Abstract
Liver is a prime organ responsible for synthesis, metabolism, and detoxification. The organ is endodermal in origin and its development is regulated by temporal, complex, and finely balanced cellular and molecular interactions that dictate its origin, growth, and maturation. We discuss the relevance of endoderm patterning, which truly is the first step toward mapping of domains that will give rise to specific organs. Once foregut patterning is completed, certain cells within the foregut endoderm gain competence in the form of expression of certain transcription factors that allow them to respond to certain inductive signals. Hepatic specification is then a result of such inductive signals, which often emanate from the surrounding mesenchyme. During hepatic specification bipotential hepatic stem cells or hepatoblasts become apparent and undergo expansion, which results in a visible liver primordium during the stage of hepatic morphogenesis. Hepatoblasts next differentiate into either hepatocytes or cholangiocytes. The expansion and differentiation is regulated by cellular and molecular interactions between hepatoblasts and mesenchymal cells including sinusoidal endothelial cells, stellate cells, and also innate hematopoietic elements. Further maturation of hepatocytes and cholangiocytes continues during late hepatic development as a function of various growth factors. At this time, liver gains architectural novelty in the form of zonality and at cellular level acquires polarity. A comprehensive elucidation of such finely tuned developmental cues have been the basis of transdifferentiation of various types of stem cells to hepatocyte-like cells for purposes of understanding health and disease and for therapeutic applications.
Anteroposterior Endoderm Patterning
The patterning of the endoderm along the anterior-posterior axis is necessary for proper development of endoderm-derived organs, such as the liver. During gastrulation and early somitogenesis, the endoderm is patterned into the foregut, midgut, and hindgut along the anterior-posterior axis by signaling factors secreted from the adjacent mesoderm. The fibroblast growth factor (FGF), Wnt, bone morphogenic protein (BMP), and retinoic acid (RA) signaling pathways are implicated in this patterning: FGF (30, 152), Wnt (41, 86), and BMP (146, 153) signaling promote posterior endoderm development, and RA (62) signaling is essential for proper foregut-hindgut boundary formation. In the posterior endoderm, FGF4 and Wnts from its surrounding mesoderm suppress foregut fate and promote hindgut fate; however, in the anterior endoderm, the suppression of these signaling results in foregut fate (30, 41, 86, 152). In Xenopus, Wnt signaling appears to be repressed by a secreted Wnt inhibitor Sfrp5 to maintain foregut fate in the anterior endoderm and to allow for subsequent liver development (73).
If foregut patterning does not occur properly, the liver that derives from the foregut endoderm does not form properly. For example, suppression of Wnt/β-catenin signaling during early somitogenesis makes posterior endodermal cells adopt foregut fate, resulting in the ectopic expression of the foregut marker Hhex and later liver markers in the intestine (86). Overactivation of Wnt/β-catenin signaling makes anterior endodermal cells adopt hindgut fate, resulting in a failure of liver formation (86). Suppression of RA signaling blocks liver formation in zebrafish, whereas enhancement of RA signaling results in the anterior expansion of the liver (133). Moreover, in zebrafish embryos deficient for the RA-synthesizing enzyme retinaldehyde dehydrogenase 2, the liver fails to form due to inappropriate endoderm patterning (133). Although RA signaling does not influence early liver development in Xenopus or chick (132), these zebrafish data reiterate the importance of proper endoderm patterning for subsequent liver development.
Hepatic Competence
Hepatic competence means an ability to respond to hepatic inducing signals, thereby inducing liver specification. Only the cells retaining hepatic competence can give rise to hepatoblasts. Hepatic competence has been principally analyzed at the level of pioneer transcription factors that can modulate the local chromatin structure (159–163) moreover, its regulation at other levels has recently been suggested (113, 122).
Pioneer transcription factors first bind and open the chromatin structure
FOXA (23, 24) and GATA (13, 23) transcription factors have been defined as pioneer transcription factors that confer hepatic competence due to their occupancy of the albumin (Alb) gene enhancer prior to liver specification and their ability to bind and open the compacted chromatin (Fig. 1). By investigating the occupancy on this enhancer in embryonic liver buds and the endoderm in which Alb is not expressed yet, it was revealed that two binding sites for FOXA and GATA factors are occupied in the foregut endoderm before Alb is expressed (13) and even in the dorsal endoderm that normally gives rise to the intestine (14). Since FOXA transcription factors have the ability to bind to their target sites in the compacted chromatin and locally open it, their binding permits subsequent binding of additional factors that cannot bind to the compacted chromatin by themselves (23). In breast cancer cells, FOXA1 binding relieves chromatin condensation and allows estrogen receptors to bind to local estrogen-responsive elements (19), providing another example to show that FOXA factors function as pioneer transcription factors. Mouse knockout data have demonstrated the essential role of FOXA transcription factors in hepatic competence (69). Liver-specific genes are not expressed in Foxa1 and Foxa2 double knockout mice, and importantly their expression was not induced in the explant cultures of the foregut endoderm isolated from the knockout embryos in the presence of FGF2, the hepatic inducing signal (69).
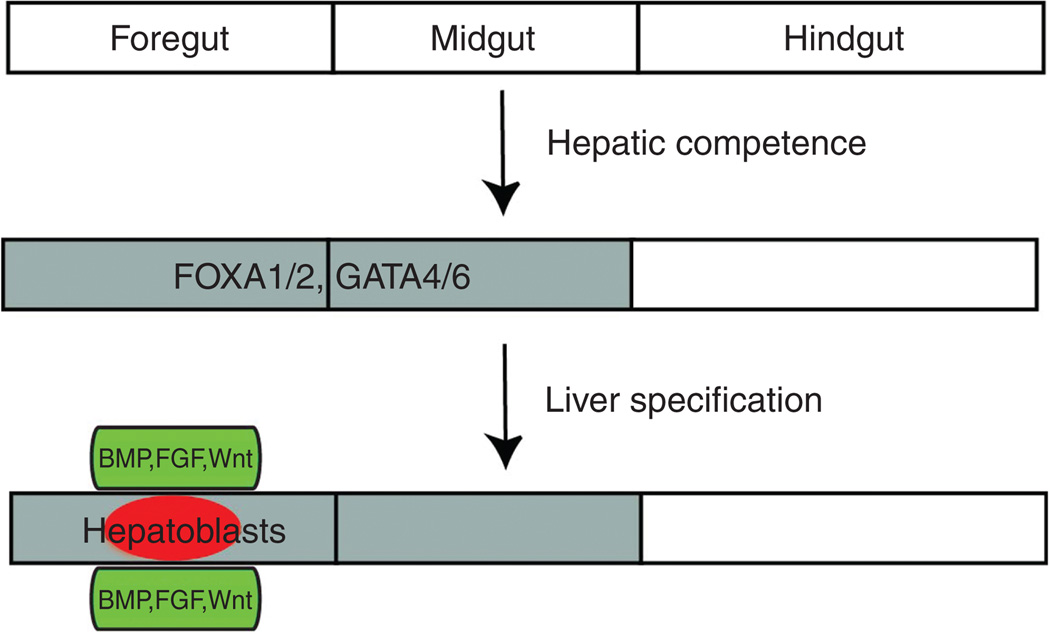
Figure 1.
