Genome-wide association studies (GWAS) have identified 12 epithelial ovarian cancer (EOC) susceptibility alleles. The pattern of association at these loci is consistent in BRCA1 and BRCA2 mutation carriers who are at high EOC risk. After imputation to the 1000 Genomes Project data, we assessed associations of 11 million genetic variants with EOC risk from 15,397 cases unselected for family history and 30,816 controls, 15,252 BRCA1 mutation carriers and 8,211 BRCA2 mutation carriers (3,096 with ovarian cancer), and combined the results in a meta-analysis. This new study design yielded increased statistical power, leading to the discovery of six new EOC susceptibility loci. Variants at 1p36 (nearest gene WNT4), 4q26 (SYNPO2), 9q34.2 (ABO) and 17q11.2 (ATAD5) were associated with EOC risk, and at 1p34.3 (RSPO1) and 6p22.1 (GPX6) specifically with the serous EOC subtype, at p<5×10−8. Incorporating these variants into risk assessment tools will improve clinical risk predictions for BRCA1/2 mutation carriers.
The risk of developing invasive EOC is higher than the population average for relatives of women diagnosed with the disease1,2, indicating the importance of genetic factors in disease susceptibility. Approximately 25% of the familial aggregation of EOC is explained by rare, high-penetrance alleles of BRCA1 and BRCA23. Furthermore, population-based GWAS have identified common variants associated with invasive EOC at 11 loci4–9 but only six have also been evaluated in BRCA1 and/or BRCA2 mutation carriers. All loci displayed associations in mutation carriers that were consistent with the associations observed in the general population10–12. In addition, the 4q32.3 locus is associated with EOC risk for BRCA1 mutation carriers only13. However, the common genetic variants explain less than 3.1% of the excess familial risk of EOC so additional susceptibility loci are likely to exist.
Women diagnosed with EOC and unaffected women from the general population ascertained through the Ovarian Cancer Association Consortium (OCAC)14 and BRCA1 and BRCA2 mutation carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA)15 were genotyped as part of the Collaborative Oncological Gene-environment Study (COGS) using the iCOGS custom array. In addition, data were available for cases and controls from three EOC GWAS. We first evaluated whether the EOC susceptibility loci at 8q21.13, 10p12.31, 17q12, 5p15.33, and 17q21.31 recently identified by OCAC7–9 also show evidence of association in BRCA1 and BRCA2 mutation carriers. Using data from >200,000 genotyped SNPs7,13,16, we performed imputation of common variants from the 1000 Genomes Project data17 and evaluated the associations of these SNPs with invasive EOC risk in OCAC and in BRCA1 and BRCA2 mutation carriers from CIMBA. Given the strong evidence for a significant overlap in loci predisposing to EOC in the general population and those associated with risk in BRCA1 and BRCA2 mutation carriers, we carried out a meta-analysis of the EOC risk associations in order to identify novel EOC susceptibility loci.
Genotype data were available for imputation on 15,252 BRCA1 mutation carriers and 8,211 BRCA2 mutation carriers, of whom 2,462 and 631, respectively, were affected with EOC13,16. From OCAC, genotyping data were available from 15,437 women with invasive EOC (including 9,627 with serous EOC) and 30,845 controls from the general population7. Imputation was performed separately for BRCA1 carriers, BRCA2 carriers, OCAC-COGS samples and the three OCAC GWAS (Supplementary Tables 1–2; Supplementary Fig. 1; Supplementary Fig. 2). The meta-analysis was based on 11,403,952 SNPs (Supplementary Fig. 3).
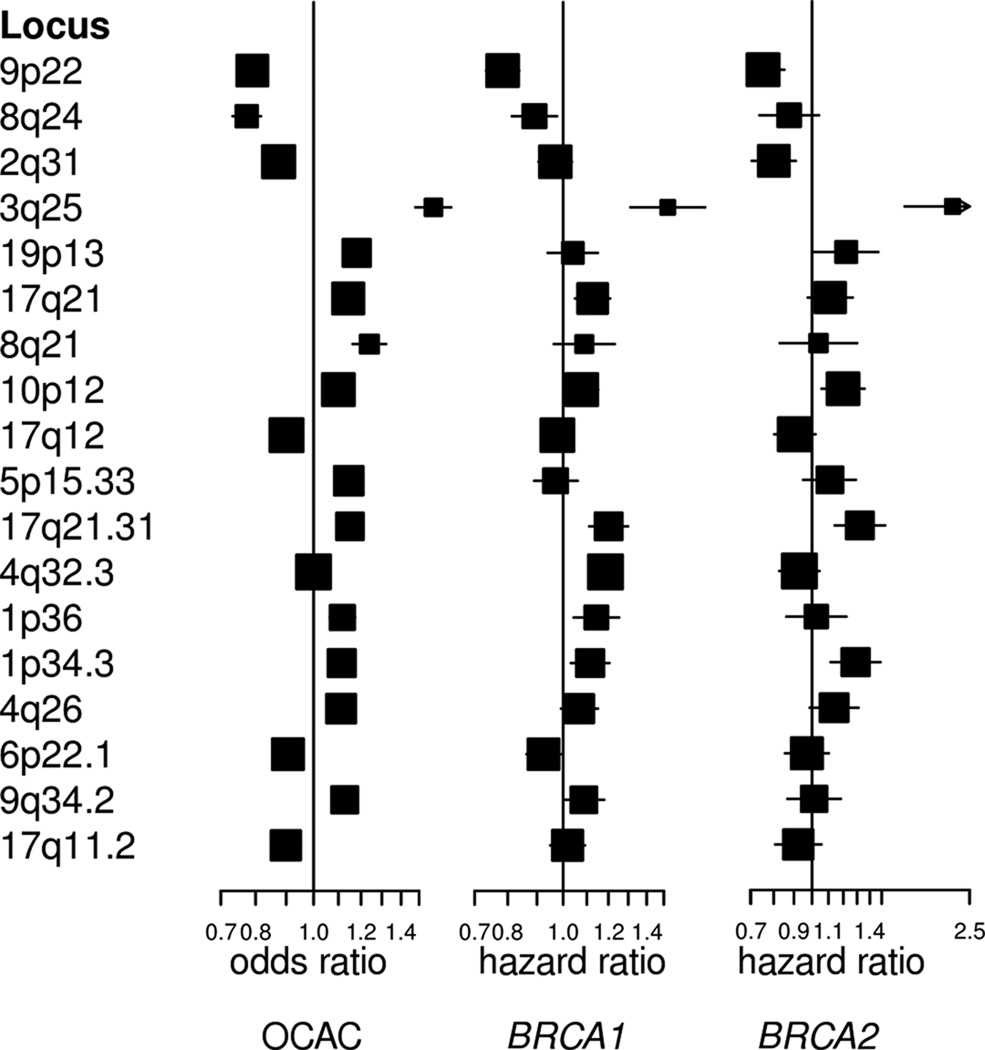
Of five EOC susceptibility loci that have not yet been evaluated in mutation carriers, two were associated with EOC risk for both BRCA1 and BRCA2 mutation carriers at p<0.05 (10p12.31 and 17q21.31) (Supplementary Table 3). Overall, seven of the twelve known EOC susceptibility loci provided evidence of association in BRCA1 mutation carriers and six were associated in BRCA2 mutation carriers. However, with the exception of 5p15.33 (TERT), all loci had hazard ratio (HR) estimates in BRCA1 and BRCA2 carriers that were in the same direction as the odds ratio (OR) estimates for serous subtype EOC from OCAC (Fig. 1). Analysing the associations jointly in BRCA1 and BRCA2 carriers and serous EOC in OCAC provided stronger evidence of association, with smaller p-values for eight of the susceptibility variants compared to the analysis in OCAC alone.
Figure 1.
