Abstract
Colorectal cancer is a heterogeneous disorder than arises via multiple distinct pathways, from tubular adenomas (TAs) and sessile serrated adenomas (SSAs), which are clinically, morphologically, and molecularly different. We examined PIK3CA amplification in colorectal precancerous legions, including TAs and SSAs. DNA was isolated from paired normal and tumoral tissues in 64 TAs and 32 SSAs. PIK3CA amplification, KRAS mutation, and BRAF mutation were analyzed by real-time PCR and pyrosequencing. PIK3CA amplification was found in 25% of TAs and 9.4% of SSAs, respectively. KRAS and BRAF mutations were mutually exclusive in both TAs and SSAs. In TAs, PIK3CA amplification was associated with left side and it was mutually exclusive with KRAS mutation. These results suggest that PIK3CA amplification may be early and important event in colorectal carcinogenesis and may drive the development of left-side TAs independently with KRAS mutation.
Keywords: colorectal cancer, mitochondria, polymorphism, sessile serrated adenomas, tubular adenomas
Introduction
Colorectal cancer (CRC) is the third most common cancer in Korea and its incidence rate has increased seriously every year 1, 2. Previous studies demonstrated that colorectal neoplasms result from the sequential accumulation of gene alterations 3-5. As a result, it is accepted that the adenoma-carcinoma sequence underlies the colorectal carcinogenesis via two distinct pathways, the chromosomal instability (CIN) pathway and microsatellite instability (MSI) 3-7. Serrated polyps are histologically classified into hyperplastic polyp, traditional serrated adenoma, sessile serrated adenoma (SSA), and mixed hyperplastic/adenomatous polyp 8. Though, SSAs have been considered to be nonneoplastic lesions without malignant potential, recent studies showed that SSAs may be the precursor of CRC with MSI 9-13. This serrated neoplastic pathway is characterized by frequent BRAF mutation and infrequent KRAS mutation 14, 15.
The phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA) gene encodes the catalytic subunit p110 alpha of phosphatidylinositol 3-kinase (PI3K) belonging to class 1A of PI3Ks. PIK3CA mutation promotes cell growth by stimulating AKT pathway in various cancers and has been reported in 10-30% of colorectal cancers 16-18. PIK3CA amplification is found more frequently than PIK3CA mutation in various cancers and it promotes another mechanism for PI3K.AKT pathway 19-21. In CRC, PIK3CA amplification was found in 38% of cancers and 25% of adenomas, suggesting its important role in the adenoma-carcinoma transformation model 22. Furthermore, their results demonstrated that it may be an independent prognostic marker for better survival. However, the role of PIK3CA amplification in colorectal carcinogenesis has not been studied using colorectal precancerous legions.
In present study, PIK3CA amplification was investigated in colorectal precursor lesions, comprising of TAs and SSAs. To contribute to better understanding on colorectal carcinogenesis, KRAS and BRAF mutations key markers in CRCs, were also studied in these lesions. Clinicopathological characteristics in these patients were analyzed according to their genetic status.
Materials and Methods
Patients and DNA Extraction
For this study, the records of colonoscopic polypectomy performed at Dongsan Medical Center between 1999 and 2003 were reviewed and 64 TAs and 32 SSAs were selected. Exclusion criteria were: previous history of surgical resection for CRCs, and evidence of hereditary non-polyposis colorectal cancer (Amsterdam criteria) or familial adenomatous polyposis. Tumor area and adjacent normal mucosa were selected from slide according to hematoxylin and eosin stained sections. Subsequently, the selected areas from paraffin embedded tissues were used for DNA extraction. DNA was isolated by using DNA extraction Kit (AbsoluteTM DNA extraction Kit, BioSewoom, Korea) according to the manufacturer's instructions.
