Abstract
The key information processing units within gene regulatory networks are enhancers. Enhancer activity is associated with the production of tissue-specific noncoding RNAs, yet the existence of such transcripts during cardiac development has not been established. Using an integrated genomic approach, we demonstrate that fetal cardiac enhancers generate long noncoding RNAs (IncRNAs) during cardiac differentiation and morphogenesis. Enhancer expression correlates with the emergence of active enhancer chromatin states, the initiation of RNA polymerase II at enhancer loci and expression of target genes. Orthologous human sequences are also transcribed in fetal human hearts and cardiac progenitor cells. Through a systematic bioinformatic analysis, we identified and characterized, for the first time, a catalog of IncRNAs that are expressed during embryonic stem cell differentiation into cardiomyocytes and associated with active cardiac enhancer sequences. RNA-sequencing demonstrates that many of these transcripts are polyadenylated, multi-exonic long noncoding RNAs. Moreover, knockdown of two enhancer-associated IncRNAs resulted in the specific downregulation of their predicted target genes. Interestingly, the reactivation of the fetal gene program, a hallmark of the stress response in the adult heart, is accompanied by increased expression of fetal cardiac enhancer transcripts. Altogether, these findings demonstrate that the activity of cardiac enhancers and expression of their target genes are associated with the production of enhancer-derived IncRNAs.
Keywords: Cardiac development, Heart failure, Gene regulation, Gene regulatory networks, Enhancers, Long noncoding RNA (IncRNAs)
1. Introduction
The development of the heart and tissue remodeling that occurs in the adult heart during the response to damage are complex biological processes modulated by the coordinated spatiotemporal execution of cardiac gene regulatory networks (GRNs) [1]. Cardiac GRNs are hardwired by groups of evolutionarily conserved cardiac transcription factors (TF) including NKX2.5, MEF2c, SRF and GATA4 [2], which interact with target cis-acting regulatory modules to drive appropriate downstream gene expression. The functional interconnections between upstream signaling pathways, cardiac TFs and their target genes direct cell specification and differentiation, and ultimately cardiac morphogenesis. Moreover, cardiac GRNs are exquisitely sensitive to genetic and environmental signals, with perturbations of these networks being responsible for the full spectrum of inherited and acquired cardiac diseases. Compensatory maladaptive mechanisms, which take place in the damaged heart, result in an increase in cardiomyocyte size and fibroblast proliferation, leading to cardiac hypertrophy and fibrosis. Importantly, hypertrophied cardiomyocytes are characterized by expression of a gene program reminiscent of that activated during embryonic development [1,3].
Although combinatorial TF binding at proximal promoters is important and has been relatively well characterized [2], the key information processing units and regulators of gene expression within the cardiac GRN are enhancers [4,5]. Enhancers are an enigmatic class of regulatory modules, which lie far from the transcriptional start sites of their target genes. They operate as information processors of temporal, spatial and environmental cues to specify and dictate GRN activity [4,6]. Enhancer function is thought to involve direct and indirect promotion of transcription at target gene promoters. Through direct interaction with the basal transcriptional machinery and indirect remodeling of the local chromatin environment at target promoters, enhancers potentiate transcriptional initiation and elongation [4]. There is also strong evidence to suggest that enhancer function requires chromatin looping to bring regulatory factors into direct physical contact with their target promoters. However, the biochemical mechanisms by which this might selectively occur are poorly characterized. Recent studies have shed light on this selectivity problem, demonstrating that active enhancers generate noncoding RNAs (ncRNAs), and that these transcripts are functionally required for enhancer activity [5,7–12]. Importantly, enhancer-associated ncRNAs are dynamically expressed in response to various stimuli including chemical, hormonal, electrophysiological, p53-dependant, and differentiation inducing signals. These dynamic expression profiles correlated with both enhancer activity and downstream target gene expression [8–11,13]. Importantly, enhancer-derived ncRNAs play fundamental roles in targeting chromatin remodeling complexes to the appropriate gene promoters [11,13–16], and thereby facilitate the formation of chromatin loops [12,13,17,18].
Enhancer-associated ncRNAs have been characterized in a limited number of cell types and contexts but evidence for function in complex developmental and pathological responses, particularly cardiogenesis and maladaptive myocardial remodeling, is lacking. Recently, the utilization of high-throughput epigenomic screens has made possible the identification of hundreds of bona fide fetal cardiac enhancers in both human and mouse [5,19–21]. Here, we provide evidence that some of these fetal cardiac enhancers are transcribed, generating ncRNAs during cardiogenesis both in vivo and in vitro. Global transcriptomic profiling reveals that hundreds of enhancers generate novel multi-exonic and polyadenylated long noncoding RNAs (IncRNAs). Interestingly, ncRNA expression correlates with that of their predicted downstream target genes. Identified transcripts are specifically enriched and differentially expressed in mouse and human cardiac progenitor cells. To highlight the functional importance of selected transcripts, we demonstrate that target gene modulation is possible via knockdown of a specific enhancer associated IncRNA. Finally, the maladaptive reactivation of the ‘fetal-gene’ program post myocardial injury is also accompanied by the re-expression of fetal enhancer-associated transcripts. The demonstration that cardiac enhancers generate functional cardiac enriched transcripts will have wide ranging consequences for our understanding of cardiac GRNs controlling cardiac development and disease.
2. Methods
For full details, see online supplement.
2. 1. ChIP sequencing from mouse and human embryonic and adult tissues
For ChIP-Seq analysis of human and mouse fetal and adult hearts we utilized previously published data sets [19,20]. Data can be found and analyzed on the GEO website (GEO accession numbers GSE32587 and GSE22549).
2.2. Transgenic mouse enhancer assay
Mouse transgenic enhancer assays were previously executed and described [19,20].
2.3. Flow cytometry
Mouse ES cells and EBs were dissociated using FACS medium and filtered through a 40-µm cell strainer. Live cells were gated on the basis of side scatter, forward scatter and propidium iodide exclusion. Flow cytometry gates were set using control wild type ES cells not containing the Nkx2.5–EmGFP cassette. Plates were analyzed for EmGFP expression using the BD FACScan (BD Biosciences). Nkx2.5–EmGFP positive cells were sorted from EBs using BDFACS Aria 1 (BD Biosciences).
2.4. Mice
For enhancer derived RNA and marker/target gene expression profiling in embryonic and adult mouse hearts’ post-cardiac injury, C57BL/6J and CD-1 background mice were used. Animal experiments were approved by the Government Veterinary Office (Lausanne, Switzerland) and performed according to the University of Lausanne Medical School institutional guidelines.
2.5. Cardiac injury models – microsurgery
Transverse aortic constriction – Chronic pressure overload was induced in 12-week old mice by transverse aortic constriction (TAC).
Ligation of the left anterior descending artery – Myocardial infarction in mice was induced as previously described. See extended experimental procedures.
2.6. Echocardiography
Transthoracic echocardiographies were performed using a 30-MHz probe and the Vevo 770 Ultrasound machine (VisualSonics, Toronto, ON, Canada).
2.7. Embryonic stem cell culture and differentiation
Nkx2.5-EmGFP BAC reporter ES cell line (129/OlaHsd strain, subline E14Tg2A.4) was kindly provided by Edward C Hsiao (Gladstone Institute of Cardiovascular Research, San Francisco) and maintained and cultured as previously described [22]. Cells were cultured on mouse embryonic fibroblast feeders or on gelatinized plates in standard ES cell medium supplemented with 1000 U/ml of LIF. Cardiac differentiation of ES cells was induced by aggregating aliquots containing 1000 cells in hanging drops to form embryoid bodies [23].
2.8. Primary cell cultures
Human fetal heart chambers and cardiac progenitor cells were isolated as previously described [24].
2.9. RNA isolation, reverse transcription, end-point PCR and quantitative PCR
RNA was isolated using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions, using on column DNase treatment. Complimentary DNA was generated using the SuperScript III kit (Invitrogen) with random hexamer primers. qRT-PCR was carried out using the Applied Biosystems SYBR Green PCR kit and an ABI Prism 7500 cycler and analyzed using the ΔΔCt method.
2.10. Cell culture and transfection
P19CL6 cells (RCB2318, RIKEN Cell Bank, Japan) were cultured in DMEM with 10% FCS and antibiotics. Transfection of P19CL6 cells with pLK0.1-puro-UbC-Tag635™ (containing shRNAi, Sigma Aldrich) was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. EomesER P19CL6 was a kind gift of Dr. Elizabeth Robertson, University of Oxford, UK. To induce differentiation in EomesER P19CL6,1 µg/ml tamoxifen was added to the cell culture medium for 3 days.
2.11. RNA sequencing and analysis
Total RNA was isolated from adult mouse hearts and differentiating mouse ESCs using the RNeasy isolation kit (Qjagen). Sequencing libraries were prepared according to the Illumina RNA Seq library kit instructions with Poly(A) selection. Libraries were sequenced with the Illumina HiSeq2000 (2 × 100 bp). Paired end RNAseq reads were mapped to mouse genome build mm9 using TopHat v2.0.5 essentially according to the protocols outlined in [25]. Using the mapped reads, transcript models were constructed using Cufflinks v2.0.2 for each individual sequencing library, masking genes from the UCSC gene set. The resulting Cufflinks models were merged using Cuffmerge, along with the UCSC gene set to create the main transcript annotation (Gene Expression Omnibus (GEO) accession number: GSE60097).