Diagrams depicting hepatic competence and liver specification. Once the endoderm is properly patterned anteroposteriorly, the foregut and midgut endoderm possess hepatic competence, shown here by FOXA1/2 and GATA4/6 expression (gray). Endodermal cells in the foregut endoderm receive hepatic inducing signals, bone morphogenic protein (BMP), fibroblast growth factor (FGF), and Wnt, from adjacent mesodermal tissues (green), and subsequently give rise to hepatoblasts (red).
GATA4 can bind to the compacted chromatin and open it, but it is less effective than FOXA1 (23). Since FOXA genes are expressed earlier than Gata genes in the endoderm during development (3, 63, 92), it appears that FOXA factors first bind to their target site in the compacted chromatin, and help GATA factors bind to the adjacent target site, thereby stably opening the compacted chromatin (23). Gata4 and Gata6 are expressed in the foregut endoderm, suggesting that these two genes redundantly regulate hepatic competence. Liver specification is not compromised in Gata4 (150) or Gata6 (168) single knockout mice although the liver bud fails to expand in these mutant embryos. The double knockout data are needed to determine whether GATA factors are required for hepatic competence. In zebrafish, single knockdown of Gata4 or Gata6 greatly reduced the size of transferrin-positive liver buds, whereas double knockdown of these genes resulted in no expression of transferrin (46). These zebrafish data also suggest the redundant roles of GATA factors in liver specification.
Broader endodermal regions retain hepatic competence
The FOXA and GATA hepatic competence factors are expressed not only in the ventral foregut endoderm but also in the dorsal endoderm, which is posterior to the foregut endoderm and normally does not give rise to the liver (3, 67, 93). These expression patterns and data from mouse explant culture studies suggest that the dorsal endoderm also retain hepatic competence (14, 43). When the dorsal endoderm was isolated between embryonic days (E) 8.5 and E11.5, and cultured free from its surrounding mesoderm, it could be induced to express Alb (14, 43). These data suggest that the dorsal endoderm retains hepatic competence and that mesodermal tissues adjacent to the dorsal endoderm suppress liver specification. The dorsal mesoderm eventually appears to lose hepatic competence as it could not be induced to express Alb when isolated at E13.5 and beyond (14). Recent zebrafish studies also demonstrated that endodermal regions posterior to the liver-forming region, which is equivalent to the murine dorsal endoderm, retain hepatic competence during development (122). When Wnt8a, a hepatic inducing signal, was overexpressed in entire tissues under the control of the heat-shock promoter, ectopic hepatoblasts, and later hepatocytes were present in the posterior endoderm. The extent of ectopic hepatocyte formation upon Wnt8a overexpression is gradually reduced as embryos develop, and the posterior endoderm eventually loses hepatic competence (122). Both the mouse explant culture studies and the zebrafish in vivo studies indicate that broader endodermal regions, in particular the dorsal endoderm, possess hepatic competence.
Signaling pathways regulate hepatic competence
In addition to the pioneer transcription factors, signaling pathways are implicated in hepatic competence. GATA4 expression in mouse foregut endoderm explants was greatly reduced by the BMP inhibitor noggin (112), suggesting that BMP signaling plays a positive role in hepatic competence. In addition, BMP signaling appears to regulate hepatic competence in zebrafish (22). Bmp2b is expressed in the lateral plate mesoderm (LPM) surrounding the endoderm before liver specification occurs in zebrafish (Fig. 2). Bmp2b signaling represses Pdx1 expression in endodermal cells adjacent to the LPM, thereby permitting Pdx1-negative endodermal cells to give rise to hepatoblasts (22). Intriguingly, BMP signaling was recently reported to be implicated in the regulation of histone modifications. BMP signaling via SMAD4 recruits histone acetyltransferase P300, results in histone acetylation at liver target elements, and enhances liver gene activation (154). Although FOXA1 can bind to the compacted chromatin, it does not bind most of its target sites in the chromatin, suggesting that certain histone modifications may prevent or allow FOXA1 binding to the sites (77). Thus, BMP signaling may regulate hepatic competence, in part, by enhancing histone acetylation, which can influence FOXA1 binding.
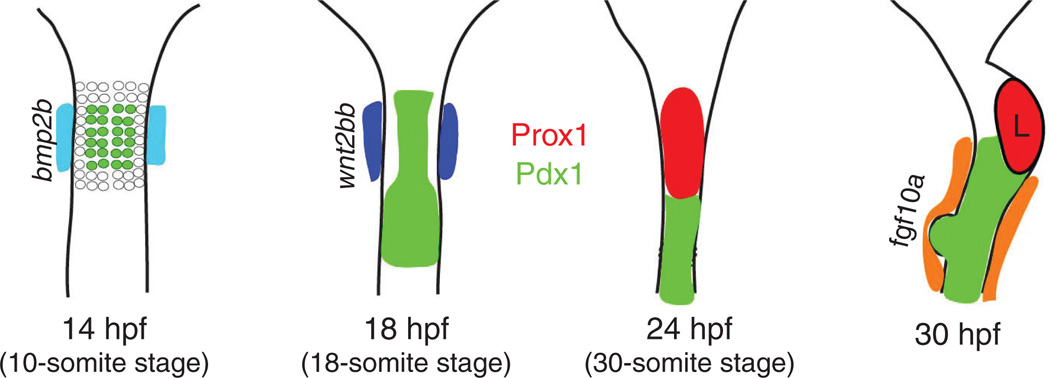
Figure 2.
Diagrams of zebrafish early liver development. bmp2b is expressed in the lateral plate mesoderm (LPM) from 14 h postfertilization (hpf), and appears to suppress Pdx1 expression (green) in the neighboring endodermal cells (22). Some of these Pdx1− endodermal cells give rise to hepatoblasts. wnt2bb is expressed in the LPM from 18 hpf (dark blue) (101); the early hepatoblast marker Prox1 starts to be expressed in endodermal cells (red) anterior to the Pdx1+ domain from 24 hpf and is continuously expressed in the liver (L) (34). fgf10a is expressed in the LPM surrounding the Pdx1+ domain from 30 hpf (orange), and appears to repress hepatic competence (122). Ventral views of the endoderm and endoderm-derived organs, anterior up.
In addition to BMP signaling, FGF signaling has been recently implicated in the regulation of hepatic competence. The extent of ectopic hepatocyte formation upon Wnt8a overexpression in zebrafish is gradually reduced as embryos develop, but this reduction is greatly delayed in fgf10a mutant embryos (122). fgf10a is expressed in the LPM surrounding the posterior endoderm in which ectopic hepatoblasts are induced upon Wnt8a overexpression. Blocking Ras activity resulted in the same phenotype as seen in fgf10a mutants, indicating Ras is downstream of FGF10a signaling in the regulation of hepatic competence (122).