Hazard ratios for the association with EOC of 12 previously reported epithelial ovarian cancer susceptibility variants and the six novel susceptibility variants for OCAC, BRCA1 mutation carriers and BRCA2 mutation carriers
Using the imputed genotypes, we observed no novel associations at p<5×10−8 in the analysis of associations in BRCA1 or BRCA2 mutation carriers separately. However, we identified seven previously unreported associations (p-values<5×10−8) in either OCAC alone, the meta-analysis of EOC associations in BRCA1, BRCA2 carriers and OCAC, or in the meta-analysis in BRCA1 and BRCA2 carriers and serous EOC in OCAC (Supplementary Fig. 4; Supplementary Tables 4–5). SNPs in six of these loci remained genome-wide statistically significant after re-imputing genotypes with imputation parameters set to maximise accuracy (Table 1; Fig. 1). SNPs at 17q11.2 (near ATAD5) were found to be associated with invasive EOC in OCAC (p<5×10−8) (Table 1). For the lead SNP, chr17:29181220:I, the estimated HR estimate for BRCA1 mutation carriers was significantly different from the estimate in OCAC (p=0.005); the association for BRCA2 carriers was consistent with the OCAC OR estimate (BRCA2-OCAC meta-analysis p=2.6×10−9). SNPs at four loci were associated at p<5×10−8 with risk of all invasive EOC in the meta-analysis (Supplementary Fig. 5): 1p36, 1p34.3, 4q26, and 9q34.2. At 1p34.3, the most strongly associated SNP, rs58722170, displayed stronger associations in the meta-analysis of serous EOC for OCAC (p=2.7×10−12). In addition, SNPs at 6p22.1 were associated at genome-wide significance level in the meta-analysis of associations with serous EOC (p=3.0×10−8), but not in the meta-analysis of all invasive EOC associations (p=6.8×10−6).
Table 1.
Association test results for loci associated at p<5×10−8 in the second imputation stage. Results reported for ovarian cancer in BRCA1 and BRCA2 mutation carriers, ovarian cancer as well as serous subtype in OCAC, the meta-analysis for ovarian cancer, and for the meta-analysis for all tumour histologies in BRCA1 and BRCA2 and serous ovarian cancer in OCAC. SNP with smallest p-value reported for each locus
| OCAC all histologies | OCAC serous | BRCA1 carriers | BRCA2 carriers | MA all histologies1 | MA serous2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locn | Nearest gene |
rs# | Ref6 | Eff6 | EAF7 | r2* | OR (95%CI) | P | OR (95%CI) | P | r2* | HR (95%CI) | P | r2* | HR (95%CI) | P | P | P |
| 1p36 | WNT4 | rs56318008 | C | T | 0.15 | 0.98 | 1.11 (1.07–1.16) |
3.9×10−7 | 1.12 (1.07–1.18) |
3.1×10−6 | 0.98 | 1.15 (1.05–1.26) |
3.1×10−3 | 0.98 | 1.03 (0.86–1.23) |
0.74 | 7.6×10−9 | 5.7×10−8 |
| 1p34.3 | RSPO1 | rs58722170 | G | C | 0.23 | 0.85 | 1.08 (1.04–1.12) |
9.7×10−5 | 1.12 (1.08–1.18) |
1.1×10−7 | 0.83 | 1.14 (1.05–1.23) |
1.5×10−3 | 0.83 | 1.35 (1.17–1.57) |
5.2×10−5 | 1.6×10−8 | 2.7×10−12 |
| 4q26 | SYNPO2 | rs17329882 | A | C | 0.24 | 0.95 | 1.09 (1.06–1.13) |
5.9×10−7 | 1.11 (1.07–1.16) |
6.4×10−7 | 0.93 | 1.08 (1.00–1.17) |
0.042 | 0.93 | 1.15 (1.00–1.33) |
0.06 | 1.4×10−8 | 1.6×10−8 |
| 6p22.1 | GPX6 | rs1161331104 | T | C | 0.31 | 0.99 | 0.93 (0.91–0.97) |
9.0×10−5 | 0.91 (0.87–0.94) |
2.6×10−7 | 0.99 | 0.92 (0.86–0.99) |
0.023 | 0.99 | 0.97 (0.85–1.10) |
0.64 | 6.8×10−6 | 3.0×10−8 |
| 9q34.2 | ABO | rs635634 | C | T | 0.19 | 0.85 | 1.11 (1.07–1.16) |
1.1×10−7 | 1.12 (1.08–1.18) |
1.0×10−6 | 0.83 | 1.11 (1.02–1.21) |
0.012 | 0.83 | 1.05 (0.89–1.23) |
0.55 | 4.4×10−9 | 4.2×10−8 |
| 17q11.2 | ATAD5 | chr17:29181220:I5 | A | AT | 0.28 | 0.95 | 0.91 (0.88–0.94) |
5.4×10−9 | 0.91 (0.87–0.94) |
8.1×10−7 | 0.94 | 1.01 (0.94–1.08) |
0.88 | 0.93 | 0.92 (0.80–1.05) |
0.23 | 2.6×10−9*3 | 3.9×10−7*3 |
Imputation accuracy r2 estimate
P-value from the meta-analysis association test for ovarian cancer in OCAC and BRCA1 and BRCA2 carriers
P-value from the meta-analysis association test for ovarian cancer in BRCA1 and BRCA2 carriers and serous ovarian cancer in OCAC
meta-analysis of ovarian cancer associations in BRCA2 carriers and OCAC only
rs116133110 listed as rs6456822 on dbSNP
chr17:29181220:I listed as rs199661266 on dbSNP
Reference and effect allele
Effect allele frequency
The most significantly associated SNP at each of the six novel loci had high imputation accuracy (r2≥0.83). At the 1p34.3, 1p36, and 6p22.1 loci, there was at least one genome-wide significant genotyped SNP correlated with the lead SNP (pairwise r2≥0.73) (Supplementary Table 6; Supplementary Fig. 5; Supplementary Note). We genotyped the leading (imputed) SNPs of the three other loci in a subset of the samples using iPLEX (Supplementary Note). The correlations between the expected allele dosages from the imputation and the observed genotypes for the variants at 4q26 and 9q34.2, (r2=0.90 and r2=0.84, respectively) were consistent with the estimated imputation accuracy (0.93 and 0.83 for CIMBA samples). The lead SNP at 17q11.2 failed iPLEX design. However, the risk allele is highly correlated with the AA haplotype of two genotyped variants on the iCOGS array (rs9910051 and rs3764419). This haplotype is strongly associated with ovarian cancer risk in the subset of samples genotyped using iCOGS (BRCA2-OCAC meta-analysis p=8.6×10−8 for haplotype, and p=1.8×10−8 for chr17:29181220:I) (Supplementary Table 7).
None of the regions contained additional SNPs that displayed EOC associations at p<10−4 in OCAC, BRCA1 carriers or BRCA2 carriers in multi-variable analyses adjusted for the lead SNP in each region, indicating that they each contain only one independent set of correlated highly associated variants (iCHAV). Relative to the 1000 Genomes Project data, we had genotyped or imputed data covering 91% of the genetic variation at 1p36, 84% at 1p34.3 and 83% at 4q26. The other three novel loci had coverage of less than 80% (Supplementary Note). There was evidence for heterogeneity at p<0.05 in the associations with histological subtype in OCAC for the lead SNPs at 1p34.4 and 6p22.1, but not for at 1p36, 4q26, 9q34.2 and 17q11.2 (Table 2).
Table 2.