2.12. Encode consortium histone modification and RNA sequencing data
For tissue specific histone modifications and RNA-Seq data, we used the ENCODE associated histone mark and RNA-Seq data sets (tracks are publicly available on the UCSC browser). For histone marks in differentiation mouse ES cells, we used custom tracks kindly provided by Benoit Bruneau (UCSF) https://b2b.hci.utah.edu/gnomex/gnomexFlex.jsp [5].
2.13. Statistical analysis
Data throughout the paper are expressed as mean ± SEM. One way ANOVA was used to test significance of data comparisons between experimental groups, with p values < 0.05 were considered significant.
3. Results
3.1. Fetal cardiac enhancers are dynamically expressed during cardiac morphogenesis
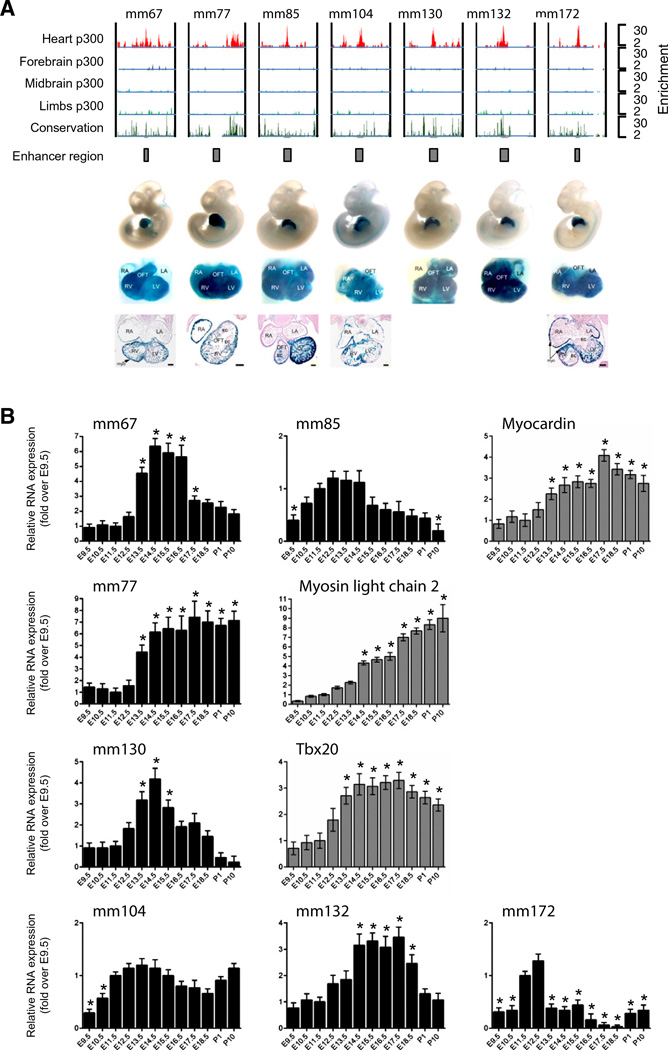
To determine whether cardiac enhancers generated ncRNAs, we took advantage of a genome-wide epigenomic screen that identified 3000 mouse fetal cardiac enhancers in the embryonic day (E) 11.5 hearts based on cardiac specific enrichment of the co-factor p300 (Fig. 1A). Of the 3000 enhancers, approximately 130 were tested in vivo for β-galactosidase reporter activity, confirming that enhancer activity drives expression in the heart [20]. Eleven enhancers, named Mus musculus (mm)67; mm73; mm76; mm77; mm85; mml03; mml04; mml30; mml32; mm172 and mm256 were then selected based on several relevant criteria. The main criteria used for the selection were the proximity of these enhancers to key protein coding genes involved in cardiac biology, the cardiac-specific activity of enhancers when tested in a reporter transgenic assay, and finally whether enhancers confer subregion specificity within the heart. Therefore, six of these enhancers were proximal to genes encoding cardiac regulatory proteins such as Myocardin, Myosin light chain-2v, Tbx20 and Endothelin-1 (Edn1; Supplemental Fig. 1). Most importantly, these enhancers drove robust cardiac-specific activity in mouse embryos (Fig. 1A). Embryo images obtained with each construct are available using the enhancer ID at the Vista Enhancer Browser (http://enhancer.lbl.gov/; [19,20]). Finally, the selected enhancers also exhibited subregion specificity (Fig. 1A). For example, mm67 exhibited activity only within the ventricles (LV and RV) and outflow tract (OFT), and not in adjacent atria (RA and LA), as visualized by β-galactosidase activity in sections of representative embryos. Primers for RT-PCR were then designed in the enhancers as demarcated by the p300 peak to generate products of at least 100 nucleotides. RT-PCR of enhancer sequences using total RNA isolated from E11.5 mouse heart, forebrain and limbs demonstrated the presence of enhancer-associated ncRNAs specifically within the heart (Supplemental Fig. 1B). Importantly, expression did not appear to be a general feature of all active cardiac enhancers since additional enhancers (mm73, mm76, mm103 and mm256) were not able to produce a transcript in cardiac precursor cells (Supplemental Fig. 1C).
Fig. 1.
Fetal cardiac enhancers are expressed in the developing heart. A. ChIP-Seq profiles of p300 occupancy at cardiac enhancers. Coverage by extended p300 reads in heart (red), forebrain (dark blue), midbrain (light blue) and limb (green). Vertebrate conservation plots (black) were obtained from the UCSC genome browser. Gray boxes correspond to candidate enhancer region. Numbers at the right indicate overlapping extended reads. Below boxes are LacZ-stained embryos and isolated hearts with in vivo enhancer activity at E11.5. B. Total RNA was extracted from embryonic (E9.5–E18.5) and neonatal (P1–P10) mouse hearts and subjected to reverse transcription followed by quantitative RT-PCR. Enhancer-associated transcripts: mm67, mm85, mm77, mm104, mml 30, mm132 and mm172, and their putative target genes: Myocardin, Myosin light chain 2 and Tbx20. Results are normalized for expression in the E11.5 heart Mean ± SEM; n = 6–8. * indicates statistical significance, p < 0.05.
To determine whether enhancer-derived transcripts were produced in a developmentally regulated manner, we proceeded via quantitative (q)RT-PCR to measure enhancer expression in the whole heart during development from E9.5 to post-natal day 10 (P10). This developmental period encapsulates all the major morphogenetic and cell fate determining events, including cardiac chamber specification (E9), maturation (E10 to E18), septation (E11–E18), terminal differentiation and myocyte exit from cell cycle (P10) [2]. Expression profiling demonstrated that cardiac differentiation and maturation markers, Myh7 and MS1/ STARS were upregulated as expected during cardiac development (Supplemental Fig. 1D) [26,27]. All selected ncRNAs were dynamically expressed during development of the heart (Fig. 1B). Enhancer expression coincided with different cardiac morphogenetic processes occurring at specific developmental stages. For example, mm77 and mm130 expression is associated with cardiac maturation and septation respectively. Putative cardiac target genes were also dynamically expressed with expression kinetics correlating with enhancer-associated ncRNAs (Fig. 1B). The induction of the ncRNAs typically is coupled to expression of target genes. In some cases, enhancer expression appeared to be repressed once target gene reached maximal levels (see for instance mm67, −85, −130). Overall, these data demonstrated that enhancer-associated ncRNAs were dynamically expressed during cardiac development coinciding with both target genes and cardiac morphogenetic processes.
