Abstract
Chromosome condensation and the global repression of gene transcription1 are features of mitosis in most eukaryotes. The logic behind this phenomenon is that chromosome condensation prevents the activity of RNA polymerases. In budding yeast, however, transcription was proposed to be continuous during mitosis2. Here we show that Cdc14, a protein phosphatase required for nucleolar segregation3 and mitotic exit4, inhibits transcription of yeast ribosomal genes (rDNA) during anaphase. The phosphatase activity of Cdc14 is required for RNA polymerase I (Pol I) inhibition in vitro and in vivo. Moreover Cdc14-dependent inhibition involves nucleolar exclusion of Pol I subunits. We demonstrate that transcription inhibition is necessary for complete chromosome disjunction, because ribosomal RNA (rRNA) transcripts block condensin binding to rDNA, and show that bypassing the role of Cdc14 in nucleolar segregation requires in vivo degradation of nascent transcripts. Our results show that transcription interferes with chromosome condensation, not the reverse. We conclude that budding yeast, like most eukaryotes, inhibit Pol I transcription before segregation as a prerequisite for chromosome condensation and faithful genome separation.
In most eukaryotic cells, a dramatic structural reorganization of the genetic material into highly condensed chromosomes and the global silencing of gene transcription1 accompany entry into mitosis. This might reflect an incompatibility between transcription and chromosome condensation and/or segregation processes. The most highly transcribed regions in eukaryotic genomes are the ribosomal gene arrays (rDNA), which require a dedicated polymerase named RNA Pol I. A study in budding yeast using cell size as an indicator of cell-cycle stage and cellular RNA transcript levels established that transcription, including rDNA, is not inhibited at any stage during mitosis2. This is surprising because rDNA becomes hyper-condensed during anaphase5,6. We thus revisited whether transcription is inhibited during yeast mitosis using more sensitive assays.
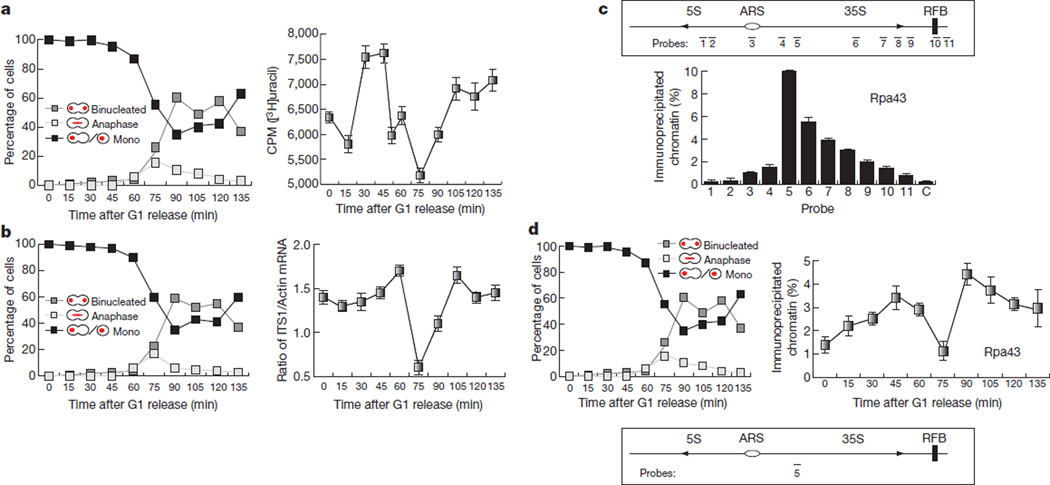
We measured total RNA synthesis in synchronised yeast cultures undergoing an entire cell cycle using incorporation of [3H]uracil into total RNA and found that cells downregulate RNA synthesis during anaphase (Fig. 1a). Analysis of nascent 35S rRNA transcripts also showed significant reduction during anaphase (Fig. 1b; 75 min and Supplementary Fig. 1). The anaphase inhibition of rRNA transcription correlates with the exclusion of the Pol I subunit Rpa43 from the 35S gene region (Fig. 1d and Supplementary Figs 2 and 3). Therefore, yeast cells, like most eukaryotes, inhibit transcription during mitosis; however, whereas transcription inhibition in most eukaryotic cells takes place in metaphase, in yeast it occurs during anaphase.
Figure 1. Transcription is inhibited in budding yeast during anaphase.
a, [3H]uracil incorporation into total RNA in wild-type yeast cells released from a G1 block. The mean (n = 3) and s.d. are shown. b, Primary rRNA transcript levels in cells released from a G1 block. qPCR using primers to the internal transcribed sequence 1 (ITS1) (Supplementary Fig. 1) were used to determine levels of intact 35S rRNA transcript. An average (n = 3) and s.d. are shown. c, ChIP analysis of Rpa43–9myc binding to ribosomal repeats in exponentially growing cells. An average (n = 2) and s.d. are shown. d, ChIP analysis of Rpa43–9myc binding to the 5′ end region of the 35S rRNA gene in wild-type yeast cells released from a G1 block. An average (n = 2) and s.d. are shown.