Hepatic competence can be regulated at multiple levels
Although hepatic competence has been mainly understood at the level of pioneer transcription factors, it can be regulated at multiple levels. To respond to hepatic inducing signals, the ligands should first bind to their receptors in responding endodermal cells. The regulation of the expression of the receptors and the secreted inhibitors can influence the ligand-receptor binding, suggesting the regulation of hepatic competence at the ligand-receptor binding level. Upon the binding, signals are transferred from the membrane to the nucleus through the intracellular signaling components. The regulation of these components could also influence hepatic competence. In the nucleus, the local chromatin structure of regions containing hepatoblast genes can influence binding of transcription factors/cofactors. If they cannot bind to their target sites in the enhancer/promoter regions, liver specification will not occur. Thus, histone modifications and DNA methylation that affect chromatin structure could regulate hepatic competence. Histone acetylation mediated by P300 has recently been implicated in liver gene activation (154). Pioneer transcription factors also change chromatin structure by opening compacted chromatin. Furthermore, hepatic competence can be regulated at the level of the corepressors, coactivators, and other transcription factors that can regulate the expression of hepatoblast genes. The GRG3 corepressor can repress hepatic competence through binding to FOXA transcription factors, thereby recruiting the corepressor to FOXA target genes and creating a compacted chromatin structure (113).
Liver Specification (Hepatic Induction)
Endodermal cells that possess hepatic competence can give rise to hepatoblasts only if they receive hepatic inducing signals (99, 159–161, 163, 164). Three hepatic inducing signals, FGF, BMP, and Wnt, have been identified so far (Fig. 1). These signals appear to be expressed in the mesoderm adjacent to the ventral foregut endoderm. The mesoderm that secretes hepatic inducing signals is the cardiac mesoderm and septum transversum mesenchyme (STM) in mice, and the LPM in zebrafish. Although mesodermal tissues that secrete hepatic inducing signals are not the same between mouse and zebrafish, they are adjacent to the ventral foregut endoderm during liver specification. Therefore, specification occurs in a specific, discrete region of the endoderm although a much broader endodermal region retains hepatic competence.
FGF signaling plays an inductive role in liver specification
Classic chick transplantation experiments suggested that the cardiac mesoderm possesses hepatic inducing signal(s) (68). Transplanted foregut endoderm developed into the liver when it was close to the cardiac mesoderm. However, it did not develop into the liver when it was close to the other mesodermal tissues. About 25 years later, Dr. Zaret’s group identified that FGF is the hepatic inducing signal from the cardiac mesoderm (53). FGF ligands, FGF1 and FGF2, but not FGF8, could replace the cardiac mesoderm to induce Alb expression from the foregut endoderm in mouse explant cultures. Moreover, the soluble extracellular domains of FGF receptors efficiently blocked Alb induction (53). The role of FGF signaling in liver specification was confirmed in chick (166) and zebrafish (123). Chick explant culture studies showed that several FGFs including FGF1 and FGF2 could substitute for the cardiogenic mesoderm to induce HHEX and ALB expression from the anterior lateral endoderm and that the treatment of the FGF receptor inhibitor could block ALB induction (166). In zebrafish, blocking FGF signaling via overexpression of the dominant-negative form of Fgfr1 resulted in the great reduction or absence of the expression of the hepatoblast markers hhex and prox1 in the liver-forming region (123). Among several FGF downstream signaling pathways, the mitogen-activated protein kinase (MAPK) pathway is essential for liver specification (18). Chemical inhibitors that suppress the MAPK pathway could block Alb induction in mouse tissue explant and whole-embryo cultures, whereas inhibitors that suppress the PI3K pathway could not do it (18).
Mouse explant culture studies have further suggested that different thresholds of FGFs are used to induce hepatoblasts and lung progenitor cells from the ventral foregut endoderm (121). A low concentration of FGF2 (5 ng/mL) induces Alb expression, whereas a high concentration (50–500 ng/mL) induces the expression of the lung progenitor marker Nkx2.1, but not Alb. Based on the induction timing of liver and lung markers and the intensity of FGF expression in the heart, it was hypothesized that endodermal cells close to the cardiac mesoderm at the 6 to 7 somite stage are exposed to a low concentration of FGF and give rise to hepatoblasts due to the low level of FGF expression in the cardiac mesoderm at this stage. However, at the 7 to 8 somite stage, the cardiac mesoderm expresses more FGF, so that endodermal cells close to the cardiac mesoderm are exposed to a high concentration of FGF and give rise to lung progenitor cells (58).
In spite of explant culture and some in vivo studies, FGF ligands and receptors responsible for liver specification have not been identified yet. So far, there is no report about any single or compound FGF ligand or receptor mutants, which show a defect in liver specification. Since several FGF ligands and receptors are expressed in the cardiac mesoderm and the foregut endoderm (27, 53, 135, 137, 166, 169), the absence of one or two FGF ligands or receptors could be compensated by the presence of other FGF ligands or receptors. Therefore, the analysis of multiple compound mutants is needed to identify FGF ligands and receptors essential for liver specification.
BMP signaling is also required for liver specification
In addition to FGF, BMP emanating from the STM is also required for liver specification. Initial explant culture studies (53) did not identify BMP as a hepatic inducing signal because dissected foregut tissues always contained septum transversum mesenchymal tissues. Subsequent explant culture studies (112) showed that noggin treatment blocked Alb induction in the foregut endoderm cocultured with the cardiac mesoderm, revealing the crucial role of BMP signaling in liver specification. BMP2, 4, and 7 rescued the induction failure, suggesting that these BMP ligands can induce liver specification. Bmp2 and Bmp4 are expressed in the STM, but not in the foregut endoderm, when liver specification occurs in mice (112), indicating the STM as a BMP source. The essential role of BMP signaling in liver specification was confirmed in chick (166) and zebrafish (123). Pellets of noggin-expressing cells implanted into chick embryos blocked HHEX expression in endodermal cells close to the pellets, whereas BMP2-containig beads ectopically induced HHEX expression in the endoderm adjacent to beads implanted just posterior or lateral to the endogenous HHEX expression domain, but not other endodermal regions (166). Blocking BMP signaling in zebrafish, by the overexpression of the dominant-negative BMP receptor after early somitogenesis, abolished or greatly reduced hhex and prox1 expression (123).
A liver bud does not form in Bmp4 mutant mice; however, endodermal thickening, the first morphological indication of liver specification, occurs with a significant delay (112). In addition, Alb is expressed in the mutant ventral foregut endoderm although its level is much lower in the mutants than wild-type (112). These data indicate that liver specification does occur in Bmp4 mutants but subsequent proliferation and migration are blocked. Since Bmp2 expression pattern is quite similar to Bmp4 (78) and BMP4 can induce Alb expression in vitro (112), Bmp2 could potentially compensate for the lack of Bmp4 in the mutants. The analysis of Bmp2 and Bmp4 double mutant mice is needed to test this hypothesis.
Wnt is a novel hepatic inducing signal
An unbiased forward-genetic screen in zebrafish led to the identification of wnt2bb mutants that have very small or no liver buds (101) (Fig. 2). Prox1 expression is greatly reduced in this mutant, and overexpression of the dominant-negative Tcf blocked liver formation, suggesting the positive role of Wnt/β-catenin signaling in liver specification (101). The presence of hhex and Prox1 expression, although delayed and greatly reduced, as well as liver recovery in this mutant suggested that other Wnt ligands might compensate for the absence of Wnt2bb. Wnt2 knockdown in wnt2bb mutants blocked liver recovery and importantly resulted in no hhex expression in the liver-forming region (107), indicating the essential role of Wnt signaling in liver specification. Both wnt2 (107) and wnt2bb (101) are expressed in the LPM adjacent to the liver-forming region. wnt2bb is expressed earlier than wnt2, which may explain delayed liver formation in wnt2bb mutants.