3.2. Enhancer-derived transcripts are enriched in cardiac progenitor cells
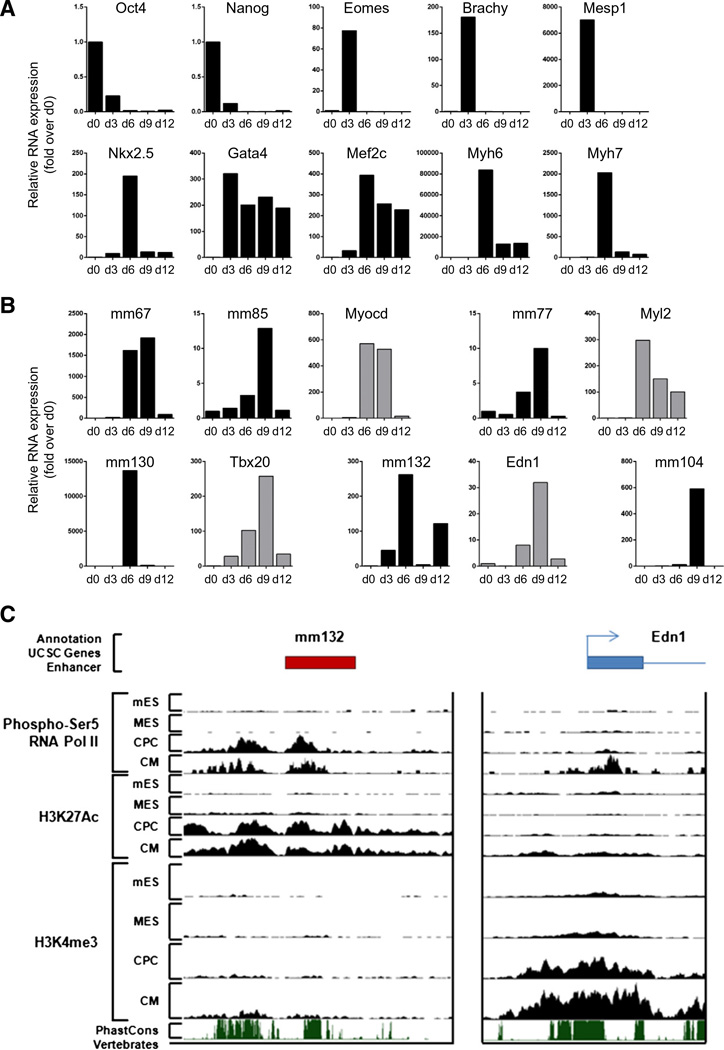
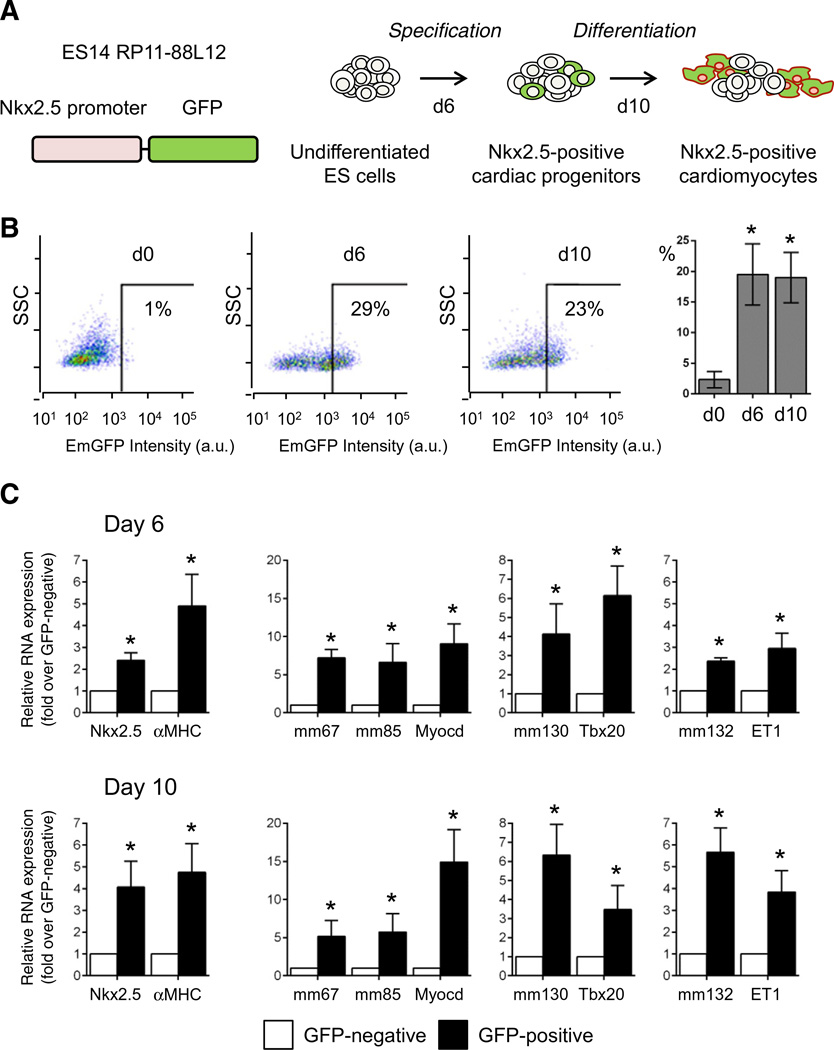
To evaluate enhancer expression during cardiogenesis, mouse embryonic stem (ES) cells were induced to differentiate using the hanging drop model [28] (Fig. 2A and B; Supplemental Fig. 2A). This model recapitulates embryonic cardiac development in vitro, generating all appropriate cardiac lineages (Supplemental Fig. 2B). We first examined the temporal gene expression patterns associated with pluripotency, cardiac mesoderm, cardiac progenitors and differentiated cardiomyocytes (Fig. 2A). Upon differentiation, the pluripotency markers Oct-3/4 and Nanog were rapidly downregulated. This down-regulation occurred concomitantly with expression of Brachyury, Eomes and Mesp1. The three core cardiac transcription factors Nkx2.5, Mef2C and GATA4, which specify cardiac progenitors and drive the cardiac gene program, were significantly upregulated by days 3 to 6. This was followed by robust expression of the cardiac differentiation and structural proteins, Myh6 and Myh7. Six fetal enhancers were expressed in differentiating embryoid bodies (EBs), and demonstrated dynamic expression profiles correlating with cardiac differentiation. Enhancer ncRNAs were predominantly induced at day 6, corresponding to CPC stage and is associated with putative target gene expression (Fig. 2B). To confirm that enhancers were transcribed in cardiac progenitor cells, we took advantage of the EmGFP–Nkx2.5 ES cell line [22], which expresses EmGFP under the control of the endogenous Nkx2.5 promoter, allowing the purification of cardiac progenitor cells by fluorescence-activated cell sorting (FACS) (Figs. 3A and B). EmGFP–Nkx2.5 ES cells were differentiated and Nkx2.5 expressing cells were isolated at days 6 and 10 (Fig. 3B). As expected, Nkx2.5 expression and Myh6 expression were enriched in EmGFP–Nkx2.5 expressing cells (Fig. 3C). In addition, enhancer-derived transcripts and their associated cardiac genes were also significantly enriched in EmGFP–Nkx2.5-positive CPCs (Fig. 3C).
Fig. 2.
Fetal cardiac enhancers are expressed during embryonic stem cell differentiation into the cardiogenic lineage. RNA was isolated on d0, d3, d6, d9 and d12 of embryoid body formation. A. Relative RNA levels of stage-specific markers of cardiac differentiation. B. Relative RNA levels of enhancer-associated ncRNAs and putative target genes during cardiac differentiation. C. UCSC genome browser views of ChIP-Seq data at the mm132 and Edn1 loci for mES, MES, CPC and CM representing different stages of cardiac differentiation.
Fig. 3.
Fetal cardiac enhancer-derived transcripts are enriched in Nkx2.5-positive cardiac progenitor cells. A. Schematic of ES cell differentiation, and isolation of Nkx2.5-positive cardiac progenitors and cardiomyocytes. B. Time course of EmGFP expression during embryoid-body formation and cardiac differentiation at d0, d6 and d10. Bars indicate mean percentages of EmGFP (Nkx2.5)-positive cells at d0, d6 and d10 of differentiation (n = 3); C. Relative RNA levels of enhancer-associated ncRNAs and putative target genes in sorted EmGFP (Nkx2.5)-positive (black bar) and EmGFP (Nkx2.5)-negative (white bar) cells. Mean ± SEM; n = 3. * indicates statistical significance, p < 0.05.
To precisely determine stage-specific activation of our cardiac enhancers, we took advantage of publicly available chromatin state maps generated using chromatin immunoprecipitation followed by sequencing (ChIP-Seq) in differentiating ES cells [5]. Analyses have been executed in pluripotent mouse ES cells (mES, i.e. Oct4-positive cells), at the cardiac mesoderm stage (MES, i.e. Mesp1-positive cells), at the cardiac progenitor stage (CPC, i.e. Nkx2.5-positive cells and GATA-4-positive cells) and in differentiated cardiomyocytes (CM, i.e. αMHC-positive cells), corresponding to d0, d3, d3–6 and d6–12 in the hanging drop differentiation protocol (Supplemental Fig. 2B). Assessment of H3K27me3 and H3K4me3 (associated with inactive and active canonical promoters respectively), and H3K4mel and H3K27ac (associated with poised and active enhancers) allowed us to analyze chromatin state transitions at the transcribed fetal enhancers during cardiogenic differentiation (Supplemental Figs. 3A – D). All enhancers were induced at the CPC stage, corresponding with enhancer transition from a poised (H3K4me1) to an active state (H3K4me1, H3K27ac). None of the expressed enhancers were associated with canonical active promoter states (H3K4me3), confirming that they represent distal regulatory elements and not previously unannotated promoters. Importantly enhancer-associated expression was coupled with enrichment of initiating RNA polymerase II (RNAP2; phosphorylated at serine 5), supporting the notion that active enhancers undergo transcription. In addition, we analyzed the presence of activating marks, i.e. H3K4me3 and initiating RNAP2, at the promoter of Endothelin 1, the predicted target gene of mm132 (Fig. 2C). Interestingly, enhancer activation and transcription appeared to precede induction of its target gene.
The enrichment of enhancer-derived transcripts specifically in cardiac progenitor cells led us to postulate that these enhancers were under the control of cardiac-specific transcription factors. We therefore executed a pair-wise sequence comparison between mouse and human sequences to identify evolutionary conserved transcription factor binding sites (TFBS). All fetal enhancers were enriched with conserved cardiac TFBS, including GATA4, Nkx2.5, Mef2 and SRF motifs (Supplemental Fig. 4). Two enhancers, mm130 and mm172, also contained additional T-Box motifs, which can be bound by the key cardiac mesoderm-specifying transcription factor Eomesodermin (Eomes). To evaluate whether these enhancers were sensitive to activation by transcription factors, we took advantage of P19CL6 mouse embryonic carcinoma cells [29], containing an Eomes gene fused to sequences encoding a mutated estrogen receptor binding domain (EomesER) for tamoxifen-induced nuclear translocation [30]. Upon tamoxifen activation, Eomes induces Mesp1 and Lhx1 expression and initiates a cardiogenic gene program in P19CL6 cells (Supplemental Fig. 4A). In addition, mm130, mm172 and their putative target genes Tbx20 and Ednra were transcriptionally induced in the presence of tamoxifen, suggesting that these enhancers are downstream of an Eomes-dependant transcriptional axis (Supplemental Fig. 4B). Conversely, none of the other cardiac enhancers, not containing T-Box motifs, were induced in the presence of tamoxifen (Supplemental Fig. 4D).