Associations with ovarian cancer subtypes in OCAC samples for loci associated with ovarian cancer at p<5×10−8 in the meta-analysis
| All histologies | Serous | Endometrioid | Clear cell | Mucinous | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | rs# | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | p-het* |
| 1p36 | rs56318008 | 1.11 (1.06–1.15) |
8×10−7 | 1.12 (1.06–1.17) |
6×10−6 | 1.09 (1.00–1.19) |
0.05 | 1.24 (1.10–1.39 |
5×10−4 | 1.03 (0.91–1.17) |
0.65 | 0.22 |
| 1p34.3 | rs58722170 | 1.07 (1.03–1.11) |
2×10−4 | 1.12 (1.07–1.17) |
4×10−7 | 0.94 (0.87–1.02) |
0.16 | 1.00 (0.89–1.12) |
0.98 | 1.08 (0.97–1.21) |
0.17 | 0.001 |
| 4q26 | rs17329882 | 1.09 (1.06–1.13) |
3×10−7 | 1.11 (1.07–1.16) |
3×10−7 | 1.09 (1.01–1.18) |
0.020 | 1.06 (0.96–1.18) |
0.26 | 1.11 (0.99–1.23) |
0.06 | 0.88 |
| 6p22.1 | rs116133110 | 0.94 (0.91–0.97) |
9×10−5 | 0.91 (0.87–0.94) |
3×10−7 | 0.95 (0.89–1.02) |
0.16 | 1.05 (0.95–1.15) |
0.34 | 1.03 (0.94–1.14) |
0.53 | 0.008 |
| 9q34.2 | rs635634 | 1.12 (1.08–1.16) |
9×10−9 | 1.13 (1.08–1.18) |
2×10−7 | 1.12 (1.03–1.21) |
0.007 | 1.03 (0.92–1.16) |
0.58 | 1.23 (1.10–1.38) |
3×10−4 | 0.23 |
| 17q11.2 | chr17:29181 220:I |
0.90 (0.87–0.93) |
1×10−9 | 0.90 (0.87–0.94) |
2×10−7 | 0.88 (0.82–0.95) |
5×10−4 | 0.88 (0.80–0.98) |
0.020 | 1.01 (0.91–1.12) |
0.84 | 0.18 |
p-value for the heterogeneity in associations with different tumour subtypes
We carried out a competing risks association analysis in BRCA1 and BRCA2 mutation carriers in order to investigate whether these loci are also associated with breast cancer risk for mutation carriers (Supplementary Note). We used the most strongly associated genotyped SNPs for this purpose because the statistical method requires actual genotypes18. The EOC HR estimates were consistent with the estimates from the main analysis for all SNPs (Supplementary Table 8). None of the SNPs displayed associations with breast cancer risk at p<0.05.
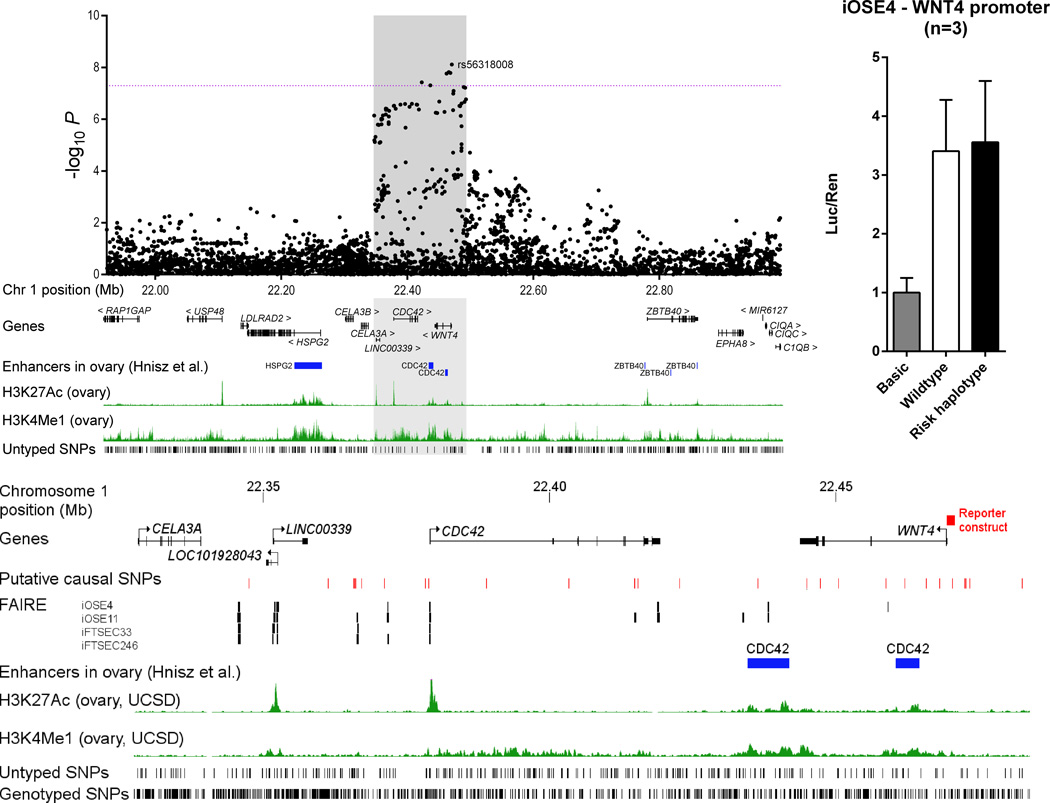
At each of the six loci, we identified a set of SNPs with odds of less than 100 to 1 against being the causal variant; most are in non-coding DNA regions (Supplementary Table 9). None were predicted to have likely deleterious functional effects although some lie in or near chromatin biofeatures in fallopian tube and ovarian epithelial cells which may represent the functional regulatory targets of the risk SNPs (Table 3; Supplementary Table 10). We also evaluated the protein coding genes in each region for their role in EOC development, and as candidate susceptibility gene targets. Molecular profiling data from 496 HGSOCs performed by The Cancer Genome Atlas (TCGA) indicated frequent loss/deletion at four risk loci (1p36, 4q26, 9q34.2 and 17q11.2) (Supplementary Table 11). Consistent with this, WNT4 and ABO were significantly down-regulated in ovarian tumours while ATAD5 was up-regulated. Somatic coding sequence mutations in the six genes nearest the index SNPs were rare. We performed expression quantitative trait locus (eQTL) analysis in a series of 59 normal ovarian tissues (Supplementary Table 12) to evaluate the gene nearest the top ranked SNP at each locus. For the five genes expressed in normal cells, we found no statistically significant eQTL associations for any of the putative causal SNPs at each locus; neither did we find any significant tumour-eQTL associations for these genes based on data from TCGA (Supplementary Table 12). At the 1p36 locus, the most strongly associated variant, rs56318008, is located in the promoter region of WNT4 which encodes a ligand in the WNT signal transduction pathway, critical for cell proliferation and differentiation. Using a luciferase reporter assay we found no effect of these putatively causal SNPs on WNT4 transcription in iOSE4 normal ovarian cells (Fig. 2). Some of the putative causal SNPs at 1p36 are located in CDC42 and LINC00339, and several are in putative regulatory domains in ovarian tissues (Supplementary Table 10; Fig. 2). CDC42 is known to play a role in migration and signalling in ovarian and breast cancer19,20. SNPs at 1p36 are also associated with increased risk of endometriosis and WNT4, CDC42 and LINC00339 have all been implicated in endometriosis21, a known risk factor for endometrioid and clear cell EOC22.
Table 3.
Summary of data on SNPs, closest gene and all genes in 1 MB region for each locus
| Loci | Position of top SNP |
# putatively causal SNPs |
Genes in window of putatively causal SNPs |
# SNPs aligned w/ biofeatures♭ |
Normal eQTL closest gene |
Tumour DNA copy number |
Significant expression difference in tumour vs normal |
Known role of gene in cancer |
# Genes in 1MB region |
Other known cancer genes in 1MB region |
|---|---|---|---|---|---|---|---|---|---|---|
| 1p36 | promoter region of WNT4 |
39 |
WNT4, CDC42, LINC00339 |
11 | NS | loss | Yes | 11 | RAP1GAP, CDC42 | |
| 1p34.3 | intron 3 of RSPO1 |
15 | RSPO1 | 0 | NS | gain | Yes | 22 | C1orf109, FHL3 | |
| 4q26 | intron 3 of SYNPO2 |
4 | SYNPO2 | 2 | NS** | loss | down | Yes | 12 | none |
| 6p22.1 | intron 1 of GPX6 |
22 | GPX6, GPX5 | 1 | N/A | gain | 23 | ZKSCAN3, TRIM27 | ||
| 9q34.2 | 4.3kb upstream of ABO |
18 | ABO, SLC2A6* | 1 | NS | loss | down | Yes | 32 |
TSC1, RALGDS, RPL7A, VAV2 |
| 17q11.2 | intron 6 of ATAD5 |
16 |
ATAD5, TEFM, ADAP2, CRLF3, SUZ12P1 |
0 | NS | loss | up | Yes | 17 | NF1 |
Proximal promoter regions were defined as 1kb upstream of the transcription start site
N/A indicated no expression of GPX6 in normal tissues. NS, not significant.
