Abstract
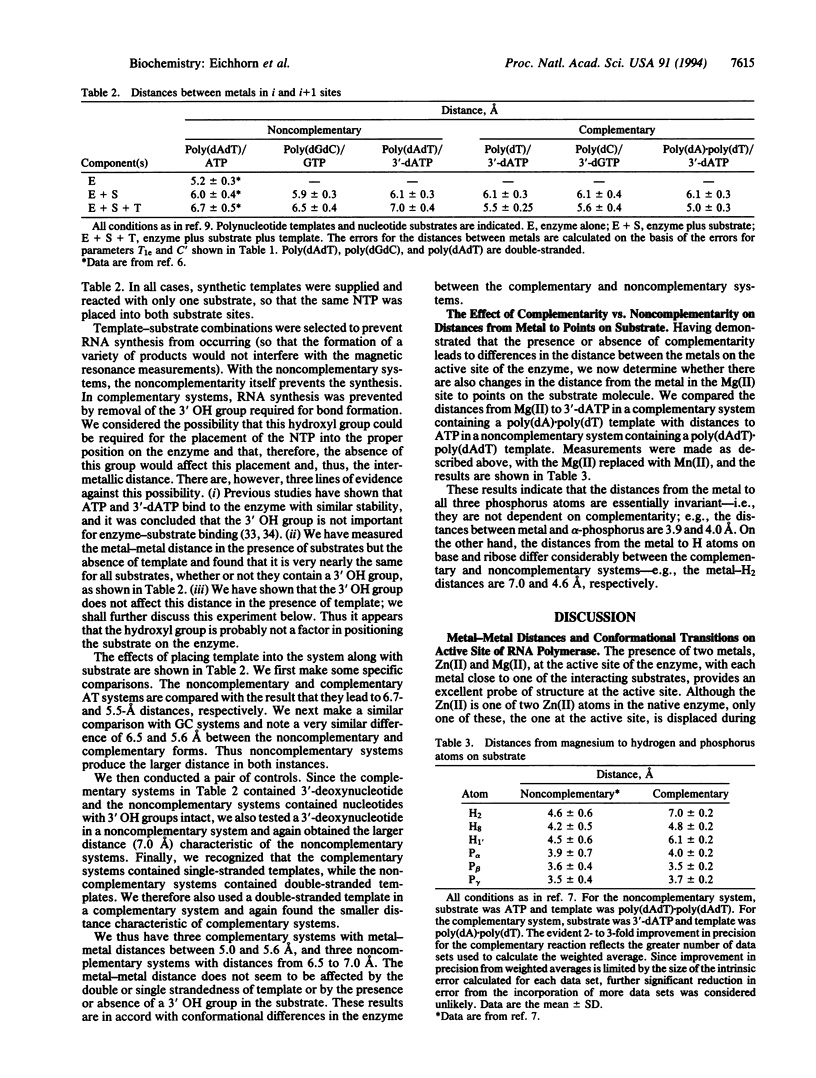
Distances between the metal ions bound to the product terminus i site and the substrate i + 1 site of Escherichia coli RNA polymerase range from 5.0 to 5.6 A when the substrate is complementary to a template base and from 6.5 to 7.0 A for a noncomplementary relationship. The metal bound to the substrate at the i + 1 site exhibits a constant distance to the three phosphates on the substrate regardless of complementarity, but the distance to base and ribose protons changes. The differences in these geometric parameters are explained by the ability of the enzyme to assume two conformations, one to place correct nucleotide substrates in optimal position for bond formation and the other to prevent incorrect nucleotides from assuming such a position. In this scheme a metal-triphosphate complex can move toward or away from the terminal 3' OH group of the growing RNA chain, to assure fidelity of transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong V. W., Yee D., Eckstein F. Mechanistic studies on deoxyribonucleic acid dependent ribonucleic acid polymerase from Escherichia coli using phosphorothioate analogues. 2. The elongation reaction. Biochemistry. 1979 Sep 18;18(19):4120–4123. doi: 10.1021/bi00586a010. [DOI] [PubMed] [Google Scholar]
- Beal R. B., Pillai R. P., Chuknyisky P. P., Levy A., Tarien E., Eichhorn G. L. Structural studies on the active site of Escherichia coli RNA polymerase. 2. Geometrical relationship of the interacting substrates. Biochemistry. 1990 Jun 26;29(25):5994–6002. doi: 10.1021/bi00477a017. [DOI] [PubMed] [Google Scholar]
- Bean B. L., Koren R., Mildvan A. S. Magnetic resonance studies of the conformation of enzyme-bound adenylyl(3' leads to 5')uridine and adenosine 5'-triphosphate on RNA polymerase from Esherichia coli. Biochemistry. 1977 Jul 26;16(15):3322–3333. doi: 10.1021/bi00634a007. [DOI] [PubMed] [Google Scholar]
- Blank A., Gallant J. A., Burgess R. R., Loeb L. A. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986 Oct 7;25(20):5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Butzow J. J., Stankis R. G. Identification of a component separated on Mono Q purification of Escherichia coli RNA polymerase as an NTPase. FEBS Lett. 1992 Mar 23;300(1):71–72. doi: 10.1016/0014-5793(92)80166-e. [DOI] [PubMed] [Google Scholar]
- Chatterji D., Wu C. W., Wu F. Y. Nuclear magnetic resonance studies on the role of intrinsic metals in Escherichia coli RNA polymerase. Effect of DNA template on the nucleotide-enzyme interaction. J Biol Chem. 1984 Jan 10;259(1):284–289. [PubMed] [Google Scholar]
- Chatterji D., Wu F. Y. Direct coordination of nucleotide with the intrinsic metal in Escherichia coli RNA polymerase. A nuclear magnetic resonance study with cobalt-substituted enzyme. Biochemistry. 1982 Sep 14;21(19):4657–4664. doi: 10.1021/bi00262a022. [DOI] [PubMed] [Google Scholar]