3.3. Orthologous human enhancers are transcribed and functional
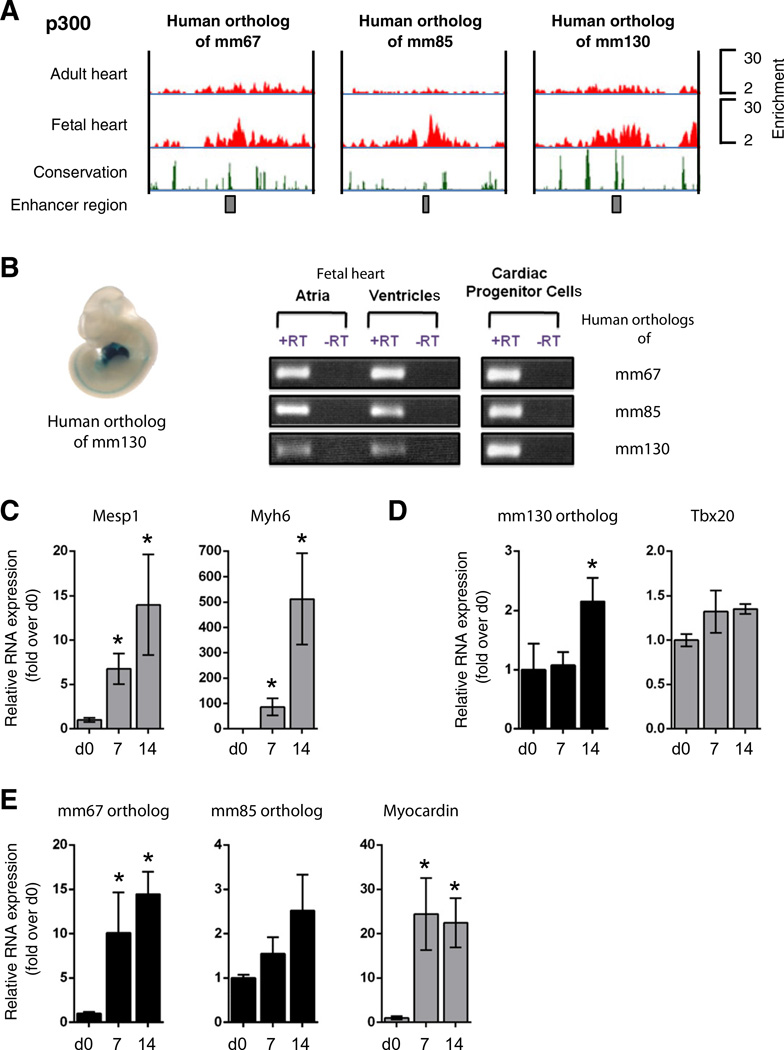
Multispecies vertebrate and mouse conservation plots suggested that four of the seven fetal cardiac enhancers (mm67, −85, −130 and −132) appeared to be evolutionarily conserved in humans (Supplemental Fig. 5A). We therefore utilized a previously executed genome-wide ChIP-Seq screen [19], and determined the occupancy profiles of enhancer-associated co-activator proteins at orthologous human enhancers in fetal and adult human hearts. We observed a significant enrichment of p300/CBP at human orthologs of mm67, −85 and −130 specifically within the fetal heart indicating that these enhancers were active during development (Fig. 4A). To demonstrate regulatory conservation of orthologous human enhancers, the human mm130 sequence was isolated and tested in a mouse transgenic enhancer assay. The orthologous human sequence recapitulated the enhancer activity of the mouse sequence, demonstrating that human orthologous enhancers were functionally conserved (Fig. 4B). In order to confirm that human orthologous enhancers were expressed, RT-PCR was carried out on total RNA extracted from human fetal ventricles and atria as well as from human Nkx2.5-positive CPCs isolated at gestational week 12 (Fig. 4B; Supplemental Fig. 5B) [24]. Primers for RT-PCR were designed in regions contained within the enhancer as demarcated by the p300/ CBP peak in order to generate products of at least 100 nucleotides. Transcripts for conserved human enhancers were present in atria, ventricles and isolated CPCs (Fig. 4B).
Fig. 4.
Orthologous human enhancer sequences are expressed in the fetal heart and in differentiating cardiac progenitor cells. A. ChIP-Seq profiles of p300/CBP occupancy in genomic regions of human orthologous enhancer sequences are indicated by red peaks. Black boxes in the lower panel correspond to the enhancer sequence. Vertebrate conservation plots (black) were obtained from the UCSC browser. B. In vivo activity of human orthologous mml30 enhancer in E11.5 transgenic mice (left panel). Fetal cardiac enhancers are expressed in both cardiac chambers and isolated cardiac progenitor cells (CPCs)(right panel); C. Relative RNA levels of cardiac differentiation markers in differentiating human CPCs; D and E. Relative RNA levels of enhancer-associated ncRNAs and putative target genes during cardiac differentiation of human CPCs. RNA was isolated on d0, d7 and d14 of differentiation. Mean ± SEM; n = 4. * indicates statistical significance, p < 0.05.
Considering the enrichment of enhancer-associated transcripts in mouse ES cell-derived CPCs, we proceeded to determine the expression of human orthologs of mm67, −85 and −130 in differentiating human CPCs [24] (Supplemental Fig. 5B). CPC differentiation was accompanied by a robust induction in the cardiac regulatory transcription factors Mesp1 and the cardiac structural protein Myh6 at 7 and 14 days following induction (Fig. 4C). Consistently, human enhancer transcripts were significantly upregulated at day 7 (mm67) or day 14 (mm85 and −130) of cardiac differentiation (Figs. 4D, E). This activation is also associated with significant upregulation of Myocardin. Therefore, human orthologous enhancer-derived ncRNAs appeared to be functionally conserved during differentiation of isolated cardiac progenitors.
3.4. Global discovery of enhancer-associated IncRNAs during cardiac differentiation
Enhancer-associated ncRNAs are currently known to exist in several forms: a) bidirectional, unspliced, non-polyadenylated eRNAs [9]; b) unidirectional, intergenic, spliced and polyadenylated long noncoding RNAs with a canonical promoter chromatin signature (H3K4me3) [10]; and c) unidirectional, intra- and intergenic, spliced and polyadenylated multi-exonic long noncoding RNAs with enhancer-associated chromatin signatures (H3K4me1 and H3K27Ac) [11,31]. Since reverse transcription reactions were primed with random hexamers in initial experiments, we were not able to readily discern the nature of our enhancer-derived ncRNAs. To address this issue, we assessed transcription of the Poly(A)+ fraction of the transcriptome using high-throughput sequencing (RNA-Seq). Total RNA was isolated from differentiating ESCs at day 0 and day 6 of cardiac differentiation, corresponding to pluripotent (d0) and cardiac precursor cells (CPCs, d6) (Supplemental Figs. 2A and B). These temporal points were selected to allow the identification of ncRNAs being transcribed from developmental cardiac enhancers during cardiac differentiation. Furthermore, day 6 of ESC differentiation corresponds approximately to E11.5 in the developing heart, the temporal point at which our fetal enhancers were identified. Furthermore, enhancer-derived transcripts were maximally expressed at day 6 during ESC cardiac differentiation (Fig. 2).
