Abstract
Transducer Like Proteins (Tlps), also known as methyl accepting chemotaxis proteins (MCP), enable enteric pathogens to respond to changing nutrient levels in the environment by mediating taxis toward or away from specific chemoeffector molecules. Despite recent advances in the characterization of chemotaxis responses in Campylobacter jejuni, the impact of Tlps on the adaptation of this pathogen to disparate niches and hosts is not fully characterized. The latter is particularly evident in the case of C. jejuni 81-176, a strain that is known to be highly invasive. Furthermore, the cytoplasmic group C Tlps (Tlp5, 6, and 8) were not extensively evaluated. Here, we investigated the role of C. jejuni 81-176 Tlps in chemotaxis toward various substrates, biofilm formation, in vitro interaction with human intestinal cells, and chicken colonization. We found that the Δtlp6 and Δtlp10 mutants exhibited decreased chemotaxis toward aspartate, whereas the Δtlp6 mutant displayed a decreased chemotaxis toward Tri-Carboxylic Acid (TCA) cycle intermediates such as pyruvate, isocitrate, and succinate. Our findings also corroborated that more than one Tlp is involved in mediating chemotaxis toward the same nutrient. The deletion of tlps affected important phenotypes such as motility, biofilm formation, and invasion of human intestinal epithelial cells (INT-407). The Δtlp8 mutant displayed increased motility in soft agar and showed decreased biofilm formation. The Δtlp8 and Δtlp9 mutants were significantly defective in invasion in INT-407 cells. The Δtlp10 mutant was defective in colonization of the chicken proximal and distal gastrointestinal tract, while the Δtlp6 and Δtlp8 mutants showed reduced colonization of the duodenum and jejunum. Our results highlight the importance of Tlps in C. jejuni's adaptation and pathobiology.
Keywords: chemotaxis, methyl accepting chemoreceptors, colonization, chicken, motility, organic acids, amino acids, virulence
Introduction
Campylobacter jejuni is a Gram-negative, thermophilic, and obligate microaerophilic bacterium that accounted for 7.5 million disability adjusted life years (DALYs) globally in 2010 (Murray et al., 2012). C. jejuni infects approximately 1 million people in the USA each year (Scallan et al., 2011). In 2012, 14.3 out of every 100,000 people were infected with Campylobacter, surpassing the 12.3 person goal set by the Centers for Disease Control and Prevention (CDC, 2013). These data highlight the need to control the incidence of food-borne C. jejuni outbreaks.
Chickens are a major source of C. jejuni, while a small proportion of campylobacteriosis cases can be attributed to other farm animals, and environmental sources (Wilson et al., 2008). However, the factors that contribute to successful colonization (in poultry) and virulence (in humans) remain not well defined. The ability to adapt to disparate environments is a key to C. jejuni's diverse lifestyles in different hosts and its effective colonization of the gut (Hermans et al., 2011). These observations emphasize the need for a better understanding of the factors that contribute to C. jejuni's adaptation.
Chemotaxis allows motile bacteria to travel toward a favorable niche or away from unfavorable conditions. Transducer Like Proteins (Tlp), also known as Methyl Accepting Chemotaxis Proteins (MCP), are chemoreceptors which mediate chemotaxis toward or away from chemoeffector molecules in an environment (Wadhams and Armitage, 2004). Although the basic components of chemotaxis are conserved among bacteria (Wadhams and Armitage, 2004), the number of Tlps/MCPs varies and has a weak correlation with bacterial genome size (Lacal et al., 2010). However, bacteria possessing genomes with higher number of Tlps such as C. jejuni are characterized by a complex lifestyle and a remarkable ability to interact with the host and other bacteria (Lacal et al., 2010). Notably, the genome sequence of C. jejuni NCTC11168 revealed homologs for ten putative chemotactic sensory receptors (Marchant et al., 2002). In C. jejuni, Tlps are classified into groups A, B and C, based on predicted domain structure and homology to chemoreceptors of other bacteria (Marchant et al., 2002). The group A Tlps (Tlp1, 2, 3, 4, 7, and 10) are integral membrane proteins with two transmembrane domains, a periplasmic ligand binding domain, and a conserved cytoplasmic signaling domain (Marchant et al., 2002). Group B Tlps [Tlp9 (CetA)] have two transmembrane domains and assist in energy taxis along with the signal sensing protein CetB (Marchant et al., 2002; Vegge et al., 2009). The group C Tlps (Tlp5, 6, and 8) possess cytoplasmic signaling domains but lack the transmembrane and periplasmic binding domain (Marchant et al., 2002). Previous research (e.g., Korolik and Ketley, 2008; Vegge et al., 2009; Tareen et al., 2010; Reuter and van Vliet, 2013; Li et al., 2014; Rahman et al., 2014) and the occurrence of multiple Tlps in the relatively small genome of C. jejuni strongly suggest that investigating these proteins might reveal crucial insights into the pathobiology of this pathogen.
C. jejuni has been shown to be chemotactic toward L-fucose, L-aspartate, L-cysteine, L-glutamate, L-serine, organic acid salts (pyruvate, succinate, fumarate, citrate, malate, and alpha-ketoglutarate), bile (from beef and chicken), and mucin (bovine gallbladder, and chicken and hog gastric mucin) (Hugdahl et al., 1988; Korolik and Ketley, 2008; Rahman et al., 2014). Nevertheless, our understanding of the role of Tlps in C. jejuni pathogenesis is still limited, especially with respect to the biological functions of the cytoplasmic group C Tlps. It is important to note that most of the previous studies on Tlps have been conducted using C. jejuni NCTC11168 or its variants (Table 1). For example, Tlp1, 3, 4, 7, 8, and 9 have been previously studied using C. jejuni NCTC11168 or NCTC11168-O (Vegge et al., 2009; Tareen et al., 2010; Reuter and van Vliet, 2013; Li et al., 2014; Rahman et al., 2014). However, the expression of tlp genes can vary based on growth conditions, isolation source, and strains (Day et al., 2012). Importantly, the overall contributions of Tlps to the success of highly invasive strains such as C. jejuni 81-176 have not received equal attention. C. jejuni 81-176 was originally isolated from the stool samples collected from a campylobacteriosis outbreak, which was associated with the consumption of raw milk (Korlath et al., 1985). C. jejuni 81-176 is known to have a relatively high invasion capacity in human epithelial intestinal cells in vitro (Hu and Kopecko, 1999), while this strain was experimentally shown to cause inflammatory diarrhea in human trials (Black et al., 1988). Although C. jejuni 81-176 and C. jejuni NCTC11168 are said to have the same complement of Tlp genes (Rahman et al., 2014), these strains are also known to have a marked difference in the genes encoding two Tlps. Specifically, tlp3 has a naturally occurring mutation in C. jejuni 81-176 (Korolik and Ketley, 2008; Day et al., 2012), while tlp7, encoded by two genes (cj0951c and cj0952) in C. jejuni NCTC11168, is encoded by a single gene (cjj81176-0975) in C. jejuni 81-176 (Tareen et al., 2010). It was reported that the tlp7 mutant was deficient in chemotaxis toward formic acid in C. jejuni B2 which, similar to NCTC11168, also has two genes encoding Tlp7 (Tareen et al., 2010). However, complementation using the corresponding gene from C. jejuni 81-176 restored the phenotype (Tareen et al., 2010). Subsequently, the functional significance of the difference in tlp7 between the strains is not currently known. Furthermore, the impairment of tlp3 in C. jejuni NCTC11168-O significantly reduced sodium deoxycholate-induced chemotaxis, colonization of jejunal mucosa in mice (Li et al., 2014), and motility and invasion of epithelial cell line, Caco-2 but had no significant impact on chicken colonization (Rahman et al., 2014). In another study, a C. jejuni NCTC11168 tlp3 mutant also showed a decreased invasion of human Colo 205 epithelial cells and chicken embryo intestinal cells in vitro but the mutant's motility in a rich medium was not affected (Vegge et al., 2009). However, although C. jejuni 81-176 inconsistently and inefficiently colonized wild-type C3H mice (Chang and Miller, 2006), which is consistent with the aforementioned tlp3 mutation, the strain is still capable of infecting humans and colonizing chickens (Black et al., 1988; Hendrixson and Dirita, 2004). Regardless, it is plausible that, in terms of specific tissue infection, high invasive capabilities might expose the pathogen and its Tlps to otherwise inaccessible niches and cognate substrates. Therefore, in this study, we investigated Tlps in the highly invasive C. jejuni 81-176 strain for their role in chemotaxis, biofilm formation, in-vitro interaction with human intestinal epithelial cells, and tissue specific colonization in the chicken host. For this purpose, we created isogenic mutants targeting all three Tlp groups, specifically tlp4 and tlp10 (Group A), tlp9 (Group B), and tlp6 and tlp8 (Group C). A mutant targeting the last group C Tlp (tlp5) was not included, because we were unsuccessful in deleting the gene, which corroborated the observations of Hendrixson et al. (2001). Our data confirmed that more than one Tlp can mediate chemotaxis toward a particular substrate. Furthermore, certain Tlps contributed substantially to persistence (biofilm formation), interaction with human intestinal cells (INT 407), and chicken tissue colonization by C. jejuni 81-176.