Biofeatures defined as open chromatin, H3K4me3 or H3K27ac marks detected in normal ovarian and/or fallopian cells
There are 16 genes in this region; ABO, SURF6, MED22, RPL7A, SNORD24, SNORD36B, SNORD36A, SNORD36C, SURF1, SURF2, SURF4, C9orf96, REXO4, ADAMTS13, CACFD1, SLC2A6, however all SNPs are within or upstream of ABO or upstream of SLC2A6.
Trend p=0.067
Figure 2.
The 1p36 epithelial ovarian cancer susceptibility locus
A) The Manhattan plot depicts the strength of association between all imputed and genotyped SNPs across the region bound by hg19 co-ordinates chr1:21922893-22991643. The dotted line represents the genome-wide significance level 5×10−8. Additional tracks show genes and enhancers in ovary as described in Hnisz et al38. Positions of SNPs for which imputation r2<0.3 and/or minor allele frequency <0.005 are shown in the bottom track as ‘untyped’ SNPs.
B) The shaded iCHAV from (A) is shown depicting the genes and the location of the WNT4 promoter construct as a red box. Red ticks show the positions of the putative causal variants following likelihood ratio testing. Signals from FAIRE-seq data derived from ovarian cells are represented by black marks, and the locations of predicted CDC42 enhancers38 as blue boxes. The positions of genotyped SNPs, and those that were neither genotyped nor well imputed (‘untyped’), are shown.
C) Normalised luciferase reporter activity following triplicate transfections of wildtype and risk haplotype WNT4 promoter constructs in iOSE4 cells. Error bars represent standard error from three independent experiments.
The strongest associated variant at 1q34, rs58722170, is located in RSPO1, which encodes R-spondin 1, a protein involved in cell proliferation (Supplementary Fig. 6). RSPO1 is important in tumorigenesis and early ovarian development23,24, and regulates WNT4 expression in the ovaries25. SYNPO2 at 4q26 encodes myopodin which is involved in cell motility and growth26 and has a reported tumour suppressor role27–30. rs635634 is located upstream of the ABO gene (Supplementary Fig. 7). A moderately correlated variant (rs505922, r2=0.52) determines ABO blood group and is associated with increased risk of pancreatic cancer31,32. Previous studies in OCAC also showed a modestly increased risk of EOC for individuals with the A blood group33. The moderate correlation between rs635634 and rs505922 and considerably weaker EOC association of rs505922 (p=1.2×10−5) suggests that the association with blood group is probably not driving the association with risk. The indel, 17:29181220:I, at 17q11.2 is located in ATAD5 which acts as a tumour suppressor gene34–36 (Supplementary Fig. 8). ATAD5 modulates the interaction between RAD9A and BCL2 in order to induce DNA damage related apoptosis. Finally, rs116133110, at 6p22.1, lies in GPX6 which has no known role in cancer.
The six novel loci reported in this study increase the number of genome-wide significant common variant loci so far identified for EOC to 18. Taken together, these explain approximately 3.9% of the excess familial relative risk of EOC in the general population, and account for approximately 5.2% of the EOC polygenic modifying variance in BRCA1 mutation carriers and 9.3% in BRCA2 mutation carriers. The similarity in the magnitude of associations between BRCA1 and BRCA2 carriers and population-based studies suggests a general model of susceptibility whereby BRCA1 and BRCA2 mutations and common alleles interact multiplicatively on the relative risk scale for EOC37. This model predicts large differences in absolute EOC risk between individuals carrying many alleles and individuals carrying few risk alleles of EOC susceptibility loci for BRCA1 and BRCA2 mutation carriers13,16. Incorporating EOC susceptibility variants into risk assessment tools will improve risk prediction and may be particularly useful for BRCA1 and BRCA2 mutation carriers.
METHODS
Study populations
We obtained data on BRCA1 and BRCA2 mutation carriers through CIMBA. Eligibility in CIMBA is restricted to females 18 years or older with pathogenic mutations in BRCA1 or BRCA2. The majority of the participants were sampled through cancer genetics clinics15, including some related participants. Fifty-four studies from 27 countries contributed data. After quality control, data were available on 15,252 BRCA1 mutation carriers and 8,211 BRCA2 mutation carriers, of whom 2,462 and 631, respectively, were affected with EOC (Supplementary Table 1).
Data were available for the stage 1 of three population-based EOC GWAS. These included 2,165 cases and 2,564 controls from a GWAS from North America (“US GWAS”)39, 1,762 cases and 6,118 controls from a UK-based GWAS (“UK GWAS”)6, and 441 cases and 441 controls from the Mayo GWAS. Furthermore, 11,069 cases and 21,722 controls were genotyped using the iCOGS array (“OCAC-iCOGS” stage data). Overall, 43 studies from 11 countries provided data on 15,347 women diagnosed with invasive epithelial EOC, 9,627 of whom were diagnosed with serous EOC, and 30,845 controls from the general population.
All subjects included in this analysis were of European descent and provided written informed consent as well as data and blood samples under ethically approved protocols. Further details of the OCAC and CIMBA study populations as well as the genotyping, quality control and statistical analyses have been described elsewhere7,13,16.
Genotype data
Genotyping and imputation details for each study are shown in Supplementary Table 1.
Confirmatory genotyping of imputed SNPs
To evaluate the accuracy of the imputation of the SNPs we found to be associated with EOC risk, we genotyped rs17329882 (4q26) and rs635634 (9q34.2) in a subset of 3,541 subjects from CIMBA using Sequenon’s iPLEX technology. The lead SNP at 17q11.2, chr17:29181220:I failed iPLEX design. We performed quality control of the iPLEX data according to the CIMBA guidelines. After quality control, we used the imputation results to generate the expected allele dosage for each genotyped sample and computed the Pearson product-moment correlation coefficient between the expected allele dosage and the observed genotype. The squared correlation coefficient was compared to the imputation accuracy as estimated from the imputation.
Quality control of GWAS and iCOGS genotyping data
We carried out quality control separately for BRCA1 carriers, BRCA2 carriers, the three OCAC GWAS, and OCAC-iCOGS samples, but quality criteria were mostly consistent across studies. We excluded samples if they were not of European ancestry, if they had a genotyping call rate < 95%, low or high heterozygosity, if they were not female or had ambiguous sex, or were duplicates (cryptic or intended). In OCAC studies, one individual was excluded from each pair of samples found to be first-degree relatives and duplicate samples between the iCOGS stage and any of the GWAS were excluded from the iCOGS data. SNPs were excluded if they were monomorphic, had call rate<95%, showed evidence of deviation from Hardy-Weinberg equilibrium or had low concordance between duplicate pairs. For the Mayo GWAS and the UK GWAS, we also excluded rare SNPs (MAF<1% or allele count <5, respectively). We visually inspected genotype cluster plots for all SNPs with P<10−5 from each of the newly identified loci. We used the R GenABEL library version 1.6.7 for quality control40.
Genotype data were available for analysis from iCOGS for 199,526 SNPs in OCAC-iCOGS, 200,720 SNPs in BRCA1 mutation carriers, and 200,908 SNPs in BRCA2 mutation carriers. After QC, for the GWAS, data were available on 492,956 SNPs for the US GWAS, 543,529 SNPs for the UK GWAS and 1,587,051 SNPs for the Mayo GWAS (Supplementary Table 2).