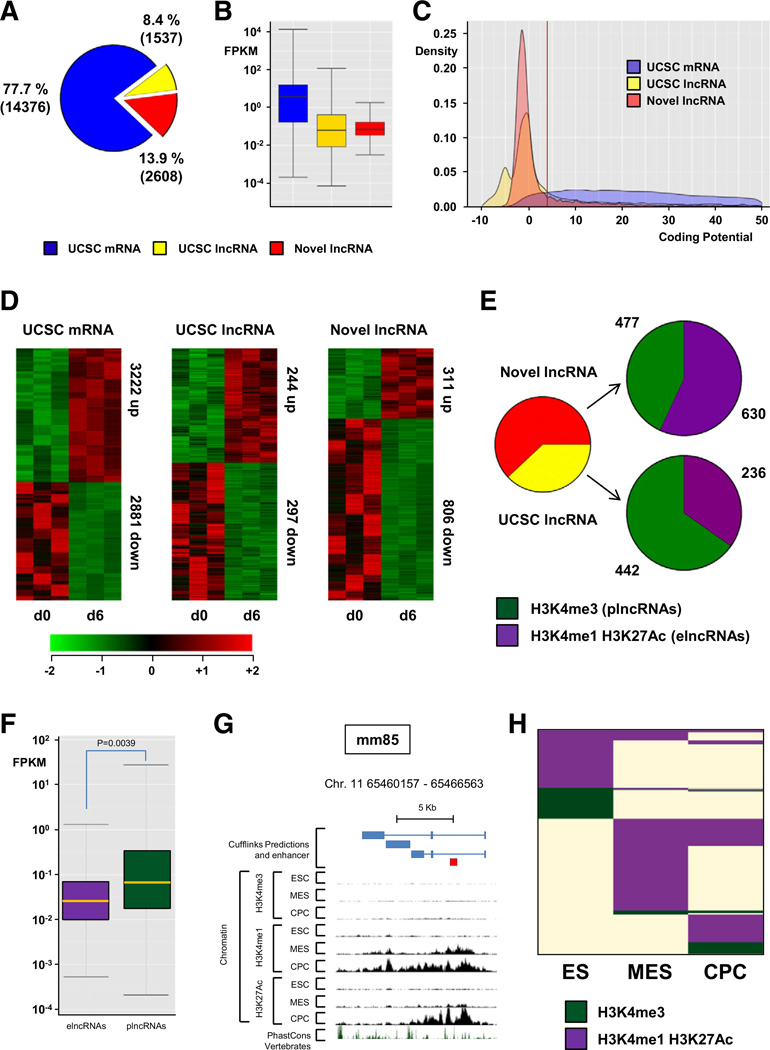
We also integrated ChIP-Seq data generated in a comparable directed differentiation system that recapitulated the step-wise differentiation of mESCs (ES) to cardiac precursor cells (CPCs) (Supplemental Fig. 2) [5]. This facilitates the annotation of chromatin states at underlying genomic sequence and assigns ncRNAs as either promoter-(plncRNA, H3K4me3) or enhancer-associated (elncRNA, H3K4me1/ H3K27Ac) IncRNAs. Furthermore, this allows us to identify enhancers that are activated during cardiac differentiation (i.e. between d0 and d6), and couple activation with differential expression of associated elncRNAs. We generated paired-end 100 bp RNA-Seq reads of Poly(A)+ selected RNA and, using TopHat [25], we mapped a total of >2 billion RNA-Seq reads to the mouse genome (Supplemental Fig. 6A). Transcripts were reconstructed de novo from these data using Cufflinks [25], and compared with UCSC gene annotations. To identify IncRNAs with high confidence, we considered only multi-exonic transcripts greater than 200 nucleotides in size, and discarded any that overlap with known mRNA exons on the same strand or that have predicted coding potential (Supplemental Fig. 6; Coding potential score <4, Fig. 5C). This analysis reconstructed 18,521 multi-exonic transcripts, of which 14,376 (3222 upregulated and 2881 downregulated comparing undifferentiated vs. differentiated ESCs) correspond to University of California Santa Cruz (UCSC) annotated protein coding genes (Figs. 5A, D, Supplemental Tables 3, 4). Our IncRNA annotation pipeline identified 4145 multi-exonic IncRNAs. There were 1537 (244 upregulated and 297 downregulated) UCSC annotated IncRNAs and 2608 (311 upregulated and 806 downregulated) novel previously unannotated IncRNAs. Novel IncRNAs and UCSC IncRNAs were expressed at significantly lower levels than coding genes (Fig. 5B). Furthermore, to verify the non-coding nature of our novel IncRNA candidates, we calculated the Gene ID coding potential score for each transcript and found that these novel transcripts have minimal protein-coding potential, comparable to UCSC annotated IncRNAs (Fig. 5C).
Fig. 5.
Global discovery of enhancer associated IncRNA expression in differentiating mESCs. RNA-Seq was performed on RNA samples isolated from undifferentiated mESC (d0) and from differentiating mESc at the cardiac precursor stage (d6 after induction of differentiation) to characterise the differentiation-associated transcriptome. (A) Pie chart showing composition of the Poly(A)+ transcriptome, UCSC mRNAs (blue), UCSC IncRNAs (yellow) and novel IncRNAs (red). (B) Box plot of transcript abundance (fragments per kilobase per million reads [FPKM]) of UCSC mRNAs (blue), UCSC IncRNAs (yellow) and novel IncRNAs (red). (C) Kernel density plot of coding potential (Gene ID score). (D) Heat maps showing hierarchical clustering of differentially expressed transcripts within the three RNA classes during ESC differentiation. (E) Pie charts showing distribution of USCS IncRNAs (yellow) and novel IncRNAs (red) associated with a canonical promoter signature (H3K3me3, green) or active enhancer state (H3K4mel/H3K27Ac) during ESC differentiation. (F) Box plot of transcript abundance (FPKM) of enhancer-templated (purple) and canonical promoter-associated (green) IncRNAs. (G) The mm85-templated IncRNA is associated with acquisition of active enhancer state during ESC differentiation. (H) Chromatin state map of all IncRNAs associated with either canonical promoter or enhancer state in at least one lineage, ES cell (ES), mesodermal precursors (MES) and cardiac precursor cells (CPCs). Rows are recursively clustered by these marks in these lineages.
To classify our identified novel and annotated IncRNAs, we examined their overlap with specific chromatin states characterized in differentiating ESCs at the pluripotency (ES), mesodermal (MES) and cardiac precursor (CPC) stages of cardiac differentiation [5]. LncRNAs were classified as being associated with either a canonical promoter (plncRNA, H3K4me3) or an active enhancer (elncRNA, H3K4mel/ H3K27Ac). Interestingly, novel IncRNAs were more associated with active enhancers (57% elncRNAs) when compared to previously annotated UCSC IncRNAs (34% elncRNAs) (Fig. 5E). These findings demonstrate that globally enhancers active during development are commonly associated with the production of Poly(A)+ multiexonic IncRNAs, supporting the notion that enhancer-associated transcription is a common feature of cardiac developmental enhancers. Recent studies have demonstrated that elncRNAs and plncRNAs exhibit significant differences in transcript abundance with plncRNAs typically more highly expressed [31]. We also find that plncRNAs are more expressed in differentiating ESCs (p = 0.003 vs. elncRNA) than elncRNAs (Fig. 5F).
We then determined if cardiac enhancers identified in E11.5 fetal hearts were associated with novel multi-exonic Poly(A)+ IncRNAs in ES cells. We identified one enhancer, mm85 that was associated with a novel IncRNA (Fig. 5G), while other enhancers were not. Considering that these enhancers generate a transcript, we suggest that they are likely to encompass the bidirectional non-polyadenylated eRNA class [9]. Since enhancer-associated transcription correlates with enhancer activity, we therefore proceeded to visualize chromatin state transitions occurring at all IncRNAs identified in this systematic analysis (Fig. 5H). A significant fraction of IncRNAs with active enhancer states exhibit exquisite stage specific chromatin state transitions during cardiac differentiation. This finding further supports the notion that cardiac developmental enhancer activity is correlated with the expression of associated ncRNAs. Finally, we find that expression of a significant proportion of elncRNA correlates with the expression of their proximal coding genes, in agreement with previous studies (Supplemental Table 5) [31]. These findings support the notion that enhancer-associated IncRNAs regulate the expression of target protein coding genes in cis during the cardiac differentiation process. To summarize, our integrated genomic analysis of differentiating ESCs has identified hundreds of IncRNAs associated with cardiac developmental enhancers. These enhancer-associated IncRNAs are differentially expressed in a coordinated manner with enhancer state transitions and correlate in expression with putative target genes.
3.5. Fetal enhancers are active and transcribed in the adult heart
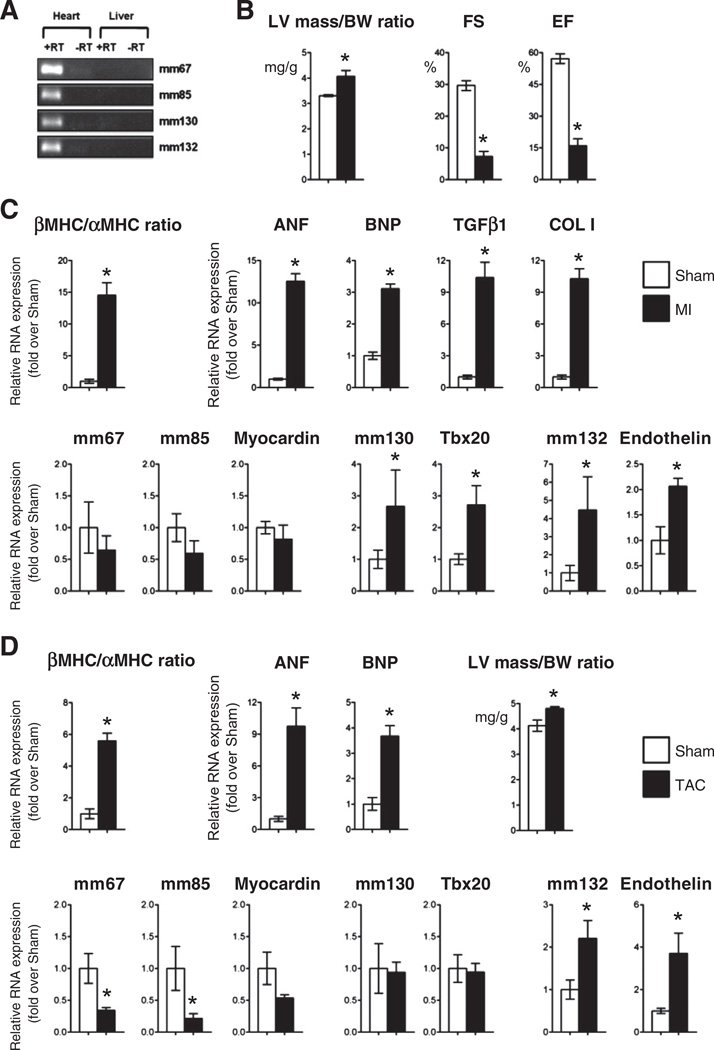
Pathological cardiac remodeling is accompanied by the reactivation of a fetal gene program in the adult heart. Therefore, we determined whether this reactivation manifested at the level of fetal enhancer expression. Utilizing ENCODE ChIP-Seq data [32,33], we first demonstrated that four of the mouse fetal enhancers (mm67, −85, −130 and −132) were associated in the adult mouse heart with epigenetic marks identifying active chromatin, i.e. enrichment with p300, H3K4me1 and H3K27ac (Supplemental Fig. 7A). We also observed heart-specific occupancy by the transcriptional machinery (RNAP2 and DNasel hyper-sensitivity) at all four fetal enhancers. These enhancer signatures were only present in the heart and not in the liver (Supplemental Fig. 7A) or other tissues (not shown), confirming the cardiac-specific nature of these regulatory sequences. To confirm that these epigenetically active enhancers were expressed, total RNA was isolated from the adult mouse heart and liver. RT-PCR analysis confirmed that these four enhancer produced ncRNA in the adult heart (Fig. 6A). In contrast, mml04 and mm172 were not expressed. Furthermore, mm77 was excluded of the analysis due the possible confounding expression of Myl2 within the mm77 locus. We then determined enhancer expression in two patho-physiological models of cardiac injury. We first used a myocardial infarction model obtained by left anterior descending artery (LAD) ligation (Figs. 6A and B; Supplemental Fig. 7B). Fourteen days post-infarction, the myocardium was characterized by remodeling, i.e. increased heart weight (HW) to body weight (BW) ratio, left ventricular (LV) mass, and septal and LV wall thickness as well as decreased ejection fraction (EF) as assessed by echocardiography (Fig. 6B; Supplemental Table 2). Total RNA was isolated from the border zone of infarct-ed region and corresponding region in sham-operated mice. Expression profiling via qRT-PCR analysis demonstrated a robust reactivation of the fetal gene program (Fig. 6C) including upregulation of cardiac markers of stress (ANP, BNP, βMHC) and pro-fibrotic genes (Col1, Tgfb1). Of the expressed enhancers in the adult heart, mml30 and −132 were significantly upregulated post-infarction (Fig. 6C). This induction correlated positively with the upregulation of their target genes Tbx20 and endothelin-1. Interestingly, both Tbx20 and endothelin-1 have been implicated in pathological cardiac remodeling post myocardial injury [34,35]. In contrast, mm67 and −85, as well as their putative target gene Myocardin were not significantly regulated post injury (Fig. 6C). To evaluate whether enhancers demonstrated active transcription during the acute phase of the response to infarction, we measured expression of all enhancers on days 1 and 7 (Supplemental Fig. 8). Enhancers were minimally activated at these early time points. Significant induction of mm132 and its target gene Edn1 was nevertheless observed (Supplemental Fig. 8). Furthermore, mm85, −67 and their predicted target gene, Myocardin, were all significantly downregulated 7 days post infarction (Supplemental Fig. 8).