Since Wnt signaling promotes the posterior endodermal fate and suppresses the anterior endodermal fate during gastrulation and early somitogenesis (86), Wnt signaling should be suppressed in the anterior endoderm during these stages to permit liver formation later. However, after anteroposterior endoderm patterning is established, Wnt signaling positively regulates liver specification. When β-catenin signaling was activated in the Xenopus endoderm from Stage 11 (midgastrula) or 20 (6–7 somite stage), the expression of the liver marker for1 was greatly reduced. In contrast, when β-catenin signaling was activated from Stage 30, for1 expression was greatly expanded (86). In addition, when β-catenin signaling was repressed in the endoderm from Stage 30, for1 expression was greatly reduced (86). These opposite roles of Wnt/β-catenin signaling in liver formation during development are also observed in zebrafish (41). A recent study in mouse liver and pancreas specification showed that BMP signaling represses Hnf6 expression at the 3 to 4 somite stage, whereas it induces the expression at the 5 to 6 somite stage, (148), emphasizing the importance of developmental timing in understanding roles of signaling pathways.
Wnt/β-catenin signaling is not only necessary but also sufficient for liver specification. Gain-of-function studies in zebrafish showed that overexpression of Wnt2bb (107) or Wnt8a (122) in entire tissues induced ectopic hepatoblast and hepatocyte formation in the posterior endoderm that normally gives rise to the intestine. Xenopus studies also showed that the activation of β-catenin signaling in the endoderm resulted in ectopic liver marker expression in the posterior endoderm. Intriguingly, overexpression of Bmp2b or the constitutive-active form of Fgf receptors did not induce ectopic hepatoblasts in zebrafish (122), suggesting that BMP and FGF signaling are necessary, but not sufficient, for liver specification.
Although zebrafish and Xenopus data reveal the essential role of Wnt/β-catenin signaling in liver specification, its evidence in mice has not been reported yet. In zebrafish and Xenopus, the timing of the anteroposterior endoderm patterning event is well separated from that of the liver specification event. However, in mice, it appears that the timings are very close to each other. Liver specification occurs at the 26 somite stage in zebrafish (101), whereas it occurs at the 7 to 8 somite stage in mice (161, 167). The short interval between endoderm patterning and liver specification in mice may make it difficult to manipulate Wnt/β-catenin signaling during the short period time. A mouse model, in which Wnt/β-catenin signaling is activated or inactivated in the foregut endoderm after anteroposterior endoderm patterning but prior to liver specification, is needed to define the role of Wnt/β-catenin signaling in liver specification. Foxa3-cre-driven β-catenin deletion is evident at E9.5 in hepatoblasts and did not affect the alteration of the hepatoblast compartment at this stage (141). While this may imply that Wnt/β-catenin signaling is dispensable for hepatic induction in mice, it may also suggest the technical challenge of a short time interval between endoderm patterning and liver specification in mice that mandates β-catenin deletion temporally to draw more meaningful conclusions. In addition, the activation of β-catenin signaling in the posterior endoderm after the endoderm pattering will reveal whether Wnt/β-catenin signaling can induce ectopic hepatoblasts in mice as in zebrafish and Xenopus.
Hepatic Morphogenesis
As the hepatic induction or specification concludes several dynamic cellular and molecular mechanisms are set into place for an orderly progress of what is termed as the phase of hepatic morphogenesis. This phase is characterized by the growth and development of hepatic bud, which is cellularly quite heterogeneous in mammals. The major cell type is the hepatoblast or the bipotential stem cell of the liver, which migrates, expands and then differentiates into either an immature hepatocyte or a cholangiocyte to the further undergo maturation to a functional cell type of either lineage (Fig. 3). In addition to these cells though, the growing liver bud contains hematopoietic elements, endothelial cells and developing stellate cells as other major cell types that actively participate in hepatic morphogenesis through cell-cell interactions and signaling. We will first discuss the development of epithelial cells followed by a discussion on the development of nonepithelial compartment and how the nonparenchymal cells may be modulating the process of hepatic morphogenesis.
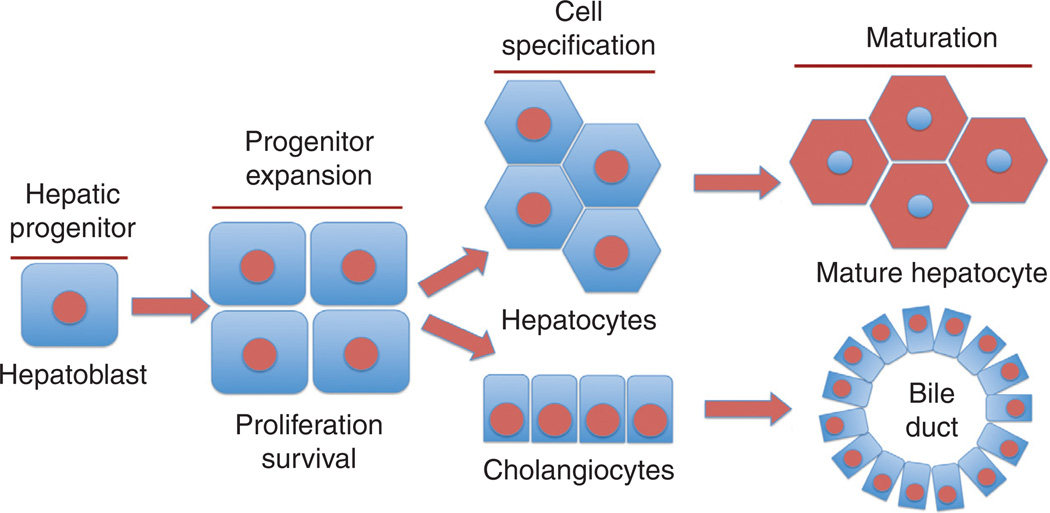
Figure 3.
Representation of hepatic morphogenesis. Hepatoblasts, which are the hepatic progenitors, undergo expansion via balanced cell proliferation and survival. These bipotential stem cells then differentiate into hepatocytes or cholangiocytes (biliary epithelial cells). As the morphogenesis continues, these cells undergo maturation by acquiring additional characteristics such as polarity and now are primed to perform key cellular functions of the liver.
Epithelial compartment
Hepatoblasts: Characteristics and migration
The hepatoblasts are the bipotential stem cells contained with the hepatic thickening. These cells express hepatic genes such as α-fetoprotein, transthyretin, and hepatocyte nuclear factor-4α, followed by Alb. At this stage (~E9.0) in mouse liver development, the hepatic diverticulum is evident which abuts septum transversum (47, 68). The basal lamina of endoderm physically demarcates the hepatic endoderm from the septum transversum. The migration of these cells into the septum transversum requires the secretion of matrix metalloproteinases, which are known to degrade the extracellular matrix and facilitating the migration (82). Here, the endothelial cells present in the area, which are known to secrete factors that are not fully characterized but include Neurturin, which may have chemotactic properties for the hepatoblasts (145). The role of endothelial cells here is of clear significance since Flk-1 deletion leads to absence of endothelial cells, which in turn impairs hepatoblast migration (85).
