrid purity | Nandakumar et al. (2004) |
| Indels | Agarose | R4M17,R4M43,R4M50,R5M13,R5M30,R6M14,R10M30,R11M23 and R12M27 | Basmati varieties | Steele et al. 2008 |
| SCAR | Agarose | B1,B7,B43,B43a,E30,F6,G22,M2CG,M11,S13 and Wka9 | Koshihikari in processed rice products | Ohtsubo and Nakamura (2007) |
| SNP | Dot-blot | 8 SNPs | 43 rice cultivars | Shirasawa et al. (2006) |
| STS | Agarose | A7, B43, M11 and G22 | Cultivars of rice wine | Ohtsubo et al. (2008) |
| STS | Agarose | PRR | Rice specific primer | Nakamura and Ohtsubo (2010) |
The fluorescent labelled microsatellite markers provide more accurate allele sizes when run in combination with either slab-gel or capillary electrophoresis. In a first attempt, Bligh (2000) used the fluorescent simple sequence length polymorphisms (SSLPs) to distinguish between known Basmati rice cultivars and likely adulterants by providing good distinction and separation levels in blind test samples using competitive nature of the fluorescence. Shortly, Nagaraju et al. (2002) used the fluorescent labelled microsatellite and ISSR (Inter-Simple- Sequence-Repeat) markers to reveal genetic relationships in traditional (TB) and evolved Basmati (EB) and semidwarf non-Basmati (NB) rice varieties. The traditional Basmati (TB) and semidwarf NB rice varieties were clearly delineated by both marker assays. Subsequently, the genetic relationships among Indian aromatic and quality rice germplasm were evaluated by Jain et al. (2004) using thirty fluorescently labelled rice microsatellite markers. Of them, a subset of eight markers viz., RM1, RM5, RM103, RM135, RM171, RM174, RM222, RM252 could differentiate the premium traditional Basmati, cross-bred Basmati, and non-Basmati rice varieties having different commercial value in the market-place. Also, Pal et al. (2004) evaluated thirteen rice cultivars viz., 4 commercial traditional Basmati, 6 cross-bred Basmati and 3 non-Basmati varieties using 35 SSR markers. Some SSRs viz., RM60, RM84, RM252, RM171, and RM257 were found unique among the closely related traditional Basmati rice varieties. Traditional Basmati rice varieties could be differentiated from one or more of the cross-bred Basmati rice varieties by allelic polymorphism at 27 of the 35 SSR loci; the most useful markers being RM171, RM1, RM44, RM110,RM229,RM235,RM242, and RM255. A capillary electrophoresis-based multiplex microsatellite marker assay for detection as well as quantification of adulteration in Basmati rice samples has been developed. The single-tube assay multiplexes eight microsatellite loci viz., RM1, RM44, RM55, RM72, RM171, RM202, RM241 and RM348 to generate variety-specific allele profiles that can detect adulteration from 1 % upwards with accuracy of quantification ±1.5 % Archak et al. (2007) and Vemireddy et al. (2007).
Recently, a DNA-based multiplex detection tool, the padlock probe ligation and microarray detection (PPLMD) tool was developed which is extended to a 15-plex traceability tool with a focus on products of wheat and Basmati rice (Voorhuijzen et al. 2012). One nucleotide difference in target sequence was sufficient for the distinction between the presence or absence of a specific target. At least 5 % Basmati rice was detected in mixtures with non-fragrant rice. PPLMD has been shown to be a useful tool for the detection of fraudulent/intentional admixtures in premium foods and is ready for the monitoring of correct labelling of premium foods worldwide.
Fragment separation methods
In addition to the molecular marker system, the fragment separation methods also greatly influence the accuracy of detection as well as quantification of the adulterants in the samples (Spaniolas et al. 2006; Tang et al. 2004). Although a consensus was made in using microsatellites for detection of adulteration, an ambiguity exists in different laboratories across the world in choosing the best fragment separation method. For instance, Archak et al. (2007) used fluorescently labelled microsatellite markers for amplification and capillary electrophoresis for fragment separation, whereas Bligh (2000) used slab-gel for fragment separation. Afterwards, it has been demonstrated that capillary electrophoresis was quite reproducible among fragment separation methods, and showed relatively less error in the estimation of allele sizes (±0.73 bp and <0.50 bp deviation) compared to slab-gel (±1.59 bp and 1 bp) and agarose gel (±8.03 bp and >3 bp) electrophoresis methods (Vemireddy et al. 2007).
Authenticity of processed foods
In rice authenticity, much attention should be paid to avoid contamination in the initial stages of the marketing i.e., at brown rice form because undetected adulterants debase the quality and price of processed food products where identification of adulterant is difficult even with DNA based markers also due to disintegration of most of the DNA. However, recently, protocols have been developed for identification of cultivars in the processed foods and bread containing both wheat and rice flours employing PCR based methods with improved DNA isolation protocol (Ohtsubo and Nakamura, 2007; Nakamura and Ohtsubo 2010). The protocol contains the preparation of the template DNA for PCR by the CTAB method followed by the extraction of DNA with 70 % ethyl alcohol (Nakamura and Ohtsubo 2010).
Hybrid seed purity
Another important area of rice authenticity where the intervention of DNA based methods required is hybrid seed production. Hybrid technology is gaining popularity worldwide due to its yield advantage over the conventionally developed rice varieties. The success of hybrid seed production is mainly depends on the purity of their parental lines. It has been estimated that 1 % contamination of hybrid seed causes yield loss of approximately 100 kg/ha (Mao et al. 1996). Maintaining of seed purity is an important aspect of hybrid seed production. Traditionally used to grow out tests (GOT) to test the seed purity based on morphological traits are time consuming and laborious. To address this issue, Nandakumar et al. (2004) and Sundaram et al. (2008) have been identified informative microsatellite markers capable of distinguishing hybrid rice parental lines and used for assessing the hybrid seed purity employing two dimensional bulked DNA sampling strategy (Sundaram et al. 2008).
GMO detection
Since the global rise of genetically modified organisms (GMOs) or transgenic plants cultivation, it is necessary to detect the GMO levels in the food products. As of now, only DNA-based methods have been used to detect the GMO content in the rice products. M¨ade et al. (2006) developed a method for detection of Bt rice varieties and quantification of the Bt content in imported rice products. They have used field-tested Bt rice (‘Anti-pest Shanyou 63’ and ‘Anti-pest Jinyou 63’) as reference material to determine transgenic DNA sequences. The cryIA(b) and cryIA(c) fusion gene and the nopaline synthase terminator (nos) sequence was used to develop a construct-specific real-time PCR based detection method. This Bt rice specific detection system along with quantitative real-time PCR method for the rice-specific reference gene gos9 could able to quantify the Bt content with a limit of quantification of below 0.1 %.