Fig. 6.
Fetal enhancer expression is induced in response to stress in the adult heart A. Fetal enhancers are expressed in the adult mouse heart B. Cardiac dimensions and function in mice 14 days after myocardial infarction. C. Relative RNA levels of cardiac stress markers, enhancer-associated ncRNAs and target genes in sham-operated (Sham; white bar) and myocardial infarction (M; black bar) groups. Ratio of β over α Myosin heavy chain expression is also indicated. Results are normalized to levels measured in sham-operated mice. Mean ± SEM; n = 4–6. * indicates statistical significance, p < 0.05. D. Left ventricular mass to body weight ratio, relative RNA levels of cardiac stress markers, mm132 enhancer-associated ncRNAs and Endothelin in sham-operated (Sham; white bar) and transaortic constriction (TAC; black bar) groups. Ratio of β over α Myosin heavy chain expression is also indicated. Results are normalized to levels measured in sham-operated mice. Mean ± SEM; n = 3–5. * indicates statistical significance, p < 0.05.
In a second series of experiments, we used a model of cardiac pressure overload obtained by transaortic constriction (TAC; Supplemental Fig. 7C). Seven days after surgery, the myocardium exhibited cardiac hypertrophy and reactivation of the fetal gene program (Fig. 6D). Again, pressure overload-induced activation of fetal gene expression was associated with induction of mm132 and its target gene endothelin-1. Endothelin-1 is incidentally one of the best characterized hypertrophic agonists post cardiac injury, responding to numerous cardiac stresses and modulating the maladaptive pathological response. In contrast, mm130 was not induced, and mm67 and mm85 were even downregulated after TAG
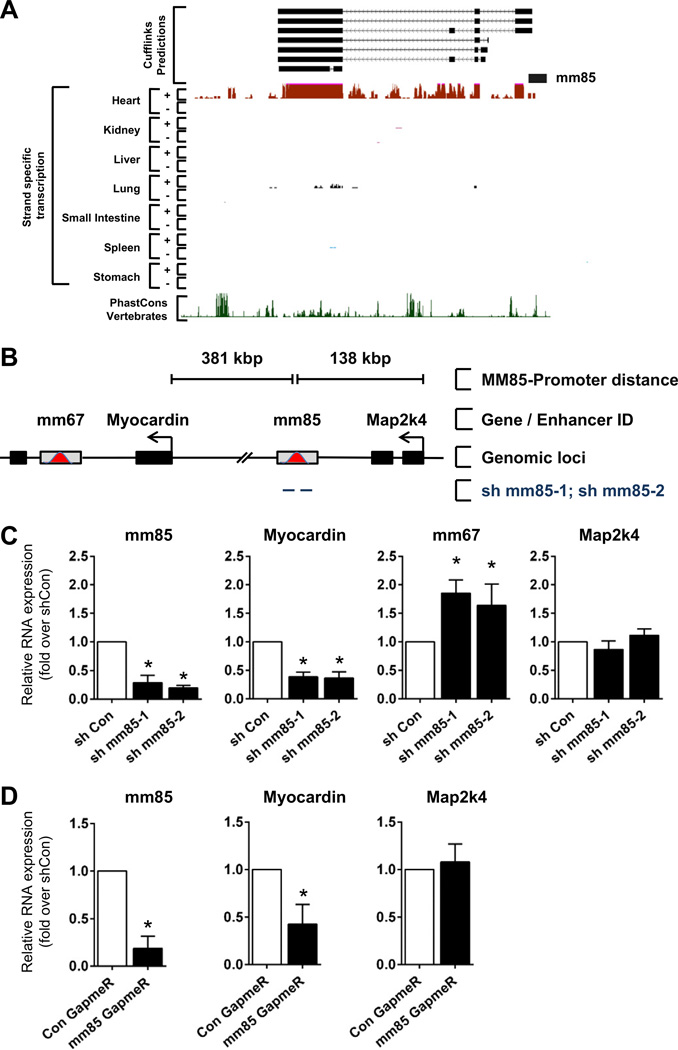
To determine the transcript structure and nature of the fetal enhancer-derived ncRNAs expressed in the adult heart, we utilized a recently published adult heart-specific IncRNA data set [36]. This study executed 100 bp paired-end RNA-sequencing of Poly(A)+ RNA in the heart of 8 week-old C57BL/6 mice followed by de novo IncRNA identification and characterization. Utilizing this data, we found that in the adult mouse heart, two enhancers, mm85 and mm77, were associated with unidirectional Poly(A)+ multi-exonic IncRNAs (Fig. 7A; Supplemental Figs. 9A – E). These ncRNAs exist as multiple iso-forms and are derived from the plus (mm77) and minus (mm85) strands. These data suggest that the other expressed enhancers were likely generating non-polyadenylated enhancer-derived ncRNAs. Delineating the intragenic structure of the multi-exonic ncRNAs associated with mm67 and mm132 was difficult to assess as our analysis is compromised by the presence of the parent coding gene transcripts (Supplemental Fig. 9C). Altogether, these results demonstrate that fetal cardiac enhancers are differentially expressed in the adult heart post injury, potentially contributing to the global reactivation of the fetal gene program.
Fig. 7.
The IncRNA associated to the mm85 fetal enhancer is required for transcription of its target gene. A. UCSC genome browser views of strand-specific RNA-Seq data at mm85 genomic locus for adult heart, kidney, liver, lung, small intestine, spleen and stomach. B. Schematic illustrating the relative genomic location and distance of fetal mm85 enhancer, fetal enhancer mm67, Myocardin proximal target genes and Map2k4 gene. Black bars indicate protein coding genes, gray bars with red peak indicate fetal enhancers. Tailed lines describe relative distances between mm85 and proximal gene transcription start sites. C. P19CL6 cells were transfected with the indicated shRNAs directed against mm85. Relative levels of mm85, Myocardin, mm67 and Map2k4 RNAs are normalized to levels found in cells transfected with control shRNA (shCon). Mean ± SEM; n = 3. * indicates statistical significance, p < 0.05. D. Mouse neonatal CMs were transfected with GapmeRs targeting mm85 IncRNA or random scrambled sequence. Cells were harvested 48 h post transfection and assayed for mm85 IncRNA, Myocardin and Map2k4 expression by qRT-PCR. Bars represent mean ± SEM; n = 2. * indicates statistical significance, p < 0.05.
3.6. Fetal enhancer-associated noncoding RNAs are functional
To assess the functional importance of enhancer-associated ncRNAs, we designed small interfering RNAs (shRNAi) to target the mm85 derived IncRNA (Figs. 7A and B). Knockdown experiments were performed in P19CL6 mouse embryonic carcinoma cells. Transfection of P19CL6 cells with shMM85-1 and shMM85-2, but not control shRNAi, reduced the enhancer transcript by approximately 80% (Fig. 7C). The predicted target gene of mm85, Myocardin, was also significantly downregulated (Fig. 7C), demonstrating that mm85 enhancer derived IncRNA was required for Myocardin expression. Since many protein-coding genes are known to be regulated by multiple enhancers, we also characterized the mm67 enhancer, which is located within the Myocardin gene (Fig. 7B). Upon mm85 ncRNA knockdown, the mm67 ncRNA was upregulated approximately two fold (Fig. 7C). Importantly, the more proximal protein coding gene, Map2k4, which is not a predicted target of mm85 based on its poor cardiac specificity, was not affected by mm85 ncRNA knockdown. Considering that the mm85-associated IncRNA is also expressed in the adult heart and modulated concomitantly with myocardin 7 days post infarction, we wanted to confirm that the mm85 IncRNA regulated myocardin in differentiated cardiomyocytes (CMs). As an experimental model, we used isolated neonatal mouse CMs. Cells were transfected with modified antisense oligonucleotides (GapmeRs) targeting mm85 (Fig. 7D). The associated transcript was reduced by greater than 80%. Myocardin was again significantly downregulated in a specific manner whereas Map2k4 remained unaffected. These results indicate that the mm85 enhancer-associated IncRNA is required for cis-activation of Myocardin and this regulation occurs with high specificity.