At the same time, several key transcriptional factors are expressed and their importance in this phase of liver development is clear through loss of function experiments. Gata4 and Gata6 are zinc transcription factors with proven roles in extraembryonic tissue, also have role in hepatoblast migration and in maintenance of expression of hepatic transcription factors (150, 168). In fact through in vitro and in vivo approaches, these factors have been shown to be critical in hepatic morphogenesis in zebrafish, xenopus and mice. At least one mechanism suggested by which these factors regulate hepatoblast development is via directly regulating expression of Hhex by binding to its promoter. Hhex is another extremely important homeobox transcription factors, which is required for multiple events in liver development (12, 46, 123). In fact Hhex null embryos lack liver and gall bladder and more detailed characterization has revealed its role in hepatic pseudostratification, which is the first step before the hepatic endoderm delaminates and hepatoblasts begin to migrate into septum transversum. In addition, Hhex ablation also leads to failure of migration of hepatoblasts. Interestingly, even when embryos go past that stage owing to conditional deletion in early hepatoblasts, Hhex loss impairs the differentiation of these bipotential progenitors to the hepatocytes (49).
Prox-1 is another homeobox domain transcription factor expressed in the hepatoblasts at this time (16). The deletion of this factor impedes the migration as well as expansion of hepatoblasts (128). It has been speculated that Prox1 may be responsible for regulating the expression of E-cadherin in developing hepatoblasts within the hepatic diverticulum. In fact, downregulation of E-cadherin is a mechanism by which hepatoblasts are able to begin their migration into septum transversum stroma after disruption of basal lamina. Another complementary set of transcription factors includes the Onecut-1 (HNF6) and Onecut-2. Loss-of-function studies for these factors demonstrate importance in the migration of hepatoblasts similar to GATA and Hhex factors albeit at a later stage. This appears to be via degradation of basal lamina (81). However, Onecut-1 and Onecut-2 loss appears to be functionally compensated by other factors as the hepatoblast migration eventually does occur, but the phenotype is know consistent with failure of the progenitors to expand. Thus, the liver size is dramatically smaller and displays additional defects.
Hepatoblast expansion and growth of primitive liver bud
The next several days mark the growth and expansion of the hepatic bud through mostly paracrine mechanisms where by mesenchymal cells secrete multiple growth factors and cytokines whose receptors and downstream effectors are richly expressed in hepatoblasts. Concurrently, these cells are receiving spatiotemporal signals that dictate their differentiation into hepatocytes or cholangiocytes.
Many signaling cascades are implicated in hepatoblast expansion through in vitro and in vivo studies. The signals emanate from mesenchyme, which consists of endothelial cells, stellate cells, and hematopoietic elements within the developing livers (Fig. 4). Some of the key signaling pathways include BMP, FGF, hepatocyte growth factor (HGF), Wnt, and RA signaling. While these signaling mechanisms can be broadly classified as receptor tyrosine kinases and others such as transforming growth factor β (TGFβ) and Wnt signaling, there is significant cross-talk that has been reported within various signaling cascades making it precarious to draw definitive linear conclusions. However, it will be sufficient to say that PI3 kinase, β-catenin, smad, MAPK, and JNK signaling are prudent for various aspects of hepatoblast development during the phase of hepatic morphogenesis.
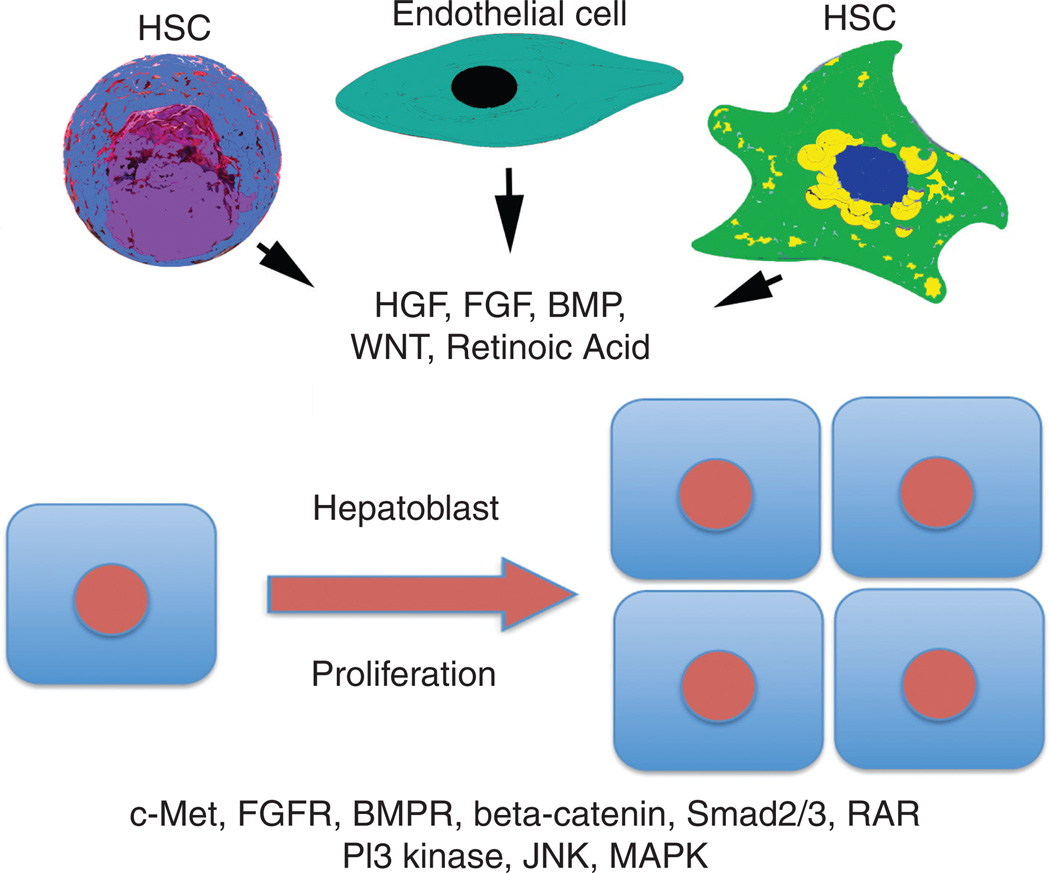
Figure 4.
Cellular and molecular basis of hepatoblast expansion. Various mesenchymal elements including endothelial cells, hepatic stellate cells (HSCs) and even hematopoietic stem cells (HSCs) are a source of key growth factors that act via paracrine mechanisms to induce proliferation of hepatoblasts that expresses receptors as well as key downstream effectors of these signaling mechanisms. Abbreviations: HGF-hepatocyte growth factor; FGF(R)-fibroblast growth factor (receptor); BMP(R)-bone morphogenic protein (receptor); RAR-retinoic acid receptor; JNK-JUN NH2-terminal kinase; PI3K–phosphoinositide 3-kinase; MAPK-mitogen activated protein kinase.