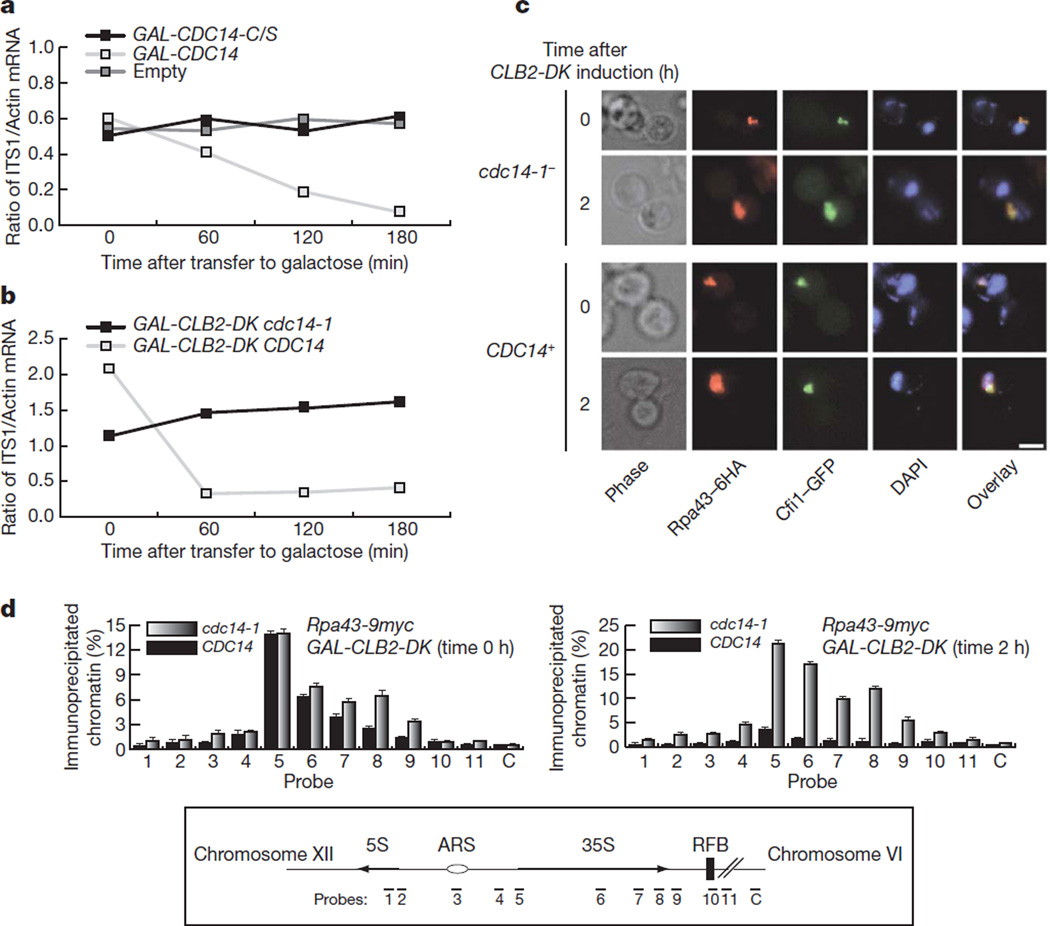
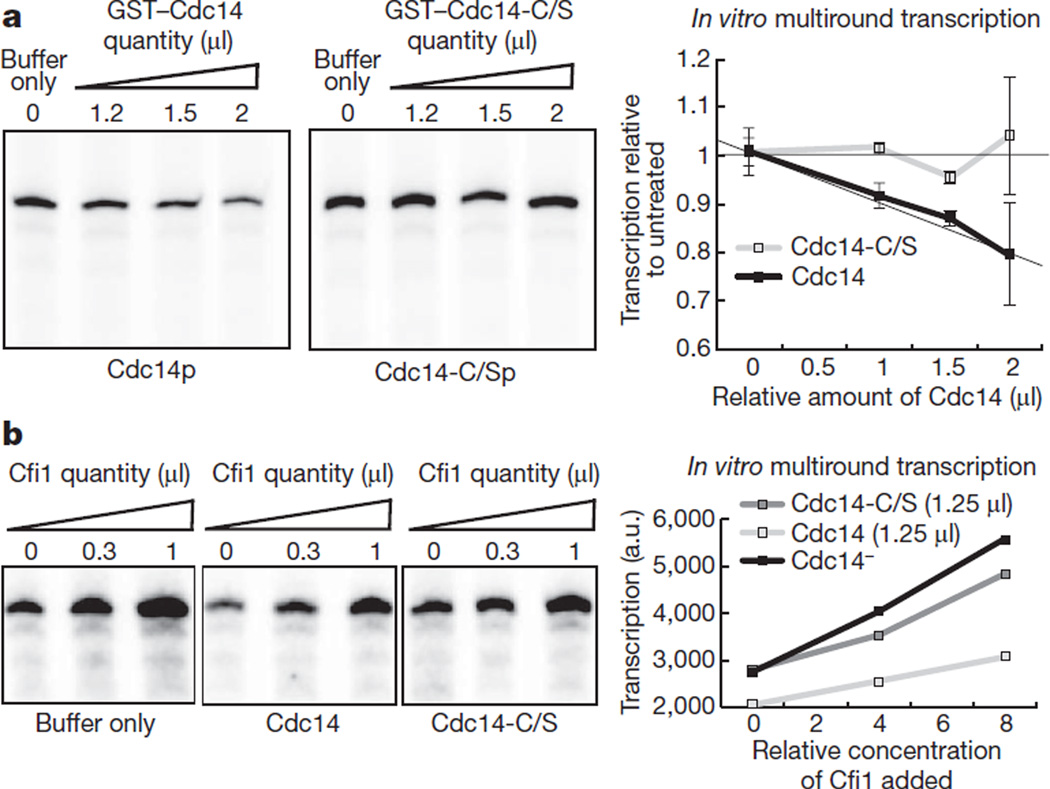
In early anaphase the conserved phosphatase Cdc14 becomes activated7. Cdc14 is required for the resolution of transcription-dependent linkages in the ribosomal gene array8,9. Expression of CDC14 from the GAL1-10 promoter in metaphase cells causes a fourfold reduction in rRNA synthesis (Fig. 2a) and the effect is dependent on its phosphate activity (Fig. 2a). Cfi1 (also known as Net1), the nucleolar inhibitor of Cdc14 and a CDK1 target10–12, interacts directly with Pol I and stimulates transcription in vitro13. Expression of a stabilized form of cyclin B2, CLB2-DK14, causes a mitotic arrest where Cdc14 is maintained at the fully released and active stage12. CLB2-DK expression also causes reduction in rRNA synthesis in the presence of wild-type Cdc14 (Fig. 2b), confirming that rDNA transcription inhibition is directly dependent on the phosphatase and not Cfi1. Moreover, CLB2-DK expression in the presence of Cdc14 causes delocalization of the essential Pol I subunit Rpa43 from the nucleolus (Fig. 2c, d and Supplementary Fig. 4). However, this is not the case for the entire Pol I holocomplex because the Rpa190 subunit is not delocalized when CLB2-DK is expressed in the presence of Cdc14 (Supplementary