Table 1.
Major and relevant findings on C. jejuni Tlps from previous studies.
| Tlp protein | Protein designation | Ligand | Mutant phenotype | This study (C. jejuni 81-176) | ||
|---|---|---|---|---|---|---|
| Motility | In vivo colonization of chickens | Other | ||||
| Tlp4 - 81-176 (Hendrixson and Dirita, 2004) - NCTC11168 (Vegge et al., 2009) -NCTC11168-O (Li et al., 2014) | Tlp4 (DocC) | - Sodium deoxycholate (Li et al., 2014) - No chemotaxis defect (Vegge et al., 2009) | - Decreased (Vegge et al., 2009) | - Decreased (Hendrixson and Dirita, 2004) | - Decreased invasion (Human Colo 205 cells, chicken embryo intestinal cells) | - No significant deficiency in the phenotypes was observed |
| Tlp5 - 81-176 (Hendrixson et al., 2001) | Tlp5 | - Unknown | - Mutant could not be generated | - Mutant could not be generated | - Mutant could not be generated | - Mutant could not be generated |
| Tlp6 - 81-176 (Hendrixson and Dirita, 2004) | Tlp6 | - Unknown | - Unknown | - No effect (cecum) | - Unknown | Decreased chemotaxis toward aspartate, isocitrate, succinate, and propionate - Decreased colonization of the chicken duodenum and jejunum |
| Tlp8 - NCTC11168 (Reuter and van Vliet, 2013) | CetZ | - Unknown (Energy taxis) | - No effect | No effect | - Opposing role in energy taxis | - Increased motility - Decreased biofilm formation - Decreased colonization of the chicken duodenum and jejunum - Decreased invasion in INT 407 cells |
| Tlp9 - NCTC11168 (Vegge et al., 2009; Reuter and van Vliet, 2013) - 81-176 (Hendrixson et al., 2001) | CetA | - Energy taxis - Decreased chemotaxis toward pyruvate | - Decreased (Vegge et al., 2009; Reuter and van Vliet, 2013) - Decreased in defined medium (Hendrixson et al., 2001) | - No effect | - Unknown | - Decreased invasion in INT 407 cells |
| Tlp10 - 81-176 (Hendrixson and Dirita, 2004) - NCTC11168 (Vegge et al., 2009) | DocB | - Unknown | - No effect | - Decreased in ceca and in proximal and distal intestinal tract | - Decreased invasion (NCTC11168 in Colo 205 cells, chicken embryo intestinal cells) (Vegge et al., 2009) | - Decreased chemotaxis toward aspartate and fumarate -Colonization defect in the chicken cecum, duodenum and jejunum |
Materials and methods
Ethics statement
Animal experiments were conducted according to the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The animal studies were approved by the Institutional Animal Care and Use Committee (IACUC), the Ohio State University. Chickens were housed at the Food Animal Health Research Program Animal Care Facility, which is fully accredited by AAALAC and the animals were supervised by a senior veterinarian. Infectious agents were administered using manual restraint for less than 1 min to minimize distress. Before necropsy, chickens were euthanized by carbon dioxide inhalation. This method is consistent with the recommendations of the panel on euthanasia of the American Veterinary Medical Association and by the Ohio State University Institutional Laboratory Animal Care and Use Committee.
Bacterial strains, media, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table S1. C. jejuni 81–176 (WT) was used to generate the tlp deletion mutants. C. jejuni strains were routinely grown on Mueller-Hinton agar (MH; Oxoid, Hampshire, United Kingdom) microaerobically [(85% N2 (v/v), 10% CO2 (v/v) and 5% O2 (v/v)] in a DG250 Microaerophilic Workstation (Microbiology International, Frederick, Maryland, USA) at 42°C. E. coli DH5α was used for cloning purposes and was routinely cultured using the Luria-Bertani (LB) medium at 37°C overnight. Growth media was supplemented with appropriate antibiotics, chloramphenicol (20 μg/ml for E. coli; 10 μg/ml for Campylobacter), kanamycin (30 μg/ml), and zeocin (50 μg/ml) where necessary.
Targeted deletion of tlp genes
Deletion of target genes (tlp4:cjj_0289; tlp6:cjj_0473; tlp8:cjj_1128; tlp9:cjj_1205; and tlp10: cjj_0046) was achieved by double crossover homologous recombination using a suicide vector containing homologous sequences on either side of the respective tlp gene as described previously (Gangaiah et al., 2009; Rajashekara et al., 2009). Briefly, a 1 kb flanking region on either side of the target gene along with coding regions of the gene were amplified by PCR using the specific tlp F and R primers and C. jejuni 81–176 genomic DNA (Table S2). The PCR product was ligated into pZErO-1 to generate the plasmid pZErO-1-tlp using Fastlink™ DNA ligation kit (Epicentre, Madison). Inverse PCR was performed on pZErO1-tlp using the specific tlp INV F and INV R primers (Table S2) to amplify the plasmid without the majority of the tlp coding sequence; except for approximately 50 bp at the ends of the original tlp gene sequence. The kanamycin cassette from pUC4K was then cloned into the inverse PCR product to generate the suicide vectors pZErO-1-inv-tlp. The resulting suicide vectors were electroporated into C. jejuni 81–176 (Wilson et al., 2003). Recombinants were selected on MH agar plates containing the appropriate antibiotics. The deletion of the specific tlp gene was confirmed by PCR.
Complementation of the Δtlp mutants
The respective tlp coding sequences along with their potential promoter regions (~100–200 bp upstream of the target gene) were amplified by PCR using specific primers and cloned into pRY111, an E. coli-Campylobacter shuttle vector (Yao et al., 1993) (Table S2). Since potential promoter regions for tlp6 and tlp10 are located further upstream, sequence encoding the open reading frame (ORF) with the ribosomal binding site was cloned into pRY111 followed by directional cloning of the potential promoter region upstream of the ORF [For tlp6 (cjj_0473): intergenic region between cjj_0474 and rpmB; for tlp10 (cjj_0046): region between ccpA (cjj_0047) and cjj_0048]. The recombinant plasmids were then transformed into E. coli DH5α, purified, and subjected to restriction analysis to confirm that they carried the desired sequence. The plasmids were introduced into the appropriate tlp mutant by biparental conjugation as described previously (Miller et al., 2000). Transconjugants were selected on MH agar plates containing kanamycin and chloramphenicol and one transconjugant for each target was selected, further confirmed by PCR to verify the presence of WT copy of the cognate gene, designated as tlp comp, and used in complementation studies.