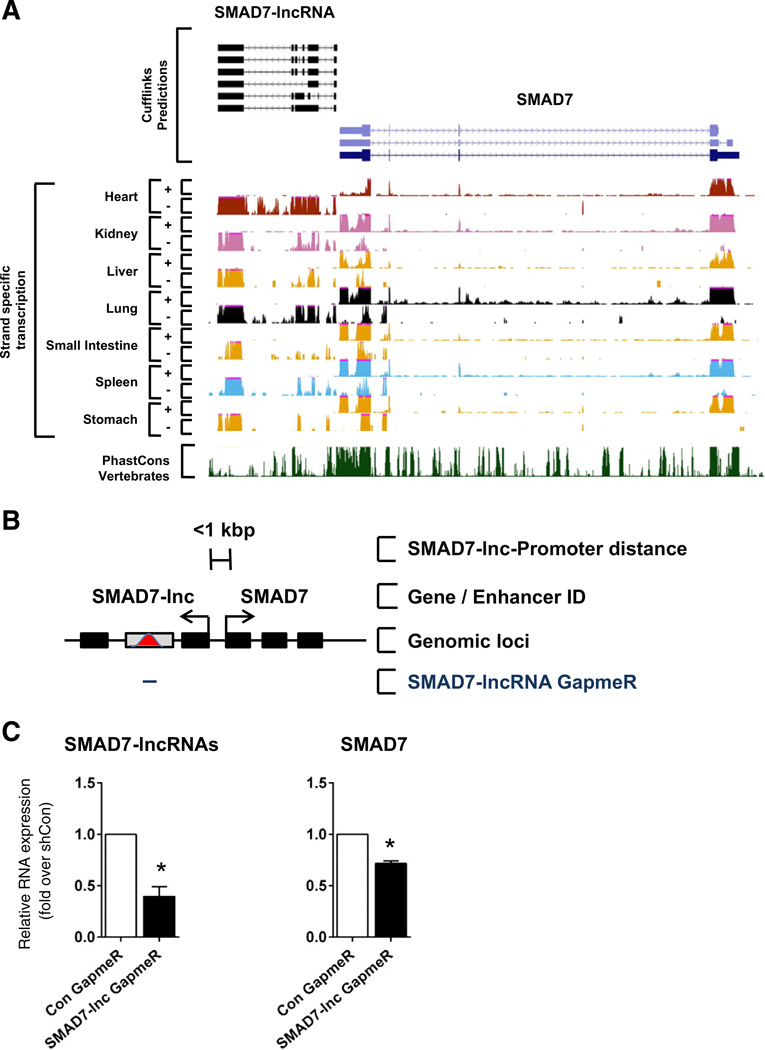
To further exemplify the functional importance of fetal enhancer-derived IncRNAs in the adult heart, we selected a novel IncRNA identified in the RNA-Seq analysis of differentiating embryonic stem cells. This IncRNA, herein named SMAD7-lncRNA, is proximal to Smad7 and well expressed in a number of differentiated adult tissues, including the heart (Fig. 8A). In preliminary experiments, we determined furthermore that this particular IncRNA was highly expressed in cardiac fibroblasts. Therefore, we used isolated neonatal mouse cardiac fibroblasts, which were transfected with GapmeR targeting the last exon of this particular IncRNA (Fig. 8B). Significant knockdown of the IncRNA (>50%, *p < 0.05) resulted in a significant decrease in the predicted target gene, Smad7(>30%, *p< 0.05) (Fig. 8C). These data further support a requirement of enhancer-derived IncRNAs for the expression of proximal target genes in cardiac fibroblasts.
Fig. 8.
The SMAD7-associated enhancer-derived IncRNA is required for its transcription. A. UCSC genome browser views of strand-specific RN A-Seq data at SMAD7 genomic locus for adult heart, kidney, liver, lung, small intestine, spleen and stomach. B. Schematic illustrating the relative genomic location and distance of the SMAD7-lncRNA and SMAD7. Black bars indicate coding and noncoding exons, gray bars with red peak indicate fetal enhancers. Tailed lines describe relative distances between IncRNA and SMAD7 transcriptional start sites. C. Neonatal cardiac fibroblasts were transfected with GapmeRs targeting SMAD7-lncRNA or random scrambled sequence. Cells were harvested 48 h post transfection and assayed for SMAD7-lncRNA and SMAD7 expression by qRT-PCR Bars represent mean ± SEM; n = 4. * indicates statistical significance, p < 0.05.
4. Discussion
Enhancers are the key information processing units within the cardiac GRN. Identification and characterization of cardiac enhancers are therefore essential to decipher the molecular basis of cardiogenesis and pathological remodeling [37]. A number of important recent studies have attempted to address this issue, utilizing genome-wide epigenomic screens to identify fetal and adult cardiac enhancers [19, 20,37,38]. Despite this progress, a physiologically relevant annotation and analysis is still lacking. Considering the recent identification of a novel class of enhancer-associated regulatory ncRNAs that are essential for enhancer function [8–10,12,13,18], we set out to determine whether cardiac enhancers are transcribed and generate cardiac-specific ncRNAs. Here, we provide first evidence that bona fide fetal cardiac enhancers are transcribed dynamically during cardiac development and disease. This highly regulated expression, both in vivo and in vitro, is consistent with expression kinetics of enhancer associated ncRNAs in other cellular systems [8]. It also supports the notion for these transcripts being functionally important rather than artifacts or transcriptional noise. Fetal enhancer ncRNAs are expressed in a cell and tissue specific manner, correlating with the activity of the enhancer and the expression of the predicted cardiac enriched target genes [19,20]. Furthermore, we demonstrate that both mouse and human enhancer-associated ncRNAs are highly enriched in fetal cardiac progenitor cell populations and that a significant fraction of the intergenic multi-exonic poly(A)+ IncRNAs identified in these cells are derived from active developmental enhancers. Importantly, very-deep sequencing allowed us, to probe the association and presence of such enhancer-derived transcripts at an unprecedented level in differentiating ESCs. Importantly, we have included in this analysis only transcripts with no predicted coding potential based on in silico determination (Gene ID score < 4). Nevertheless, we cannot formally exclude that some of these IncRNAs might be translated into small peptides under particular conditions. This warrants further investigation. Finally, we demonstrate that the reactivation of the fetal gene program post myocardial stress is also accompanied by the re-expression of fetal enhancers. Interestingly our group has recently profiled and characterized the long noncoding transcriptome in the adult mouse heart following myocardial infarction [36]. Notably, the vast majority of novel multi-exonic unidirectional IncRNAs that were identified are associated with adult heart-specific cardiac enhancers, comparable to our observations here in ESCs differentiating towards the cardiogenic lineage. Furthermore, the differentially modulated fraction of IncRNAs post infarction, was even more enriched with cardiac enhancer states, suggesting that cardiac enhancer IncRNAs are required for the global transcriptional reprogramming that underpins maladaptive remodeling and subsequent transition to heart failure.
Many evolutionary conserved adult cardiac enhancers exhibit RNAP2 occupancy in the mouse heart. Furthermore, thousands of recently discovered novel cardiac enhancers identified in differentiating mES cells (at cardiac progenitor and cardiomyocyte stage) are associated with initiating RNAP2 when enhancers are in an active state (H3K4me1 and H3K27ac), supporting the notion that enhancer-associated transcription is a common feature of cardiac enhancers. Previously characterized enhancer ncRNAs have been demonstrated to compose two functionally and structurally diverse ncRNA classes. The so-called enhancer-RNA (eRNA) class are unspliced, non-polyadenylated and bidirectionally transcribed [8,9]. The long non-coding RNA (IncRNA) class are RNAP2 transcribed, polyadenylated and multi-exonic [10,11]. To determine which class the enhancer-associated ncRNAs described in the present study represent, we executed RNA-Seq on Poly(A)+ RNA derived from differentiating mouse ESCs and integrated this with publicly available ChlP/RNA-Seq data sets [5,33]. Ab initio transcript reconstruction identified novel, unidirectional, Poly(A)+ and multi-exonic ncRNAs mapping to mm85 and mm77. Interestingly, both transcripts were associated with different chromatin states. Mm77 was enriched with H3K4me3 at its transcription start site, rendering it similar to intergenic lincRNAs [39] and enhancing IncRNAs [10]. This is also consistent with mm77 potentially representing an alternative TSS for the Myl2 gene itself. Mm85 was of particular interest as it was not associated with H3K4me3 but enriched with a purely active enhancer-associated chromatin state (H3K4mel and H3K27ac) rendering it comparable to the recently described elncRNAs [31]. Extending this conclusion, we demonstrate globally that numerous developmental enhancers are associated with unidirectional multi-exonic IncRNAs. The very-deep sequencing approach allowed us to identify hundreds of previously unannotated novel IncRNAs derived from these active enhancers. Interestingly, the novel IncRNAs were more associated with active enhancers as compared to UCSC IncRNAs, comparable with recent observation in the adult heart [36].