PIK3CA Amplification
Copy number of PIK3CA gene was analyzed by quantitative real-time (qRT) PCR. For the quantitative determination of PIK3CA content relative to nDNA, primers for specific amplification of exon 20 in PIK3CA gene and nDNA-encoded ß-actin gene were selected according to previous study 21. Real-time PCR was then carried out on an LightCycler 480 II system (Roche Diagnostics, Germany) with a total volume of 20 µl reaction mixture containing 10 µl SYBR Green Master MIX (Takara, Japan), 8 pmol of each primers, and DNA (50 ng). The PCR conditions were 95°C for 1 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 30 s. The threshold cycle number (Ct) values of the ß -actin gene and PIK3CA gene were determined. The copy number of PIK3CA in each tested specimen was then normalized against that of ß -actin gene to calculate the relative PIK3CA copy number. Each measurement was repeated in triplicate and 5 serially diluted control samples were included in each experiment. Copy amplification of PIK3CA gene was defined by a copy number ≥ 3. Samples with a PIK3CA/ ß -actin ratio between 1.0 and 3.0 were classified as having PIK3CA gain.
Microsatellite instability (MSI)
MSI was analyzed with two microsatellite markers, BAT25 and BAT 26 based on previous studies demonstrated that these analyses can accurately detect MSI without the need for additional markers. Briefly, polymerase chain reaction (PCR) was performed using a thermal cycler (Applied Biosystems, USA). The PCR products were also denatured in formamide loading buffer (95% formamide, 20 mmol EDTA, 10 mmol NaOH, 0.05% bromophenol blue, 0.05% xylene cyanol) and electrophoresed through 7.5% and 10% polyacrylamide gels. Silver stain was performed to develop bands. MSI was defined as either a band shift or the appearance of a novel band in DNA from precancerous lesions. All experiments were repeated at least twice to rule out any artifacts. Direct DNA sequencing was performed on those PCR products that showed altered band mobility in the above analysis using the ABI 3730 DNA sequencer (Bionics Inc, Korea).
KRAS and BRAF Mutations
KRAS mutations in codons 12 and 13, and BRAF V600E mutation were analyzed by pyrosequencing (PyroMark Q24, Sweden). Primers for amplification and pyrosequencing were designed as previously described 23. The pyrosequencing reaction was performed on a PyroMark Q24 instrument using the Pyro Gold Q24 Reagents (Qiagen, Netherlands). The pyrosequencing primers were used in a final concentration of 0.3 µmol/L. Resulting data were analyzed and quantified with the PyroMark Q24 software version 2.0.6 (Qiagen, Netherlands).
Statistical Analysis
SPSS software for Windows was used. Statistical comparisons for significance were made with Wilcoxon signed-rank test for paired samples. Chi-square, Fischer' exact tests and Mann Whitney U test were used to analyze the relationship between variables. A p value < 0.05 was considered statistically significant.
Result
Precursors of CRCs were comprised of 64 tubular adenomas (TAs) and 32 sessile serrated adenomas (SSAs). Clinicopathological characteristics of TAs and SSAs were presented in Table 1. Higher frequency of KRAS mutation was found in TA than that in SSAs (p = 0.012). However, BRAF mutation was shown only SSAs (p < 0.001), therefore, KRAS and BRAF mutations were mutually exclusive in TAs and SSAs. Other clinicopathological characteristics were not associated with KRAS and BRAF mutations. Using real-time PCR methods, the expression level of PIK3CA gene were analyzed in paired TAs and SSAs. The average of PIK3CA expression level between tumors and normal tissues was not significantly different in both TAs and SSAs (Figure 1). In normal and tumor tissues, higher expression of PIK3CA gene was found in TAs than SSAs (p < 0.05). With a gene copy number of 3 or more defined as amplification, we found the incidence of PIK3CA amplification in colorectal precancerous legions was 19.8% (19/96) in the present study (Table 2). Higher frequency of PIK3CA amplification was found in TAs (25.0%, 16/64) than SSAs (9.4%, 3/32), however, it did not reach statistical significance (p = 0.07). Similar frequency of PIK3CA gain was found in TAs and SSAs. Clinicopathological characteristics of PIK3CA amplification was presented in Table 3. In TAs, PIK3CA amplification was associated with left-side (p = 0.025). And mutual exclusivity between PIK3CA amplification and KRAS mutation was found in TAs, though it did not get to statistical significance (p = 0.06). In SSAs, PIK3CA amplification did not have any clinical significance.