Syringe capillary assay for chemotaxis
To quantify chemotaxis, we adapted a modified capillary chemotaxis assay that quantitatively measured bacterial tactic responses (Figure S1). Capillary assays are the most commonly used methods to assess bacterial chemotaxis (Lewus and Ford, 2001). The assay was previously used for quantifying chemotaxis in subsurface microaerophilic bacteria (Mazumder et al., 1999) and other Epsilonproteobacteria, namely Helicobacter pylori (Cerda et al., 2003, 2011). Bacterial culture from each C. jejuni strain grown microaerobically at 42°C for 18 h on MH agar were harvested using a sterile disposable loop, washed and resuspended by gentle pipetting in chemotaxis buffer (Phosphate Buffered Saline, pH 7.4), and OD600 was adjusted to 0.5. A 100 μl of a solution containing 100 mM of the compound to be tested for chemotactic response (buffer alone served as a control) was aspirated through a 22 G stainless-steel needle (0.25 mm diameter × 20 mm long) into a 1 ml tuberculin syringe. The concentration of the compound was selected based on previous studies and a series of preliminary experiments that showed that 100 mM resulted in the strongest chemotaxis response (Vegge et al., 2009; Tareen et al., 2010). A 100 μl of the adjusted bacterial suspension was drawn into a 200 μl disposable pipette tip, which was then sealed from one end. The needle-syringe system was fitted to a pipette tip in such a way that most of the needle immersed into the bacterial suspension. The system was positioned horizontally and incubated at 42°C for 1 h. Finally, the needle-syringe system was detached and bacterial suspension in the syringe was 10-fold serially diluted in the chemotaxis buffer. Dilutions were plated onto MH agar plates, incubated for 24 h at 42°C microaerobically, and the CFU were counted. Each assay was performed in triplicate. Results were expressed as the mean of three independent assays. To express the taxis toward a test compound, Relative Chemotaxis Ratio (RCR) was calculated as the ratio between the numbers of bacteria entering the test needle-syringes and those in the control (buffer only) needle-syringes. A test compound was considered as an attractant if the RCR was significantly = 2 (P < 0.05) (Adler, 1973; Moulton and Montie, 1979; Mazumder et al., 1999; Cerda et al., 2003). A mutant was considered deficient in chemotaxis toward a substrate if both the corresponding RCR-value was significantly < 2 (P < 0.05) and the CFU of the mutants were significantly lower (P < 0.05) than those of the wildtype. A C. jejuni 81-176 cheY mutant which is incapable of directional movement (negative control; Yao et al., 1997) and 0.1% porcine gastric mucin (positive control; Sigma) were also used to evaluate the integrity of the assay. To test the response to repellents, C. jejuni cultures were mixed with a repellent and the bacteria that entered the syringe, which in this instance contained only buffer, to escape the repellent were quantified as described above. To account for any methodological bias, capillary chemotaxis results were further verified by using the disc method (Vegge et al., 2009; Tareen et al., 2010) for selected compounds.
Motility and biofilm assays
To determine motility, C. jejuni mutants and the wild type (WT) were grown microaerobically at 42°C for 18–24 h on MH agar and resuspended in MH broth to an OD600 of 0.05. Bacterial cells were stabbed (2 μl) with pipette tips into semisolid MH agar (0.4% agar) plates. Plates were incubated for 48 h at 42°C microaerobically. Motility was assessed by measuring the diameter of the zone of motility after 48 h (Rajashekara et al., 2009). A C. jejuni 81-176 cheY mutant which is deficient in motility and the RY213 strain (diploid for cheY) which exhibits increased motility (Yao et al., 1997) were used as controls.
Static biofilm formation was assessed in borosilicate tubes by incubating 1 ml of 0.05 OD600 adjusted MH broth culture at 42°C microaerobically for 72 h without shaking (Gangaiah et al., 2009). The biofilms were stained with 1% crystal violet for 15 min, washed to remove excess stain, and quantified by measuring the absorbance (OD570) after incubation in 1 ml DMSO (80%) for 48 h.
Quantitative reverse transcriptase PCR (qRT-PCR) of biofilm associated genes
The C. jejuni WT and Δtlp8 strains were examined for changes in expression of biofilm related genes in 3 day old biofilms and overnight grown planktonic shaking cultures. The cultures were adjusted to an initial OD600 of 0.05 in MH broth and incubated at 42°C microaerobically either in standing condition for 3 days (biofilm growth) or overnight under shaking at 100 rpm (planktonic). Total RNA was extracted using RNeasy Mini Kit (Qiagen) and cDNA was synthesized using SuperScript® III First-Strand Synthesis SuperMix (Invitrogen). RNA and cDNA concentrations and purity were determined using NanoDrop ND-2000c spectrophotometer (Wilmington, DE).
qRT-PCR analysis was performed on genes involved in biofilm formation. These included: kpsM, capsular polysaccharide synthesis; pglH, protein glycosylation; neuB, lipo-oligosaccharide; fliS, flagella protein synthesis; cj0688, phosphate acetyltransferase; maf5, flagella formation and luxS, quorum sensing (Joshua et al., 2006; Reeser et al., 2007). Gene specific primers (Table S2) used in this analysis have been described previously (Drozd et al., 2014). qRT-PCR was performed using SensiMixPlus® SYBR RT-PCR Kit (Quantace, Norwood, MA) in a Realplex2 Mastercycler (Eppendorf, Westbury, NY). Nuclease free water (no DNA) was used as negative control. The relative levels of expression of target genes were normalized to 16S rRNA gene expression of the same strain. The relative fold changes in gene expression was calculated using the comparative threshold cycle (CT) method to yield fold-difference in transcript level compared to WT (Livak and Schmittgen, 2001). The qRT-PCR was performed three times with duplicate samples in each assay.
Adherence and invasion assay
As described in Kassem et al. (2012) and Gaynor et al. (2005), each well of a 24-well tissue culture plate was seeded with 1.4 × 105 INT 407 cells (human embryonic intestine cells, ATCC CCL 6) in MEM with 10% (v/v) fetal bovine serum (FBS) and incubated for 18 h at 37°C under 5% CO2. Following incubation, cells were infected with C. jejuni WT and mutants at a multiplicity of infection 100:1 in both adherence and invasion assays. To assay for adherence, the infected monolayers were incubated for 3 h followed by washing the INT 407 cells 3 times with MEM. Cells were then lysed using 0.1% (v/v) Triton X-100, serially diluted (10-fold) in MEM and 100 μl of each dilution were spread on MH agar plates. The plates were incubated for 48 h at 42°C under microaerobic conditions after which CFU were counted. To assess invasion, INT 407 cells were infected for 3 h at 37°C after which cells were treated with gentamicin (150 μg/ml) and incubated for an additional 2 h. Subsequently, infected cells were rinsed with MEM three times, lysed with 0.1% Triton-X 100, serially diluted in MEM, and spread on MH agar plates to determine the number of CFU. In parallel, we also cultured the supernatant of gentamicin treated monolayers to ensure the quality of the gentamicin protection assay.
Chicken colonization studies
Three day-old specific pathogen free chicks (n = 6/group) from our hatching facility (Food Animal Health Research Program, OARDC, Wooster, OH) were used in this study. Birds in each group were housed in a separate cage, and the birds were further confirmed to be Campylobacter free by testing cloacal samples. The birds were then inoculated orally with 104 CFU of the C. jejuni WT and Δtlp6, 8, 9, and 10 mutants in 200 μl of 1X PBS (pH 7.4). Chicks were euthanized 7 days post-inoculation. The ceca, duodenum, jejunum, liver, spleen, and bursa were collected aseptically, weighed, homogenized, serially diluted in 1X PBS (pH 7.4), and 100 μl of the homogenates were spread onto MH agar containing Campylobacter selective supplement with or without kanamycin. Plates were incubated at 42°C microaerobically and CFU/gram of tissues was determined.
Statistical analysis
Data analysis was performed using One-Way analysis of variance (ANOVA) with Tukey's post-test. Statistical significance of data was also determined using Student's t-test in cases where only two data sets were compared. Data from the chicken colonization experiment were analyzed using the Mann-Whitney test. P = 0.05 (α level) was considered statistically significant.