FGF signaling, which is relevant at earlier stages as discussed in the preceding section is also of essence in hepatic morphogenesis via not only being upstream of PI3K/AKT but also through effect on β-catenin signaling (10, 18, 53, 117). Likewise, BMP signaling mediates its effect not only through Smad1/5/8 and Smad4-dependent target gene expression during hepatic morphogenesis, but also by regulating other effectors including β-catenin (2, 120, 172). Wnt/β-catenin signaling is also a major mediator of hepatoblast expansion. Cytoplasmic and nuclear localization of β-catenin was observed at times of high hepatoblast proliferation (88, 139). The first study to demonstrate its direct role in hepatoblast proliferation was using antisense oligonucleotides against β-catenin gene in embryonic liver cultures (96). The decreased cell proliferation was evident due to impaired cyclin-D1 expression, which was also validated in vivo through hepatoblast-specific ablation of β-catenin gene expression (141). This led to severely hypoplastic livers that also showed additional abnormalities in differentiation and maturation as discussed in forthcoming sections. Studies in zebrafish have also demonstrated a positive role of β-catenin in hepatic morphogenesis, where lack of APC led to enhanced hepatoblast proliferation and larger livers during liver development (41). Recent studies have shown that direct upstream effectors of β-catenin including Wnt9 and Wnt2 may be produced by mesoderm, endothelial, and stellate cells to induce hepatoblast expansion during hepatic development (60, 84, 101).
The role of TGFβ is through activation of smad2/3 and smad4 heterotrimeric complex-dependent gene regulation, which may activate MAP kinase signaling among others. Because of redundancy of TGFβ/Smads, and due to plethora of events beings regulated by this signaling mechanism, it is cumbersome to address specific impact on hepatic morphogenesis. The best-known effect of TGFβ on hepatic morphogenesis is evident in embryos that are Smad2+/−; Smad 3+/− double heterozygotes, which display hypoplastic liver due to defective β1-integrin and defective β-catenin signaling (151). Loss of embryonic liver fodrin, a β-spectrin, and a scaffolding protein that interacts with Smad3 and Smad4 leads to defective hepatic morphogenesis, which again highlights the role of TGFβ (144).
It is also relevant to emphasize that different laboratories have utilized different strategies to successfully isolate hepatoblasts during different stages of liver development. HBC-3 cells were isolated from E9.5 stage of murine development and stains positively for α-FP, Alb, and cytokeratin 14 (CK14), and while these cells retain an undifferentiated hepatoblast phenotype on feeders, their culture in dimethyl sulfoxide induces hepatocyte and in matrigel induces a biliary differentiation (111). Similarly, another group showed that cells from E13.5 livers that are CD45− TER119− c-Kit− CD29− CD49f+ and CD45− TER119− c-Kit− c-Met+ CD49f+/low and from E11.5 that are CD45− TER119− c-Kit− c-Met+ CD49f+/low had high capacity to form hepatic colony-forming units (140). Another report showed at E11 that CD45−, TER119− c-Kitlow fraction contained hepatoblasts as well (89). Other two important surface molecules used for identification and sorting hepatoblasts include Delta-like protein 1 (Dlk1) and epithelial cell adhesion molecule (EpCAM) [reviewed in reference (143)]. In fact cells positive for these two markers from E11.5 livers showed robust liver stem cells activity. It should be noted that EpCAM expression diminishes over developmental stages and eventually in adults only bile ducts express this marker, whereas Dlk1 expression is temporal only in hepatoblasts and is lost upon differentiation to hepatocytes or cholangiocytes.
Hepatoblast survival during hepatic morphogenesis
As the hepatoblast expansion occurs, not only do they proliferate but also resist cell death. A recurring theme to this end has been a major cytoprotective role of NF-kappa B (κ) signaling during hepatic morphogenesis. There are several models where owing to genetic deletion of components of this signaling pathway has exhibited as massive hepatic apoptosis. Disruption of p65, a subunit of κ led to lethality at E15-E16 (7). Similarly, loss of T2K, a kinase that associates with tumor necrosis factor receptor associated factor-2 leads to embryonic lethality associated with hepatic degeneration at around E14.5 (11). Glycogen synthase kinase-3β (GSK3β), a kinase known to phosphorylate β-catenin to induce its recognition by the ubiquitin proteosome has an independent function in NF-κB signaling, where it has been shown to be required for NF-κB activity (45). Genetic disruption of GSK3β led to mid gestational lethality due to excessive hepatocyte apoptosis that in vitro showed hypersensitivity to TNFα-mediated cell death and reduced NF-κB activation.
Necrotic death due to oxidative stress or other mechanisms has also been observed due to loss of certain genes during hepatic morphogenesis. Nrf1, a transcription factor critical for regulating redox balance, when knocked out led to fetal lethality associated with enhanced fetal hepatocyte apoptosis and high oxidative stress (20). Similarly, metal-responsive transcriptional activator MTF-1, which regulates the basal and inducible expression of metallothioneins, when genetically ablated leads to embryonic lethality at E14 due to massive hepatic degeneration mostly due to necrosis (44). The expression of metallothionein I and II genes was absent, which in turn led to decreased glutathione biosynthesis. In fact, the mouse embryo fibroblasts from the mutant embryos were more sensitive to cadmium or hydrogen peroxide. C-Jun, an AP-1 transcription factor, also appears critical in hepatoblast survival as well as proliferation (32).
While several of these observations are similar, several of these pathways are distinct and the mode of death is disparate as well. For example, while NF-κB disruption led to mostly apoptosis of hepatoblasts, loss of MTF-1 led to predominantly necrosis. While more studies are essential to clarify systematically the requirement of survival pathways during hepatoblast development, it is quite clear that there is a need of multitude of survival signals in the developing hepatoblasts in counteracting the cellular stresses that appear innate to the stage of hepatic morphogenesis.
Hepatoblast differentiation to hepatocyte or cholangiocyte
Hepatoblasts are the bipotential progenitors that are destined to become either a hepatocyte or a cholangiocyte to form intrahepatic bile ducts (Fig. 3). As the hepatoblasts expand in numbers, they simultaneously, after a certain stage, begin to undergo lineage specification. This stage corresponds to around E13.5 in mouse liver development. A hepatoblast or hepatic progenitor is a cell with high nuclear to cytoplasmic ratio and is interspersed between hematopoietic cells and other nonparenchymal cells in the developing livers. These cells express unique markers (Dlk1), certain markers of hepatocytes (Alb, HNF4α) and of immature hepatocytes (α-FP). As the liver development continues, these cells make a commitment to being either a hepatocyte or a cholangiocyte (CK19) and the processes that determine their fate are being increasingly understood. The hepatoblasts that acquire the hepatocyte cell fate do so through gradual acquisition of liver-enriched transcription factors that in turn regulate the expression of genes that make it a polarized, cuboidal, and functioning cell [reviewed in reference (126)]. There is interplay between various growth factors and cytokines concurrent with cell-cell interactions that enable upregulation of certain transcription factors at the expense of others within a subset of hepatoblasts, which will guide them along a specific-cell type differentiation. Once the decision is made, similar factors or additional signaling cascades then promote the maturation, organization, and functioning of these cell types.