Molecular profiling or DNA barcoding
While protein-based methods could differentiate the rice varieties as per their subspecies/groups, the DNA-based methods has the potential to differentiate at the inter-varietal and intra-varietal level. Earlier, Archak et al. (2007) developed cultivar-specific allele profiles using 8 microsatellite markers viz., RM1, RM55, RM44, RM72, RM348, RM241, RM202 and RM171 to differentiate the traditional basmati, evolved basmati and non-basmati varieties. Very recently, Vemireddy et al. (2014) developed DNA barcodes/fingerprints for 90 high yielding rice varieties using a panel of eight hypervariable microsatellite (hvRM) markers viz., RM11313, RM13584, RM15004, RM5844, RM22250, RM22565, RM24260 and RM8207 and validated in survey samples of Sona Mahsuri variety.
Quantification of adulteration
In addition to detection of the adulterant, accurate quantification of the adulterants is also a key aspect in rice authenticity (Vlachos and Arvanitoyannis 2008). Since rice is a farm product, a certain amount of inadvertent mixture is expected during post harvesting period of cultivation. In view of the unavoidable adulteration, in 2005, a code of practice was developed for the control of basmati rice in the UK and it recommended 7 % as the ceiling for inadvertent mixtures in Basmati imports to United Kingdom (www.riceassociation.org.uk/). Unlike the methods of detection of adulteration where a range of markers and fragment detection systems available, very few methods are available for accurate quantification of the adulterants especially DNA-based microsatellite markers coupled with capillary electrophoresis or real time PCR method found suitable. Quantification of adulteration has been a global issue and several methods/protocols have been established.
Non-DNA-based methods
Of non-DNA based methods, physico-chemical methods have been used for the quantification of adulteration. Especially, the quality trait, alkali spreading value has been used to detect grains of RD 23 and Chai Nat 1 (CN1; high amylose content, intermediate gelatinization temperature) in Thai rice industry. However, their accuracy is low, especially when Jasmine rice is adulterated with similar amylose content rice such as Pathum Thani 1 (PTT1) variety, or at low adulteration level (Pitiphunpong et al. 2011). In a study by Grimm et al. (2001) on jasmine rice, adulteration level was estimated based on the textural and pasting measurements using Principal Component Analysis (PCA), Multiple Linear Regression and Multivariate Analysis. The alkaline oxidation value has also been used as an indicator of adulteration of Basmati with scented as well as non-scented rice. American long grain rice and Basmati rice were separated by treating with a 2 % solution of sodium bisulphite. When 5 % v/v HCl was added, the Basmati kernels turned chalky in about 20 min, while 40 min or more were required for all kernels of American rice to become chalky (Singhal et al. 1997).
DNA-based methods
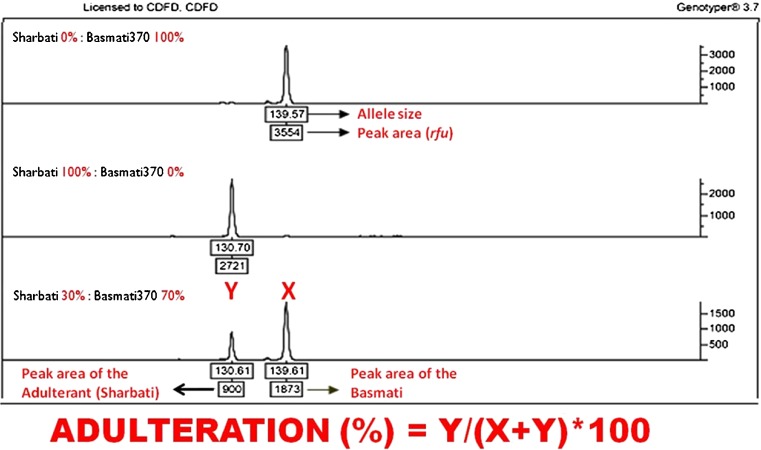
For quantification of adulteration, combination of microsatellite markers and either capillary electrophoresis or real time PCR have been widely employed (Table 3). The principle of quantification of the adulterant by DNA-based method is rely on the relative quantities of the amplified allelic product at a common locus between competing DNA templates of the genuine rice and the adulterant mixed in progressive proportions (Archak et al. 2007) (Fig. 2). In order to normalize quantification, TB grains were mixed with a NB (adulterant) in a progressive dilution. PCR was carried out with diagnostic microsatellite markers and the product resolved on capillary electrophoresis. The ratio of peak area of the two alleles of that of the adulterant and the Basmati will be plotted against the progressive proportion of adulteration to develop a standard curve using Curve Expert version 1.38 (Fig. 3). The ratio between the quantities of the amplicons (ratio of peak areas of TB and NB variety) would reveal the ratio of the quantities of competing DNA template in a PCR mixture. This method can quantify the adulteration even at 1 % level. However, the model used for construction of calibration curve is one of the factors which influence the accuracy of the quantification of the adulteration. For instance, when three blind samples at 4, 8 and 12 % adulteration were genotyped using capillary electrophoresis, Archak et al. (2007) estimated standard errors as ±1.94, ±0.17 and ±0.93 %, while Bligh (2000) reported higher standard errors as ±1.06, ±0.77 and ±10.07 at an actual adulteration of 39, 44 and 25 % in slab-gel, respectively. Recently, Colyer et al. (Colyer et al. 2008) compared different calibration curves using slab-gel and a single microsatellite marker RM222. This group concluded that the linear regression of ratio of peak areas to the ratio of content proportions as the most precise method for accurate quantification of the adulteration. By employing this method, it is possible to arrive at lowest standard error within the agreeable limits of adulteration (7 % non Basmati).
Table 3.
Methods used for quantification of the adulterants
| Marker type | Fragment separation method | DNA Markers | Method of quantification | Reference |
|---|---|---|---|---|
| Microsatellites | Slab-gel | RM1,RM19,RM208,RM55,RM201,RM219,RM225,RM229,RM202,M2 and M16 | Calibration curve | Bligh (2000) |
| Microsatellites | Capillary electrophoresis | RM1,RM171,RM202,RM44,RM55,RM348,RM72 and RM241 | Calibration curve-Logistic model | Archak et al. (2007) |
| Real time PCR | Fgr and RM348 | Cycle number | ||
| Microsatellites | Slab-gel, Capillary electrophoresis | RM1,RM171,RM202,RM44,RM55,RM348,RM72 and RM241 | Ratio of peak area of adulterant and total peak area | Vemireddy et al. (2007) |
| Microsatellites | Slab-gel | RM222 | Calibration curve-Linear regression | Colyer et al. 2008 |
| Microsatellites and Indel | Real time PCR | RM241 and BAD2 gene | High resolution melting profile | Ganopoulos et al. (2011) |
| SCAR | Real time PCR | Wka9q, S13q, E30q, and F6q | Standard curve- Cycle number | Okunishi et al. (2005) |
| Gene specific primer | Real time PCR | gos9 sequence | Cycle number | Herńandez et al. (2005) |
Fig. 2.