Results
Contribution of tlps to chemotaxis in response to amino acids and organic acids
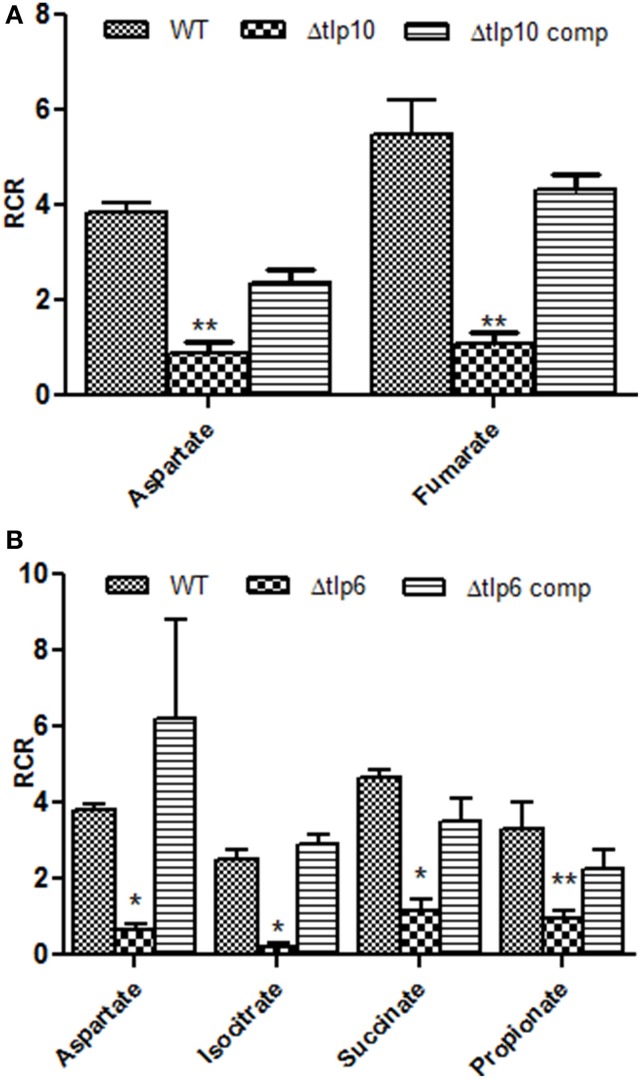
To determine the contribution of Tlps toward chemotaxis, we generated deletion mutants of tlp4, 6, 8, 9, and 10. The mutants did not exhibit a growth defect while incubated in MH broth (data not shown). Our preliminary analysis indicated that RCR-values for WT C. jejuni toward known chemoattractants were >2 and for chemorepellants such as bile acids (cholic acid and deoxycholic acid) was < 0.1 (Hugdahl et al., 1988; Cerda et al., 2003), which further validated the usefulness of the syringe-capillary method to assess chemotaxis in C. jejuni. Furthermore, using the capillary assay we show that C. jejuni 81-176 exhibited strong chemotaxis toward 0.1% porcine gastric mucin (RCR = 9.0), while a cheY mutant (non-motile) had an RCR below the detection limit (~ 0), because no or very few colonies were retrieved from the syringe containing tested chemoattractants (aspartate, fumarate, propionate, and pyruvate). All compounds tested in this chemotaxis assay were used at a concentration of 100 mM which was determined based on our preliminary studies using varying concentrations of compounds from 10 to 100 mM (Figure S2A). Figure 1 shows tlp mutants with significantly defective chemotaxis against various substrates tested in this study. Chemotactic responses of tlp mutants to various substrates are shown in Table 2. Furthermore, the response of the mutants to chemorepellents (Cholic acid and Deoxycholic acid) was not significantly different compared to those of the wildtype. Specifically, all strains exhibited an RCR of <0.1 in response to the repellents.
Figure 1.
The Δtlp mutants that are defective in chemotaxis toward essential nutrients. Chemotaxis of tlp10 (A) and tlp6 (B) was assessed by determining the relative chemotactic ratio (RCR) which is the ratio of bacterial numbers migrating toward the chemical in the syringe to the bacterial numbers migrating toward the chemotaxis buffer. Chemotaxis was determined using the capillary method and significant results were further confirmed using the disc diffusion method (Table S3). Mucin and a ΔcheY mutant were used as a positive and negative control, respectively. An RCR-value of 2 or above indicates chemotaxis toward the test chemical. Only those Δtlp mutants with a significant defect in chemotaxis are shown and the complete chemotaxis results for all compounds tested are listed in Table 2. The data represent the average and SE of three experiments with 3 replicates in each experiment. *P ≤ 0.05, **P ≤ 0.01.
Table 2.
RCR-values for the WT and chemotaxis mutants for all compounds tested.
| Compound | WT | Δtlp4 | Δtlp6 | Δtlp8 | Δtlp9 | Δtlp10 |
|---|---|---|---|---|---|---|
| Average CFU | Average CFU | Average CFU | Average CFU | Average CFU | Average CFU | |
| (RCR)a | (RCR) | (RCR) | (RCR) | (RCR) | (RCR) | |
| Aspartate | (9.76 ± 0.85)*105 (3.81 ± 0.3) | (2.21 ± 0.63)*106 (8.65 ± 2.46) | (1.74 ± 0.59)*105 (0.68 ± 0.23) | (6.17 ± 1.56)*105 (2.41 ± 0.61) | (8.69 ± 3.33)*105 (3.39 ± 1.3) | (2.30± 1.05)*105 (0.90 ± 0.41) |
| L-glutamine | (3.79 ±0.48)*106 (4.19 ± 0.5) | (1.26 ± 0.14)*107 (13.98 ± 1.48) | (1.64 ± 0.44)*106 (1.81 ± 0.48) | (3.13 ± 0.84)*106 (3.46 ± 0.93) | (3.3 ± 0.78)*106 (3.65 ± 0.87) | (1.56 ± 0.17)*106 (1.73 ± 0.19) |
| L-serine | (1.21± 0.23)*106 (4.03 ± 0.76) | (2.29 ± 0.23)*106 (7.62 ± 0.75) | (1.6± 0.64)*106 (5.32 ± 2.12) | (1.33 ± 0.36)*106 (4.42 ± 1.21) | (1.15 ± 0.32)*106 (3.82 ± 1.08) | (1.19 ± 0.52)*106 (3.95 ± 1.73) |
| Fumarate | (2.47 ±0.58)*106 (5.48 ± 1.28) | (4.64 ± 0.58)*106 (10.32 ± 1.29) | (1.44 ± 0.34)*106 (3.19 ± 0.76) | (1.31 ± 0.14)*106 (2.90 ± 0.31) | (1.89 ± 0.65)*106 (4.21 ± 1.44) | (5.09 ± 2.16)*105 (1.13 ± 0.48) |
| Isocitrate | (7.05 ± 1.15)*105 (2.51 ± 0.41) | (1.17 ± 0.64)*106 (4.17 ± 2.28) | (6.18 ± 3.65)*104 (0.22 ± 0.13) | (1.43 ± 0.45)*106 (5.08 ± 1.59) | (1.59 ± 0.33)*106 (5.65 ± 1.18) | (7.56 ± 2.05) *105 (2.69 ± 0.73) |
| Formate | (6.02 ±0.18)*106 (4.41 ± 0.13) | (8.43 ± 2.57)*106 (6.18 ± 1.88) | (3.29 ± 1.24)*106 (2.41 ± 0.91) | (3.11 ± 0.93)*106 (2.28 ± 0.68) | (6.55 ± 1.66)*106 (4.8 ± 1.22) | (2.32 ± 0.34)*106 (1.7 ± 0.25) |
| Succinate | (1.95 ±0.13)*106 (4.68 ± 0.32) | (3.68 ± 2.32)*106 (8.81 ± 5.56) | (4.88 ± 1.96)*105 (1.17 ± 0.47) | (2.29 ± 0.65)*106 (5.48 ± 1.56) | (2.14 ± 1.75)*106 (5.13 ± 4.18) | (7.81 ± 2.51)*105 (1.87 ± 0.60) |
| Pyruvate | (1.85 ± 0.04)*106 (3.46 ± 0.08) | (2.95 ± 0.21)*106 (5.52 ± 0.39) | (1.07± 0.18)*106 (2.01 ± 0.32) | (1.13± 0.34)*106 (2.11 ± 0.63) | (6.95 ± 0.59)*105 (1.3 ± 0.11) | (1.27 ± 0.3)*106 (2.37 ± 0.55) |
| Propionate | (4.04 ± 1.47)*106 (3.33 ± 1.21) | (3.86 ± 0.17)*106 (3.19 ± 0.14) | (1.14 ± 0.45)*106 (0.94 ± 0.37) | (6.8 ± 2.33)*106 (5.6 ± 1.92) | (6.03 ± 1.15)*106 (4.97 ± 0.95) | (4.38 ± 3.64)*106 (3.61±3) |
The RCR was calculated by taking the ratio of bacterial numbers migrating toward the chemical in the syringe to the bacterial numbers migrating toward the chemotaxis buffer. The results show the means of three independent assays. An RCR-value of 2 and above indicates chemotaxis toward the test chemical (Mazumder et al., 1999). RCR-values that were significantly lower than 2 (P < 0.05) are indicated in bold font.