Fig. 5), despite the fact that Pol I transcription is inhibited (Fig. 2b). Therefore Cdc14 probably inhibits Pol I transcription by destabilization of specific subunits. These findings suggest that Cdc14 is a Pol I transcriptional repressor. Indeed, purified Cdc14 inhibits Pol I transcription in vitro (Fig. 3a) whereas the phosphatase-dead mutant does not (Fig. 3a). Cdc14 does not prevent stimulation Pol I transcription by Cfi1 (Fig. 3b). Therefore the activities of Cfi1 and Cdc14 in the activation and repression of Pol I transcription are independent.
Figure 2. Cdc14 phosphatase inhibits rRNA transcription and prevents binding of Pol I subunits to ribosomal genes.
a, Primary rRNA transcript levels in metaphase-arrested cells with expression of wild-type CDC14 or phosphatase-dead CDC14-C/S. b, Primary rRNA transcript levels upon expression of a stabilized form of cyclinB2 (CLB2-DK) in the presence of a wild-type CDC14 or temperature-sensitive mutant cdc14-1. c, Localization of Rpa43 and the nucleolar marker Cfi1 in CLB2-DK-expressing cells with wild-type CDC14 or cdc14-1. Scale bar 5 µm. d, ChIP analysis of Rpa43 binding to 35S DNA in cells before (left) and after (right) expression of CLB2-DK in the presence of a wild-type CDC14 or cdc14-1. An average (n = 2) and s.d. are shown.
