Abstract
We understand little about photopreference and the molecular mechanisms governing vision-dependent behavior in vector mosquitoes. Investigations of the influence of photopreference on adult mosquito behaviors such as endophagy and exophagy and endophily and exophily will enhance our ability to develop and deploy vector-targeted interventions and monitoring techniques. Our laboratory-based analyses have revealed that crepuscular period photopreference differs between An. gambiae and An. stephensi. We employed qRT-PCR to assess crepuscular transcriptional expression patterns of long wavelength-, short wavelength-, and ultraviolet wavelength-sensing opsins (i.e., rhodopsin-class G-protein coupled receptors) in An. gambiae and in An. stephensi. Transcript levels do not exhibit consistent differences between species across diurnal cycles, indicating that differences in transcript abundances within this gene set are not correlated with these behavioral differences. Using developmentally staged and gender-specific RNAseq data sets in An. gambiae, we show that long wavelength-sensing opsins are expressed in two different patterns (one set expressed during larval stages, and one set expressed during adult stages), while short wavelength- and ultraviolet wavelength-sensing opsins exhibit increased expression during adult stages. Genomic organization of An. gambiae opsins suggests paralogous gene expansion of long wavelength-sensing opsins in comparison with An. stephensi. We speculate that this difference in gene number may contribute to variation between these species in photopreference behavior (e.g., visual sensitivity).
Keywords: Anopheles gambiae, Anopheles stephensi, photopreference, crepuscular behavior, rhodopsin
Among deployable malaria control and prevention techniques, those targeting the primary host of Plasmodium—the vector mosquito—continue to constitute our most effective methods of intervention. The use of long-lasting insecticide-treated bed nets (Mittal et al. 2012) and indoor residual spraying (Kim et al. 2012), along with environmental management (Imbahale et al. 2012), have led to significant reductions in malaria-related morbidity and mortality in a number of disease-endemic countries (Fullman et al. 2013). However, we must be attentive to impacts on vector-targeted interventions of insecticide resistance (Weill et al. 2000, Reimer et al. 2008). In addition, the inexorable genesis of resistance and extended clearance times of malaria parasites following treatment with drugs, such as chloroquine, mefloquine, and most recently artemisinin, continue to compromise the utility of antimalarial drug-based interventions (Bray et al. 1998, Djimde et al. 2001, Dondorp et al. 2009, Alonso and Tanner 2013).
Creation of next-generation vector-targeted interventions that focus on aspects of the mosquito life cycle that are not targeted by present interventions (indoor residual spraying or IRS, and insecticide-treated bednets or ITNs) will depend, in part, on development of a broader understanding of the behaviors of vector mosquitoes. Many mosquito behaviors—including resting, foraging and feeding behaviors, olfactory responses, flight activity, and flight patterns—have been studied to identify prospective points of attack for next-generation vector-targeted interventions. Toward that end, we have begun to investigate illumination preferences of Anopheline mosquitoes.
Light traps are often used to monitor vector mosquito population compositions and densities (Overgaard et al. 2012, Tchouassi et al. 2012), and we anticipate that light sources could be incorporated into push-pull strategies (Takken 2010) for deflecting vector mosquitos from human dwellings. Still, light traps used to monitor biting rates have been known to provide conflicting results that can vary based on study methods, species observed, and geographical location (Mathenge et al. 2005, Mala et al. 2011). By understanding mosquito light preference in greater depth, we will expand our grasp of vector bionomics, and contribute to improvements in the use of light-based tools for monitoring vector populations and for the development of next-generation interventions that will contribute to decreasing the malaria burden in disease-endemic regions.
Anopheles funestus, Anopheles stephensi, and Aedes aegypti (L.) exhibit increased flight activity in dim-light settings compared with a setting of complete darkness, and the illumination intensities that stimulate flight vary among these species (Ribbands 1946, Manouchehri et al. 1976, Rowland 1989, Kawada et al. 2005). For instance, An. stephensi biting rates increase during nighttime hours, and house-entering behavior of An. funestus increases on moonlit nights (Ribbands 1946, Manouchehri et al. 1976, Rowland 1989). Mosquito house-entering and resting behaviors have been shown to be dependent on temperature microclimates, inside and outside of dwellings (Paaijmans and Thomas 2011). These resting preferences and illumination-influenced behaviors can impact malaria transmission by vector mosquitos and determine how accurately mosquito-monitoring techniques will reflect species prevalence. Integrative consideration of such bionomic factors has begun to influence the development of multiple interventions, including exposure to surface-applied malathion and fungal biocontrol agents, based on more extensive understanding of mosquito resting and flight behaviors (Perich et al. 2000, Mnyone et al. 2012).
While many innate behaviors have been well-characterized in many vector species, illumination preference is a mosquito behavior that has proven difficult to assay in lab and field settings. We have little molecular insight into possible mechanisms underlying illumination-dependent behavioral differences. For instance, multiple studies have reported conflicting results regarding the attractiveness to mosquitoes of blue and green wavelengths of light. Field studies of Culex spp. have reported attraction toward blue light, albeit the least intense of the visible wavelengths with regard to brightness in the study (Ali et al. 1989). Other field studies have concluded that a majority of mosquito species (among the genera Anopheles, Aedes, Coquillettidia, Mansonia, Psorophora, and Uranotaenia) prefers green wavelengths, although Culex nigripalpus females are reported to prefer blue wavelengths (Bentley et al. 2009).
On the other hand, laboratory-based experiments have shown that Culex nigripalpus feed for longer periods of time under illumination of 500 and 600 nm, within the green range of the visible spectrum (Burkett et al. 2012). Other species such as Mansonia perturbans are said to prefer wavelengths of 400–600 nm (blue–green range), while An. stephensi is said to be attracted to near-UV and incandescent light rather than to specific wavelengths (Wilton and Fay 1972, Browne and Bennett 1981). At present, we do not understand whether light preference differences among species, or potentially within species, depend on intrinsic genetic and molecular mechanisms, or on features of life history that engender habituation and learned preferences for specific wavelengths.
Within the order Diptera, molecular mechanisms underlying phototransduction and circadian rhythm have been investigated most extensively in Drosophila melanogaster, given the genetic and molecular tools available in this model organism (Montell 2012). We speculate that circadian variation in the expression of mosquito phototransduction genes may underlie diurnally variable mosquito behaviors. In the Drosophila head, over 150 genes associated with a variety of biological processes exhibit circadian oscillation in expression (Claridge-Chang et al. 2001). Hymenoptera, such as Apis mellifera, exhibit circadian fluctuations in expression of a green-sensitive opsin gene and an arrestin gene, each of which encodes phototransduction components, and their circadian rhythms may be controlled by a mechanism other than that mediated by Cryptochrome-2 (Sasagawa et al. 2003, Yuan et al. 2007).