- Chuknyisky P. P., Rifkind J. M., Tarien E., Beal R. B., Eichhorn G. L. Structural studies on the active site of Escherichia coli RNA polymerase. 1. Interaction of metals on the i and i + 1 sites. Biochemistry. 1990 Jun 26;29(25):5987–5994. doi: 10.1021/bi00477a016. [DOI] [PubMed] [Google Scholar]
- Erie D. A., Hajiseyedjavadi O., Young M. C., von Hippel P. H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993 Nov 5;262(5135):867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- Ferrin L. J., Mildvan A. S. NMR studies of conformations and interactions of substrates and ribonucleotide templates bound to the large fragment of DNA polymerase I. Biochemistry. 1986 Sep 9;25(18):5131–5145. doi: 10.1021/bi00366a023. [DOI] [PubMed] [Google Scholar]
- Kahn J. D., Hearst J. E. Reversibility of nucleotide incorporation by Escherichia coli RNA polymerase, and its effect on fidelity. J Mol Biol. 1989 Jan 20;205(2):291–314. doi: 10.1016/0022-2836(89)90342-2. [DOI] [PubMed] [Google Scholar]
- Knight W. B., Dunaway-Mariano D., Ransom S. C., Villafranca J. J. Investigations of the metal ion-binding sites of yeast inorganic pyrophosphatase. J Biol Chem. 1984 Mar 10;259(5):2886–2895. [PubMed] [Google Scholar]
- Koren R., Mildvan S. Magnetic resonance and kinetic studies of the role of the divalent cation activator of RNA polymerase from Escherichia coli. Biochemistry. 1977 Jan 25;16(2):241–249. doi: 10.1021/bi00621a013. [DOI] [PubMed] [Google Scholar]
- Libby R. T., Nelson J. L., Calvo J. M., Gallant J. A. Transcriptional proofreading in Escherichia coli. EMBO J. 1989 Oct;8(10):3153–3158. doi: 10.1002/j.1460-2075.1989.tb08469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A., Kunkel T. A. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Lowe P. A., Hager D. A., Burgess R. R. Purification and properties of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase. Biochemistry. 1979 Apr 3;18(7):1344–1352. doi: 10.1021/bi00574a034. [DOI] [PubMed] [Google Scholar]
- Maggio E. T., Kenyon G. L., Mildvan A. S., Hegeman G. D. Mandelate racemase from Pseudomonas putida. Magnetic resonance and kinetic studies of the mechanism of catalysis. Biochemistry. 1975 Mar 25;14(6):1131–1139. doi: 10.1021/bi00677a006. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H., Kassavetis G. A., Geiduschek E. P. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J Mol Biol. 1994 Jan 28;235(4):1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- Mildvan A. S., Gupta R. K. Nuclear relaxation measurements of the geometry of enzyme-bound substrates and analogs. Methods Enzymol. 1978;49:322–359. doi: 10.1016/s0076-6879(78)49017-2. [DOI] [PubMed] [Google Scholar]
- Ninio J., Bernardi F., Brun G., Assairi L., Lauber M., Chapeville F. On the mechanism of nucleotide incorporation into DNA and RNA. FEBS Lett. 1975 Sep 15;57(2):139–144. doi: 10.1016/0014-5793(75)80702-2. [DOI] [PubMed] [Google Scholar]