Figure 3. Cdc14 inhibits RNA Pol I transcription in vitro.
a, Multiround in vitro RNA Pol I transcription assays varying the relative amount of purified wild-type Cdc14 or phosphatase-dead Cdc14 (Cdc14-C/S) added to the reaction (concentration, 160 µgml−1 of Cdc14 or Cdc14-C/S). Addition of Cdc14, but not Cdc14-C/S, inhibits RNA Pol I transcription. An average (n = 2) and s.d. are shown. b, Multiround in vitro RNA Pol I transcription assays varying the relative amount of Cfi1 added (concentration as in ref. 19) to reactions containing wild-type Cdc14, phosphatase-dead Cdc14-C/S or buffer only. a.u., arbitrary units.
Phosphorylation of Pol I has been linked to active transcription15. In metaphase-arrested cells at least two phospho-forms of Rpa43 can be detected (Supplementary Fig. 6). Expression of Cdc14 causes the loss of one of the Rpa43 phospho-bands (Supplementary Fig. 6). Therefore Cdc14 promotes dephosphorylation of Rpa43. Deletions in the carboxy-terminal region of Rpa43, where most phospho-sites are located16, prevent Cdc14-dependent nucleolar delocalization but do not affect transcription inhibition (Supplementary Fig. 7). Therefore additional Cdc14 targets in the regulation of Pol I transcription repression must exist. Nonetheless, Rpa43 seems to be a key factor because a phospho-mimicking mutant of Rpa43 shows defects in transcription inhibition (Supplementary Fig. 8).
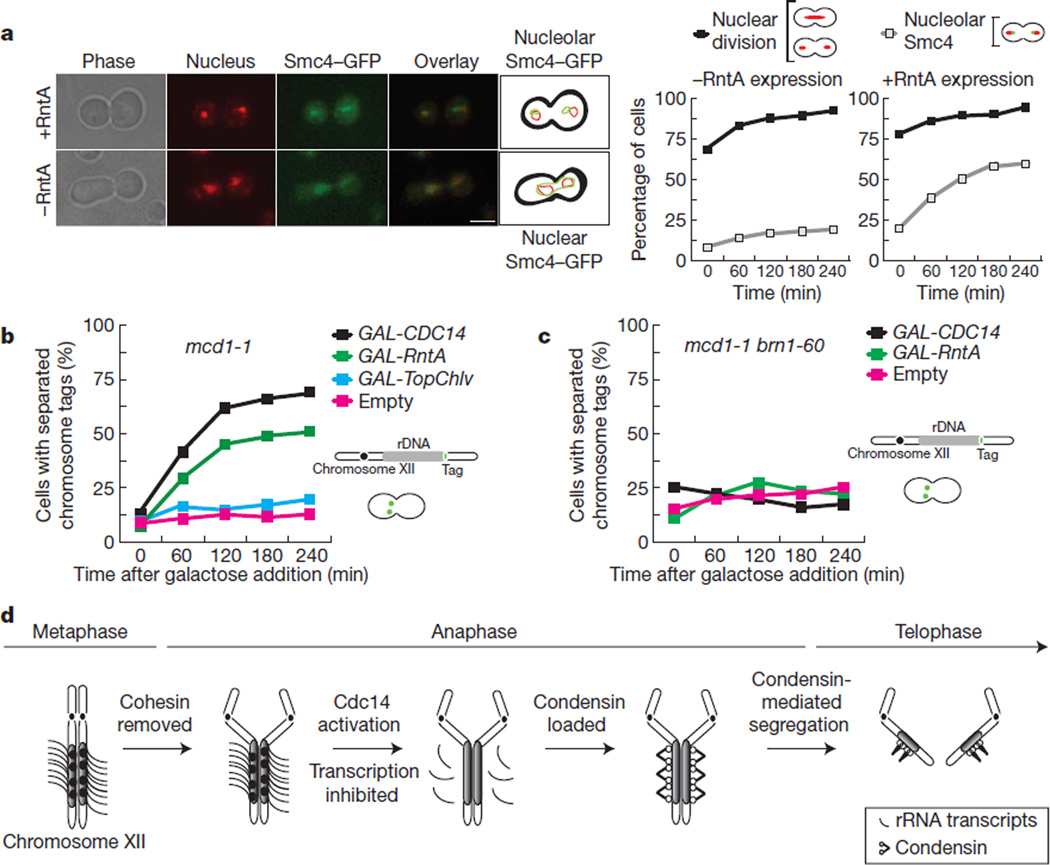
The requirement of Cdc14 for rDNA condensation17,18 and segregation17–20 is thought to stem from a defect in condensin localization17,20. Condensin binding is incompatible with rDNA transcription21, and inactivation of Pol I allows rDNA segregation in the absence of Cdc14 (refs 8 and 9). Therefore transcription inhibition by Cdc14 is probably sufficient to promote condensin binding to rDNA. Indeed, in the absence of Cdc14 removal of rRNA transcripts by expression of the RntA RNase T1 from Aspergillus oryzae22 (Supplementary Fig. 9) promotes condensin binding to rDNA (Fig. 4a).