Given the presence of 11 annotated opsin genes in the An. gambiae genome, An. gambiae has the largest number of opsin genes of any of the insects for which genome assemblies exist at present (Hill et al. 2002, Holt et al. 2002). This expanded opsin gene set has arisen, in part, due to an early duplication of long wavelength-sensitive opsin genes to create a set comprising six long wavelength-sensitive (λmax >500 nm) genes (GPROP1, GROP3-7)—in combination with one UV wavelength-sensitive (λmax <400 nm) opsin gene (GPROP8), one short wavelength-sensitive (λmax 400–500 nm) opsin gene (GPROP9), one functionally undefined opsin gene (GPROP10), and two pteropsin genes (GPROP11, GRPOP12; Spaethe and Briscoe 2004). To date, none of these An. gambiae opsin genes has been shown to exhibit statistically significant circadian variation in expression, although a number do vary in level over the 24-h circadian cycle (Rund et al. 2011). Behavioral analyses of An. gambiae have shown that manipulation of light can influence the timing of blood-feeding behavior (Das and Dimopoulos 2008). Finally, it has been proposed that variation between An. gambiae and Ae. aegypti in the localization of opsin2 and opsin8 expression within the compound eye may underlie species-specific behavioral patterns (e.g., photopreference in low light settings) that differ between these two vector mosquito species (Hu et al. 2009).
In this study we have developed a simple, laboratory-based assay to assess photopreference of An. gambiae and An. stephensi. We have employed these photopreference assays to determine that An. gambiae and An. stephensi exhibit different photopreferences, depending on the time of day and the illumination zone into which they are introduced. Subsequent qRT-PCR analysis fails to reveal significant diurnal differences in opsin gene expression, when comparing the two species. RNAseq analysis of An. gambiae opsins during four life stages indicates that one-half of the long wavelength-sensing opsins are expressed predominantly during larval stages and the other half during adult life-stages, while ultraviolet wavelength- and short wavelength-sensing opsins are expressed predominantly during adult stages. Further analysis of the organization of the long wavelength-sensitive opsin genes in the two species reveals that An. gambiae possess two more long wavelength-sensing opsins than An. stephensi, and we speculate that this difference in gene number may contribute to the differences in photopreference that we observe in the two species.
Methods
Colony
An. gambiae G3 colony (courtesy of Dr. Flaminia Catteruccia, Harvard School of Public Health, Boston, MA) and An. stephensi Sind-Kasur strain Nijmegen (courtesy of Dr. Maria Mota, University of Lisbon, Lisbon, Portugal) were used for all experiments. All experiments were performed on mosquitoes 7–10 d post emergence, which were also aged 3–5 d post blood feeding and 1–3 d post egg laying. A photoperiod of 11:11 (L:D) h was maintained with 1 h dawn:dusk transitions between light and dark periods, with a constant temperature of 27°C and 80% relative humidity. Mosquitoes were fed 10% glucose solution ad libitum and were kept in the presence of the opposite sex throughout their life cycle.
Photopreference Assays
Photopreference assays were performed during the dawn:dusk and dusk:dawn transition periods. Assays were conducted using the arenas illustrated in Supp File 1 (online only). A 60” long, clear, plexiglass tube with a 2” interior diameter was used for the containment portion of the apparatus. For the trinary assays, photic zones were approximately 20” in length and were illuminated with 0 Lux, 100 Lux, or 400 Lux. Illumination levels were based on lux values of a lit room (Yu et al. 2007), and lux values obtained from observations outdoors during dawn and dusk hours in Chestnut Hill, MA. Binary assays consisted of a 30” dark zone (0 Lux) and a 30” illuminated zone (400 Lux). There was no temperature change within the tube throughout the course of the experiment, and the dark and illuminated zones of the tube remained at the same temperature. For each experimental run, approximately 50–75 mosquitoes were aspirated from the colony and introduced to the end of the tube employed for that run. A set of three biological replicates was completed for each pattern of introduction (i.e., illuminated end or dark end introduction). After mosquitoes were allowed to move throughout the tube for 20 min, mosquitoes were asphyxiated quickly by rapid exposure to high-concentration CO2, to avoid alteration of resting patterns, and counts of male and female mosquitoes within each photic zone were then performed. The length of time used for each assay (20 min) was chosen as mosquito activity, i.e., the movement of mosquitoes among regions within the tube, did not change further beyond 20 min following the introduction of mosquitoes (data not shown).
Statistical Analysis
Statistical comparisons for the assessment of photopreference were performed using a Chi-Squared test to determine whether observed distributions deviated significantly from a random distribution. Statistical analyses were performed using Prism 5.0 software.
Collection of Samples and qRT-PCR of Selected Phototransduction Pathway Genes
All gene sequences, nomenclature, and identifiers are according to VectorBase VB-2013-12 (https://www.vectorbase.org; Last accessed 25 February 2015, Megy et al. 2012). qRT-PCR was performed for genes associated with known functions, including light detection and phototransduction pathways in both An. gambiae and An. stephensi. Samples were collected over a 48-h time period in order to encompass two complete diurnal L:D cycles. Collections were made every 4 h and consisted of approximately 10–15 female mosquitoes. Mosquito heads were immediately removed, and RNA was extracted using TriReagent (Sigma: St. Louis, MO), for use in subsequent analyses.
RPS7 (AGAP010592) gene expression was used as a reference for both species. Long wavelength-sensing (AGAP012982), short wavelength-sensing (AGAP010089), and ultraviolet wavelength-sensing (AGAP006126) genes were assayed for expression patterns, as compared to control genes, in both species. Sequences and concentrations of primers used for qRT-PCR can be found in Supp File 2 (online only). An. stephensi genes orthologous to those in An. gambiae were identified using local BLAST and manual annotation of the of the An. stephensi genome (VectorBase VB-2013-12). USB VeriQuest SYBR Green One-Step qRT-PCR Master Mix 2X (Affymetrix: Santa Clara, CA) was used to perform qRT-PCR. Cycling conditions were 50°C for 10 min, 95°C for 10 min, 40 cycles of 95°C for 15 s and 58°C for 30 s for An. gambiae (61°C for 30 s for An. stephensi). Reactions were run on a 7500 Fast Real-Time PCR System (Applied Biosystems: Grand Island, NY). qRT-PCR reaction products were subsequently sequenced to verify amplification of correct target sequences. All values were normalized to the highest expression value obtained for the given gene, for visualization purposes.