Based on the RCR indices and CFU numbers, the Δtlp6 and Δtlp10 mutants were defective in chemotaxis toward aspartate compared to WT (Figures 1A,B). The Δtlp10 mutant showed decreased chemotaxis toward fumarate, while the Δtlp6 mutant showed a diminished chemotaxis toward succinate, isocitrate, and propionate (Figure 1B). Chemotaxis defects observed in the mutants was restored to WT levels in the complemented strains; except in Δtlp10 where there was partial complementation for chemotaxis to aspartate (Figure 1). However, this partial complementation observed in case of tlp10 was not due to a polar effect, because the relative expression of a downstream gene (cjj_0045) was similar to the WT (ΔΔCt fold change for cjj_0045 was 1.068). Chemotaxis results observed above for the Δtlp mutants were further confirmed by assessing chemotaxis using the disc method for selected substrates (Table S3).
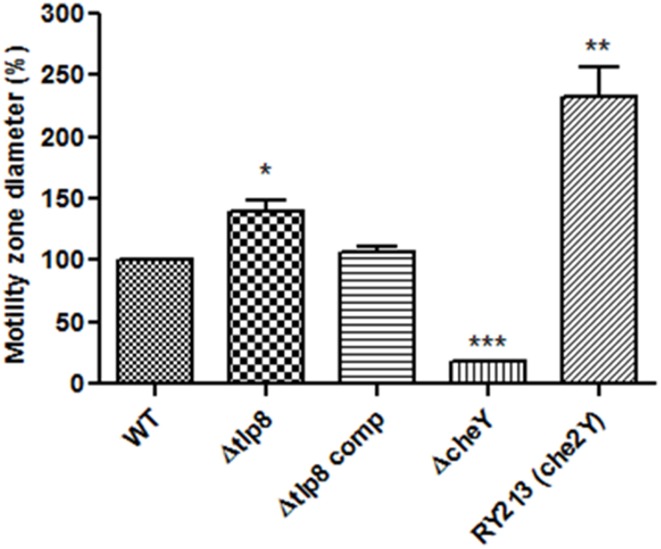
Contribution of tlps towards motility and biofilm formation
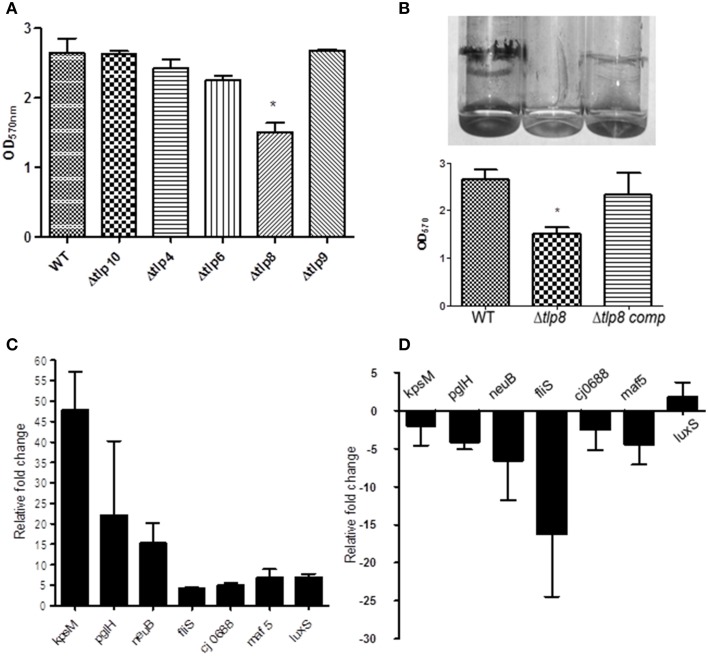
Most of the Δtlp mutants demonstrated either a slight or no defect in motility compared to the WT strain. However, the Δtlp9 mutant showed the most significant defect (a 54.4% decrease, P = 0.0131) (Figure S2B). The Δtlp8 (ΔcetZ) mutant exhibited an increased motility phenotype (Figure 2). Under bright field microscopy, no visible differences were observed in the motility of Δtlp8 mutant compared to the WT (data not shown). However, the Δtlp8 mutant revealed a defect in biofilm formation under microaerobic conditions (Figures 3A,B). To further investigate this observation, we analyzed the expression of genes that have been previously implicated in biofilm formation in the Δtlp8 biofilm and planktonic cultures. The kpsM, pglH, neuB, fliS, maf5, and cj0688 genes were upregulated in 3 day old Δtlp8 biofilms, but they were downregulated in its planktonic culture. However, the luxS gene was upregulated in the Δtlp8 mutant in both the biofilm and the planktonic phases (Figures 3C,D).
Figure 2.
Role of chemoreceptors in motility. Histogram showing the diameter of the zone of motility expressed in percentages relative to the WT. The zone of motility was determined after stabbing 2 μL of C. jejuni (OD600 of 0.05) in the middle of a semi-solid (0.4%) MH agar plate. The agar plates were incubated microaerobically for 48 h and motility was assessed by measuring the diameter of the zone of motility. The Δtlp8 mutant showed increased motility. Motility was restored to values similar to the WT levels in the complemented strain. A C. jejuni 81-176 cheY mutant and the RY213 strain (diploid for cheY) were used as controls. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 3.
Contribution of tlps to biofilm formation. (A) Biofilm was assessed by crystal violet staining. One ml of C. jejuni cultures (OD600 of 0.05) was incubated in borosilicate tubes microaerobically for 72 h, the tubes were washed with distilled water and stained using (1%) crystal violet. Extra stain was removed by washing and the biofilm were suspended in 1 mL DMSO (80%) for quantification by a spectrophotometer. Each bar represents the mean ± SE of 3 independent experiments. *P ≤ 0.05. (B) The Δtlp8 mutant shows decreased biofilm formation and this defect was restored in the complemented strain. Each bar represents the mean ± SE of 3 independent experiments, *P ≤ 0.05. (C,D) Transcriptional changes in biofilm associated genes in the Δtlp8 mutant measured by qRT-PCR using 3 day old biofilm cultures (C) and overnight grown planktonic culture (D). The difference in gene expression relative to the WT was determined by the threshold cycle (CT) method. The assay was repeated three times. The data represent the mean relative fold change in expression ± SE.
Role of tlps in adhesion to and invasion of INT 407 cells
To investigate if Tlps affect the interaction with human cells, we examined the Δtlp mutants' adherence to and invasion of INT 407 human intestinal epithelial cells. In comparison to WT, there were no significant differences (P > 0.05) in the mutants' CFU numbers that adhered to the INT 407 cells (Figure S3). The Δtlp8 and the Δtlp9 mutants exhibited a significant (P ≤ 0.05) defect in invasion of INT 407 cells (Figure 4) with 1–2 logs less bacteria recovered compared to WT, respectively.