Imputation
We performed imputation separately for BRCA1 carriers, BRCA2 carriers, OCAC-iCOGS samples and each of the OCAC GWAS. We imputed variants from the 1000 Genomes Project data using the v3 April 2012 release17 as the reference panel. For OCAC-iCOGS, the UK GWAS and the Mayo GWAS, imputation was based on the 1000 Genomes Project data with singleton sites removed. To improve computation efficiency we initially used a two-step procedure, which involved pre-phasing in the first step and imputation of the phased data in the second. We carried out pre-phasing using the SHAPEIT software41. We used the IMPUTE version 2 software for the subsequent imputation42 for all studies with the exception of the US GWAS for which the MACH algorithm implemented in the minimac software version 2012.8.15, mach version 1.0.18 was used. To perform the imputation we divided the data into segments of approximately 5Mb each. We excluded SNPs from the association analysis if their imputation accuracy was r2<0.3 or their minor allele frequency (MAF) was <0.005 in BRCA1 or BRCA2 carriers or if their accuracy was r2<0.25 in OCAC-iCOGS, the UK GWAS, UK GWAS or Mayo GWAS.
We performed more accurate imputation for the regions around the novel EOC loci from the joint analysis of the data from BRCA1 and BRCA2 carriers and the general population (any SNP with P<5×10−8). The boundaries of these regions were set +/− 500kb from any significantly associated SNP in the region. As in the first run, the 1000 Genomes Project data v3 were used as the reference panel and the software IMPUTE2 was applied. However, for the second round of imputation, we imputed genotypes without pre-phasing in order to improve accuracy. To further increase the imputation accuracy we changed some of the default parameters in the imputation procedure. These included an increase of the MCMC iterations to 90 (out of which the first 15 were used as burn-in), an increase of the buffer region to 500kb and an increase of the number of haplotypes used as templates when phasing observed genotypes to 100. These changes were applied consistently for all data sets.
Statistical analyses
Association analyses in the unselected ovarian cancer cases and controls from OCAC
We evaluated the association between genotype and disease using logistic regression by estimating the associations with each additional copy of the minor allele (log-additive models). The analysis was adjusted for study and for population substructure by including the eigenvectors of the first five ancestry specific principal components as covariates in the model. We used the same approach to evaluate the SNP associations with serous ovarian cancer after excluding all cases with any other or with unknown tumour subtype. For imputed SNPs we used expected dosages in the logistic regression model to estimate SNP effect sizes and p-values. We carried out analyses separately for OCAC-iCOGS and the three GWAS and pooled thereafter using a fixed effects meta-analysis. We carried out the analysis of re-imputed genotypes of putative novel susceptibility loci jointly for the OCAC-iCOGS and GWAS samples. All results are based on the combined data from iCOGS and the three GWAS. We used custom written software for the analysis.
Associations in BRCA1 and BRCA2 mutation carriers from CIMBA
We carried out the ovarian cancer association analyses separately for BRCA1 and BRCA2 mutation carriers. The primary analysis was carried out within a survival analysis framework with time to ovarian cancer diagnosis as the endpoint. Mutation carriers were followed until the age of ovarian cancer diagnosis, or risk-reducing salpingo-oophorectomy (RRSO) or age at last observation. Breast cancer diagnosis was not considered as a censoring event. In order to account for the non-random sampling of BRCA1 and BRCA2 mutation carriers with respect to their disease status we conducted the analyses by modelling the retrospective likelihood of the observed genotypes conditional on the disease phenotype18. We assessed the associations between genotype and risk of ovarian cancer using the 1 degree of freedom score test statistic based on the retrospective likelihood18,43. To account for the non-independence among related individuals in the sample, we used an adjusted version of the score test statistic, which uses a kinship adjusted variance of the score44. We evaluated associations between imputed genotypes and ovarian cancer risk using a version of the score test as described above but with the posterior genotype probabilities replacing the genotypes. All analyses were stratified by the country of origin of the samples.
We carried out the retrospective likelihood analyses in CIMBA using custom written functions in Fortran and Python. The score test statistic was implemented in R version 3.0.145.
We evaluated whether there is evidence for multiple independent association signals in the region around each newly identified locus by evaluating the associations of genetic variants in the region while adjusting for the SNP with the smallest meta-analysis p-value in the respective region. This was done separately for BRCA1 carriers, BRCA2 carriers and OCAC.
For one of the novel associations, it was not possible to confirm the imputation accuracy of the lead SNP chr17:29181220:I at 17q11.2 through genotyping. Therefore, we inferred two-allele haplotypes for rs9910051 and rs3764419, highly correlated with the lead SNP (r2=0.95), using an in-house program. These variants were genotyped on the iCOGS array and therefore this analysis was restricted to 14,733 ovarian cancer cases and 9,165 controls from OCAC-COGS, and 8,185 BRCA2 mutation carriers that had available genotypes for both variants based on iCOGS. The association between the AA haplotype and risk was tested using logistic regression in OCAC and using Cox regression in BRCA2 mutation carriers.
Meta-analysis
We conducted a meta-analysis of the EOC associations in BRCA1, BRCA2 carriers and the general population for genotyped and imputed SNPs using an inverse variance approach assuming fixed effects. We combined the logarithm of the per-allele hazard ratio estimate for the association with EOC risk in BRCA1 and BRCA2 mutation carriers and the logarithm of the per-allele odds ratio estimate for the association with disease status in OCAC. For the associations in BRCA1 and BRCA2 carriers, we used the kinship adjusted variance estimator44 which allows for inclusion of related individuals in the analysis. We only used SNPs with results in OCAC and in at least one of the BRCA1 or the BRCA2 analyses. We carried out two separate meta-analyses, one for the associations with EOC in BRCA1 carriers, BRCA2 carriers and EOC in OCAC, irrespective of tumour histological subtype, and a second using only the associations with serous EOC in OCAC. The number of BRCA1 and BRCA2 samples with tumour histology information was too small to allow for subgroup analyses. However, previous studies have demonstrated that the majority of EOCs in BRCA1 and BRCA2 mutation carriers are high-grade serous49–53. Meta-analyses were carried out using the software “metal”, 2011-03-25 release54.
Candidate causal SNPs in each susceptibility region
In order to identify a set of potentially causal variants we excluded SNPs with a likelihood of being causal of less than 1:100, by comparing the likelihood of each SNP from the association analysis with the one of the most strongly associated SNP46. The remaining variants were then analysed using pupasuite 3.1 to identify potentially functional variants (Supplementary Table 9).
Functional analysis
Expression quantitative trait locus (eQTL) analysis in normal OSE and FTSE cells
Early-passage primary normal ovarian surface epithelial cells (OSECs) and fallopian tube epithelial cells were harvested from disease-free ovaries and fallopian tubes. Normal ovarian epithelial cells were collected by brushing the surface of the ovary with a sterile cytobrush, and were cultured in NOSE-CM55. Fallopian tube epithelial cells were harvested by Pronase digestion as previously described56, plated onto collagen-coated plastics (Sigma) and cultured in DMEM/F12 (Sigma-Aldrich) supplemented with 2% Ultroser G (BioSepra) and 1× penicillin/streptomycin (Lonza). By the time of RNA harvesting, fallopian tube cultures tested consisted of PAX8 positive fallopian tube secretory epithelial cells (FTSECs), consistent with previous observations that ciliated epithelial cells from the fallopian tube do not proliferate in vitro.