Figure 4. Condensin localization to rDNA and nucleolar segregation requires removal of rRNA transcripts.
a, Localization of the condensin subunit Smc4 in cdc14-1 metaphase-arrested cells expressing the RntA RNase22 (+RntA) or not (−RntA). Smc4 locates throughout the nucleoplasm (nuclear) or at the nucleolus (nucleolar). Representative micrographs and quantification of the experiment are shown. Scale bar, 5 µm. Two hundred cells were scored per time point. b, rDNA segregation in the cohesin mutant mcd1-1 arrested in metaphase at 37 °C and expressing CDC14, RntA RNase, TopChlv or empty vector. Two hundred cells were scored per time point. c, rDNA segregation in the double cohesin condensin mutant mcd1-1/brn1-60 arrested in metaphase at 37 °C and expressing CDC14, RntA or empty vector. Two hundred cells were scored per time point. d, Schematic representation of CDC14-dependent rRNA transcription inhibition during yeast anaphase.
Inactivation of cohesin in metaphase-arrested cells causes separation of all genomic regions except the rDNA because Cdc14 is inactive at this stage17,18. Expression of RntA in metaphase cells where cohesin has been inactivated (or a cdc14-1 arrest; Supplementary Fig. 10) causes separation of nucleolar regions (Fig. 4b). Recent work has shown that expression of topoisomerase II fromParamecium bursaria Chlorella virus 1 (TopChlv), suppresses nucleolar segregation defects in condensin mutants23. In contrast, TopChlv does not promote separation of rDNA regions in metaphase cells inactivated for cohesin (Fig. 4b). This result is unexpected because Cdc14 and condensin were anticipated to be in the same pathway for rDNA segregation; thus the physical nature of rDNA linkages in the absence of either of these two factors should be identical. This is clearly not the case, as expression of RntA in metaphase cells where both cohesin and condensin are inactive does not rescue rDNA segregation (Fig. 4c).
An important feature of eukaryotic chromosomes is the pairing of replicated chromatids until metaphase and their separation in anaphase. In budding yeast, it was known that rDNA regions require Cdc14 (refs 17–20) and condensin24 to segregate, in addition to cohesin cleveage17,18. However, the specific roles of these factors during segregation were not clear. Here, we have shown that Cdc14 function during rDNA segregation is the inhibition of transcription. Moreover, Cdc14 is a direct inhibitor of Pol I (Fig. 3a). We have shown that condensin localization to rDNA regions is dependent on this transcriptional inhibition, thus indirectly dependent on Cdc14. Condensin binding to rDNA is essential for compaction of the locus5,24, a requirement for rDNA segregation6. Indeed, reducing the number of rDNA units partially bypasses the segregation requirement for Cdc148 (and hence downstream condensation). But are there additional requirements to transcription inhibition and condensation for rDNA segregation? Topological stress produced ahead of replication forks can be transferred to behind the forks by allowing replisome rotation, which generates precatenanes that become catenated daughter molecules after replication. In budding yeast, depletion of Top2, the enzyme that resolves catenations, causes severe segregation defects and ‘cut phenotypes’25. In principle, DNA catenations within rDNA could also prevent its segregation. Indeed, some studies have suggested links between condensin and yeast Top2 (ref. 26); thus after transcription inhibition, condensin binding could mediate rDNA segregation by promoting not only condensation but also decatenation through Top2. Whether condensin function at rDNA includes Top2-mediated decatenation is an interesting question for the future.
Inhibition of transcription activity during mitosis was discovered many years ago1. The common explanation behind this phenomenon was that chromosome condensation limits the activity, although not the accessibility, of RNA polymerases to DNA. Our results show the opposite: the condensation complex (condensin) cannot access chromatin until rRNA transcription is significantly reduced. We show that in yeast rDNA this occurs during anaphase and is a process dependent on the mitotic exit phosphatase Cdc14 (Fig. 4d). Finally, we demonstrate that failure to reduce rRNA synthesis interferes with chromosome segregation. Therefore we conclude that mitotic inhibition of transcription is critical to maintain genome integrity through cell division.