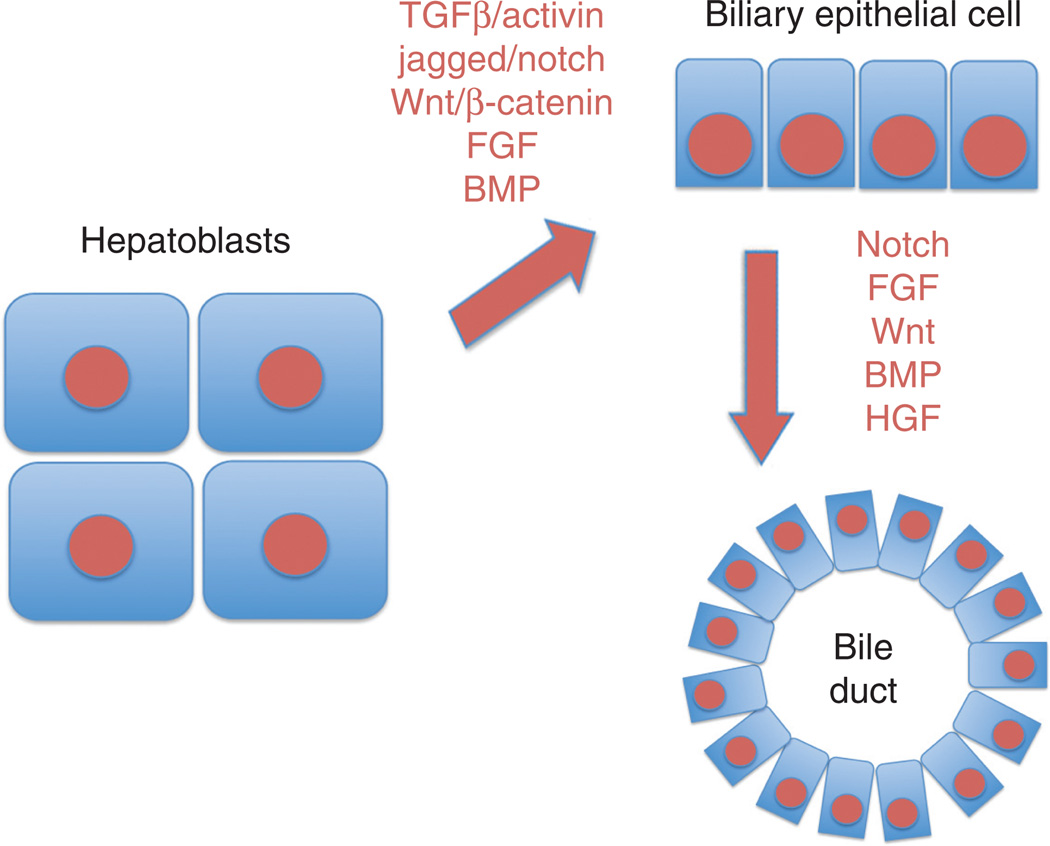
Several signaling molecules are implicated in lineage determination of a hepatoblast to cholangiocyte (Fig. 5). A key cellular interaction heralding this process is between the developing portal vein and hepatoblasts surrounding this structure at around E13 stage during murine hepatic development. In fact there exists a gradient of activin/TGFβ with the highest exposure limited to the cells around portal mesenchyme (25). This allows for differentiation of hepatoblasts to cholangiocytes. Similarly, Jagged1 expression seems to be apparent in the periportal mesenchyme and biliary cells, while Notch2 is present in biliary cells (39, 170). The function of Notch signaling in biliary differentiation is also substantiated by phenotype observed upon hepatic inactivation of RBP-Jκ, a transcriptional effector of Notch signaling, which results in diminished biliary differentiation of hepatoblasts (170). It should be noted that even before the discovery of temporal expression and regulation of Notch signaling during intrahepatic bile duct development, Human Alagille syndrome, which is associated with paucity of bile ducts, was identified to be due to mutations in Jagged-1 or Notch-2 (NOTCH2) and hence due to deficient Notch signaling (72, 102, 109, 171). Some of the key transcription factors that allow for biliary differentiation include Sox9, whereas expression of Onecut transcription factors HNF6 and OC-2 counteract the activin/TGFβ signaling away from the portal mesenchyme where hepatoblasts differentiate into hepatocytes and acquire distinct hepatocyte-enriched transcription factors such as HNF4α (25). It is relevant to note that in response to mesenchyme signals, the hepatoblasts located adjacent to portal vein not only upregulate biliary factors such as Sox9, HNF6, OC2, and HNF1β but at the same time downregulate hepatocyte-enriched transcription factors such as HNF4α. In addition, there is a rigorous role of HNF6 and OC2 in modulating hepatoblast differentiation to hepatocytes and cholangiocytes and any change in their expression could jeopardize the timing and extent of hepatobiliary differentiation. Here, an important role of C/EBPα in repressing the gene expression of both HNF6 and HNF1β has also been shown in hepatocytes in the liver parenchyma, whereas suppression of its expression in periportal hepatoblasts allows for a higher expression of both HNF6 and HNF1β (125, 155). The role of C/EBPα in negatively regulating cholangiocyte fate of hepatoblasts was further ascertained by observation that when C/EBPα-null liver samples were transplanted into scid mice testis, mostly biliary structures were observed (155). Similarly, genetic interactions between HNF6 and Notch signaling have been identified in playing a critical role in development of intrahepatic bile ducts (147). Additional factors have shown to assist in biliary differentiation include FGF and BMP signaling pathways although exact mechanism remains to be elucidated (156).
Figure 5.
Molecular basis of cholangiocyte differentiation and maturation. Various instructive signals, either paracrine or autocrine, stimulate specification of cholangiocytes from the hepatoblasts and these signals induce specific transcription factor programs within the cholangiocytes. Once these cells are specified, they continue to respond to various growth factors, again in an autocrine or paracrine fashion, to mature and organize as ductal structures with specific functions. Abbreviations: TGFβ-transforming growth factor β; HGF-hepatocyte growth factor; FGF-fibroblast growth factor; BMP-bone morphogenic protein.
Wnt signaling has also been shown to play a dramatic role in assisting hepatoblasts differentiate to cholangiocyte (66, 97). The first evidence came from a study where ex vivo knockdown of β-catenin in embryonic liver cultures that consisted of hepatoblasts led to failure of CK-19-positive cells to form duct like structures (96). When the same organoid cultures were maintained in Wnt3a-conditioned media, it led to survival of only biliary epithelia, whereas HGF and Wnt3a was sufficient to sustain both hepatocytes and cholangiocytes (50). These studies were also confirmed in vivo when Foxa3-cre mediated deletion of β-catenin led to failure of proper bile duct differentiation of hepatoblasts and there was a lack of formation of primitive bile ducts (141). When β-catenin was prematurely stabilized due to loss of APC in hepatoblasts, these β-catenin overexpressing cells formed immature cholangiocytes, which matured when transplanted in vivo (29). What are the downstream mechanisms that may be playing a role in lineage specification of hepatoblasts to cholangiocytes still remains to be elucidated (66). It is intriguing to note that very recently two forms of β-catenin were identified during late hepatic development (65). While the predominant truncated form of β-catenin that lacks aminoterminal was observed in developing hepatocytes, full-length β-catenin was exclusively localized to developing bile ducts at E17. It would be critical to identify how this form of β-catenin may be playing an important role in bile duct morphogenesis.