Detection and Quantification of the adulteration. A representative profile of Sharbati (a common adulterant) mixed with Basmati 370 (a traditional basmati) in 0:100 (pure basmati), 100:0 (pure adulterant) and30:70 (30 % adulteration) is shown. The top box below the easch peak is allele size of the variety (bp) and the botton box is its peak area expressed in rfu (relatve fluorescent unit. Y is Peak area of the adulterant (Sharbati); X is the Peak area of the Basmati (Basmati 370)
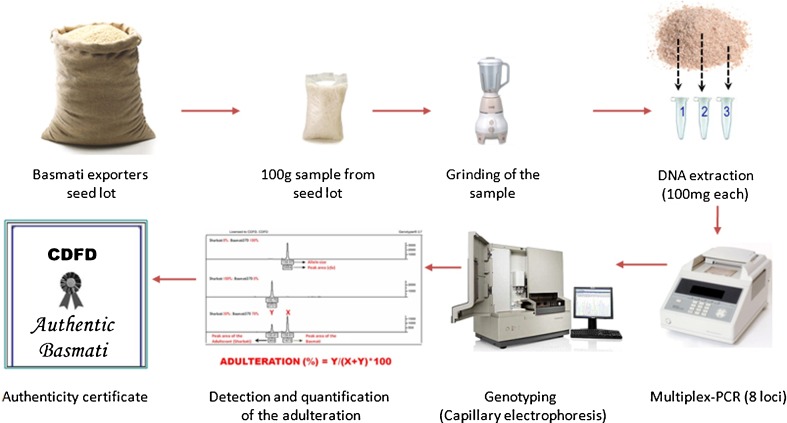
Fig. 3.
Protocol followed for Basmati authenticity at APEDA-CDFD Centre for Basmati DNA analysis. The detailed protocol was given in the main text (parts of the figure are from the following sources: Basmati exporters seed lot (Rice bag)- http://www.exportersindia.com/krk-traders/products.htm, 100g sample from seed lot (Rice polished rice) - http://www.keywordpicture.com/keyword/bag%20rice/, Genotyping(Capillary electrophoresis) - http://pag.ibis.ulaval.ca/seq/en/index.php)
The quantitative competitive PCR (QC-PCR) and real-time PCR allows simultaneous detection and conformation of fragments using specific probes or fluorescently labelled primers by PCR. The Real-time PCR analysis used in the genetically modified (GM) crops, relies on the continuous measurement of increments in the fluorescence during cycles and the PCR cycle number required to generate a signal that is significantly above noise level (cycle threshold).
As of today, real time PCR assay is regarded as the most sought after method for accurate quantification of the nucleic acids. It is a reliable as well as commonly used method to detect foreign DNA in genetically modified (GM) food samples (Baeumler et al. 2006; Leimanis et al. 2006). In case of GM food, detection and quantification of transgene has been relatively straight forward because transgenes are distinct “foreign” DNA elements in the host genome, which can be specifically amplified with high accuracy (detection) and sensitivity (quantification) by real time PCR (Nakamura et al. 2013). On the other hand, in case of Basmati adulteration, the so-called “adulterant” is another long-grain rice of cheap quality. As of now Basmati-specific sequence has not been identified to generate varietal-specific alleles. However, an 8 bp deletion in the betaine-aldehyde dehydrogenase-2 gene (bad-2) specific to aromatic rices (Bradbury et al. 2005) was exploited to amplify an 80 bp product exclusively in a non Basmati variety (i.e., adulterant) using standard method of capillary electrophoresis (Archak et al. 2007). This method showed a mean deviation of 1.76 % which is comparable to real time assay. This infers that the estimation of adulteration by capillary electrophoresis is realistically accurate for practical applications. Similarly, TaqMan based real time PCR method has also been developed for quantification of adulteration in Basmati by employing bad-2 gene specific primers (Lopez 2008; Mathure et al. 2014). Although it was possible to distinguish genuine Basmati, this assay was found unsuitable for practical applications, as many unnotified evolved Basmati also carry the same genuine Basmati allele.
In a study by Ganopoulos et al. (2011) high resolution melting (HRM) analysis has been used for detection and quantification of adulteration. The HRM analysis was reported as a rapid, cost effective DNA based method with efficiency comparable to capillary electrophoresis. With this analysis it is possible to discriminate PCR products of the same allele size with different melting profiles due to their difference in nucleotide base composition.
In addition, twenty non-transgenic rice varieties were quantitatively detected by Herńandez et al. (2005) with RTi-PCR. The real-time PCR assay was based on the gos9 sequence and was able to specifically detect and quantify DNA from the rice plants with a limit of detection of 3.3 genome copies. Whereas the quantification limit was approximately 100 haploid genomes for real-time PCR systems with 95 % confidence. Furthermore, the performance of this assay on twelve commercial food products revealed 100 % identification.
Determination of the age of the samples
Generally, the rice material stored for over long periods of time commands high price than the new one due to high chalkiness nature of the later. As it is difficult to estimate storage period of rice, dishonest traders blend rice of different storage periods. The DNA-based methods are unsuitable to address this problem as there will not be any genotypic differences between different age group samples. However, the variation between different age group samples can be witnessed at metabolite level. Widjaja et al. (1996) reported increase of the total volatile compounds of paddy, brown and white rice upon long storage in air due to accumulation of aldehydes and ketones as a result of lipid oxidation process. Recently, a simple, fast and non-destructive approach was developed to classify rice seeds of different storage time with discrimination accuracy of 97.5 % employing near infrared reflectance spectroscopy (NIR) (Dengsheng and Xiaoli 2007).
Determination of the geographical area of the samples
Geographical area also fetches sometime additional price to some speciality rices especially Basmati as its quality traits particularly exquisite aroma and cooking traits are influenced by the environment where they have been cultivated. Determination of the geographical origin of rice is important feature especially Basmati rice as unscrupulous producers/traders attempts to increase their profits by mislabelling the inferior/low quality products grown outside their regular cultivated area of foot hills of Himalayas. Earlier, grain proteins (Lan et al. 2002; Montalvan et al. 1998) and isozymes (Li and Rutger 2000) were used for discrimination of rice cultivars grown in different geographical regions. The principle underlying this method is that trace element and isotope composition of the rice grains reflect the soil and latitude of the area in which it is grown. The trace elements are labile in the soil and easily transported into plants and can be good indicators of geographical origin. Further, the concentrations of the trace elements depends on topography and soil characteristics. Currently, multi-isotopic and multi-elements analyses are being used to determine the geographical area of the samples (Kelly et al. 2002, 2005; Suzuki et al. 2008) (Table 4). The amounts of P, K, Mg, Ca, Mn, Zn, Fe, and Cu were determined with Inductively Coupled Plasma Atomic Emission Spectrometry (ICP-AES); while those of Rb, Mo, Ba, Sr, Ni, Cd, Cs, Pb, Al, Cr, and Co were determined with High Resolution Mass Spectrometry (HRMS). These analyses could distinguish rice samples cultivated in different (Tohoku/Kanto area from those of Hokuriku) geographic regions. For instance, Kawasaki et al. (2002) determined strontium isotope ratio (Sr-87/Sr-86) with Multiple Collectors Inductively Coupled Plasma Mass Spectrometry (MC-ICP-MS) in order to identify provenance of brown rice. After the removal of rubidium from the sample solutions, the Sr isotope ratios were determined with a precision of <0.01 %. The Sr isotope ratios of the Japanese rice samples ranged from 0.706 to 0.709, while those of the Chinese and Vietnamese samples ranged from 0.710 to 0.711, slightly higher than those of almost all the Japanese samples. Kelly et al. (2005) analysed rice samples cultivated in the USA, Europe, and Basmati grown regions (India/Pakistan) with Isotope Ratio Mass Spectrometry (IRMS) and Inductively Coupled Plasma Mass Spectrometry (ICP-MS). The nine key variables: carbon-13, oxygen-18, boron, holmium, gadolinium, magnesium, rubidium, selenium, and tungsten were used for the proper identification of rice samples by discriminate analysis. High levels of boron (>2,500 ppb) were associated with rice samples from America while high levels of holmium were found in the samples from Arkansas. European rice samples generally contained relatively high levels of magnesium, while Basmati rice (Indian/Pakistani samples) were characterized by relatively low oxygen-18 (18O) abundance. Also, for rapid discrimination of Chinese rice wines of different geographical origin, particularly Shaoxing wines from non-Shaoxing wines, near-infrared spectroscopy (NIRS) was used (Shen et al. 2010; Yu et al. 2007). Also, fourier transform near-infrared spectroscopy (FT-NIR) was used to discriminate Chinese rice wine of different geographical origins as well as different vintage years. A clear separation between the different vintage years was reported using tartaric acid as a discriminating descriptor (Yu et al. 2007).