LncRNAs have well characterized roles in gene regulation [15,16, 40–43]. They have been shown to be master cell-specific cis- and trans-modulators of gene expression, via direct regulation of target gene promoter activity or indirect interaction with coding and noncoding regulatory networks [15,41–45]. In particular, IncRNAs are especially important for regulating the epigenome, which is established as a major dynamic determinant of cardiac gene expression [5,15,44]. LncRNAs are more cell-specific in their expression compared to protein coding mRNAs [46]. They function as molecular scaffolds, targeting epigenetic and chromatin remodeling complexes to their correct geno-mic loci [15,44]. Analogous to the enhancer-derived IncRNA HOTTIP [13] and to activating IncRNAs [12], the cardiac enriched IncRNAs described herein could potentiate chromatin looping between enhancer sequences and cardiac-specific gene promoters. Interestingly, many IncRNAs have been shown to interact with the Polycomb Repressive Complex 2 (PRC2), known to regulate histone methylation [47,48]. Two recent studies have implicated this complex in epigenetic programming in cardiac precursor cells, being critical for cardiac specification, development and adult myocardial adaptation to stress [49,50]. It would be of interest to determine what proportion of our newly identified enhancer associated IncRNAs are also able to interact with and target PRC2 in a cardiac developmental specific manner, akin to the regulatory roles conferred by the cardiogenic IncRNAs, Braveheart and Fendrr [51,52].
We also demonstrate that enhancer-associated ncRNA expression is regulated in pathophysiological models of heart disease in vivo. These data provide a foundation for the functional annotation of the cardiac-enriched IncRNA transcriptome and suggests that many of these noncoding transcripts may encompass the enhancer ncRNA class. Furthermore, the cardiac enhancers analyzed in this study are highly specific to temporal and spatial cues. This is evidenced by their spatial domains of activity in the developing heart as well as their temporal activation kinetics during in vitro cardiogenesis and in response to pathophysiological stimuli. Since cardiac enhancer ncRNAs exhibit significant cell-specific and context-dependant expression [46], this renders them ideal candidates for therapeutic approaches in the heart. Indeed, as demonstrated for mm85 and SMAD7-lncRNA, it is possible to modulate target gene expression via enhancer-associated ncRNA manipulation. In this context, quantitative profiling of cardiac enhancer-associated ncRNAs can provide exquisite insights into cardiac enhancer activity and more globally into the regulatory state of the cardiac GRN during various cardiogenic processes. Finally, it is important to note that the existence of functional enhancer-associated ncRNAs radically alters also the way we conceptualize genetic variations. Indeed, the vast majority of single nucleotide variants (SNVs) associated with cardiac pathologies and cardiovascular risks reside in noncoding sequences [53–56]. It is assumed that SNVs within noncoding regulatory elements impact upon transcription factor binding motifs [57], However, a number of SNVs were actually found in the human orthologs of some of the mouse cardiac enhancers characterized in the present study (data not shown). Our findings therefore demonstrate that we should also consider the impact of these noncoding SNVs on the expression, structure and function of enhancer-derived IncRNAs.
Acknowledgements
We are grateful to Keith Harshman and Genomic Technologies Facility, University of Lausanne, Switzerland facility for support and sequencing. We thank loannis Xenarios, Swiss Institute of Boinformatics, Lausanne, Switzerland and Roderic Guigό, Center for Genomic Regulation, Barcelona, Spain for his help in bioinformatic analysis. We thank Benoit Bruneau, Gladstone Institute, San Francisco, CA for providing access to custom ChlP-Seq tracks. In the process of this analysis, we took advantage of CvDC data produced as part of the Bench-to-Bassinet Program. EomesER P19CL6 was a kind gift of Dr. Elizabeth Robertson, University of Oxford, UK.
Sources of funding
This work was supported by grants from the Swiss National Science Foundation (T.P., grant no 406340-128129) within the frame of the National Research Program 63 on “Stem Cells and Regenerative Medicine”. LAP. was supported by NHGR1 grant R01HG003988. L.A.P., D.M. and MJ.B. performed work at Lawrence Berkeley National Laboratory and at the United States Department of Energy Joint Genome Institute, Department of Energy Contract DE-AC02-05CH11231, University of California.
Abbreviations
- ncRNAs
noncoding RNAs
- GRN
gene regulatory networks
- CPC
cardiac progenitor cell
- TF
transcription factor
- EB
embryoid body
- RNAP2
RNA polymerase II
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.yjmcc.2014.08.009.
Disclosure statement
None declared.
References
- 1.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313(5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res. 2002;90(5):509–519. doi: 10.1161/01.res.0000013072.51957.b7. [DOI] [PubMed] [Google Scholar]
- 3.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451(7181):943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 4.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151(1):206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, et al. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148(3):473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11(19):2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):el000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45(4):447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20(2):142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31(1):51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 17.Kandarian B, Sethi J, Wu A, Baker M, Yazdani N, Kym E, et al. The medicinal leech genome encodes 21 innexin genes: different combinations are expressed by identified central neurons. Dev Genes Evol. 2012;222(1):29–44. doi: 10.1007/s00427-011-0387-z. [DOI] [PubMed] [Google Scholar]
- 18.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 19.May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Aldyama JA, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2012;44(1):89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChlP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42(9):806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108(14):5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsiao EC, Yoshinaga Y, Nguyen TD, Musone SL, Kim JE, Swinton P, et al. Marking embryonic stem cells with a 2A self-cleaving peptide: a NKX2-5 emerald GFP BAC reporter. PLoS ONE. 2008;3(7):e2532. doi: 10.1371/journal.pone.0002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wobus AM, Kaomei G, Shan J, Wellner MC, Rohwedel J, Ji G, et al. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J Mol Cell Cardiol. 1997;29(6):1525–1539. doi: 10.1006/jmcc.1997.0433. [DOI] [PubMed] [Google Scholar]
- 24.Gonzales C, Ullrich ND, Gerber S, Berthonneche C, Niggli E, Pedrazzini T. Isolation of cardiovascular precursor cells from the human fetal heart. Tissue Eng Part A. 2012;18(1–2):198–207. doi: 10.1089/ten.tea.2011.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25(8):3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuwahara K, Teg Pipes GC, McAnally J, Richardson JA, Hill JA, Bassel-Duby R, et al. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J Clin Invest. 2007;117(5):1324–1334. doi: 10.1172/JCI31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuegemann CJ, Samraj AK, Walsh S, Fleischmann BK, Jovinge S, Breitbach M. Differentiation of mouse embryonic stem cells into cardiomyocytes via the hanging-drop and mass culture methods. Curr Protoc Stem Cell Biol. 2010 doi: 10.1002/9780470151808.sc01f11s15. http://dx.doi.org/10.1002/9780470151808.sc01f11s15 [Chapter 1: Unit 1F 11 ] [DOI] [PubMed] [Google Scholar]
- 29.Habara-Ohkubo A. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct Fund. 1996;21(2):101–110. doi: 10.1247/csf.21.101. [DOI] [PubMed] [Google Scholar]
- 30.Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ. The T-box transcription factor Eomesodermin acts upstream of Mespl to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol. 2011;13(9):1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 2013;14(11):R131. doi: 10.1186/gb-2013-14-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, et al. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9(4):el001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugden PH. An overview of endothelin signaling in the cardiac myocyte. J Mol Cell Cardiol. 2003;35(8):871–886. doi: 10.1016/s0022-2828(03)00153-6. [DOI] [PubMed] [Google Scholar]
- 35.Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121(12):4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J. doi: 10.1093/eurheartj/ehu180. 4/30/2014 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A. 2013;110(50):20164–20169. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visel A, Blow MJ, Li Z, Zhang T, Aldyama JA, Holt A, et al. ChlP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattick JS. The double life of RNA. Biochimie. 2011;93(11):viii–ix. doi: 10.1016/S0300-9084(11)00355-5. [DOI] [PubMed] [Google Scholar]
- 43.Derrien T, Guigo R, Johnson R. The long non-coding RNAs: a new (p)layer in the “dark matter”. Front Genet. 2011;2:107. doi: 10.3389/fgene.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res. 2012;110(3):406–415. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, et al. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012;44(3):343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grote P, Wittier L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific IncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bown MJ, Braund PS, Thompson J, London NJ, Samani NJ, Sayers RD. Association between the coronary artery disease risk locus on chromosome 9p21.3 and abdominal aortic aneurysm. Circ Cardiovasc Genet. 2008;1(1):39–42. doi: 10.1161/CIRCGENETICS.108.789727. [DOI] [PubMed] [Google Scholar]
- 54.Samani NJ, Schunkert H. Chromosome 9p21 and cardiovascular disease: the story unfolds. Circ Cardiovasc Genet. 2008;1(2):81–84. doi: 10.1161/CIRCGENETICS.108.832527. [DOI] [PubMed] [Google Scholar]
- 55.Shah S, Nelson CP, Gaunt TR, van der Harst P, Barnes T, Braund PS, et al. Four genetic loci influencing electrocardiographic indices of left ventricular hypertrophy. Circ Cardiovasc Genet. 2011;4(6):626–635. doi: 10.1161/CIRCGENETICS.111.960203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward LD, Kellis M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science. 2012;337(6102):1675–1678. doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]