RNA Sequencing and Analysis
Male and female whole body RNAseq data sets from An. gambiae (GASUA strain) mosquitoes were obtained from Dr. Larry Zweibel and Dr. Jason Pitts (Vanderbilt University, Memphis, TN; Pitts et al. 2011). Those mosquitoes, which were reared with a photoperiod of 12:12 (L:D) h in 75% humidity, were collected for sequencing at Zeitgeber time 10–12, and were therefore exposed to illumination preceding collection of RNA. We collected two biological replicates at the same time points as Pitts et al. (2011), i.e., first (L1) and third (L3) instar larvae, as well as single biological replicates of adult males and females (whole body) of An. gambiae G3 to compliment the Vanderbilt University data set. We collected only single adult replicates as our goal was to validate expression levels reported by Pitts et al. (2011), rather than define statistically significant differences in transcriptional expression among life stages. RNAseq data sets have been deposited in the European Nucleotide Archive under the SRA accession PRJEB5712. RNA extraction and sequencing of these collections were performed by Otogenetics Corp. (Norcross, GA) and the Broad Institute (Cambridge, MA). All RNA-seq data were aligned to An. gambiae P3 assembly, from VectorBase VB-2013-12, using Tophat2 (Kim et al. 2013). FPKM values and comparisons between samples were performed using Cufflinks-Cuffdiff2, and the subsequent heatmap was visualized using CumberBund (Trapnell et al. 2013). Genes analyzed included all long wavelength-sensing opsins GPROP1 (AGAP013149), GPROP3 (AGAP012982), GPROP4 (AGAP012985), GPROP5 (AGAP001162), GPROP6 (AGAP001161), GPROP7 (AGAP002462), ultraviolet wavelength-sensing opsin GPROP8 (AGAP006126), short wavelength-sensing opsin GPROP9 (AGAP010089), an unknown wavelength-sensing opsin GPROP10 (AGAP007548), and the two pteropsins GPROP11 (AGAP002443) and GPROP12 (AGAP002444).
Results and Discussion
Determination of Photopreferences in An. gambiae and An. stephensi
First, we measured photopreference characteristics of An. gambiae and An. stephensi to determine whether there are distinctions between the two species. We developed an assay that assesses the photopreference of An. gambiae using a binary choice arena (0 Lux vs. 400 Lux, Fig. 1, Table 1, Supp File 1 [online only]). Introduction of mosquitos into the illuminated end of the apparatus during either dawn or dusk crepuscular periods reveals that females exhibit a significant preference for darkness, while males exhibit no reference between illumination and darkness (Fig. 1A andE). Binary choice assays in which An. gambiae was introduced into the darkened end of the apparatus reveal that males and females exhibit a significant preference for resting in darkness (Fig. 1C andG).
Fig. 1.
An. gambiae and An. stephensi binary photopreference. Bar graphs depict percent of mosquitos resting in specific photic regions (±SEM, N = 3) for each experiment. Left and right columns depict An. gambiae and An. stephensi resting patterns for each condition, respectively, with males and females being depicted within each column. Dawn and dusk refer to relative crepuscular period. Right hand titles indicate introduction site followed by relative crepuscular period. Black bars represent mosquitos resting in the 0 Lux region of the tube at the end of the experiment, and open bars represent those resting in the 400 Lux region. (A and B) Introduction into 400 Lux region at dawn. (C and D) Introduction into 0 Lux region at dawn. (E and F) Introduction into 400 Lux region at dusk. (G and H) Introduction into 0 Lux region at dusk. ★P < 0.05, ★★P < 0.01, ★★★P < 0.001. Tabulations can be found in Table 1.
Table 1.
An. gambiae and An. stephensi binary photopreference data
| Zeit. time | Int Site |
An. gambiae |
An. stephensi |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Female | Male | Female | Male | Female | Male | ||
| Dawn | 400 | 76.1 ± 3.3 | 68.7 ± 8.7 | 25.1 ± 2.9 | 38.0 ± 7.7 | 37.6 ± 2.6 | 70.5 ± 3.3 | 63.0 ± 2.8 | 30.5 ± 3.2 |
| Dawn | 0 | 69.8 ± 4.3 | 75.4 ± 2.6 | 32.09 ± 4.71 | 25.3 ± 2.6 | 48.2 ± 4.2 | 78.3 ± 2.0 | 53.2 ± 4.1 | 22.3 ± 2.1 |
| Dusk | 400 | 74.6 ± 0.7 | 57.3 ± 5.0 | 25.5 ± 0.7 | 44.6 ± 5.0 | 29.7 ± 4.1 | 50.7 ± 2.0 | 73.3 ± 5.8 | 49.6 ± 2.0 |
| Dusk | 0 | 62.0 ± 1.3 | 62.2 ± 0.5 | 38.2 ± 1.2 | 37.8 ± 0.5 | 34.1 ± 7.5 | 44.3 ± 7.6 | 72.3 ± 8.0 | 59.9 ± 6.6 |
| 0 Lux | 400 Lux | 0 Lux | 100 Lux | ||||||
| Photic preference zone | |||||||||
Tabulation of results presented in Figure 1. Zeitgeber time and Introduction site are presented in the left-hand columns, with photic regions represented with 0 Lux and 400 Lux. Values are percent resting in respective region ± SEM.
Analogous experiments with An. stephensi reveal that females prefer the illuminated portion of the apparatus when added to the illuminated end of the apparatus at dawn, while males prefer darkness (Fig. 1B). When introduced into the illuminated end of the apparatus, females exhibit a preference for illumination at dusk, while males no longer display any illumination preference (Fig. 1F). When added to the darkened portion of the apparatus at dawn, An. stephensi females lack any discernible photopreference, while males display a preference for darkness (Fig. 1D). When introduced into darkened end of the apparatus at dusk, An. stephensi males exhibit no preference, while females exhibit a preference for the illuminated portion of the apparatus (Fig. 1H).
The differences we observed between An. gambiae and An. stephensi photopreferences are consistent with differences observed in past studies of each species in other physical settings (Jones et al. 1967, Rowland 1989). Female An. gambiae generally exhibit a significant preference for a darkened photic zone, which can be attributed to an active avoidance of increased illumination. The active avoidance of illumination by An. gambiae females, when they are introduced to the 400 Lux end of the arena (Fig. 1A and E), indicates an avoidance of the light rather than a simple, consistent preference toward the end of the apparatus into which the mosquitos are introduced. Given that previous studies of An. gambiae indicate that peak flight activity occurs at the dawn and dusk hours, the possibility that An. gambiae are not actively moving within our apparatus is unlikely (Jones et al. 1967).
Interestingly, An. stephensi photopreference differs greatly from that of An. gambiae. Female An. stephensi prefer the 400 Lux region of the apparatus in all conditions, except when introduced into 0 Lux at dawn, when no significant preference was observed. This suggests a requirement for increased illumination to perform visual-based behaviors, such as identifying a feeding source, an oviposition site, or a mating swarm, or for achieving increased visual acuity. Male An. stephensi exhibit a preference for darkness or no preference, for all patterns of introduction, similar to findings for An. gambiae males. This suggests that light preference may be less important for Anopheline males in the processes of finding mates and food sources. In order to further validate the distinctions in photopreferences we observe between the two species in a binary choice assay, we subsequently conducted trinary choice assays.