Table 4.
List of methods used for determination of the geographical area of the cultivated rice
| Geographical based methods | Isotope Ratio Mass Spectrometry and ICP MS | Grains | Rice from USA, Europe, Basmati regions of India/Pakistan | Carbon 13, Oxygen 18, and other elements | Kelly et al. 2005 |
| Inductively Coupled Plasma Atomic Emission Spectrometry | Grains | Oryza sativa | Amount of P, K, Mg, Ca, Mn, Zn, Fe, Cu | Yashui and Shindoh 2000 | |
| Inductively Coupled Plasma High Resolution Mass Spectrometry (ICP-HRMS-MS) | Unhulled rice | Oryza sativa Tohoku/Kanto and Hokuriku | Amount of Rb, Mo, Ba, Sr, Ni, Cd, Cs, Pb, Al, Cr, Co | Yashui and Shindoh 2000 | |
| Inductively coupled Plasma Optical Emission Spectrometry | Grains | Basmati ricce | Trace elements | Kelly et al. 2002 | |
| Multiple Collector Inductively Coupled Plasma Mass Spectrometry (MC-ICP-MS) | Grain | Brown rice, Japanese, Chinese and Vietnamese rice | Provenance based on Strontium isotope Sr87/Sr86 | Kawasaki et al. 2002 | |
| Atomic Fluorescence Spectroscopy (AFS) | Polished rice | Pllished rice from China | Selenium Concentration | Chen et al. 2002 |
Case study: Basmati rice
Basmati is a long-grain type of rice (Oryza sativa L.) cultivated exclusively in the foot hills of the Himalayas in the regions of Haryana, Uttar Pradesh and the Punjab province of India. Basmati, “king of rices”, carved out a special place among all aromatic rice cultivars and also commands a premium price at international markets. Although Basmati is bestowed with unique qualities like extra long slender grain, soft texture of the cooked rice and exquisite fragrance, it possesses undesirable characters like tall culms, photoperiod sensitivity and susceptibility to diseases. This prompted the breeders to develop hybrids with traditional Basmati, which has eventually led to the existence of TB, EB and relatively inferior non Basmati (NB) long-grain rice varieties (Archak et al. 2007; Nagaraju et al. 2002; Vemireddy et al. 2007). Existence of obvious price difference, zero percent import duty and difficulty in distinguishing TB, EB and NB varieties offers a room to the dishonest traders to look for adulteration to earn higher profits. Moreover, a survey conducted by the Food Standards Agency (FSA), United Kingdom on Basmati rice consignments from India and Pakistan revealed that such practices are prevalent.
In India, authorized centre for certifying Basmati samples is “APEDA-CDFD Centre for Basmati DNA analysis” housed at “Centre for DNA Fingerprinting and Diagnostics”, Hyderabad. The rice samples received from Export Inspection Council (EIC) are tested for their purity using the DNA protocol developed at the centre. The protocol followed is available at http://www.cdfd.org.in/lmg/APEDA/index.php. The protocol was followed for the analysis of standard and blind samples in the ring trial during 2005 and the results were in agreement across laboratories. Generally, the centre receives rice samples from export inspection agency (EIA) and export inspection council (EIC) and some leading companies like LT Overseas, Amira Foods, Kohinoor Foods Limited, PICRIC, Best Food International Limited, Aeroplane and Hindustan Lever Limited. Approximately, 100 g of grain from each sample received for testing is powdered and the remaining sample is stored for up to 3 months. From the powdered sample, three sub-samples of 1 g each is drawn randomly, from which 100 mg of grain powder will be collected for DNA extraction. DNA is isolated using DNeasy plant mini kit (Qiagen). The quantity and quality of the DNA (at 260/230 and 260/280 absorbance) will be verified using a spectrophotometer. PCR is performed using standardized conditions (Archak et al. 2007).
The fluorescently labelled PCR products are mixed with 0.3 μl of GeneScan-500 ROX (6-Carboxy-X-Rhodamine) size standard (Applied Biosystems, USA) and 10.2 μl of Hi-Di Formamide (Applied Biosystems, USA), electrophoresed by capillary electrophoresis on an ABI PRISM 3100 genetic analyzer (Applied Biosystems, USA) according to the manufacturer’s instructions. Subsequently, fluorescent DNA fragments are resolved using the GeneScan version 3.7, and allele size and peak-area of the true peaks are determined by Genemapper version 3.7 (Applied Biosystems, USA). In case of adulterated samples, each experiment is repeated thrice, starting from the DNA extraction, to confirm the extent of adulteration. The quantity of an amplified PCR product is represented by peak area (measured in relative fluorescent units, rfu). The adulteration is expressed as a percent fraction to that of pure Basmati as
Where, A is the rfu of the adulterant and B is the rfu of the main variety.
Once the DNA analysis of given rice sample is completed, the duly signed authenticity certificate, containing details of percentage of adulterated rice in the test Basmati rice sample, is provided to the sender (Fig. 3).
The protocol has been authorized by the export development and regulatory agencies of the Government of India for issuance of certificate of purity for Basmati export samples. Employing this assay, more than 1,000 referral samples of Basmati rice destined to international market have so far been certified. Patents have been applied for the technology developed at CDFD for Basmati DNA analysis (USPTO 10/357, 488 and 11/406, 257; PCT/IN06/00254). The know-how on ready-to use “Basmati VerifilerTM Kit” licensed by CDFD has been transferred to Labindia (Indian partners of Applied Biosystems) under license transfer agreement.