- Rosenberger R. F., Hilton J. The frequency of transcriptional and translational errors at nonsense codons in the lacZ gene of Escherichia coli. Mol Gen Genet. 1983;191(2):207–212. doi: 10.1007/BF00334815. [DOI] [PubMed] [Google Scholar]
- Slepneva I. A., Weiner L. M. 31P NMR studies of the interaction of ATP with RNA polymerase from Escherichia coli. FEBS Lett. 1981 Aug 3;130(2):283–286. doi: 10.1016/0014-5793(81)81140-4. [DOI] [PubMed] [Google Scholar]
- Smagowicz W. J., Scheit K. H. The properties of ATP-analogs in initiation of RNA synthesis catalyzed by RNA polymerase from E coli. Nucleic Acids Res. 1981 May 25;9(10):2397–2410. doi: 10.1093/nar/9.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springgate C. F., Loeb L. A. On the fidelity of transcription by Escherichia coli ribonucleic acid polymerase. J Mol Biol. 1975 Oct 5;97(4):577–591. doi: 10.1016/s0022-2836(75)80060-x. [DOI] [PubMed] [Google Scholar]
- Szafrański P., 2nd, Smagowicz W. J., Wierzchowski K. L. Substrate selection by RNA polymerase from E. coli. The role of ribose and 5'-triphosphate fragments, and nucleotides interaction. Acta Biochim Pol. 1985;32(4):329–349. [PubMed] [Google Scholar]
- Villafranca J. J., Balakrishnan M. S., Wedler F. C. Determination of metal-metal distances in E. coli glutamine synthetase by EPR. Biochem Biophys Res Commun. 1977 Mar 21;75(2):464–471. doi: 10.1016/0006-291x(77)91065-8. [DOI] [PubMed] [Google Scholar]
- Volloch V. Z., Rits S., Tumerman L. A possible mechanism responsible for the correction of transcription errors. Nucleic Acids Res. 1979 Apr;6(4):1535–1546. doi: 10.1093/nar/6.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. W., Goldthwait D. A. Studies of nucleotide binding to the ribonucleic acid polymerase by equilibrium dialysis. Biochemistry. 1969 Nov;8(11):4458–4464. doi: 10.1021/bi00839a035. [DOI] [PubMed] [Google Scholar]
- Wu F. Y., Tyagi S. C. Fluorescence resonance energy transfer studies on the proximity relationship between the intrinsic metal ion and substrate binding sites of Escherichia coli RNA polymerase. J Biol Chem. 1987 Sep 25;262(27):13147–13154. [PubMed] [Google Scholar]
- Yamashita M. M., Almassy R. J., Janson C. A., Cascio D., Eisenberg D. Refined atomic model of glutamine synthetase at 3.5 A resolution. J Biol Chem. 1989 Oct 25;264(30):17681–17690. doi: 10.2210/pdb2gls/pdb. [DOI] [PubMed] [Google Scholar]
- de Mercoyrol L., Soulié J. M., Job C., Job D., Dussert C., Palmari J., Rasigni M., Rasigni G. Abortive intermediates in transcription by wheat-germ RNA polymerase II. Dynamic aspects of enzyme/template interactions in selection of the enzyme synthetic mode. Biochem J. 1990 Aug 1;269(3):651–658. doi: 10.1042/bj2690651. [DOI] [PMC free article] [PubMed] [Google Scholar]