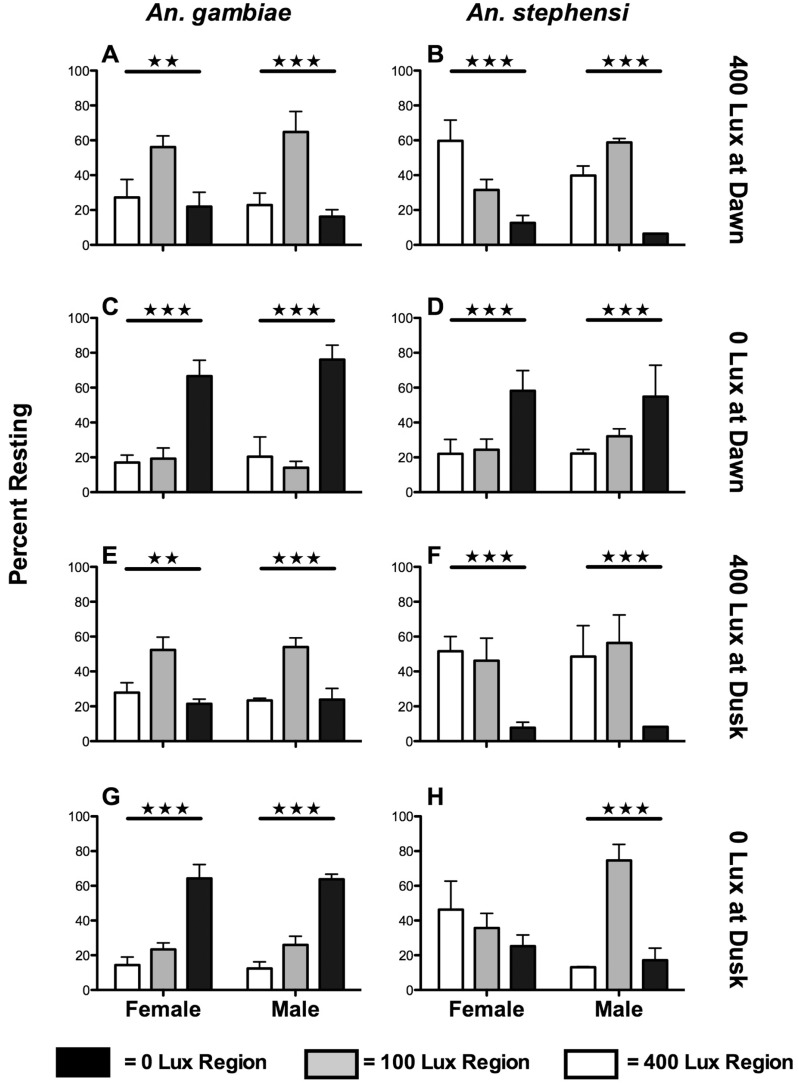
Assessment of An. gambiae photopreference in a trinary choice assay (0 Lux vs. 100 Lux vs. 400 Lux, Fig. 2, Table 2), which allows for greater delineation of photopreference, illustrates that females and males prefer 100 Lux illumination during dawn and dusk crepuscular periods, when introduced to the 400 Lux end of the apparatus (Fig. 2A andE). When the assay was repeated with the introduction of mosquitos into the 0 Lux end of the apparatus, both sexes of An. gambiae prefer to remain in the darkened end of the apparatus during both crepuscular periods (Fig. 2C andG). An. stephensi display tendencies to rest in 400 and 100 Lux regions of the apparatus, instead of the nonilluminated region, when introduced to the 400 Lux-illuminated region of the apparatus during dawn or dusk (Fig. 2B andF). Following introduction into the darkened end of the apparatus during dawn, An. stephensi males and females remain in the darkened region (Fig. 2D). Females exhibit no preference following introduction into the darkened end during dusk, and males exhibit significant preference toward the 100 Lux-illuminated region when introduced in the same manner (Fig. 2H).
Fig. 2.
An. gambiae and An. stephensi trinary photopreference. Bar graphs depict percent of mosquitos resting in specific photic regions (±SEM, N = 3) for each experiment. Left and right columns depict An. gambiae and An. stephensi resting patterns for each condition, respectively. Dawn and dusk refer to relative crepuscular period. Right hand titles indicate introduction site, followed by relative crepuscular period. Black bars represent mosquitos resting in the 0 Lux region of the tube at the end of the experiment, gray bars represent those resting in the 100 Lux region and open bars represent those resting in the 400 Lux region. (A and B) Introduction into 400 Lux region at dawn. (C and D) Introduction into 0 Lux at dawn. (E and F) Introduction into 400 Lux at dusk. (G and H) Introduction into 0 Lux at dusk. ★P < 0.05, ★★P < 0.01, ★★★P < 0.001. Tabulations can be found in Table 2.
Table 2.
An. gambiae and An. stephensi trinary photo preference data
| Zeitgeber time | Introduction site | An. gambiae | An. stephensi | ||||
|---|---|---|---|---|---|---|---|
| Females | |||||||
| Dawn | 400 | 27 ± 6.0 | 56.1 ± 3.7 | 22.0 ± 4.7 | 59.7 ± 6.9 | 31.5 ± 3.5 | 12.6 ± 2.5 |
| Dawn | 0 | 17.1 ± 2.5 | 19.3 ± 3.5 | 66.7 ± 5.2 | 22.0 ± 4.8 | 24.4 ± 3.5 | 58.2 ± 6.7 |
| Dusk | 400 | 27.9 ± 3.6 | 52.4 ± 4.2 | 21.5 ± 1.5 | 51.6 ± 4.9 | 46.2 ± 9.5 | 7.7 ± 1.8 |
| Dusk | 0 | 14.4 ± 2.6 | 23.4 ± 2.2 | 64.3 ± 4.6 | 46.3 ± 9.5 | 35.7 ± 4.9 | 25.2 ± 3.7 |
| Males | |||||||
| Dawn | 400 | 22.9 ± 4.0 | 64.8 ± 6.8 | 16.2 ± 2.3 | 39.8 ± 3.2 | 58.8 ± 1.3 | 6.5 ± 0.0 |
| Dawn | 0 | 20.4 ± 6.6 | 14.1 ± 2.1 | 76.1 ± 4.8 | 22.2 ± 1.3 | 32.1 ± 2.4 | 54.9 ± 10.4 |
| Dusk | 400 | 23.4 ± 0.7 | 54.1 ± 3.0 | 23.9 ± 3.7 | 48.6 ± 10.2 | 56.4 ± 9.3 | 8.9 ± 0.1 |
| Dusk | 0 | 12.3 ± 2.9 | 25.9 ± 2.9 | 63.8 ± 1.7 | 13.1 ± 0.2 | 74.6 ± 5.3 | 17.2 ± 4.0 |
| 400 Lux | 100 Lux | 0 Lux | 400 Lux | 100 Lux | 0 Lux | ||
| Photic preference zone | |||||||
Tabulation of results presented in Figure 3. Zeitgeber time and introduction site are presented in the left-hand columns, with photic regions represented with 0, 100, and 400 Lux. Values are percent resting in respective region ±SEM.