Conclusion
Authentication of rice cultivars is an important issue to be addressed to protect the interests of farmers as well as to provide healthy food to consumers. To this direction, each country established quality standards for agricultural products by providing a label with complete details including country of origin and chemical composition. For food authentication, a database of genuine samples is required to which the ‘suspect’ test sample can be compared to establish its authenticity. Currently, the traditional as well as sophisticated methods of analysis are available for detection and quantification of the rice adulteration. Adulteration can be effectively determined using a variety of methods based as described above. The implementation of such tests by food control laboratories depends on the accuracy of detection of adulterant, the complexity of the technique and cost factors. With increased regulation of food products, in response to consumer concern, new technologies emerge to be adopted for use in the market place. The molecular markers such as RAPDs, AFLPs, ISSR, and SSR have been successfully used in rice authentication. The selection of the most suitable molecular approach depends on different aspects, including the amount of genetic variation of the analyzed species, the time needed for the analysis, the cost/effectiveness ratio and the expertise of laboratories.
Out of the numerous technologies that were applied to detect the adulteration, DNA based method particularly, the combination of capillary electrophoresis and microsatellite markers proved to be quite amenable to solve the adulteration problem. Further, the DNA-based methods have applicability in detecting the adulterants in parental lines of the hybrid seed production and processed foods. However, they are unsuitable for detection of the age of the samples and also the geographical area of the cultivated rice. These issues are effectively tackled by the protein-based methods and multi-element and multi-isotopic analyses. Based on the existing literature on rice authenticity, it can be inferred that with a combination of methods it is quite possible to eliminate the adulteration in any form or at any stage from the rice industry. Therefore, it is time for the Governments to come forward to apply the current proven technology to cull out the adulteration from the rice industry completely. One of the noteworthy example comes from the exploitation of a high throughput capillary electrophoresis based microsatellite multiplex assay developed at CDFD to combat the Basmati rice adulteration in India. Similar technology also has potential application in hybrid purity maintenance and development of the cultivar specific molecular profiles/fingerprints. Therefore, not only the advanced technology but also clear cut policies need to be implemented in the rice industry to ensure that the products of the end user are genuine.
Abbreviations
- TB
Traditional Basmati
- EB
Evolved Basmati
- NB
Non Basmati
- CE
Capillary electrophoresis
References
- Agrawal NS, Sinha AC (1965) In Symposium on Technology of Rice and Rice Products, American Association of Cereal Chemists (1969). Approved Methods of the AACC. Methods 28–31
- Ahmed J, Ramaswamy H, Ayad A, Alli I. Thermal and dynamic rheology of insoluble starch from basmati rice. Food Hydrocoll. 2008;22:278–287. [Google Scholar]
- Archak S, Lakshminarayanareddy V, Nagaraju J. High-throughput multiplex microsatellite marker assay for detection and quantification of adulteration in Basmati rice (Oryza sativa) Electrophoresis. 2007;28:2396–2405. doi: 10.1002/elps.200600646. [DOI] [PubMed] [Google Scholar]
- Arvanitoyannis IS. Trends in food authentication in modern techniques for food authentication, Ed. Da-Wen Sun. Amsterdam: Academic (Elsevier Ltd); 2008. pp. 617–643. [Google Scholar]
- Ashfaq M, Khan AS. Genetic diversity in Basmati rice (Oryza sativa L.) germplasm as revealed by microsatellite (SSR) markers. Genetika. 2012;48:62–71. [PubMed] [Google Scholar]
- Attaviroj N, Kasemsumran S, Noomhorm A. Rapid variety identification of pure rough rice by Fourier transform near infrared spectroscopy. Cereal Chem. 2011;88:490–496. [Google Scholar]
- Baeumler S, Wulff D, Tagliani L, Song P. A real-time quantitative PCR detection method specific to widestrike transgenic cotton (event 281-24-236/3006-210-23) J Agric Food Chem. 2006;54:6527–6534. doi: 10.1021/jf0610357. [DOI] [PubMed] [Google Scholar]
- Beerh OP, Srinivas T. Some histological factors associated with Basmati rice. Oryza. 1991;28:399–401. [Google Scholar]
- Bergman CJ, Delgado JT, Bryant R, Grimm C, Cadwallader KR, Webb BD. Rapid gas chromatographic technique for quantifying 2-acetyl-1-pyrroline and hexanal in rice (Oryza sativa, L.) Cereal Chem. 2000;77:454–458. [Google Scholar]
- Bett-Garber KL, Champagne ET, McClung AM, Moldenhauer KA, Linscombe SD, McKenzie KS. Categorizing rice cultivars based on cluster analysis of amylose content, protein content and sensory attributes. Cereal Chem. 2001;78:551–558. [Google Scholar]
- Bhattacharjee P, Singhal RS, Kulkarni PR. Basmati rice: a review. Int J Food Sci Technol. 2002;37:1–12. [Google Scholar]
- Bhattacharya KR. Rice Bran: Regional Extension Service Centre (Rice Milling) Scientific Series No. 7. Department of Food, Government of India. Mysore: CFTRI; 1982. [Google Scholar]
- Blair MW, Hedetale V, McCouch SR. Fluorescent-labeled microsatellite panels useful for detecting allelic diversity in cultivated rice (Oryza sativa L) Theor Appl Genet. 2002;105:449–457. doi: 10.1007/s00122-002-0921-5. [DOI] [PubMed] [Google Scholar]
- Bligh HFJ. Detection of adulteration of Basmati rice with non-premium long-grain rice. Int J Food Sci Technol. 2000;35:257–265. [Google Scholar]
- Bradbury LM, Fitzgerald TL, Henry RJ, Jin Q, Waters DL. The gene for fragrance in rice. Plant Biotechnol J. 2005;3:363–370. doi: 10.1111/j.1467-7652.2005.00131.x. [DOI] [PubMed] [Google Scholar]
- Buttery RG, Ling LC, Juliano BO, Turnbaugh JG. Cooked rice aroma and 2-acetyl-1-pyrroline. J Agric Food Chem. 1983;31:823–826. [Google Scholar]
- Cai X, Chen T, Zhou QY, Xu L, Qu LQ, Hua XJ, Lin JX (2011) Development of Casparian strip in rice cultivars. Plant Signal Behav 6:59–65 [DOI] [PMC free article] [PubMed]
- Carter RM, Yan Y, Tomlins K. Digital imaging based classification and authentication of granular food products. Meas Sci Technol. 2006;17:235–240. [Google Scholar]