Figure 4.
Contribution of tlps to Invasion of INT 407 cells. Invasion was assessed using the gentamicin protection assay. INT 407 cells were infected with C. jejuni strains for 3 h after which the cells were treated with gentamicin (150 μg/ml) and incubated for an additional 2 h. The cells were then washed and lysed and the resulting lysate was diluted and spread (100 μL) on MH agar plates. The Δtlp8 and 9 mutants displayed an invasion defect in INT 407 human intestinal epithelial cells. The data represent the average of 2 experiments with 3 replicates in each experiment. *P ≤ 0.05.
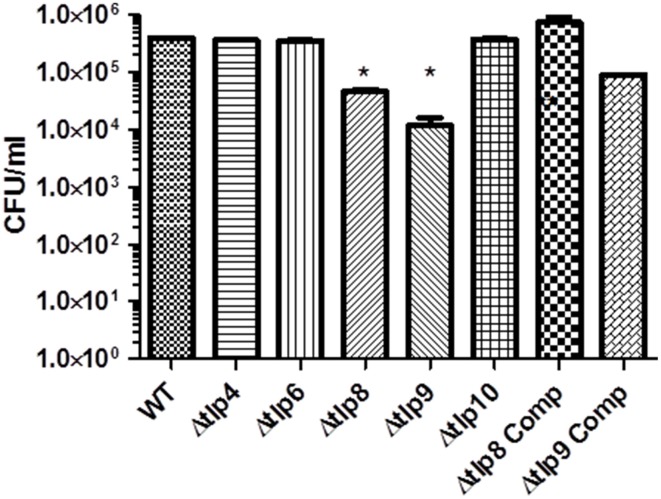
Contribution of tlps to organ specific colonization in chicken
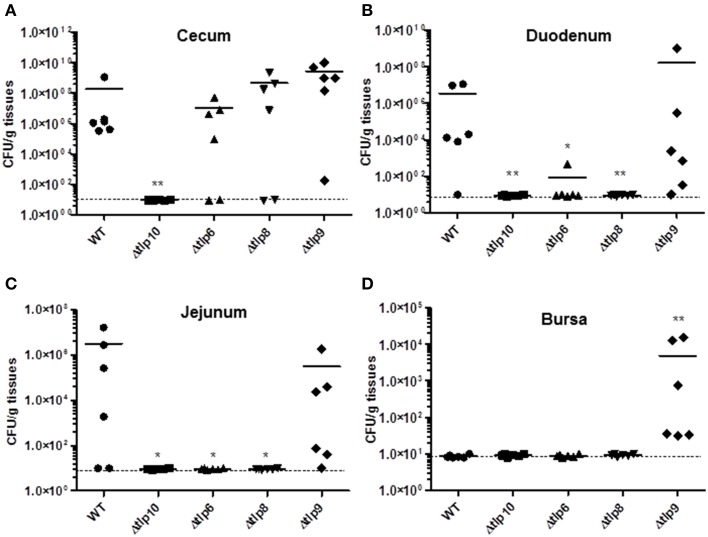
We investigated the in-vivo colonization potential of the Δtlp mutants that showed a significant defect in the phenotypes listed in the studies above. We determined the role of these tlps in organ specific colonization in the cecum, jejunum, duodenum, liver, spleen, and bursa. The Δtlp10 mutant was severely defective in colonization of the cecum, jejunum, and duodenum (Figures 5A–C) with no bacteria recovered from any of these tissues. The Δtlp6 mutant showed a 1 log decrease in cecal colonization compared to WT. Colonization potential was also affected in the Δtlp6 and 8 mutants in duodenum (2–7 log less) and jejunum (5–7 log less) compared to WT. Interestingly, only the Δtlp9 mutant was able to colonize all the examined segments of the intestine. The liver and spleen showed no colonization by C. jejuni WT or the tlp mutants. However, bursal colonization was observed only with the Δtlp9 mutant; even as no bacteria were recovered from the bursa of the WT inoculated group (Figure 5D).
Figure 5.
Colonization of the Δtlp mutants in chickens. Three day-old chicks (n = 6/group) were inoculated orally with 104 CFU of the C. jejuni WT and Δtlp6, 8, 9, and 10 mutants. Seven days post-inoculation, the ceca, duodenum, jejunum, liver, spleen, and bursa were collected, homogenized, and 100 μL of serially diluted homogenates were spread onto MH agar containing Campylobacter selective supplement. (A) The Δtlp10 mutant exhibited a significant defect in colonization of the cecum. (B) The Δtlp8 and 10 mutants exhibited a colonization defect in the duodenum. (C) The Δtlp6, 8, and 10 mutants showed a decreased colonization in the jejunum. (D) The Δtlp9 mutant exhibited an increased colonization of the bursa. Each data point represents CFU/g of tissue, *P < 0.05, **P ≤ 0.01. Dotted line represents the detection limit. The horizontal bars represent the arithmetic mean values for CFU.
Discussion
In this study, we investigated the contribution of five chemoreceptors (belonging to the three major Tlp groups A, B, and C) toward the chemotaxis of C. jejuni 81-176, motility, biofilm formation, invasion of human epithelial intestinal cells, and colonization of the chicken host. A previous study identified Tlp1 as the aspartate chemosensory receptor in C. jejuni (Hartley-Tassell et al., 2010). Here, we observed a decreased chemotaxis toward aspartate and glutamate in the Δtlp6 and Δtlp10 mutants. This corroborated that more than one chemoreceptor can mediate chemotaxis toward a particular substrate (Rahman et al., 2014). This is not uncommon in bacterial chemotaxis where heterologous receptors can generate a cooperative signaling behavior similar to that observed for the chemoreceptors of Sinorhizobium meliloti and E. coli (Meier et al., 2007; Raterman and Welch, 2013). More importantly, the association of tlp6 and tlp10 with chemotaxis toward aspartate has not been reported before. It can be thus hypothesized that the ability of more than one Tlp to sense the same ligand provides robustness to the C. jejuni chemotaxis system, which is a vital factor in the adaptation of this enteric pathogen.
Our study also provides analysis on the role of the tested tlps in chemotaxis toward physiologically relevant organic acids. The Δtlp6 and 10 mutants showed decreased chemotaxis toward organic acids (aspartate, isocitrate, succinate, propionate, and fumarate); many of which are TCA cycle intermediates (Figure 1). Interestingly, Tlp6 is a group C cytosolic chemoreceptor, and the Δtlp6 mutant showed chemotaxis defect toward isocitrate, propionate, and succinate. Previous studies of cytosolic chemoreceptors in Rhodobacter sphaeroides, S. meliloti, and H. pylori (Wadhams et al., 2002; Meier et al., 2007; Schweinitzer et al., 2008) suggest that intracellular Tlps modulate bacterial chemotaxis toward environments with optimum concentration of metabolites which affect the energy status of the bacteria (Porter et al., 2011). The latter confers an obvious competitive advantage and further highlight the importance of Tlps in the survival strategies of C. jejuni. It was notable that the RCR of certain mutants (e.g., the Δtlp4 mutant) was higher than that of the WT in response to some substrates (Table 2). While this may seem counterintuitive, it can be explained by the evolutionary pressure to seek a large variety of substrates via interaction of different Tlps to enhance adaptation and competitiveness (Sourjik and Armitage, 2010). In that regard, the absence of one Tlp from a Tlp cooperative cluster might have enhanced the interactions of the others, subsequently enhancing chemotaxis to certain substrates.