With the availability of a photic zone with intermediate illumination in which to rest, both An. gambiae and An. stephensi photopreferences are altered compared with those measured in the binary photo assay format. Female and male An. gambiae exhibit strong preferences for darkness when introduced to the 0 Lux end of the apparatus, as in the binary photo assay. However, both sexes prefer to rest in the intermediate (100 Lux) illumination zone when introduced to the 400 Lux zone (Fig. 2A andE). These results indicate An. gambiae males and females still actively avoid the most intensely illuminated region of the apparatus, but do not necessarily prefer complete darkness. Rather, the avoidance of 400 Lux illumination, as seen in the binary assays, can be achieved by resting in the 100 Lux region rather than the 0 Lux region of the arena. The differing An. stephensi trinary preference data indicate a strong preference for an illuminated area when introduced to the 400 Lux end of the arena (Fig. 2B andF), consistent with the hypothesis that An. stephensi mosquitos require more intense light in order to experience visual perception comparable with that of An. gambiae. These data are also consistent with past findings that An. stephensi exhibits increased flight activity in a dim-light setting compared with complete darkness (Rowland 1989).
The photopreference differences that we define in binary and trinary assays indicate that our simple photopreference arena—the first of its kind for vector mosquitos—is adequate for assessing differences in photopreferences between species, in a laboratory setting. The simple fabrication, low monetary cost, and ease of transportation and setup of the assay arena imply that the assay could be performed with field-captured mosquitoes in a field setting. This strategy would reduce the need to create stable laboratory colonies of field-caught mosquitoes for photopreference behavioral assays and may enable more accurate analysis of a given species’ photopreference in the field. Photopreference is of interest as it may inform how insecticides are applied in the field, in addition to expanding our understanding of vector photobiology. Better knowledge of mosquito photopreference may enable the application of insecticides to more specific areas of interest in the home and in the field, in conjunction with control efforts, rather than the use of broad-pattern application that covers many areas without biological relevance to the vector-targeted control. Current insecticide application methods, such as indoor residual spraying, often involve treating the entirety of a dwelling and leaving a residual coating of insecticide for months after treatment. A given vector mosquito population might experience minimal contact with many of these treated surfaces, depending on its resting patterns within dwellings. By understanding these resting patterns in greater depth, the amount of insecticide needed for spraying may be reduced and better allocated to increase vector contact with insecticides and thereby increase the effectiveness of residual insecticide treatment methods.
Diurnal Variation of Opsin Gene Expression
Previous studies have shown that larval swimming behavior in the ascidian Ciona intestinalis can be altered by knocking down Ci-Opsin1, which results in reduced photoresponsiveness (Inada et al. 2003). Given these findings, we chose to determine whether diurnal transcriptional expression patterns of selected opsin gene superfamily members in An. gambiae and An. stephensi are correlated with distinct diurnal photopreferences we observe in these species. The An. gambiae haploid genome contains 11 annotated opsin genes (Hill et al. 2002, Holt et al. 2002). Eight of the 11 genes have attributable functions, and are defined as long wavelength-sensing, short wavelength-sensing, and ultraviolet wavelength-sensing opsin genes. Our Reciprocal Best Blast analysis and manual annotation of the An. stephensi genome (VectorBase VB-2013-12) using An. gambiae opsin genes as query sequences led to the identification of four long wavelength-sensing opsin genes, one short wavelength-sensing opsin gene, and a single ultraviolet wavelength-sensing opsin gene within the An. stephensi genome. The organization of a subset of An. gambiae opsin genes and homologous genes in An. stephensi is depicted in Figure 3. On chromosome 2R, An. gambiae possesses four long wavelength-sensing opsin genes within a gene cluster (GPROP3, GPROP4, GPROP5, GPROP6; Fig. 3). An. stephensi contains a similar cluster that includes only three long wavelength-sensing genes. The difference between these clusters in the two genomes is an apparent opsin gene duplication and inversion of GPROP4 in An. gambiae. In other organisms, mainly primates, increased range of wavelength sensing and trichomatic color vision have been correlated with evolutionary duplications of long wavelength-sensing and medium wavelength-sensing opsin genes (Dulai et al. 1999). Therefore, the increased number of long wavelength-sensing opsin genes in An. gambiae as compared with An. stephensi may contribute mechanistically to differences in their photopreference behaviors.
Fig. 3.
Long wavelength opsin gene organization on An. gambiae chromosome ARM 2R. Five of the six long wavelength-sensing opsin genes cluster toward the telomeric end of chromosome 2R in An. gambiae. This gene number contrasts with the four orthologous long wavelength-sensing opsin genes present in An. stephensi.
We assessed only the long wavelength-sensing GPROP3 for diurnal expression variation for a number of reasons. First, previous studies by Rund et al. (2011) did not suggest diurnal variation in the expression of any opsin (Rund et al. 2011). Second, due to sequence conservation among the long wavelength-sensing opsin gene set we have defined, GPROP3 was the only long wavelength-sensing opsin gene that could be verified specifically as being expressed using qRT-PCR in An. gambiae.
The GPROP3, GPROP8, and GPROP9 genes in An. gambiae, which are predicted to detect long wavelengths, ultraviolet wavelengths, and short wavelengths, respectively, exhibit no significant diurnal variation in transcription during the 48-h time period assayed (Fig. 4 A,C andE). Among the orthologous genes in An. stephensi—annotated as LW, UV, and SW for putative long wavelength-, ultraviolet wavelength-, and short wavelength-responsive opsin genes, respectively—the LW and SW genes fail to exhibit striking diurnal variation in transcription (Fig. 4B andF). The UV gene transcript levels increase during the dusk crepuscular period compared to levels during other intervals of Zeitgeber time (Fig. 4D). As there are no significant differences in diurnal expression patterns for opsin genes we assayed, we can reject the hypothesis that variation in expression of the opsin genes assayed is correlated with variations in photopreference that we observe between these two species. Although the transcript levels do not vary throughout diurnal phases, it is possible that protein levels may vary due to translational or post-translational regulation. However, assessment of those possibilities lies beyond the scope of our analysis. Alternatively, as subcellular localization of some opsins in the photoreceptor cells of Ae. aegypti and An. gambiae has been described, changes in this subcellular localization, again beyond the scope of our analysis, may account for variability in photopreference between species (Hu et al. 2009, 2011, 2013).
Fig. 4.