For gene expression analysis, RNA was harvested from 59 early passage samples: 54 OSECs and 5 FTSECs from cell cultures harvested at ~80% confluency using the QIAgen miRNAeasy kit with on-column DNase 1 digestion. 500ng RNA was reverse transcribed using the Superscript III kit (Life Technologies). We preamplified 10ng cDNA using the TaqMan® Preamp Mastermix; the resulting product was diluted 1:60 and used to quantify gene expression using the following TaqMan® gene expression probes: WNT4, Hs01573504_m1; RSPO1, Hs00543475_m1; SYNPO2, Hs00326493_m1; ATAD5, Hs00227495_m1 and GPX6, Hs00699698_m1. Four control genes were also included: ACTB, Hs00357333_g1; GAPDH, Hs02758991_g1; HMBS, Hs00609293_g1 and HPRT1 Hs02800695_m1 (all Life Technologies). Assays were run on an ABI 7900HT Fast Real-Time PCR system (Life Technologies).
Data Analysis
Expression levels for each gene were normalized to the average of all four control genes. Relative expression levels were calculated using the δδCt method. Genotyping was performed on the iCOGs chips, as described above. Where genotyping data were not available for the most risk-associated SNP, the next most significant SNP was used: rs3820282 at 1p36, rs12023270 at 1p34.3, rs752097 at 4q26, rs445870 at 6p22.1, rs505922 at 9q34.2 and rs3764419 at 17q11.2. Correlations between genotype and gene expression were calculated in ‘R’. Genotype specific gene expression in the normal tissue cell lines (eQTL analysis) was compared using the Jonckheere-Terpstra test. IData were normalized to the four control genes and we tested for eQTL associations, grouping OSECs and FTSECs together. Secondly, OSECs were analysed alone. eQTL analyses were performed using 3 genotype groups, or two groups (with the rare homozygote samples grouped together with the heterozygote samples).
eQTL analysis in primary ovarian tumours
eQTL analysis in primary tumours was based on the publicly available data available from The Cancer Genome Atlas (TCGA) project, which includes 489 primary high grade serous ovarian cancers. The methods have been described elsewhere57. Briefly, we determined the ancestry for each case based on the germ line genotype data using EIGENSTRAT software with 415 HapMap genotype profiles as a control set. Only populations of Northern and Western European ancestries were included. We first performed a cis-eQTL analyses using a method we described previously, in which the association between 906,600 germline genotypes and the expression levels of mRNA or miRNA (located within 500Kb on either side of the variant) were evaluated using linear regression model with the effects of somatic copy number and CpG methylation being deducted (For miRNA expression, the effect of CpG methylation is not adjusted for since the data are not available). To adjust for multiple tests, we adjusted the test P values using Benjamini-Hochberg method. A significant association was defined by a false discovery rate (FDR) of less than 0.1.
Having established a genome-wide cis-eQTL associaitions in this series of tumours, we then evaluated cis-eQTL associations for the top risk associations between each of the six new loci and the gene in closest proximity to the risk SNP. For each risk locus, we retrieved the genotype of all SNPs in ovarian cancer cases based on the Affymetrix 6.0 array. Using these genotypes and the impute2 March 2012 1000 Genomes Phase I integrated variant cosmopolitan reference panel of 1,092 individuals (Haplotypes were phased via SHAPEIT), we imputed the genotypes of SNPs in the 1000 Genomes Project in the target regions for TCGA samples58. For each risk locus where data for the most risk-associated variant were not available, we retrieved the imputed variants tightly correlated with the most risk-associated variant. We then tested for association between imputed SNPs and gene expression using the linear regression algorithm described above, where each imputed SNP was coded as an expected allele count. Again, significant associations are defined by a false discovery rate (FDR) of less than 0.1.
Regulatory profiling of normal ovarian cancer precursor tissues
We performed genome-wide formaldehyde assisted regulatory element (FAIRE) and ChIP seq with histone 3 lysine 27 acetylation (H3K27ac) and histone 3 lysine 4 monomethylation (H3K4me) for two normal OSECs, two normal FTSECs and two HGSOC cell lines (UWB1.289 and CAOV3) [Shen et al. in preparation]. These datasets annotate epigenetic signatures of open chromatin, and collectively indicate transcriptional enhancer regions. We analysed the FAIRE-seq and ChIP-seq datasets and publically available genomic data on promoter and UTR domains, intron/exon boundaries, and positions of non-coding RNA transcripts to identify SNPs from the 100:1 likely causal set that align with biofeatures that may provide evidence of SNP functionality.
Candidate Gene Analysis Using Genome Wide Profiling of Primary Ovarian Cancers
Data Sets
The Cancer Genome Atlas (TCGA) Project and COSMIC Datasets
TCGA has performed extensive genomic analysis of tumours from a large number of tissue types including almost 500 high-grade serous ovarian tumours. These data include somatic mutations, DNA copy number, mRNA and miRNA expression and DNA methylation. COSMIC is the catalogue of somatic mutations in cancer that collates information on mutations in tumours from the published literature59. They have also identified The Cancer Gene Census, which is a list of genes known to be involved in cancer. Data are available on a large number of tissue types, including 2,809 epithelial ovarian tumours.
Somatic coding sequence mutations
We analysed all genes for coding somatic sequence mutations generated from either whole exome or whole genome sequencing. In TCGA, whole exome sequencing data were available for 316 high-grade serous EOC cases. In addition, we determined whether mutations had been reported in COSMIC59 and whether the gene was a known cancer gene in the Sanger Cancer Gene Census.
mRNA expression in tumour and normal tissue
Normalized and gene expression values (Level 3) gene expression profiling data were obtain from the TCGA data portal for three different platforms (Agilent, Affymetrix HuEx and Affymetrix U133A). We analysed only the 489 primary serous ovarian tumour samples included in the final clustering analysis58 and eight normal fallopian tube samples. The boxplot function in R was used to compare ovarian tumour samples to the fallopian tube for 91 coding genes with expression data on any platform within a 1MB region around the most significant SNP at the six loci. A difference in relative expression between EOC and normal tissue was carried out using the Wilcoxon rank-sum test.
DNA copy number analysis
Serous EOC samples for 481 tumours with log2 copy number data were analysed using the cBio portal for analysis of TCGA data60,61. For each gene in a region the classes of copy number; homozygous deletion, heterozygous loss, diploid, gain, and amplification were queried individually using the advanced onco query language (OQL) option. The frequency of gain and amplification were combined as “gain”, and homozygous deletion and heterozygous loss were combined as “loss”.
Analysis of copy number vs mRNA expression
Serous EOC samples for 316 complete tumours (those with CNA, mRNA and sequencing data) were analysed. Graphs were generated using the cBio portal for analysis of TCGA data and the setting were mRNA expression data Z-score (all genes) with the Z-score threshold of 2 (default setting) and putative copy number alterations (GISTIC). The Z-score is the number of standard deviations away from the mean of expression in the reference population. GISTIC is an algorithm that attempts to identify significantly altered regions of amplification or deletion across sets of patients.
Luciferase Reporter Assay
The putative causal SNPs at the 1p36 locus lie in the WNT4 promoter and so we tested their effect on transcription in a luciferase reporter assay (Fig. 2D). Wild-type and risk haplotype (comprising five correlated variants) sequences corresponding to the region bound by hg19 co-ordinates chr1:22469416-22470869 were generated by Custom Gene Synthesis (GenScript Corporation), and then sub-cloned into pGL3-basic (Promega). Equimolar amounts of luciferase constructs (800 ng) and pRL-TK Renilla (50 ng) were co-transfected into ~8 × 104 iOSE462 normal ovarian cells in triplicate wells of 24 well plates using LipoFectamine 2000 (Life Technologies). Independent transfections were repeated three times. The Dual-Glo Luciferase Assay kit (Promega) was used to assay luciferase activity 24 hours post transfection using a BioTek Synergy H4 plate reader. The iOSE-4 cell line (derived by K. Lawrenson) was maintained under standard conditions and routinely tested for Mycoplasma and short tandem repeat profiled.