- Chen L, Yang F, Xu J, Hu Y, Hu Q, Zhang Y, Pang G. Determination of selenium concentration of rice in china and effect of fertilization of selenite and selenate on selenium content of rice. J Agric Food Chem. 2002;50:5128–5130. doi: 10.1021/jf0201374. [DOI] [PubMed] [Google Scholar]
- Choudhury PR, Kohli S, Srinivasan K, Mohapatra T, Sharma RP. Identification and classification of aromatic rices based on DNA fingerprinting. Euphytica. 2001;118:243–251. [Google Scholar]
- Choudhury G, Ranjitkumar N, Malathi S, Deborah DAK, Abhilash V, Anuradha G, Siddiq EA, Vemireddy LR. Molecular genetic diversity of major Indian rice cultivars over decadal periods. Plos ONE. 2013;8(6):e66197. doi: 10.1371/journal.pone.0066197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn JR, Temnykh S, Paul E, McCouch SR. Design and application of microsatellite marker panels for semiautomated genotyping of rice (Oryza sativa L) Crop Sci. 2002;42:2092–2099. [Google Scholar]
- Colyer A, Macarthur R, Lloyd J, Hird H. Comparison of calibration methods for the quantification of Basmati and non-Basmati rice using microsatellite analysis. Food Addit Contam Part A: Chem Anal Control Expo Risk Assess. 2008;25:1189–1194. doi: 10.1080/02652030802040141. [DOI] [PubMed] [Google Scholar]
- Dengsheng Z, Xiaoli L. Determination of storage time of rice seed using ANN Based on NIRS lecture notes in computer. Sci Adv Neural Netw. 2007;4493:1043–1048. [Google Scholar]
- Dhanya K, Sasikumar B. Molecular marker based adulteration detection in traded food and agricultural commodities of plant origin with special reference to spices. Curr Trends Biotechnol Pharm. 2010;4:454–489. [Google Scholar]
- Findlay ITA, Quirke P, Frazier R, Urquhart A. DNA fingerprinting from single cells. Nature. 1997;389:555–556. doi: 10.1038/39225. [DOI] [PubMed] [Google Scholar]
- Gangidi RR, Proctor A, Meullenet JF. Milled rice surface lipid measurement by diffuse reflectance Fourier transform infrared spectroscopy (DRIFTS) J Am Oil Chem Soc. 2002;79:7–12. [Google Scholar]
- Ganopoulos I, Argiriou A, Tsaftaris A. Adulterations in Basmati rice detected quantitatively by combined use of microsatellite and fragrance typing with high resolution melting (HRM) analysis. Food Chem. 2011;129:652–659. doi: 10.1016/j.foodchem.2011.04.109. [DOI] [PubMed] [Google Scholar]
- Glaszman JC. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74(1):21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- Grimm CC, Bergman C, Delgado JT, Bryant R. Screening for 2-Acetyl-1-pyrroline in the Headspace of Rice Using SPME/GC-MS. J Agric Food Chem. 2001;49:245–249. doi: 10.1021/jf0008902. [DOI] [PubMed] [Google Scholar]
- Hamada JS. Separation and molecular mass distribution of rice proteins by size-exclusion high-performance liquid chromatography in a dissociating buffer. J Chromatogr A. 1996;734:195–203. [Google Scholar]
- Herńandez M, Esteve T, Pla M. Real-time polymerase chain reaction based assays for quantitative detection of barley, rice, sunflower, and wheat. J Agric Food Chem. 2005;53:7003–7009. doi: 10.1021/jf050797j. [DOI] [PubMed] [Google Scholar]
- Himmelsbach DS, Barton FE, McClung AM, Champagne ET. Protein and apparent amylase contents of milled rice by NIRFT/ Raman spectroscopy. Cereal Chem. 2001;78:488–492. [Google Scholar]
- Hirannaiah BV, Bhashyam MK, Ali SZ. An improved cooking quality test for Basmati rice. J Food Sci Technol. 2001;38:116–119. [Google Scholar]
- Huebner FR, Bietz JA, Webb BD, Juliano BO. Rice cultivar identification by high performance liquid chromatography of endosperm proteins. Cereal Chem. 1990;67:129–135. [Google Scholar]
- Jain S, Jain RK, McCouch SR. Genetic analysis of Indian aromatic and quality rice (Oryza sativa L) germplasm using panels of fluorescently-labeled microsatellite markers. Theor Appl Genet. 2004;109:965–977. doi: 10.1007/s00122-004-1700-2. [DOI] [PubMed] [Google Scholar]
- Jha JS. Spectrophotometric studies of rice bran oil and mustard oil mixtures: I. J Am Oil Chem Soc. 1980;57:283–285. [Google Scholar]
- Kanno K, Kawamura Y, Kato K. J Agric Chem. 1989;63:1207–1211. [Google Scholar]
- Kawasaki A, Oda H, Hirata T. Determination of strontium isotope ratio of brown rice for estimating its provenance. Soil Sci Plant Nutr. 2002;486:635–640. [Google Scholar]
- Kelly S, Baxter M, Chapman S, Rhodes C, Dennis J, Brereton P. The application of isotopic and elemental analysis to determine the geographical origin of premium long grain rice. Eur Food Res Technol. 2002;214:72–78. [Google Scholar]
- Kelly S, Heaton K, Hoogewerff J. Tracing the geographical origin of food: The application of multi-element and multi-isotope analysis. Trends Food Sci Technol. 2005;16:555–567. [Google Scholar]
- Kim SS, Jo JS, Kim YJ, Sung NK. Authentication of rice by three-sided image analysis of kernels using two mirrors. Cereal Chem. 1997;74:212–215. [Google Scholar]
- Kim SS, Rhyu MR, Kim JM, Lee SH. Authentication of rice using near-infrared reflectance spectroscopy. Cereal Chem. 2003;80:346–349. [Google Scholar]
- Lai VMF, Lu SN, He WH, Chen HH. Non-starch polysaccharide compositions of rice grains with respect to rice variety and degree of milling. Food Chem. 2007;101:1205–1210. [Google Scholar]
- Lan Y, Fang Q, Kocher MF, Hanna MA. Detection of fissures in rice grains using image enhancement. Int J Food Prop. 2002;5:205–215. [Google Scholar]
- Largo-Gosens A, Hernandez-Altamirano M, Garcia-Calvo L, Alonso-Simon A, Alvarez J, Acebes JL. Fourier transform mid infrared spectroscopy applicatios for monitoring the structural plasticity of plant cell walls. Front Plant Sci. 2014 doi: 10.3389/fpls.2014.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimanis S, Hernandez M, Fernandez S, Boyer F, Burns M, Bruderer S, Glouden T, Harris N, Kaeppeli O, Philipp P, Pla M, Puigdomenech P, Vaitilingom M, Bertheau Y, Remacle J. A microarray-based detection system for genetically modified (GM) food ingredients. Plant Mol Biol. 2006;61:123–139. doi: 10.1007/s11103-005-6173-4. [DOI] [PubMed] [Google Scholar]
- Li Z, Rutger JN. Geographic distribution and multilocus organization of isozyme variation of rice (Oryza sativa L) Theor Appl Genet. 2000;101:379–387. [Google Scholar]
- Li X, Bai H, Wang X, Li L, Cao Y, Wei J, Liu Y, Liu L, Gong X, Wu L, Liu S, Liu G. Identification and validation of rice reference proteins for western blotting. J Exp Bot. 2011;14:4763–4772. doi: 10.1093/jxb/err084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockley K, Bardsley RG. DNA-based methods for food authentication. Trends Food Sci Technol. 2008;11:67–77. [Google Scholar]