Table 1.
Clinicopathological characteristics of TAs and SSAs in present study
| TA (n = 64, %) | SSA (n = 32, %) | p | |
|---|---|---|---|
| Age (mean ± SD) | 60.36 ± 10.57 | 59.03 ± 9.56 | 0.412 |
| Sex | 0.877 | ||
| Male | 43 (67.2) | 22 (68.8) | |
| Female | 21 (32.8) | 10 (31.2) | |
| Region | 0.51 | ||
| Proximal | 18 (28.1) | 7 (21.9) | |
| Distal | 46 (71.9) | 25 (78.1) | |
| MSI | 1.00 | ||
| (+) | 7 (10.9) | 4 (12.5) | |
| (-) | 57 (89.1) | 28 (87.5) | |
| Kras mutation | 0.012 | ||
| (+) | 15 (23.4) | 1 (3.1) | |
| (-) | 49 (76.6) | 31 (96.9) | |
| Braf mutation | < 0.001 | ||
| (+) | 0 (0) | 6 (18.8) | |
| (-) | 64 (100) | 26 (81.2) |
Figure 1.
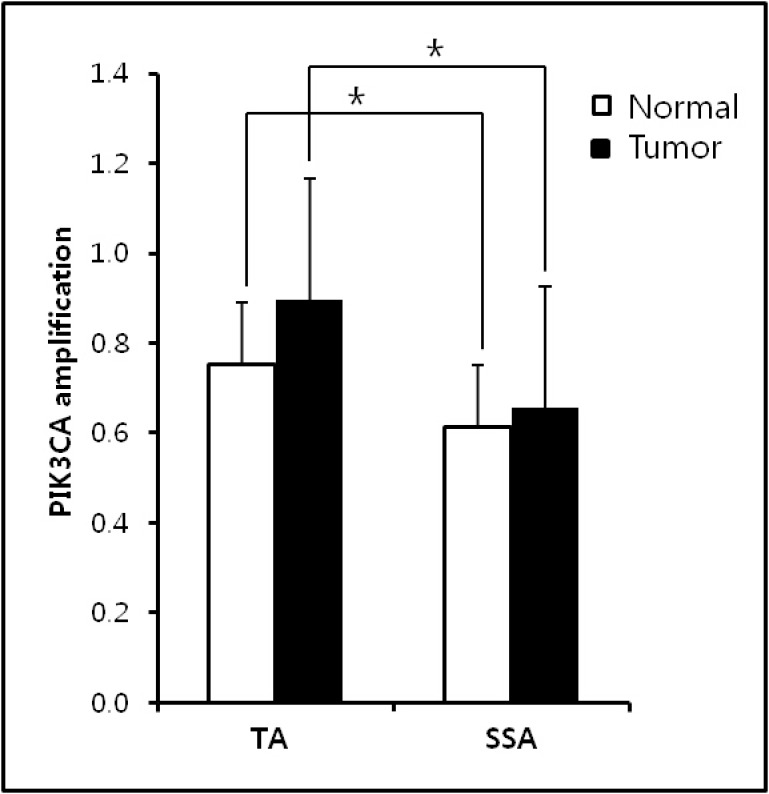
PIK3CA expression level in TAs and SSAs. No significant difference between normal and tumor samples was found in both TAs and SPs. Higher PIK3CA amplification was observed in normal and tumor samples of TAs than SSAs.
Table 2.