METHODS
Yeast strains and plasmids
All yeast strains used are shown in Supplementary Table 1. Chromosome tags have been described elsewhere6. C-terminal epitope tagging of Cdc14, Smc4 and Rpa43 was performed using PCR allele replacement methods29. Plasmids for overexpression of Cdc14 and Top2 have been described previously17. GAL-NLS-RntA-MYC plasmid was created from pERT-1 (ref. 22; a gift from K. Kitamoto), adding an NLS sequence at the N terminus and a myc epitope at the C terminus (details available upon request). The glutathione S-transferase (GST)–Cdc14 and GST–Cdc14 phosphatase-dead mutant (CDC14-C283S/R289; Cdc14-C/S) were gifts from A. Amon and D. Morgan. Top2 from Chlorella virus was amplified from genomic PBCV-1 (a gift from J. Van Etten). An NLS sequence at the N terminus was added to the Top2 gene and a GAL1-10 promoter for expression in yeast cells was cloned upstream of the ORF (details available upon request). Deletion of the C-terminal region of Rpa43 and point mutations were done using PCR allele replacement methods29.
Cell-cycle experiments
Cells arrested in G1 were treated with α-factor as described previously8. To release cells from α-factor blocks, we transferred them to fresh media plus pronase E (0.1 mg ml−1). For releases at non-permissive temperatures, we exposed cells to 37 °C for 30 min before their transfer to fresh media (also at 37 °C). Metaphase arrests using nocodazole were done incubating cells for 3 h with 15 µgml−1 nocodazole. For the experiments of RntA overexpression in cdc14 mutants, cells were inoculated overnight at 25 °C in –ura medium with 2% raffinose. Cells were transferred to medium containing 2% raffinose at 37 °C for 3 h to induce the arrest due to the inactivation of Cdc14. After microscopic confirmation of telophase arrest, the culture was incubated in medium containing 2% galactose at 37 °C to express RntA. For the experiments of RntA, CDC14, CDC14-C/S and Top2 overexpression in mcd1-1 or mcd1-1 brn1-60 metaphase-arrested cells, cultures were arrested in metaphase (nocodazole) at 25 °C in 2% raffinose before shifting the temperature to 37 °Cto inactivate cohesin (mcd1-1) and condensin (brn1-60) and adding 2% galactose to the media.
RNA methods
For RT–PCR analysis, RNAs were isolated with the RNeasy Mini Kit (Qiagen) using RNase-Free DNase Set (Qiagen) to digest DNA during RNA purification. One microgram of total RNA was used for the complementary DNA (cDNA) synthesis, using the Omniscript Reverse Transcription Kit (Qiagen). Finally, cDNA amplification was done using Go Taq polymerase (Promega). Primer pairs and probes to the intergenic spacer region 1 (ITS1) were used to detect primary 35S rRNA (intergenic spacer sequences are removed from the primary 35S rRNA transcripts during the initial processing steps). Quantifications were done using a Stratagene MX3000P Real-time PCR System and were confirmed by serial dilution analysis of RT–PCR reactions.
Microscopy and determination of segregation defects
Yeast cells with GFP-tagged proteins were analysed by fluorescence microscopy after 4,6-diamidino-2-phenylindole (DAPI) or propidium iodide staining. Images were collected on a Leica IRB using a Hamamatsu C4742-95 digital camera and OpenLab software (Improvision). Imaging was done in antifade medium (Molecular Probes) at room temperature. In each chromosome tag segregation/separation experiment, an average of 200 cells were counted for each time point and condition.
Cell extracts and western blotting
Collected cells were resuspended in 20 µl of RIPA buffer (10 mM sodium phosphate, 1% Triton X-100, 0.1% SDS, 10 mM EDTA, 150 mM NaCl, pH 7). Cells were disrupted in a Fast Prep machine (QBiogene). The total cell extract was recovered washing the beads with 200 µl of RIPA buffer. The soluble proteins were obtained by centrifugation for 5 min at 12,000 r.p.m. (4 °C). The protein concentration was measured using the BCA Protein Assay (Sigma). For SDS–PAGE gel electrophoresis, 30 µg of total cell extract was loaded per lane in 8% polyacrilamide gels (running in Tris-glicine buffer; Tris 3 g l−1, glycine 14.4 g l−1, SDS 1 g l−1). Gels were transferred to Hybond-P membranes (Amersham-Bioscience) in Tris-glicine buffer without SDS. Membranes were blocked with 5% milk in PBS, 0.1% Tween-20 (Sigma). The Myc epitope was detected with an anti-Myc 9E10 monoclonal antibody (Roche). The haemagglutinin (HA) epitope was detected with anti-HA 12CA5 monoclonal antibody (Roche). For phosphatase assays, 30 µg of total cell extracts were incubated for 60 min at 30 °C with λ-phosphatase (New England Biolabs) before electrophoresis. For in vitro dephosphorylation assay, Rpa43-6HA was immunoprecipitated from cell extract containing 500 µg of total protein using µMACS™ Epitope Tag Protein Isolation Kit. Immunoprecipitates were incubated in phosphatase buffer (25 mM HEPES-NaOH; 150 mM NaCl; 0.1 mg ml−1 BSA; 2 mM MnCl2) with GST–Cdc14, GST–Cdc14(C283S/R289A) (both purified from Escherichia coli) or λ-phosphatase. Proteins were analysed by immunoblotting with anti-HA antibodies.