Supplementary Material
Acknowledgments
We thank all the individuals who took part in this study, and all the researchers, clinicians and technical and administrative staff who made possible the many studies contributing to this work (a full list is provided in the Supplementary Note), including Xiao Qing Chen for iPLEX genotyping. The COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 - HEALTH-F2-2009-223175). The CIMBA data management and data analysis were supported by Cancer Research–UK grants C12292/A11174 and C1287/A10118. The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). The scientific development and funding for this project were in part supported by the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19-CA148112). This study made use of data generated by the Wellcome Trust Case Control consortium. Funding for the project was provided by the Wellcome Trust under award 076113. The results published here are in part based upon data generated by The Cancer Genome Atlas Pilot Project established by the National Cancer Institute and National Human Genome Research Institute (dbGap accession number phs000178.v8.p7). The cBio portal is developed and maintained by the Computational Biology Center at Memorial Sloan-Kettering Cancer Center. SH is supported by an NHMRC Program Grant to GCT. Details of the funding of individual investigators and studies are provided in the Supplementary Note. This study made use of data generated by the Wellcome Trust Case Control consortium, funding for which was provided by the Wellcome Trust under award 076113. The results published here are, in part, based upon data generated by The Cancer Genome Atlas Pilot Project established by the National Cancer Institute and National Human Genome Research Institute (dbGap accession number phs000178.v8.p7). A full list of the investigators who contributed to the generation of the data is available on the website (see URL). The cBio portal was developed and is maintained by the Computational Biology Center at Memorial Sloan-Kettering Cancer Center.
Footnotes
Author Contributions
Writing group: K.B.K., A.C.A., G.C-T., S.J.R, J.Beesley, P.P.P., S.G. Performed statistical analyses for CIMBA: K.B.K. Performed statistical analyses for OCAC: J.T. Performed the meta-analyses: K.B.K. CIMBA database management: L.McG. and D.B. Supervised CIMBA statistical analyses and CIMBA data management: A.C.A. Supervised OCAC statistical analyses: P.P.P. Initiated and coordinated CIMBA: G.C-T. Coordinated OCAC: A.Berchuck and P.P.P. Conceived and coordinated the synthesis of the iCOGS array: D.F.E. Co-ordinated iCOGS genotyping : J.S., K.Offit, F.J.C. iCOGS genotyping, calling and quality control: J.M.Cunningham, J.D., A.Lee, P.S., D.F.E., G.C-T., Provided DNA samples and/or phenotypic data: S.J.R., J.T., A.Lee, H.C.S., K.L., S.Healey, J.M.L., T.J.S., Y.G.L., T.P., Y.B., Q.L., S.C., D.H., A.Miron, M.Southey, M.B.T., D.E.G., S.S.B., R.J., C.M.D., E.J.vanR., S.L.N., Y.C.D., T.V.O.H, L.J., A-M.G., B.E., J.D., J.Benitez, A.O., M.J.G., I.Komenka, J.N.W., P.G., P.P., L.Bernard, A.V., B.B., B.P., S.Manoukian, P.R., L.P., L.O., F.F., I.Konstantopolous, J.Garber, D.F., J.Perkins, R.P., S.E., EMBRACE, A.K.G., R.K.S., A.Meindl, C.E., C.S., O.M.S., GEMO, F.D., S.Mazoyer, D.S-L., K.Claes, K.D.L., J.Kirk, G.C.R., M.Piedmonte, D.M.O'M., M.de la H., T.C., K.A., H.Nevalinna, J.M.Collee, M.A.Rookus, J.C.O., F.B.L.H., HEBON, E.O., O.D., I.B., J.Brunet, C.L., M.A.P., A.Jakubowska, J.Gronwald, J.Lubinksi, G.S., R.B.B., M.Plante, J.S., P.S., M.M., S.Tognazzo, M.R.T., kConFab, V.S.P., X.Wang, N.L., C.I.S., N.K., J.V., C.A.A., G.P., A.Berger, C.F.S., M-K.T., C.M.P., M.H.G., P.L.M., G.R., A.M.M., S.Tchatchou, I.L.A., G.G., A.E.T., U.B.J., T.A.K., M.T., A.Bojesen, J.Z., E.F., Y.L., M.Soller, A.Liljegren, B.A., Z.E., M.S-A., O.I.O., R.L.N., T.R.R., K.L.N., S.M.D., K.H.L., B.Y.K., C.W., J.Lester, ACS, AOCS, P.W., A.H., A.B.E., M.W.B., P.A.F., D.Lambrechts, E.V.N., I.V., S.Lambrechts, E.D., J.A.D., K.G.W., M.A.Rossing, A.R., J.C-C., S.W-G., U.E., K.B.M., K.Odunsi, L.S., S.Lele, L.R.W., M.T.G., P.J.T., Y.B.S., I.B.R., M.D., P.Hillemans, T.D., N.A., N.B., A.Leminen, L.M.P., R.B., F.M., J.L.K., R.P.E., R.B.N., A.du B., F.H., I.S., P.Harter, K.M., S.Hosono, S.O., A.Jensen, S.K.K., E.H., H.N.H., M.A.N.A., S-H.T., Y-L.W., B.L.F., E.L.G., J.M.Cunningham, R.A.V., F.B., G.G.G., D.Liang, M.A.T.H., X.Wu, D.A.L., M.B., A. Berchuck, E.S.I., J.M.S., P.C., R.P.W., D.W.C., K.L.T., E.M.P., S.S.T., E.V.B., I.O., S.H.O., C.K., H.B.S., I.L.T., L.Bjorge, A.M.van A., K.K.H.A., L.A.K., L.F.A.G.M., M.K., A.B-W., L.E.K., L.S.C., N.D.L., C.C., H.Y., J.Lissowska, L.A.B., N.W., C.H., L.L., L.N., H.B., H.S., D.E., I.G.C., I.McN., J.Paul, K.Carty, N.S., R.G., A.S.W., J.H.R., V.McG., W.S., B-T.J., W.Z., X-O.S., Y-T.G., B.R., H.A.R., J.R.McL., S.A.N., A.N.M., A.C., H-Y.L., J.P-W., T.A.S., Y-Y.T., Z.C., A.Z., H.A-C., A.G-M., U.M., P.Harrington, A.W.L., A.H.W., C.L.P., G.C., M.C.P., A.D-M., A.T., I.K.R., J.Kupryjanczyk, M.F., H.Noushmehr, D.F.E., K.Offit, F.J.C., S.G., P.P.P., A.C.A., G.C-T. All authors read and approved the final manuscript.