Prevalence of PIK3CA content changes in TAs and SSAs
| Total | Gain (N, %) | Amplification (N, %) | |
|---|---|---|---|
| TAs | 64 | 19 (29.7) | 16 (25.0) |
| SSAs | 32 | 11 (34.4) | 3 (9.4) |
| Total | 96 | 30 (31.2) | 19 (19.8) |
Table 3.
Clinicopathological characteristics of PIK3CA amplification in TAs and SSAs
| TA (n = 64) | p | SSA (n = 32) | p | |
|---|---|---|---|---|
| Total * | 25.0% (16/64) | 9.4% (3/32) | ||
| Sex | 0.28 | 0.22 | ||
| Male | 20.9% (9/43) | 13.6% (3/22) | ||
| Female | 33.3% (7/21) | 0% (0/10) | ||
| Side | 0.025 | 0.34 | ||
| Right | 5.6% (1/18) | 0% (0/7) | ||
| Left | 32.6% (15/46) | 12.0% (3/25) | ||
| MSI | 1.00 | 1.00 | ||
| (+) | 28.6% (2/7) | 0% (0/4) | ||
| (-) | 24.6% (14/57) | 10.7% (3/28) | ||
| KRAS mutation | 0.06 | 0.74 | ||
| (+) | 6.7% (1/15) | 0% (0/1) | ||
| (-) | 30.6% (15/49) | 9.7% (3/31) | ||
| BRAF mutation | - | 0.50 | ||
| (+) | - | 16.7% (1/6) | ||
| (-) | 25.0% (16/64) | 7.7% (2/26) |
*p = 0.07 between TA and SSA
Discussion
This article demonstrates that PIK3CA amplification is important and early event in colorectal precancerous legions, especially in TAs. PIK3CA amplification was found in 19.8% (19/96) of precancerous legions and this result was in agreement with previous studies 19, 21, 23-27. PIK3CA amplification has been reported in 30% of ovarian cancer, 33.1% of lung squamous cell carcinomas, 32.3% of head and neck squamous cell carcinomas, 12.2% of endometrial carcinomas, 37.9% of CRC, 15% of anaplastic thyroid cancer, and 67% of gastric cancer. Jehan et al. 22 showed that PIK3CA amplification was observed in 25.0% of colorectal adenomas and CRCs with PIK3CA amplification is an independent prognostic marker for better survival. So, we studied clinicopathological characteristics of PIK3CA amplification in precancerous legions, comprising of TAs and SSAs. Higher frequency of PIK3CA amplification was found in TAs more than SSAs, though it did not reach statistical significance. In SSAs, PIK3CA amplification was not associated with any clinicopatholotical characteristics, however, PIK3CA amplification had various significance in TAs. Higher frequency of PIK3CA amplification was found in left-side TAs and KRAS wild type. It indicates that PIK3CA amplification may play an important role in the development of left-side TA independently with KRAS mutation. In many cancers, PIK3CA amplification was associated with poor prognosis and cancer development, by activating the PI3K/Akt signaling pathway aberrantly. Thus, specific genotype-based targeting against the PI3K/Akt signaling pathway may be an effective therapeutic strategy for TAs. Its molecular mechanisms should be studied further to clarify its classification.
Mutual exclusivity between PIK3CA amplification and KRAS mutation was introduced firstly in previous study by Konopka et al. 24. It has been reported inverse relationship between PIK3CA amplification and PIK3CA mutation in various cancers 19, 21. Previous study indicated that PIK3CA mutation may play important role in benign and early stage of colorectal neoplasm 28. We studied PIK3CA mutation in 20 patients with TAs, as a preliminary study, however, PIK3CA mutation was not found (data not shown). Therefore, we did not include the analysis of PIK3CA mutation in present study. Previous studies also demonstrated rare PIK3CA mutation in Korean populations with CRCs 29, 30. Besides the ethnic factor, the absence of PIK3CA mutation might be affected by the morphology of the adenoma. Chang, Chiu 31 demonstrated that the frequency of the PIK3CA mutation was lower in non-polypoid colorectal neoplasm; however, we had no record of the morphology of colorectal lesion. Nevertheless, these data and our result indicated that PIK3CA amplification is more important factor than PIK3CA mutation in colorectal carcinogenesis.