- Lopez SJ. TaqMan based real time PCR method for quantitative detection of Basmati rice adulteration with non-Basmati rice. Eur Food Res Technol. 2008;227:619–622. [Google Scholar]
- Lorieux M, Petrov M, Huang N, Guiderdoni E, Ghesquiere A. Aroma in rice: Genetic analysis of a quantitative trait. Theor Appl Genet. 1996;93:1145–1151. doi: 10.1007/BF00230138. [DOI] [PubMed] [Google Scholar]
- M¨ade D, Degner C, Grohmann L. Detection of genetically modified rice: a construct-specific real-time PCR method based on DNA sequences from transgenic Bt rice. Eur Food Res Technol. 2006;224:271–278. [Google Scholar]
- Mackill DJ, Zhang Z, Redona ED, Colowit PM. Level of polymorphism and genetic mapping of AFLP markers in rice. Genome. 1996;39:969–977. doi: 10.1139/g96-121. [DOI] [PubMed] [Google Scholar]
- Mao CX, Virmani SS, Kumar I. Technological innovations to lower the costs of hybrid rice seed production. In: marker for fragrance genotyping in rice. Mol Breed. 1996;16:279–283. [Google Scholar]
- Mariotti M, Sinelli N, Catenacci F, Pagani MA, Lucisano M. Retrogradation behavior of milled and brown rice pastes during ageing. J Cereal Sci. 2009;49:171–177. [Google Scholar]
- Mathure SV, Jawali N, Thengane RJ, Nadaf AB. Comparative quantitative analysis of headspace volatiles and their association with BADH2 marker in non-Basmati scented, Basmati and non-scented rice (Oryza sativa L.) cultivars of India. Food Chem. 2014;142:383–391. doi: 10.1016/j.foodchem.2013.07.066. [DOI] [PubMed] [Google Scholar]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L) DNA Res. 2002;9:257–279. doi: 10.1093/dnares/9.6.257. [DOI] [PubMed] [Google Scholar]
- Monakhova YB, Rutledge DN, Robmann A, Waiblinger, Mahler M, Ilse M, Kuballa T, Lachenmeier DW (2013) Determination of rice type by 1H NMR spectroscopy in combination with different chemometric tools. Chemometics doi:10.1002/cem.2576
- Montalvan R, Ando A, Echeverrigaray S. Use of seed protein polymorphism for discrimination of improvement level and geographic origin of upland rice cultivars. Genet Mol Biol. 1998;21:531–535. [Google Scholar]
- Nadaf AB, Krishnan S, Wakte KV. Histochemical and biochemical analysis of major aroma compound (2-acetyl-1-pyrroline) in Basmati and other scented rice (Oryza sativa L) Curr Sci. 2006;91:1533–1536. [Google Scholar]
- Nagaraju J, Kathirvel M, Kumar RR, Siddiq EA, Hasnain SE. Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proc Natl Acad Sci U S A. 2002;99:5836–5841. doi: 10.1073/pnas.042099099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Ohtsubo K. PCR method for the detection and identification of cultivars of rice flours used in yeast leavened breads containing both wheat and rice flours. J Cereal Sci. 2010;52:16–21. [Google Scholar]
- Nakamura K, Akiyama H, Kawano N, Kobayashi T, Yoshimatsu K, Mano J, Kitta K, Ohmori K, Noguchi A, Kondo K, Teshima R. Evaluation of real-time PCR detection methods for detecting rice products contaminated by rice genetically modified with a CpTI-KDEL-T-nos transgenic construct. Food Chem. 2013;141:2618–2624. doi: 10.1016/j.foodchem.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Nandakumar N, Singh AK, Sharma RK, Mohapatra T, Prabhu KV, Zaman FU. Molecular fingerprinting of hybrids and assessment of genetic purity of hybrid seeds in rice using microsatellite markers. Euphytica. 2004;136:257–264. [Google Scholar]
- Narshimulu G, Jamaloddin M, Vemireddy LR, Anuradha G, Siddiq EA. Potentiality of evenly distributed hypervariable microsatellite markers in marker-assisted breeding of rice. Plant Breed. 2011;130:314–320. [Google Scholar]
- Ohtsubo K, Nakamura S (2007) Cultivar identification of rice (Oryza sativa L.) by PCR method and its application to processed rice products. J Appl Glicosci 54:235–243 [DOI] [PubMed]
- Ohtsubo K, Suzuki K, Haraguchi K, Nakamura S. Novel method for preparation of the template DNA and selection of primers to differentiate the material rice cultivars of rice wine by PCR. J Biochem Biophys Methods. 2008;70:1020–1028. doi: 10.1016/j.jbbm.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Okunishi T, Nakamura S, Ohtsubo K. Quantitative identification of rice cultivars by Real-Time PCR. Food Sci Technol Res. 2005;11:344–348. [Google Scholar]
- Osborne BG, Mertens B, Thompson M, Fearn T. The authentication of Basmati rice using near infrared spectroscopy. J Near Infrared Spectrosc. 1993;1:77–83. [Google Scholar]
- Osborne BG, Mertens B, Thompson M, Fearn T. The authentication of Basmati rice using near-infrared spectroscopy. J Near Infrared Spectrosc. 1997;1:77–83. [Google Scholar]
- Pal S, Jain S, Saini N, Aarti N, Jain RK. Identification of microsatellite markers for differentiating some Basmati and non-Basmati rice varieties. Indian J Biotechnol. 2004;3:519–526. [Google Scholar]
- Pitiphunpong S, Champangern S, Suwannaporn P. The Jasmine rice (KDML 105 variety) adulteration detection using physico-chemical properties. Chiang Mai J Sci. 2011;38:105–115. [Google Scholar]
- Primrose S, Woolfe M, Rollinson S. Food forensics: methods for determining the authenticity of foodstuffs. Trends Food Sci Technol. 2010;21:582–590. [Google Scholar]
- Rahman SN, Islam MS, Nasiruddin KM. Genetic polymorphism in rice (Oryza sativa L) through RAPD analysis. Indian J Biotechnol. 2007;6:224–229. [Google Scholar]
- Rao RSP, Muralikrishna G. Non-starch polysaccharide-phenolic acid complexes from native and germinated cereals and millet. Food Chem. 2004;84:527–531. [Google Scholar]
- Rittiron R, Saranwong S, Kawano S. Detection of variety contamination in milled Japanese rice using a single kernel near infrared technique in transmittance mode. J Near Infrared Spectrosc. 2005;13:19–25. [Google Scholar]
- Saini N, Jain N, Jain S, Jain RK. Assessment of genetic diversity within and among Basmati and non-Basmati rice varieties using AFLP, ISSR and SSR markers. Euphytica. 2004;140:133–146. [Google Scholar]
- Shen F, Yang D, Ying Y, Li B, Zheng Y, Jiang T (2010) Discrimination between Shaoxing wines and other Chinese rice wines by near-infrared spectroscopy and chemometrics. Food Bioprocess Technology doi:10.1007/s11947-010-0347-z
- Shirasawa K, Shiokai S, Yamaguchi M, Kishitani S, Nishio T. Dot-blot-SNP analysis for practical plant breeding and cultivar identification in rice. Theor Appl Genet. 2006;113:147–155. doi: 10.1007/s00122-006-0281-7. [DOI] [PubMed] [Google Scholar]