Protein purification
E. coli cultures carrying the expression plasmids were pre-grown overnight (50 ml of lysogeny broth (LB) plus ampicillin) at 37 °C. One litre of LB medium was inoculated using the overnight culture and allowed to reachOD600 nm of 1.0. Cells were induced with 0.5MIPTG for 6 h. The pellet was re-suspended in 30 ml lysis buffer (0.1% Triton X-100, PBS and 1 mg ml−1 lysozyme) and incubated at room temperature for 15 min. Cells were sonicated three times for 30 s on ice, the extract was centrifuged at 14,000 r.p.m. for 30 min and the supernatant was collected. The supernatant was further incubated with 1 ml pre-equilibrated GST beads for 30 min at room temperature, centrifuged and washed three times with 5 ml lysis buffer, then twice with 5 ml lysis buffer plus 0.5M NaCl and Triton X-100, followed by a final set of three washes with 5 ml lysis buffer. Elution was done by incubating with 5 ml of glutathione for 5 min. Purification of Cfi1 was done as in ref. 13.
Immunofluorescent staining
Cells were fixed with 3.7% formaldehyde for 1 h at room temperature. Cells were then washed and treated with 25 µgml−1 zymolase 20T (in 0.1% β-mercaptoethanol and PBS) for 15 min at 37 °C. Next, cells were washed three times in PBS (sorbitol 1.2 M) and re-suspended in PBS (sorbitol 1.2 M, 1% Triton X-100) for 30 s. The Myc epitope was detected with an anti-Myc 9E10 monoclonal antibody (Roche) (diluted 1:500) overnight at 4 °C. Anti-mouse fluorescein-conjugated rabbit antibody (AbCam) was used for secondary detection (diluted 1:500).
Chromatin immunoprecipitation
ChIP analysis was done as in ref. 27. In brief, cells were fixed by treatment with 1% formaldehyde. Fixed cells were harvested, washed and re-suspended in ice-cold buffer I (140 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) sodium deoxycholate and 50 mM HEPES/KOH at pH 7.5) supplemented with protease inhibitor cocktail (Complete, Roche). Cells were broken with a FastPrep machine (Qbiogene) for four rounds of 30 s each with intervening incubations on ice for 5 min. Cell extracts were separated from the glass beads and sonicated (in a Bioruptor) for twelve rounds of 15 s with intervals of 45 s. This procedure fragments chromosomal DNA to an average size of about 500 base pairs. Purified anti-HA 12CA5 or anti-MYC 9e10 monoclonal antibodies (Roche) were used at 5 µg per 50 ml of initial culture. The antibodies were incubated with either protein A- or G-agarose beads (Roche) for several hours before use. Sonicated cell extracts were first pre-cleared with protein A- or G-agarose beads and then incubated overnight at 4 °C with the corresponding antibody beads. The beads were then washed three times for 15 min in buffer I, followed by three washes for 15 min in buffer II (500 mM NaCl, 1 mM EDTA,1% (v/v) Triton X-100, 0.1% (w/v) sodium deoxycholate and 50 mM HEPES/KOH at pH 7.5) and three further washes also for 15 min in buffer III (250 mM LiCl, 1 mM EDTA, 0.5% (v/v) Nonidet P-40, 0.5% (w/v) sodium deoxycholate and 10 mM Tris-Cl at pH 8.0). All wash buffers were kept at 4 °C and supplemented with the above-described protease inhibitor cocktail. The beads were then transferred to a fresh tube and washed with TE (10 mM Tris at pH 7.6 and 1 mM EDTA). Immunoprecipitated material was incubated in elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl at pH 8.0) at 65 °C for 10 min. The immunoprecipitated material was then separated from the beads and further incubated overnight at 65 °C to reverse the formaldehyde crosslinks. Aliquots of total chromatin solution were similarly heat-treated (whole-cell extract). After treatment with proteinase K (Qiagen), material was then treated with RNase (Qiagen) and finally purified through a DNA binding spin mini-column (Qiagen).
PCR analysis of co-immunoprecipitated DNA
The whole-cell extract (or total input) and immunoprecipitated samples were analysed by quantitative PCR as described above. Primer sequences are available on request. The values are given as a percentage of material immunoprecipitated (ChIP/input).
In vitro transcription
Pol I was prepared as described previously28 from cells carrying His6-(HA)3 epitope tags on the N terminus of A135 (NOY2173 and NOY2174). Multiround transcription with all pure components was performed as described at room temperature for 30 min (ref. 28).