Taken together, we studied the PIK3CA amplification in colorectal precancerous legions for the first time and analyzed their clinicopathological significance. For the first time, a possible role of PIK3CA amplification in development of CRC is suggested, and it may drive the development of left-side TAs independently with KRAS mutation. This finding may have an important implication for the treatment and the prevention of CRCs.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Shim JI, Kim Y, Han MA, Lee HY, Choi KS, Jun JK. et al. Results of colorectal cancer screening of the national cancer screening program in Korea, 2008. Cancer research and treatment: official journal of Korean Cancer Association. 2010;42:191–8. doi: 10.4143/crt.2010.42.4.191. doi:10.4143/crt.2010.42.4.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 4.Peinado MA, Malkhosyan S, Velazquez A, Perucho M. Isolation and characterization of allelic losses and gains in colorectal tumors by arbitrarily primed polymerase chain reaction. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10065–9. doi: 10.1073/pnas.89.21.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kern SE, Fearon ER, Tersmette KW, Enterline JP, Leppert M, Nakamura Y. et al. Clinical and pathological associations with allelic loss in colorectal carcinoma [corrected] JAMA: the journal of the American Medical Association. 1989;261:3099–103. doi: 10.1001/jama.261.21.3099. [DOI] [PubMed] [Google Scholar]
- 6.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW. et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer research. 1998;58:5248–57. [PubMed] [Google Scholar]
- 7.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. doi:10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 8.Jass JR. Serrated adenoma and colorectal cancer. The Journal of pathology. 1999;187:499–502. doi: 10.1002/(SICI)1096-9896(199904)187:5<499::AID-PATH309>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Jass JR, Whitehall VL, Young J, Leggett BA. Emerging concepts in colorectal neoplasia. Gastroenterology. 2002;123:862–76. doi: 10.1053/gast.2002.35392. [DOI] [PubMed] [Google Scholar]
- 10.Hawkins NJ, Bariol C, Ward RL. The serrated neoplasia pathway. Pathology. 2002;34:548–55. [PubMed] [Google Scholar]
- 11.Goldstein NS, Bhanot P, Odish E, Hunter S. Hyperplastic-like colon polyps that preceded microsatellite-unstable adenocarcinomas. American journal of clinical pathology. 2003;119:778–96. doi: 10.1309/DRFQ-0WFU-F1G1-3CTK. doi:10.1309/DRFQ-0WFU-F1G1-3CTK. [DOI] [PubMed] [Google Scholar]
- 12.Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VL, Pike T. et al. High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology. 2006;131:1400–7. doi: 10.1053/j.gastro.2006.08.038. doi:10.1053/j.gastro.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 13.Jass JR, Baker K, Zlobec I, Higuchi T, Barker M, Buchanan D. et al. Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a 'fusion' pathway to colorectal cancer. Histopathology. 2006;49:121–31. doi: 10.1111/j.1365-2559.2006.02466.x. doi:10.1111/j.1365-2559.2006.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA. et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer research. 2005;65:6063–9. doi: 10.1158/0008-5472.CAN-05-0404. doi:10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 15.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nature reviews Cancer. 2009;9:489–99. doi: 10.1038/nrc2645. doi:10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 16.Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S Jr. et al. The prevalence of PIK3CA mutations in gastric and colon cancer. European journal of cancer. 2005;41:1649–54. doi: 10.1016/j.ejca.2005.04.022. doi:10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Kato S, Iida S, Higuchi T, Ishikawa T, Takagi Y, Yasuno M. et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. International journal of cancer Journal international du cancer. 2007;121:1771–8. doi: 10.1002/ijc.22890. doi:10.1002/ijc.22890. [DOI] [PubMed] [Google Scholar]