- Siddiq EA (1982) Breeding for quality improvement in rice-present state and strategy for the future. In: Golden Jubilee Symposium on increase in the Productivity of Rice, Rice Research Stations, Chinsurah and Ban Kura, West Bengal, India
- Siddiq EA, Nerkar YS, Mehta SL. Intra and inter subspecific variation in soluble proteins of Oryza sativa L. Theor Appl Genet. 1972;42(8):351–356. doi: 10.1007/BF00275360. [DOI] [PubMed] [Google Scholar]
- Siddiq EA, Vemireddy LR, Nagaraju J (2012) Basmati Rices: Genetics, Breeding and Trade. Agric Res 1(1):25–36 doi:10.1007/s40003-011-0011-5
- Singh RK, Sharma RK, Singh AK, Singh VP, Singh NK, Tiwari SP, Mohapatra T. Suitability of mapped sequence tagged microsatellite site markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica. 2004;135:135–143. [Google Scholar]
- Singhal RS, Kulkarni PR, Rege DV. Handbook of indices of food quality and authenticity. Cambridge: Woodhead Publishing Limited; 1997. [Google Scholar]
- Siwach P, Jain S, Saini N, Chowdhury VK, Jain RK. Allelic diversity among Basmati and non-Basmati long-grain Indica rice varieties using microsatellite markers. J Plant Biochem Biotechnol. 2004;13:25–32. [Google Scholar]
- Sood BC, Siddiq EA. A rapid technique for scent determination in rice. Indian J Genet Plant Breed. 1978;38:268–271. [Google Scholar]
- Spaniolas S, May ST, Bennett MJ, Tucker GA. Authentication of coffee by means of PCR-RFLP analysis and lab-on-a-chip capillary electrophoresis. J Agric Food Chem. 2006;54:7466–7470. doi: 10.1021/jf061164n. [DOI] [PubMed] [Google Scholar]
- Srikumar TS. The mineral and trace element composition of vegetables, pulses and cereals of southern India. Food Chem. 1993;46:163–167. [Google Scholar]
- Sriseadka T, Wongpornchai S, Kitsawatpaiboon P. Rapid method for quantitative analysis of the aroma impact compound, 2-acetyl-1-pyrroline, in fragrant rice using automated headspace gas chromatography. J Agric Food Chem. 2006;54:8183–8189. doi: 10.1021/jf0614490. [DOI] [PubMed] [Google Scholar]
- Srividhya A, Vemireddy LR, Hariprasad AS, Jayaprada M, Sridhar S, Ramanarao P, Anuradha G, Siddiq EA (2010) Identification and mapping of landrace derived QTL associated with yield and its components in rice under different nitrogen levels and environments. Int J Plant breed Genet 4(4):210–227
- Steele KA, Ogden R, McEwing R, Briggs H, Gorham J. InDel markers distinguish Basmatis from other fragrant rice varieties. Field Crop Res. 2008;105:81–87. [Google Scholar]
- Storck CR, Picolli da Silva L, Fagundes CA. Categorizing rice cultivars based on differences in chemical composition. J Food Compos Anal. 2005;18:333–341. [Google Scholar]
- Sundaram RM, Naveenkumar B, Biradar SK, Balachandran SM, Mishra B, IlyasAhmed M, Viraktamath BC, Ramesha MS, Sarma NP. Identification of informative SSR markers capable of distinguishing hybrid rice parental lines and their utilization in seed purity assessment. Euphytica. 2008;163:215–224. [Google Scholar]
- Suzuki Y, Ise K, Li C, Honda I, Iwai Y, Matsukura U. Volatile components in stored rice [oryza sativa (L.)] of varieties with and without lipoxygenase-3 in seeds. J Agric Food Chem. 1999;47:1119–1124. doi: 10.1021/jf980967a. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Chikaraishi Y, Ogawa NO, Ohkouchi N, Korenaga T. Geographical origin of polished rice based on multiple element and stable isotope analyses. Food Chem. 2008;109:470–475. doi: 10.1016/j.foodchem.2007.12.063. [DOI] [PubMed] [Google Scholar]
- Tang T, Huang J, Zhong Y, Shi S. High-throughput S-SAP by fluorescent multiplex PCR and capillary electrophoresis in plants. J Biotechnol. 2004;114:59–68. doi: 10.1016/j.jbiotec.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Thind GK, Sogi DS. Identification of coarse (IR-8), fine (PR-106) and superfine (Basmati-386) rice cultivars. Food Chem. 2005;91:227–233. [Google Scholar]
- Vaingankar NM, Kulkarni PR. A cooking quality parameter as an indicator of adulteration of Basmati rice. J Sci Food Agric. 1989;48:381–384. [Google Scholar]
- Vemireddy LR, Archak S, Nagaraju J. Capillary electrophoresis is essential for microsatellite marker based detection and quantification of adulteration of Basmati rice (Oryza sativa) J Agric Food Chem. 2007;55:8112–8117. doi: 10.1021/jf0714517. [DOI] [PubMed] [Google Scholar]
- Vemireddy LR, Ranjithkumar N, Vipparla A, Surapaneni M, Choudhary G, Sudhakarrao KV, Siddiq EA (2014) Molecular profiling of major indian rice cultivars using a set of eight hypervariable microsatellite markers. Cereal Res Commun (epub)
- Vlachos A, Arvanitoyannis IS. A review of rice authenticity/adulteration methods and results. Crit Rev Food Sci Nutr. 2008;48:553–598. doi: 10.1080/10408390701558175. [DOI] [PubMed] [Google Scholar]
- Voorhuijzen MM, van Dijk JP, Prins TW, Van Hoef AM, Seyfarth R, Kok EJ. Development of a multiplex DNA-based traceability tool for crop plant materials. Anal Bioanal Chem. 2012;402:693–701. doi: 10.1007/s00216-011-5534-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja R, Craske JD, Wootton M. Changes in volatile components of paddy, brown and white fragrant rice during storage. J Sci Food Agric. 1996;71:218–224. [Google Scholar]
- Woolfe M, Primrose S. Food forensics: using DNA technology to combat misdescription and fraud. Review. Trends Biotechnol. 2004;22:222–226. doi: 10.1016/j.tibtech.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Xue X, Wang Q, Li Y, Wu L, Chen L, Zhao J, Liu F. 2-Acetylfuran-3-glucopyranoside as a novel marker for the detection of honey adulterated with rice syrup. J Agric Food Chem. 2013;61:7488–7493. doi: 10.1021/jf401912u. [DOI] [PubMed] [Google Scholar]
- Yadav S, Singh A, Singh MR, Goel N, Vinod KK, Mohapatra T, Singh AK. Assessment of genetic diversity in Indian rice germplasm (Oryza sativa L.): use of random versus trait-linked microsatellite markers. J Genet. 2013;92:545–557. doi: 10.1007/s12041-013-0312-5. [DOI] [PubMed] [Google Scholar]
- Yashui A, Shindoh K. Determination of the geographic origin of brown-rice with trace-element composition. Bunseki Kagaku. 2000;49:405–410. [Google Scholar]
- Yu H, Zhou Y, Fu X, Xie L, Ying Y. Discrimination between Chinese rice wines of different geographical origins by NIRS and AAS. Eur Food Res Technol. 2007;225:313–320. [Google Scholar]
- Zhang Q, Maroof MA, Lu TY, Shen BZ. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor Appl Genet. 1992;83:495–499. doi: 10.1007/BF00226539. [DOI] [PubMed] [Google Scholar]