Opsin expression profiles across Zeitgeber time. Relative quantity (2ΔCt ± SEM) of opsin gene transcripts normalized to ribosomal protein subunit-7 transcript, respectively. Time points indicate samples taken every 4 h, with time point 0 being at the beginning of a 11:11 light:dark cycle with 1 h dusk:dawn transition periods, spanning two full diurnal cycles. Each time point consists of collections of 10 female mosquitos, with N = 3. Values are normalized so the highest level of expression is equal to one for each analysis. Filled bars represent time points sampled during the dark phase of the cycle. Open bars represent time points sampled during the light phase of the cycle. Panels A,D and E represent Anopheles gambiae long-wave (GPROP3), ultraviolet (GPROP8) and short-wave (GPROP9) gene levels, respectively. Panels B, C and F represent putative orthologous long-wave, ultraviolet and short-wave genes in Anopheles stephensi.
Developmental Expression and Evolution of Opsins in An. gambiae
The difference we observe in long wavelength-sensing opsin gene number in An. gambiae and An. stephensi led us to question the potential functional significance the existence of six long wavelength-sensing opsin genes in An. gambiae and only four long wavelength-sensing opsin genes in An. stephensi. To investigate this question in An. gambiae, we utilized RNAseq analysis to assess expression of each of the 11 opsin superfamily gene members during first and third larval instars, and in female and male adults (Fig. 5, Supp File 3 [online only]). Three annotated long wavelength-sensing opsin genes—GPROP1, GPROP3, and GPROP4—are expressed more highly during adult stages, and long wavelength-sensing opsin genes GPROP5–GPROP7 all exhibit increased expression during larval stages, consistent with previous findings from microarray-based expression analyses (Marinotti et al. 2006, Rund et al. 2011). GPROP11 and GPROP12, pteropsins, are also expressed at low levels during all life stages studied. In contrast, GPROP10, an opsin of unknown wavelength sensitivity, is expressed predominantly during adult stages. The remaining opsin genes—GPROP8 and GPROP9—which encode one ultraviolet wavelength-sensing opsin and one short wavelength-sensing opsin, respectively, each exhibit higher expression in adults as compared with first and third instar larvae.
Fig. 5.
Heatmap of An. gambiae Opsin gene expression. Expression of Opsin1, 3-12 in An. gambiae in mixed-gender first larval instars (L1), mixed-gender third larval instars (L3), adult females (FB), and adult males (MB). Color intensity scale indicates increasing expression, with yellow reflecting the highest expression, measured as FPKM, and blue reflecting the lowest expression. VectorBase ID identifiers and names are given for each transcript. All opsin genes are also grouped based on wavelength detected, PT (pteropsin), UN (unknown), SW (short wavelength), UV (ultraviolet wavelength), LW (long wavelength).
The developmental partitioning of opsin superfamily gene expression that we observe—most notably the dichotomous expression of long wavelength-sensing opsin genes between larval and adult stages—is unexpected and may have functional implications. Past studies of opsin gene expression during An. gambiae development have utilized the Plasmodium/Anopheles Genome Array, which groups long wavelength-sensing GPROP1, GPROP3, and GPROP4 genes into a single probe set (Ag.2R.268.0_CDS_s_at from VectorBase; Marinotti et al. 2006, Rund et al. 2011). Thus, the respective expression profiles for these three genes have not been defined previously. Each of the other long wavelength-sensing opsin genes (GPROP5, GPROP6, and GPROP7) is detectable with distinct probes on the array, respectively, allowing for accurate expression profiling of those three opsin genes. The use of RNAseq has allowed us to define the expression of each of these opsin genes, despite the very limited sequence variation among them, and its use will enable delineation of these paralogs in subsequent analyses.
The fact that half of long wavelength-sensing opsin genes are expressed predominantly during larval stages implies that these opsins may mediate functions specific to larval life stages. In this regard, it is notable that gene structures for the subset of long-wavelength sensing opsin genes expressed predominantly during larval stages exhibit structural similarities that distinguish them from those expressed predominantly in adults (Fig. 3). Larval-biased GPROP5, GPROP6, and GRPOP7 genes each include two exonic CDS regions, and significant 5′ UTR and 3′ UTR regions are present in GPROP5 and GPROP6. In contrast, adult-biased GPROP1, GPROP3, and GPROP4 each contain a single splice-site within the 5′-UTR of each gene and minimal 3′ UTRs and the entireties of their coding capacities reside within a single exon, respectively. These differing structures are consistent with the hypothesis that the two stage-biased opsin gene subsets arose from duplication of distinct ancestral genes, with limited subsequent divergence of coding sequences and gene organization within each subset.
However, the life stage-biased functions these long wavelength-sensing opsins mediate remain unclear. Visual acuity may play an important role during larval life stages for the detection of predators within aqueous environments (Klecka and Boukal 2012), while adults may process figures or shapes from the air in search of potential sugar sources, bloodmeal sources, resting sites, and oviposition sites (Allan et al. 1987). The predominant expression of some long wavelength-sensing opsin genes during larval stages, and the expression of other long wavelength-sensing opsin genes, and short wavelength-sensing and ultraviolet wavelength-sensing opsin genes only in adults may have arisen because of differing opsin requirements underlying visual acuity in aqueous environments as compared with atmospheric environments.
Subsets of long wavelength-sensing opsins are arranged in homologous loci, which are partially conserved between An. gambiae and An. stephensi (Fig. 3). The homologous locus in An. gambiae that contains two larval-biased genes and one adult-biased gene (i.e., GPROP4-6) is highly conserved in An. stephensi. If these gene trios are derived from a single gene cluster in the most recent common ancestor (MRCA) of An. gambiae and An. stephensi, then that MRCA may have possessed similar larval–adult variability in the expression of long wavelength-sensing opsin genes. Similarly, An. stephensi contains an ortholog of An. gambiae GPROP7, and genomic regions surrounding the orthologous gene in each species appear to be syntenic as reflected by the location of An. gambiae and An. stephensi GPROP7 orthologs next to AGAP002463 and ASTE008930, respectively, which are orthologs with homologies to ubiquitin-associated and SH3 domain-containing protein B (UBASH3B [Megy et al. 2012], Fig. 3). Taken together, these observations imply that the GPROP4-6 long wavelength-sensing opsin gene cluster and the GPROP7 orthologs were present in the MRCA of these two species. This invites the hypothesis that the gene family expansion in An. gambiae that created GPROP1 and GPROP3 occurred after divergence of the two species, and that the differing illumination preferences in the two species also arose following their divergence from a common ancestor, in conjunction with opsin gene family expansion. As GPROP1 and GPROP3 are expressed predominantly in adults, An. gambiae may have been selected during its evolutionary history for greater photosensitivity based on a mechanism mediated by adult opsin gene expression. Other organisms, such as butterflies, that exhibit increases in long wavelength-sensing opsin gene number also exhibit expanded spectral diversity for visual function (Sison-Mangus et al. 2006, Frentiu et al. 2007). Therefore, the expansion of long wavelength-sensing opsin gene number may underlie dynamic evolution of visual sensitivity across an expanded spectral range in An. gambiae, as compared with An. stephensi.