Websites
Nature Publishing Group. Nature Genetics - iCOGS, http://www.nature.com/icogs/
The Cancer Genome Atlas Project - http://cancergenome.nih.gov/
The cBio Cancer Genomics Portal - http://www.cbioportal.org/
Pupasuite 3.1 -http://pupasuite.bioinfo.cipf.es
CIMBA QC guidelines-http://ccge.medschl.cam.ac.uk/consortia/cimba/members/data%20management/CIMBA%20and%20BCAC%20Quality%20Control%20November%202008%20v2.doc
REFERENCES
- 1.Auranen A, et al. Cancer incidence in the first-degree relatives of ovarian cancer patients. Br J Cancer. 1996;74:280–284. doi: 10.1038/bjc.1996.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stratton JF, Pharoah P, Smith SK, Easton D, Ponder BA. A systematic review and meta-analysis of family history and risk of ovarian cancer. Br J Obstet Gynaecol. 1998;105:493–499. doi: 10.1111/j.1471-0528.1998.tb10148.x. [DOI] [PubMed] [Google Scholar]
- 3.Jervis S, et al. Ovarian cancer familial relative risks by tumour subtypes and by known ovarian cancer genetic susceptibility variants. J Med Genet. 2013 doi: 10.1136/jmedgenet-2013-102015. [DOI] [PubMed] [Google Scholar]
- 4.Bolton KL, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goode EL, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pharoah PD, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–370. 370e361–370e362. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Permuth-Wey J, et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat Commun. 2013;4:1627. doi: 10.1038/ncomms2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–384. 384e371–384e372. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couch FJ, et al. Common variants at the 19p13.1 and ZNF365 loci are associated with ER subtypes of breast cancer and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2012;21:645–657. doi: 10.1158/1055-9965.EPI-11-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramus SJ, et al. Ovarian cancer susceptibility alleles and risk of ovarian cancer in BRCA1 and BRCA2 mutation carriers. Hum Mutat. 2012;33:690–702. doi: 10.1002/humu.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramus SJ, et al. Genetic variation at 9p22.2 and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2011;103:105–116. doi: 10.1093/jnci/djq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couch FJ, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolton KL, Ganda C, Berchuck A, Pharaoh PD, Gayther SA. Role of common genetic variants in ovarian cancer susceptibility and outcome: progress to date from the Ovarian Cancer Association Consortium (OCAC) J Intern Med. 2012;271:366–378. doi: 10.1111/j.1365-2796.2011.02509.x. [DOI] [PubMed] [Google Scholar]
- 15.Chenevix-Trench G, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudet MM, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9:e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genomes Project, C et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnes DR, et al. Evaluation of association methods for analysing modifiers of disease risk in carriers of high-risk mutations. Genet Epidemiol. 2012;36:274–291. doi: 10.1002/gepi.21620. [DOI] [PubMed] [Google Scholar]
- 19.Bourguignon LY, Gilad E, Rothman K, Peyrollier K. Hyaluronan-CD44 interaction with IQGAP1 promotes Cdc42 and ERK signaling, leading to actin binding, Elk-1/estrogen receptor transcriptional activation, and ovarian cancer progression. J Biol Chem. 2005;280:11961–11972. doi: 10.1074/jbc.M411985200. [DOI] [PubMed] [Google Scholar]
- 20.Zuo Y, Wu Y, Chakraborty C. Cdc42 negatively regulates intrinsic migration of highly aggressive breast cancer cells. J Cell Physiol. 2012;227:1399–1407. doi: 10.1002/jcp.22853. [DOI] [PubMed] [Google Scholar]
- 21.Pagliardini L, et al. An Italian association study and meta-analysis with previous GWAS confirm WNT4, CDKN2BAS and FN1 as the first identified susceptibility loci for endometriosis. J Med Genet. 2013;50:43–46. doi: 10.1136/jmedgenet-2012-101257. [DOI] [PubMed] [Google Scholar]
- 22.Pearce CL, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol. 2012;13:385–394. doi: 10.1016/S1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomaselli S, et al. Human RSPO1/R-spondin1 is expressed during early ovary development and augments beta-catenin signaling. PLoS One. 2011;6:e16366. doi: 10.1371/journal.pone.0016366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parma P, et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38:1304–1309. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- 25.Tomizuka K, et al. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum Mol Genet. 2008;17:1278–1291. doi: 10.1093/hmg/ddn036. [DOI] [PubMed] [Google Scholar]
- 26.De Ganck A, et al. Multiple isoforms of the tumor suppressor myopodin are simultaneously transcribed in cancer cells. Biochem Biophys Res Commun. 2008;370:269–273. doi: 10.1016/j.bbrc.2008.03.086. [DOI] [PubMed] [Google Scholar]
- 27.Jing L, et al. Expression of myopodin induces suppression of tumor growth and metastasis. Am J Pathol. 2004;164:1799–1806. doi: 10.1016/S0002-9440(10)63738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin F, et al. Myopodin, a synaptopodin homologue, is frequently deleted in invasive prostate cancers. Am J Pathol. 2001;159:1603–1612. doi: 10.1016/S0002-9440(10)63006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Carbayo M, Schwarz K, Charytonowicz E, Cordon-Cardo C, Mundel P. Tumor suppressor role for myopodin in bladder cancer: loss of nuclear expression of myopodin is cell-cycle dependent and predicts clinical outcome. Oncogene. 2003;22:5298–5305. doi: 10.1038/sj.onc.1206616. [DOI] [PubMed] [Google Scholar]
- 30.Yu YP, Luo JH. Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 2006;66:7414–7419. doi: 10.1158/0008-5472.CAN-06-0227. [DOI] [PubMed] [Google Scholar]
- 31.Amundadottir L, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rummel S, Shriver CD, Ellsworth RE. Relationships between the ABO blood group SNP rs505922 and breast cancer phenotypes: a genotype-phenotype correlation study. BMC Med Genet. 2012;13:41. doi: 10.1186/1471-2350-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole EM, et al. ABO blood group and risk of epithelial ovarian cancer within the Ovarian Cancer Association Consortium. Cancer Causes Control. 2012;23:1805–1810. doi: 10.1007/s10552-012-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikdar N, et al. DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle. 2009;8:3199–3207. doi: 10.4161/cc.8.19.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell DW, et al. Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet. 2011;7:e1002245. doi: 10.1371/journal.pgen.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee KY, et al. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J Biol Chem. 2010;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wacholder S, Han SS, Weinberg CR. Inference from a multiplicative model of joint genetic effects for [corrected] ovarian cancer risk. J Natl Cancer Inst. 2011;103:82–83. doi: 10.1093/jnci/djq510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Permuth-Wey J, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71:3896–3903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GenABEL: genome-wide SNP association analysis. 2013 [Google Scholar]
- 41.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nature Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. doi: [DOI] [PubMed] [Google Scholar]
- 42.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. Plos Genetics. 2009;5 doi: 10.1371/journal.pgen.1000529. doi:ARTN e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antoniou AC, et al. RAD51 135G-->C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antoniou AC, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R: A Language and Environment for Statistical Computing v. 3.0.1. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 46.Udler MS, Tyrer J, Easton DF. Evaluating the power to discriminate between highly correlated SNPs in genetic association studies. Genet Epidemiol. 2010;34:463–468. doi: 10.1002/gepi.20504. [DOI] [PubMed] [Google Scholar]
- 47.Reumers J, et al. Joint annotation of coding and non-coding single nucleotide polymorphisms and mutations in the SNP effect and PupaSuite databases. Nucleic Acids Res. 2008;36:D825–D829. doi: 10.1093/nar/gkm979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conde L, et al. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res. 2006;34:W621–W625. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mavaddat N, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2012;21:134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lakhani SR, et al. Pathology of ovarian cancers in BRCA1 and BRCA2 carriers. Clin Cancer Res. 2004;10:2473–2481. doi: 10.1158/1078-0432.ccr-1029-3. [DOI] [PubMed] [Google Scholar]
- 51.Rubin SC, et al. Clinical and pathological features of ovarian cancer in women with germ-line mutations of BRCA1. N Engl J Med. 1996;335:1413–1416. doi: 10.1056/NEJM199611073351901. [DOI] [PubMed] [Google Scholar]
- 52.Maehle L, et al. High risk for ovarian cancer in a prospective series is restricted to BRCA1/2 mutation carriers. Clin Cancer Res. 2008;14:7569–7573. doi: 10.1158/1078-0432.CCR-08-0112. [DOI] [PubMed] [Google Scholar]
- 53.Shaw PA, et al. Histopathologic features of genetically determined ovarian cancer. Int J Gynecol Pathol. 2002;21:407–411. doi: 10.1097/00004347-200210000-00011. [DOI] [PubMed] [Google Scholar]
- 54.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li NF, et al. A modified medium that significantly improves the growth of human normal ovarian surface epithelial (OSE) cells in vitro. Lab Invest. 2004;84:923–931. doi: 10.1038/labinvest.3700093. [DOI] [PubMed] [Google Scholar]
- 56.Fotheringham S, Levanon K, Drapkin R. Ex vivo culture of primary human fallopian tube epithelial cells. J Vis Exp. 2011 doi: 10.3791/2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, et al. Expressopn QTL based analyses reveal candidate causal genes and loci across five tumor types. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu228. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10(Unit 10):11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lawrenson K, et al. Senescent fibroblasts promote neoplastic transformation of partially transformed ovarian epithelial cells in a three-dimensional model of early stage ovarian cancer. Neoplasia. 2010;12:317–325. doi: 10.1593/neo.91948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.