Studies on C. jejuni NCTC11168 have shown that the tlp1, tlp3, docB, and docC mutants displayed a decreased ability to invade human intestinal epithelial and chicken embryo cells (Vegge et al., 2009; Tareen et al., 2010). In this study, tlp8 and tlp9 were the only mutants defective in invasion of INT407 cells. Tlp8 (CetZ) and Tlp9 (CetA) are involved in energy taxis (Reuter and van Vliet, 2013), which emphasizes the importance of these two genes and the cognate acquisition of energy in virulence. Interestingly, the Δtlp8 mutant was defective in biofilm formation under microaerophilic conditions despite exhibiting an increased motility on semi-solid agar plates. Previously, it has been shown that a C. jejuni Cj1496c hypermotile mutant was defective in colonization of the chicken host (Kakuda and Dirita, 2006). It was suggested that the deletion of Cj1496c may lead to elevated levels of phosphorylated CheY, which increased the tumbling frequency. Furthermore, a diploid cheY mutant displayed non-adherent, non-invasive, and hypermotile phenotypes (Yao et al., 1997) similar to what was observed with the ΔCj1496c mutant. However, the mechanisms involved in the cheY-diploid phenotype have not been delineated. Similarly, the decreased invasiveness of the Δtlp8 mutant suggests an involvement of other flagella independent mechanisms which require further investigation. It is possible that to meet crucial energy needs in the Δtlp8 mutant, compensatory mechanisms such as increased motility might favor randomly detecting energy sources.
Biofilm formation is a complex process that is affected by quorum sensing, flagella, aerobic stress, and nutrient availability in C. jejuni (Reeser et al., 2007; Reuter et al., 2010). Additionally, biofilm formation in C. jejuni has been shown to involve both flagella dependent and independent mechanisms (Kalmokoff et al., 2006; Reuter et al., 2010). Interestingly, BLAST analysis of Tlp8 revealed a 41% identity with BdlA protein of Pseudomonas aeruginosa. BdlA is a putative MCP which mediates biofilm dispersion in P. aeruginosa, a motility independent phenotype (Morgan et al., 2006). However, unlike BdlA, Tlp8 doesn't contribute to biofilm shedding (Figure S4). Furthermore, biofilm-related genes (maf5, kpsM, fliS, neuB, and cj0688) (Joshua et al., 2006) were upregulated in the Δtlp8 mutant in biofilms as compared to planktonic culture (Figure 3D). These results likely suggest that Tlp8 may be interacting with other factors which dictate biofilm formation, highlighting potential complex interactions between Tlps, nutrient sensing, motility, and biofilm formation in C. jejuni.
We observed that tlps contribute to differential colonization of C. jejuni in different sections of the chicken gastrointestinal tract (cecum, jejunum, and duodenum). Consistent with previous reports (Hendrixson and Dirita, 2004) (Table 1), the Δtlp10 (ΔdocB) mutant was highly defective in colonization of the cecum and the Δtlp9 (ΔcetA) mutant showed no colonization defect. The Δtlp6 and 10 mutants colonized differently in the duodenum and the jejunum, which also coincided with defects in the chemotaxis of these mutants toward essential nutrients such as aspartate, pyruvate, and fumarate. These observations further suggest that a relationship between substrate-specific chemotaxis and tissue-specific colonization may exist and further investigation of these interactions may provide a better insight into C. jejuni-host interactions.
In comparison with other studies (Table1), our Δtlp4 and Δtlp8 mutants exhibited slightly different phenotypes. Specifically, our Δtlp4 mutant was not deficient in the phenotypes that we tested. However, a tlp4 mutant in C. jejuni NCTC11168 exhibited decreased motility and reduced invasion of human Colo 205 epithelial cells and chicken embryo intestinal cells in vitro (Vegge et al., 2009). Furthermore, a tlp4 mutant in C. jejuni NCTC11168-O was deficient in chemotaxis in response to sodium deoxycholate (Li et al., 2014). Hendrixson and Dirita (2004) showed that a tlp4 mutant in C. jejuni 81-176 exhibited decreased colonization of the chicken cecum; however, this was not tested in our study. Finally, a Δtlp8 mutant in NCTC 11168 did not exhibit increased motility (Reuter and van Vliet, 2013). While the reasons behind these discrepancies are not currently clear and might be due to the use of different testing methods, it is known that the expression of tlp genes can vary based on growth conditions, isolation source, and strains (Day et al., 2012). It is also important to note that Tlp4 and Tlp8 in C. jejuni 81-176 exhibit 100 and 99% protein sequence identity with their cognate homologs in C. jejuni NCTC-11168, respectively. Furthermore, it is possible that certain Tlps might confer different phenotypes to C. jejuni 81-176, which conforms to our initial predictions. In turn, the invasive properties of C. jejuni 81-176 might provide access to niches and cognate substrates that may not be available to C. jejuni NCTC-11168.
In conclusion, we delineated the contributions of Tlps and cognate chemotaxis to survival and host colonization phenotypes of C. jejuni 81-176. The effects on biofilm formation, motility, and host colonization highlight the far reaching implications of impairing the chemotaxis system in C. jejuni, a pathogen that preferentially relies on non-sugar carbon sources for energy production, adaptation, and success in disparate niches. Our data also emphasize the complex interactions that allow a bacterial pathogen to respond to its environment.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Juliette Hanson for assistance with the chicken colonization studies. Dr. Rajashekara's laboratory is supported by the funds from Ohio Agricultural Research and Development Center (OARDC), The Ohio State University, and the Agriculture and Food Research Initiative (AFRI) grant# 2012-68003-19679, U. S. Department of Agriculture. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2015.00046/abstract
References
- Adler J. (1973). A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74, 77–91. 10.1099/00221287-74-1-77 [DOI] [PubMed] [Google Scholar]
- Black R. E., Levine M. M., Clements M. L., Hughes T. P., Blaser M. J. (1988). Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157, 472–479. 10.1093/infdis/157.3.472 [DOI] [PubMed] [Google Scholar]
- CDC. (2013). Incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. Sites, 1996–2012. MMWR Morb. Mortal. Wkly Rep. 62, 283–287. [PMC free article] [PubMed] [Google Scholar]
- Cerda O. A., Nunez-Villena F., Soto S. E., Ugalde J. M., Lopez-Solis R., Toledo H. (2011). tlpA gene expression is required for arginine and bicarbonate chemotaxis in Helicobacter pylori. Biol. Res. 44, 277–282. 10.4067/S0716-97602011000300009 [DOI] [PubMed] [Google Scholar]
- Cerda O., Rivas A., Toledo H. (2003). Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224, 175–181. 10.1016/S0378-1097(03)00423-3 [DOI] [PubMed] [Google Scholar]
- Chang C., Miller J. F. (2006). Campylobacter jejuni colonization of mice with limited enteric flora. Infect. Immun. 74, 5261–5271. 10.1128/IAI.01094-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day C. J., Hartley-Tassell L. E., Shewell L. K., King R. M., Tram G., Day S. K., et al. (2012). Variation of chemosensory receptor content of Campylobacter jejuni strains and modulation of receptor gene expression under different in vivo and in vitro growth conditions. BMC Microbiol. 12:128. 10.1186/1471-2180-12-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozd M., Chandrashekhar K., Rajashekara G. (2014). Polyphosphate-mediated modulation of Campylobacter jejuni biofilm growth and stability. Virulence 5, 680–690. 10.4161/viru.34348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaiah D., Kassem I. I., Liu Z., Rajashekara G. (2009). Importance of polyphosphate kinase 1 for Campylobacter jejuni viable-but-nonculturable cell formation, natural transformation, and antimicrobial resistance. Appl. Environ. Microbiol. 75, 7838–7849. 10.1128/AEM.01603-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor E. C., Wells D. H., MacKichan J. K., Falkow S. (2005). The Campylobacter jejuni stringent response controls specific stress survival and virulence-associatedphenotypes. Mol. Microbiol. 56, 8–27. 10.1111/j.1365-2958.2005.04525.x [DOI] [PubMed] [Google Scholar]
- Hartley-Tassell L. E., Shewell L. K., Day C. J., Wilson J. C., Sandhu R., Ketley J. M., et al. (2010). Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol. Microbiol. 75, 710–730. 10.1111/j.1365-2958.2009.07010.x [DOI] [PubMed] [Google Scholar]