In vivo labelling of RNA
Labelling and analysis of RNA was performed as described30. In brief, cultures were grown in yeast peptone dextrose at 25 °C to OD600 nm = 0.2–0.3. Cultures were adjusted to OD600 nm = 0.4, pulse labelled with [3H]uracil (22 µCi ml−1 final; New England Nuclear Life Sciences/Perkin Elmer Life Sciences) for 5 min. The incorporation of [3H]uracil into RNA was measured using a scintillation counter.
Supplementary Material
Acknowledgements
We are grateful to A. Amon, D. Morgan, K. Kitamoto and J. Van Etten for providing reagents, and M. Nomura for advice. We thank members of the Aragon laboratory for discussions. We acknowledge grant support from UAB HSF-GEF to D.A.S. and from ‘La Junta de Extremadura’ to A.C.-B. The Aragon laboratory is supported by the Medical Research Council of the UK.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions F.M. made the initial observation that transcription impairs rDNA segregation in the absence of Cdc14 function. All experiments shown were performed by A.C.-B., except ChIPs which were performed by M.M.-S. and in vitro phosphatase assays which were performed by D.A.S. A.J. cloned the RNAse constructs. H.T. provided a battery of Rpa43 mutants. All experiments shown were conceived by L.A. and the person performing the specific experiment. L.A. wrote the paper.
References
- 1.Taylor J. Nucleic acid synthesis in relation to the cell division cycle. Ann. NY Acad. Sci. 1960;90:409–421. doi: 10.1111/j.1749-6632.1960.tb23259.x. [DOI] [PubMed] [Google Scholar]
- 2.Elliott SG, McLaughlin CS. Regulation of RNA synthesis in yeast. III. Synthesis during the cell cycle. Mol. Gen. Genet. 1979;169:237–243. doi: 10.1007/BF00382269. [DOI] [PubMed] [Google Scholar]
- 3.Granot D, Snyder M. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil. Cytoskeleton. 1991;20:47–54. doi: 10.1002/cm.970200106. [DOI] [PubMed] [Google Scholar]
- 4.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 5.Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machin F, Torres-Rosell J, Jarmuz A, Aragon L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J. Cell Biol. 2005;168:209–219. doi: 10.1083/jcb.200408087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 8.Machin F, et al. Transcription of ribosomal genes can cause nondisjunction. J. Cell Biol. 2006;173:893–903. doi: 10.1083/jcb.200511129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomson BN, D’Amours D, Adamson BS, Aragon L, Amon A. Ribosomal DNA transcription-dependent processes interfere with chromosome segregation. Mol. Cell. Biol. 2006;26:6239–6247. doi: 10.1128/MCB.00693-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shou W, et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 11.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 12.Azzam R, et al. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- 13.Shou W, et al. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell. 2001;8:45–55. doi: 10.1016/s1097-2765(01)00291-x. [DOI] [PubMed] [Google Scholar]
- 14.Hendrickson C, Meyn MA, III, Morabito L, Holloway SL. The KEN box regulates Clb2 proteolysis in G1 and at the metaphase-to-anaphase transition. Curr. Biol. 2001;11:1781–1787. doi: 10.1016/s0960-9822(01)00564-4. [DOI] [PubMed] [Google Scholar]
- 15.Fath S, et al. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl Acad. Sci. USA. 2001;98:14334–14339. doi: 10.1073/pnas.231181398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber J, et al. Site specific phosphorylation of yeast RNA polymerase I. Nucleic Acids Res. 2008;36:793–802. doi: 10.1093/nar/gkm1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 19.Torres-Rosell J, Machin F, Jarmuz A, Aragon L. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle. 2004;3:496–502. [PubMed] [Google Scholar]
- 20.Wang BD, Yong-Gonzalez V, Strunnikov AV. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang BD, Butylin P, Strunnikov A. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle. 2006;5:2260–2267. doi: 10.4161/cc.5.19.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii T, Yamaoka H, Gomi K, Kitamoto K, Kumagai C. Cloning and nucleotide sequence of the ribonuclease T1 gene (rntA) from Aspergillus oryzae and its expression in Saccharomyces cerevisiae and Aspergillus oryzae. Biosci. Biotechnol. Biochem. 1995;59:1869–1874. doi: 10.1271/bbb.59.1869. [DOI] [PubMed] [Google Scholar]
- 23.D’Ambrosio C, Kelly G, Shirahige K, Uhlmann F. Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr. Biol. 2008;18:1084–1089. doi: 10.1016/j.cub.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 24.Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Bhalla N, Biggins S, Murray AW. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nature Protocols. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 28.Keener J, Josaitis CA, Dodd JA, Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 29.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 30.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.