In conclusion, we have begun to investigate molecular mechanisms underlying photopreference differences we observe between An. gambiae and An. stephensi in laboratory-based assays, by assessing differences between these two species in the genomic organization and expression of opsin genes. Using binary and trinary choice assays, we determine that An. gambiae and An. stephensi prefer different illumination intensities at subjective dawn and dusk. Analyses of long wavelength-, short wavelength- and ultraviolet wavelength-sensing opsin gene expression, measured over two diurnal cycles, reveal no conclusive differences between these two species in diurnal patterns of transcript expression. RNAseq data sets in An. gambiae indicate that long wavelength-sensing opsin genes are expressed in two distinct gene sets, during either larval or adult stages. Conversely, short wavelength- and ultraviolet wavelength-sensing opsin genes are expressed more highly in adult females and males. An. stephensi possess only four long wavelength-sensing opsin genes compared with the six long wavelength-sensing opsin genes found in An. gambiae, a reflection of paralogous expansion of the opsin gene superfamily in An. gambiae. The differences we observe in the number of long wavelength-sensing opsin genes in these two species may contribute to the differences we observe between their illumination intensity preferences.
Supplementary Data
Supplementary data are available at Journal of Medical Entomology online.
Acknowledgments
We would like to thank Jason Pitts and Larry Zwiebel for sharing An. gambiae RNAseq data sets; Maria Mota and Flaminia Catteruccia for mosquito eggs and larvae; Alan Kopin for sharing in financial support for RNAseq analysis; the Boston College Biology Department for financial support. Alan Kopin, Neil Lobo, Samuel Rund, and Colleen Hitchcock for their thoughtful reading of the draft manuscript; and Rob Waterhouse and Daniel Neafsey for continuous assistance during analyses of RNAseq data sets.
References Cited
- Ali A., Nayar J. K., Knight J. W., Stanley B. H. 1989. Attraction of Florida mosquitoes (Diptera:Culicidae) to artificial light in the field. 57th Annu. Conf. Calif. Mosq. Vector Control Assoc. Inc. Los Angeles, California, USA. [Google Scholar]
- Allan S. A, Day J. F., Edman J. D. 1987. Visual ecology of biting flies. Annu. Rev. Entomol. 32: 297–316. [DOI] [PubMed] [Google Scholar]
- Alonso P. L., Tanner M. 2013. Public health challenges and prospects for malaria control and elimination. Nat. Med. 19: 150–155. [DOI] [PubMed] [Google Scholar]
- Bentley M. T., Kaufman P. E., Kline D. L., Hogsette J. A. 2009. Response of adult mosquitoes to light-emitting diodes placed in resting boxes and in the field. J. Am. Mosq. Control Assoc. 25: 285–291. [DOI] [PubMed] [Google Scholar]
- Bray P. G., Mungthin M., Ridley R. G., Ward S. A. 1998. Access to hematin: The basis of chloroquine resistance. Mol. Pharmacol. 54: 170–179. [DOI] [PubMed] [Google Scholar]
- Browne S. M., Bennett G. F. 1981. Response of mosquitoes (Diptera: Culicidae) to visual stimuli. J. Med. Entomol. 18: 505–521. [DOI] [PubMed] [Google Scholar]
- Burkett D. A., Butler J. F., The S., Entomologist F., Dec N. 2012. Laboratory evaluation of colored light as an attractant for female Aedes aegypti, Aedes albopictus, Anopheles quadrimaculatus, and Culex nigripalpus. Fla. Entomol. 88: 383–389. [Google Scholar]
- Claridge-Chang A., Wijnen H., Naef F., Boothroyd C., Rajewsky N., Young M. W. 2001. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32: 657–671. [DOI] [PubMed] [Google Scholar]
- Das S., Dimopoulos G. 2008. Molecular analysis of photic inhibition of blood-feeding in Anopheles gambiae. BMC Physiol. 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde A., Doumbo O. K., Cortese J. F., Al E. 2001. A molecular marker for Chloroquine-resistant falciparum Malaria. N. Engl. J. Med. 344: 257–263. [DOI] [PubMed] [Google Scholar]
- Dondorp A. M., Nosten F., Al E. 2009. Artemisinin resistance in plasmodium falciparum malaria. N. Engl. J. Med. 361: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulai K. S., Dornum M. Von, Mollon J. D., Hunt D. M. 1999. The evolution of trichromatic color vision by opsin gene duplication in new world and old world primates the evolution of trichromatic color vision by opsin gene duplication in new world and old world primates. Genome Res. 9: 629–638. [PubMed] [Google Scholar]
- Frentiu F. D., Bernard G. D., Sison-Mangus M. P., Brower A. V. Z., Briscoe A. D. 2007. Gene duplication is an evolutionary mechanism for expanding spectral diversity in the long-wavelength photopigments of butterflies. Mol. Biol. Evol. 24: 2016–28. [DOI] [PubMed] [Google Scholar]
- Fullman N., Burstein R., Lim S. S., Medlin C., Gakidou E. 2013. Nets, spray or both? The effectiveness of insecticide-treated nets and indoor residual spraying in reducing malaria morbidity and child mortality in sub-Saharan Africa. Malar. J. 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. A., Fox A. N., Pitts R. J., Kent L. B., Tan P. L., Chrystal M. A., Cravchik A., Collins F. H., Robertson H. M., Zwiebel L. J. 2002. G protein – coupled receptors in Anopheles gambiae. Science 298: 176–178. [DOI] [PubMed] [Google Scholar]
- Holt R., Subramanian G. M., Halpern A., Sutton G. G., Charlab R., Nusskern D. R., Wincker P., Clark A. G., Ribeiro J.M.C., Wides R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- Hu X., England J. H., Lani A. C., Tung J. J., Ward N. J., Adams S. M., Barber K. A, Whaley M. A, O’Tousa J. E. 2009. Patterned rhodopsin expression in R7 photoreceptors of mosquito retina: Implications for species-specific behavior. J. Comp. Neurol. 516: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Leming M. T., a Whaley M., O’Tousa J. E. 2013. Rhodopsin coexpression in UV photoreceptors of Aedes aegypti and Anopheles gambiae mosquitoes. J. Exp. Biol. 217: 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., a Whaley M., Stein M. M., Mitchell B. E., O’Tousa J. E. 2011. Coexpression of spectrally distinct rhodopsins in Aedes aegypti R7 photoreceptors. PLOS ONE 6: e23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbahale S. S., Githeko A., Mukabana W. R., Takken W. 2012. Integrated mosquito larval source management reduces larval numbers in two highland villages in western Kenya. BMC Public Health 12: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada K., Horie T., Kusakabe T., Tsuda M. 2003. Targeted knockdown of an opsin gene inhibits the swimming behaviour photoresponse of ascidian larvae. Neurosci. Lett. 347: 167–170. [DOI] [PubMed] [Google Scholar]