- Hendrixson D. R., Akerley B. J., DiRita V. J. (2001). Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40, 214–224. 10.1046/j.1365-2958.2001.02376.x [DOI] [PubMed] [Google Scholar]
- Hendrixson D. R., Dirita V. J. (2004). Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52, 471–484. 10.1111/j.1365-2958.2004.03988.x [DOI] [PubMed] [Google Scholar]
- Hermans D., Van Deun K., Messens W., Martel A., Van Immerseel F., Haesebrouck F., et al. (2011). Campylobacter control in poultry by current intervention measures ineffective: urgent need for intensified fundamental research. Vet. Microbiol. 152, 219–228. 10.1016/j.vetmic.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Hu L., Kopecko D. J. (1999). Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect. Immun. 67, 4171–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugdahl M. B., Beery J. T., Doyle M. P. (1988). Chemotactic behavior of Campylobacter jejuni. Infect. Immun. 56, 1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua G. W., Guthrie-Irons C., Karlyshev A. V., Wren B. W. (2006). Biofilm formation in Campylobacter jejuni. Microbiology 152, 387–396. 10.1099/mic.0.28358-0 [DOI] [PubMed] [Google Scholar]
- Kakuda T., Dirita V. J. (2006). Cj1496c encodes a Campylobacter jejuni glycoprotein that influences invasion of human epithelial cells and colonization of the chick gastrointestinal tract. Infect. Immun. 74, 4715–4723. 10.1128/IAI.00033-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmokoff M., Lanthier P., Tremblay T. L., Foss M., Lau P. C., Sanders G., et al. (2006). Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188, 4312–4320. 10.1128/JB.01975-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem I. I., Khatri M., Esseili M. A., Sanad Y. M., Saif Y. M., Olson J. W., et al. (2012). Respiratory proteins contribute differentially to Campylobacter jejuni's survival and in vitro interaction with hosts' intestinal cells. BMC Microbiol. 12:258. 10.1186/1471-2180-12-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlath J. A., Osterholm M. T., Judy A., Forgang J. C., Robinson R. A. (1985). A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152, 592–596. 10.1093/infdis/152.3.592 [DOI] [PubMed] [Google Scholar]
- Korolik V., Ketley J. (2008). Chemosensory Signal Transduction Pathway of Campylobacter jejuni. Washington, DC: ASM Press; 10.1128/9781555815554.ch20 [DOI] [Google Scholar]
- Lacal J., Garcia-Fontana C., Munoz-Martinez F., Ramos J. L., Krell T. (2010). Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 12, 2873–2884. 10.1111/j.1462-2920.2010.02325.x [DOI] [PubMed] [Google Scholar]
- Lewus P., Ford R. M. (2001). Quantification of random motility and chemotaxis bacterial transport coefficients using individual-cell and population-scale assays. Biotechnol. Bioeng. 75, 292–304. 10.1002/bit.10021 [DOI] [PubMed] [Google Scholar]
- Li Z., Lou H., Ojcius D. M., Sun A., Sun D., Zhao J., et al. (2014). Methyl-accepting chemotaxis proteins 3 and 4 are responsible for Campylobacter jejuni chemotaxis and jejuna colonization in mice in response to sodium deoxycholate. J. Med. Microbiol. 63(Pt 3), 343–354. 10.1099/jmm.0.068023-0 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Marchant J., Wren B., Ketley J. (2002). Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 10, 155–159. 10.1016/S0966-842X(02)02323-5 [DOI] [PubMed] [Google Scholar]
- Mazumder R., Phelps T. J., Krieg N. R., Benoit R. E. (1999). Determining chemotactic responses by two subsurface microaerophiles using a simplified capillary assay method. J. Microbiol. Methods 37, 255–263. 10.1016/S0167-7012(99)00072-X [DOI] [PubMed] [Google Scholar]
- Meier V. M., Muschler P., Scharf B. E. (2007). Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J. Bacteriol. 189, 1816–1826. 10.1128/JB.00883-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. G., Bates A. H., Horn S. T., Brandl M. T., Wachtel M. R., Mandrell R. E. (2000). Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66, 5426–5436. 10.1128/AEM.66.12.5426-5436.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R., Kohn S., Hwang S. H., Hassett D. J., Sauer K. (2006). BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 188, 7335–7343. 10.1128/JB.00599-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton R. C., Montie T. C. (1979). Chemotaxis by Pseudomonas aeruginosa. J. Bacteriol. 137, 274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. J., Vos T., Lozano R., Naghavi M., Flaxman A. D., Michaud C., et al. (2012). Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: a systematic analysis for the global burden of disease study 2010. Lancet 380, 2197–2223. 10.1016/S0140-6736(12)61689-4 [DOI] [PubMed] [Google Scholar]
- Porter S. L., Wadhams G. H., Armitage J. P. (2011). Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9, 153–165. 10.1038/nrmicro2505 [DOI] [PubMed] [Google Scholar]
- Rahman H., King R. M., Shewell L. K., Semchenko E. A., Hartley-Tassell L. E., Wilson J. C., et al. (2014). Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog. 10:e1003822. 10.1371/journal.ppat.1003822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashekara G., Drozd M., Gangaiah D., Jeon B., Liu Z., Zhang Q. (2009). Functional characterization of the twin-arginine translocation system in Campylobacter jejuni. Foodborne Pathog. Dis. 6, 935–945. 10.1089/fpd.2009.0298 [DOI] [PubMed] [Google Scholar]
- Raterman E. L., Welch R. A. (2013). Chemoreceptors of Escherichia coli CFT073 play redundant roles in chemotaxis toward urine. PLoS ONE 8:e54133. 10.1371/journal.pone.0054133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeser R. J., Medler R. T., Billington S. J., Jost B. H., Joens L. A. (2007). Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl. Environ. Microbiol. 73, 1908–1913. 10.1128/AEM.00740-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Mallett A., Pearson B. M., Van Vliet A. H. (2010). Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76, 2122–2128. 10.1128/AEM.01878-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., van Vliet A. H. (2013). Signal balancing by the CetABC and CetZ chemoreceptors controls energy taxis in Campylobacter jejuni. PLoS ONE 8:e54390. 10.1371/journal.pone.0054390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerging Infect. Dis. 17, 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinitzer T., Mizote T., Ishikawa N., Dudnik A., Inatsu S., Schreiber S., et al. (2008). Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J. Bacteriol. 190, 3244–3255. 10.1128/JB.01940-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V., Armitage J. P. (2010). Spatial organization in bacterial chemotaxis. EMBO J. 29, 2724–2733. 10.1038/emboj.2010.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen A. M., Dasti J. I., Zautner A. E., Gross U., Lugert R. (2010). Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behaviour towards formic acid and are important for invasion of host cells. Microbiology 156, 3123–3135. 10.1099/mic.0.039438-0 [DOI] [PubMed] [Google Scholar]
- Vegge C. S., Brondsted L., Li Y. P., Bang D. D., Ingmer H. (2009). Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl. Environ. Microbiol. 75, 5308–5314. 10.1128/AEM.00287-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams G. H., Armitage J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037. 10.1038/nrm1524 [DOI] [PubMed] [Google Scholar]
- Wadhams G. H., Martin A. C., Porter S. L., Maddock J. R., Mantotta J. C., King H. M., et al. (2002). TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol. Microbiol. 46, 1211–1221. 10.1046/j.1365-2958.2002.03252.x [DOI] [PubMed] [Google Scholar]
- Wilson D. J., Gabriel E., Leatherbarrow A. J., Cheesbrough J., Gee S., Bolton E., et al. (2008). Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. 10.1371/journal.pgen.1000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. L., Bell J. A., Young V. B., Wilder S. R., Mansfield L. S., Linz J. E. (2003). Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149, 3603–3615. 10.1099/mic.0.26531-0 [DOI] [PubMed] [Google Scholar]
- Yao R., Alm R. A., Trust T. J., Guerry P. (1993). Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130, 127–130. 10.1016/0378-1119(93)90355-7 [DOI] [PubMed] [Google Scholar]
- Yao R., Burr D. H., Guerry P. (1997). CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23, 1021–1031. 10.1046/j.1365-2958.1997.2861650.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.