- 18.Abubaker J, Bavi P, Al-Harbi S, Ibrahim M, Siraj AK, Al-Sanea N. et al. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. Oncogene. 2008;27:3539–45. doi: 10.1038/sj.onc.1211013. doi:10.1038/sj.onc.1211013. [DOI] [PubMed] [Google Scholar]
- 19.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS. et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer research. 2004;64:7678–81. doi: 10.1158/0008-5472.CAN-04-2933. doi:10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 20.Bertelsen BI, Steine SJ, Sandvei R, Molven A, Laerum OD. Molecular analysis of the PI3K-AKT pathway in uterine cervical neoplasia: frequent PIK3CA amplification and AKT phosphorylation. International journal of cancer Journal international du cancer. 2006;118:1877–83. doi: 10.1002/ijc.21461. doi:10.1002/ijc.21461. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N. et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer research. 2008;68:6913–21. doi: 10.1158/0008-5472.CAN-07-5084. doi:10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jehan Z, Bavi P, Sultana M, Abubaker J, Bu R, Hussain A. et al. Frequent PIK3CA gene amplification and its clinical significance in colorectal cancer. The Journal of pathology. 2009;219:337–46. doi: 10.1002/path.2601. doi:10.1002/path.2601. [DOI] [PubMed] [Google Scholar]
- 23.Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:790–9. doi: 10.1158/1078-0432.CCR-09-2446. doi:10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 24.Konopka B, Janiec-Jankowska A, Kwiatkowska E, Najmola U, Bidzinski M, Olszewski W. et al. PIK3CA mutations and amplification in endometrioid endometrial carcinomas: relation to other genetic defects and clinicopathologic status of the tumors. Human pathology. 2011;42:1710–9. doi: 10.1016/j.humpath.2010.01.030. doi:10.1016/j.humpath.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 25.Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G. et al. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC cancer. 2012;12:50.. doi: 10.1186/1471-2407-12-50. doi:10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou P, Liu D, Shan Y, Hu S, Studeman K, Condouris S. et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:1161–70. doi: 10.1158/1078-0432.CCR-06-1125. doi:10.1158/1078-0432.CCR-06-1125. [DOI] [PubMed] [Google Scholar]
- 27.Suda T, Hama T, Kondo S, Yuza Y, Yoshikawa M, Urashima M. et al. Copy Number Amplification of the PIK3CA Gene Is Associated with Poor Prognosis in Non-lymph node metastatic Head and Neck Squamous Cell Carcinoma. BMC cancer. 2012;12:416.. doi: 10.1186/1471-2407-12-416. doi:10.1186/1471-2407-12-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velho S, Moutinho C, Cirnes L, Albuquerque C, Hamelin R, Schmitt F. et al. BRAF, KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: Primary or secondary genetic events in colorectal carcinogenesis? BMC cancer. 2008;8:255. doi: 10.1186/1471-2407-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon MJ, Lee SE, Kang SY, Choi YL. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: Comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathology, research and practice. 2011;207:762–8. doi: 10.1016/j.prp.2011.10.002. doi:10.1016/j.prp.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Kim SY, Shim EK, Yeo HY, Baek JY, Hong YS, Kim DY. et al. KRAS Mutation Status and Clinical Outcome of Preoperative Chemoradiation With Cetuximab in Locally Advanced Rectal Cancer: A Pooled Analysis of 2 Phase II Trials. International journal of radiation oncology, biology, physics. 2013;85:201–7. doi: 10.1016/j.ijrobp.2012.03.048. doi:10.1016/j.ijrobp.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 31.Chang L-C, Chiu H-M, Shun C-T, Liang J-T, Lin J-T, Chen C-C. et al. Mutational profiles of different macroscopic subtypes of colorectal adenoma reveal distinct pathogenetic roles for KRAS, BRAF and PIK3CA. BMC gastroenterology. 2014;14:221. doi: 10.1186/s12876-014-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