- Jones M. D., Hill M., Hope A. M. 1967. The circadian flight activity of the mosquito Anopheles gambiae: Phase setting by the light regime. J. Exp. Biol. 47: 503–511. [DOI] [PubMed] [Google Scholar]
- Kawada H., Takemura S., Arikawa K. 2005. Comparative study on nocturnal behavior of Aedes aegypti and Aedes albopictus. J. Med. Entomol. 42: 312–318. [DOI] [PubMed] [Google Scholar]
- Kim D., Fedak K., Kramer R. 2012. Reduction of malaria prevalence by indoor residual spraying: A meta-regression analysis. Am. J. Trop. Med. Hyg. 87: 117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. 2013. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecka J., Boukal D. S. 2012. Who eats whom in a pool? A comparative study of prey selectivity by predatory aquatic insects. PLOS ONE 7: e37741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mala A. O., Irungu L. W., Shililu J. I., Muturi E. J., Mbogo C. M., Njagi J. K., Mukabana W. R., Githure J. I. 2011. Plasmodium falciparum transmission and aridity: A Kenyan experience from the dry lands of Baringo and its implications for Anopheles arabiensis control. Malar. J. 10: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manouchehri A. V., Djanbakhsh B., Eshghi N. 1976. The biting cycle of Anopheles dthali. A. fluviatilis and A. stephensi in southern Iran. Trop. Geogr. Med. 28: 224–227. [PubMed] [Google Scholar]
- Marinotti O., Calvo E., Nguyen Q. K., Dissanayake S., Ribeiro J. M. C., James A. A. 2006. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol. Biol. 15: 1–12. [DOI] [PubMed] [Google Scholar]
- Mathenge E. M., Misiani G. O., Oulo D. O., Irungu L. W., Ndegwa P. N., Smith T. A, Killeen G. F., Knols B.G.J. 2005. Comparative performance of the Mbita trap, CDC light trap and the human landing catch in the sampling of Anopheles arabiensis, An. funestus and culicine species in a rice irrigation in western Kenya. Malar. J. 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megy K., Emrich S. J., Lawson D., Campbell D., Dialynas E., Hughes D.S.T., Koscielny G., Louis C., Maccallum R. M., Redmond S. N., et al. 2012. VectorBase: Improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 40: D729–D734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal P. K., Sood R. D., Kapoor N., Razdan R. K., Dash A. P. 2012. Field evaluation of Icon®Life, a long-lasting insecticidal net (LLIN) against Anopheles culicifacies and transmission of malaria in District Gautam Budh Nagar (Uttar Pradesh), India. J. Vector Borne Dis. 49: 181–187. [PubMed] [Google Scholar]
- Mnyone L. L., Lyimo I. N., Lwetoijera D. W., Mpingwa M. W., Nchimbi N., Hancock P. A., Russell T. L., Kirby M. J., Takken W., Koenraadt C.J.M. 2012. Exploiting the behaviour of wild malaria vectors to achieve high infection with fungal biocontrol agents. Malar. J. 11: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. 2012. Drosophila visual transduction. Trends Neurosci. 35: 356–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard H. J., Saebø S., Reddy M. R., Reddy V. P., Abaga S., Matias A., Slotman M. A. 2012. Light traps fail to estimate reliable malaria mosquito biting rates on Bioko Island, Equatorial Guinea. Malar. J. 11: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaijmans K. P., Thomas M. B. 2011. The influence of mosquito resting behaviour and associated microclimate for malaria risk. Malar. J. 10: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perich M. J., Davila G., Turner A, Garcia A, Nelson M. 2000. Behavior of resting Aedes aegypti (Culicidae: Diptera) and its relation to ultra-low volume adulticide efficacy in Panama City, Panama. J. Med. Entomol. 37:541–546. [DOI] [PubMed] [Google Scholar]
- Pitts R. J., Rinker D. C., Jones P. L., Rokas A., Zwiebel L. J. 2011. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics 12: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer L., Fondjo E., Patchoke S., Diallo B., Lee Y., Ng A., Ndjemai H. M., Atangana J., Traore S. F., Lanzaro G., et al. 2008. Relationship between kdr mutatin and resistance to pyrethroid and DDT insecticides in natural populations of Anopheles gambiae. J. Med. Entomol. 45: 260–266. [DOI] [PubMed] [Google Scholar]
- Ribbands C. R. 1946. Moonlight and house-haunting habits of female anophelines in West Africa. Bull. Entomol. Res. 36: 395–417. [DOI] [PubMed] [Google Scholar]
- Rowland M. 1989. Changes in the circadian flight activity of the mosquito Anopheles stephensi associated with insemination, blood-feeding, oviposition and nocturnal light intensity. Physiol. Entomol. 14: 77–84. [Google Scholar]
- Rund S.S.C., Hou T. Y., Ward S. M., Collins F. H., Duffield G. E. 2011. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc. Natl Acad. Sci. USA 108: 12979–12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa H., Narita R., Kitagawa Y., Kadowaki T. 2003. The expression of genes encoding visual components is regulated by a circadian clock, light environment and age in the honeybee (Apis mellifera). Eur. J. Neurosci. 17:963–970. [DOI] [PubMed] [Google Scholar]
- Sison-Mangus M. P., Bernard G. D., Lampel J., Briscoe A. D. 2006. Beauty in the eye of the beholder: The two blue opsins of lycaenid butterflies and the opsin gene-driven evolution of sexually dimorphic eyes. J. Exp. Biol. 209: 3079–3090. [DOI] [PubMed] [Google Scholar]
- Spaethe J., Briscoe A. D. 2004. Early duplication and functional diversification of the opsin gene family in insects. Mol. Biol. Evol. 21: 1583–1594. [DOI] [PubMed] [Google Scholar]
- Takken W. 2010. Push-pull strategies for vector control. Malar. J. 9: I16. [Google Scholar]
- Tchouassi D. P., Sang R., Sole C. L., Bastos A. D. S., Cohnstaedt L. W., Torto B. 2012. Trapping of Rift Valley Fever (RVF) vectors using light emitting diode (LED) CDC traps in two arboviral disease hot spots in Kenya. Parasit. Vectors. 5: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill M., Chandre F., Brengues C., Manguin S., Akogbeto M., Pasteur N., Guillet P., Raymond M. 2000. The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol. Biol. 9: 451–455. [DOI] [PubMed] [Google Scholar]
- Wilton D. P., Fay R. W. 1972. Responses of adult Anopheles stephensi to light of various wavelengths. J. Med. Entomol. 9: 301–304. [DOI] [PubMed] [Google Scholar]
- Yu M., Kautz M. A, Thomas M. L., Johnson D., Hotchkiss E. R., Russo M. B. 2007. Operational implications of varying ambient light levels and time-of-day effects on saccadic velocity and pupillary light reflex. Ophthalmic Physiol. Opt. 27: 130–41. [DOI] [PubMed] [Google Scholar]
- Yuan Q., Metterville D., Briscoe A. D., Reppert S. M. 2007. Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24: 948–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.