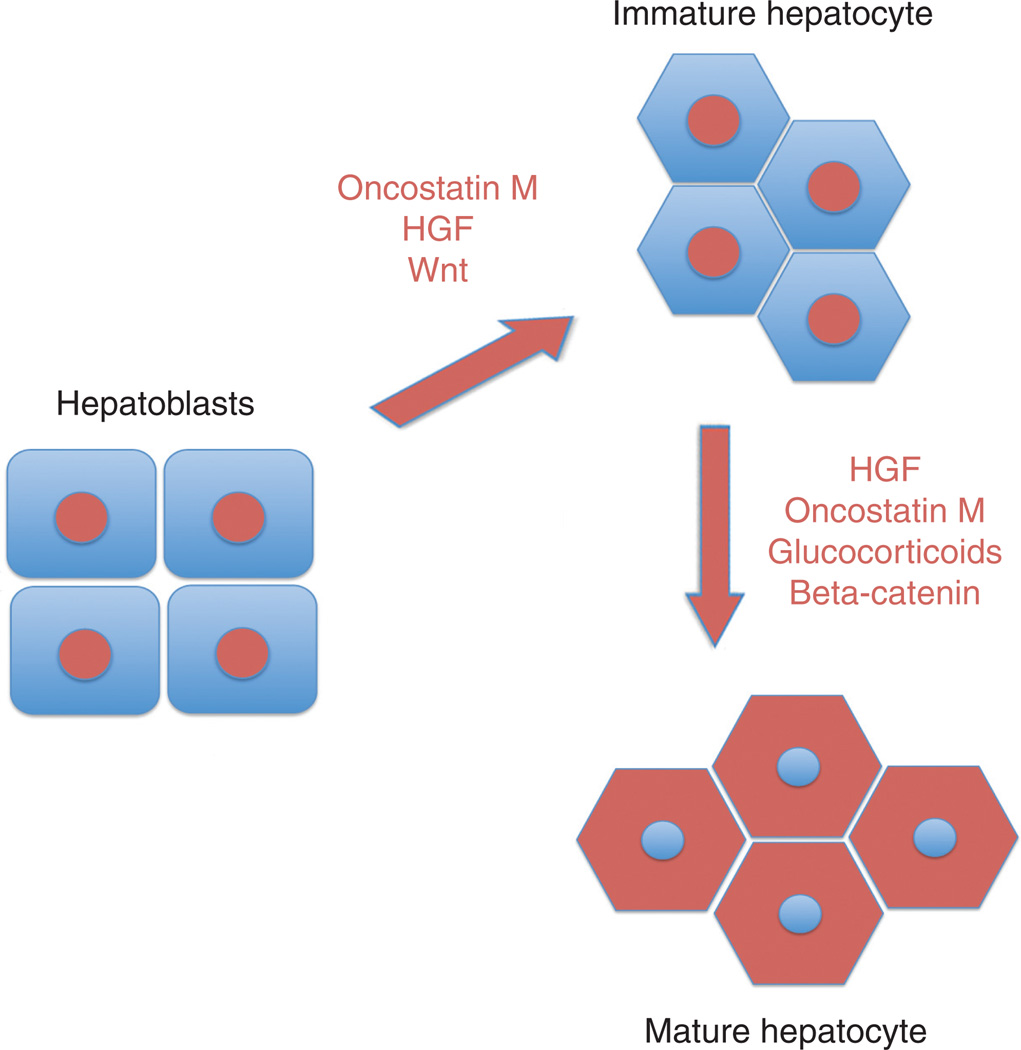
The major transcription factors that are expressed in hepatocytes as they emerge from hepatoblasts include FoxA1/2, HNF1α and β, HNF4α, and HNF6 [reviewed in reference (126)]. Other than being spatiotemporally distant from TGFβ gradient, additional factors are known to play a role in hepatoblast differentiation to hepatocytes although the exact mechanisms are not completely clear (25). However, several factors have been shown to play an important role in hepatocyte differentiation of the hepatoblast (Fig. 6). Oncostatin M is released from developing hematopoietic compartment within the liver and is an important mediator of hepatocyte differentiation (55, 56, 91). Additionally, HGF promotes hepatic morphogenesis having effects on hepatoblast expansion along with hepatocyte maturation (87, 116). Other factors that cooperate with Oncostatin M and HGF to induce hepatocyte differentiation include the glucocorticoid hormones and Wnt signaling. The Wnt signaling through β-catenin was shown to promote hepatocyte maturation. When embryonic livers were cultured in the presence of antisense to β-catenin, there was defect in maturation of hepatocytes, which was observed as sustained expression of progenitor cell markers in hepatocytes (96). Foxa3-cre-mediated β-catenin gene deletion in developing hepatoblasts also led to retarded maturation of hepatocytes which was reflected in maintained high nuclear to cytoplasmic ratio of cells, failure of hepatocytes to accumulate glycogen and these findings were associated with decrease in HNF4α and C/EBPα (141). More recently, an aminoterminal truncated form of β-catenin has been identified in maturing hepatocytes as a result of calpain-induced cleavage (65). This form of β-catenin has distinct target genes and additional studies will be necessary to elucidate the role and regulation in hepatocyte maturation.
Figure 6.
Molecular basis of hepatocyte differentiation and maturation. Various instructive signals, either paracrine or autocrine, stimulate specification of hepatocytes from the hepatoblasts and these signals induce specific transcription factor programs within the hepatocytes. Once these cells are specified, they continue to respond to various growth factors, again in an autocrine or paracrine fashion, to mature, acquire cell polarity and gain expression of genes to enable specific functions.
The downstream effectors of these various signaling cascades may directly induce target gene expression responsible for hepatocyte functions but also indirectly through induction of hepatocyte-enriched transcription factor expression. In fact a factor such as HNF4α has been shown to bind to promoters of nearly half of the genes associated with hepatocyte polarity, junctional integrity, and metabolic and xenobiotic functions (105). This characteristic of HNF4α was exploited recently when mouse fibroblasts were converted to hepatocytes by introduction of two transcription factors, one of which was HNF4α and the other one was FoxA1 or A2 or A3 (119). The generated induced-hepatocyte-like cells possessed in vivo and in vitro characteristics of near-hepatocyte-like cells.
Hepatocyte maturation
Hepatocytes that emerge from hepatoblasts look distinct from hepatocytes at later stages of hepatic development. In fact, when hepatocytes are compared for example at E15 and E17 stages, the cuboidal morphology and presence of clear cytoplasm due to glycogen is quite apparent at the latter stage (94). This indicates that once hepatocytes originate from hepatoblasts, they continue to undergo maturation under the control of factors such as Oncostatin M, glucorticoids, HGF, Wnt, and additional factors and through expression of transcription factors such as HNF4α and C/EBPα (35, 105, 149) (Fig. 6). Hepatocytes continue to change their characteristics and functional capabilities drastically both perinatally as well as postnatally under the control of several of these liver-enriched transcription factors. C/EBPα knockout mice are lethal at neonatal stage since they lack key enzymes that regulate glucose and ammonia homeostasis (59). In fact, these animals die due to defects in gluconeogenesis and ammonia detoxification since several enzymes in ornithine cycle are regulated by C/EBPα, which in turn dictate hepatocyte maturation and function.
The hepatocytes begin to acquire location-specific functions, which is termed as zonation (131). This term was initially coined by Jungermann who pointed out the distinct properties of cells within the liver based on their location in the hepatic lobule (37, 54). The hepatocytes located in close proximity to the hepatic inflow around the portal triad are referred to as being in periportal zone, while those around central vein are located in centrizonal or pericentral zone. The functions of hepatocytes in periportal, mid-, or pericentral zone are distinct and are a function of interplay between Wnt/β-catenin signaling pathway and HNF4α signaling (134). β-Catenin signaling is active in the pericentral area and many of the genes downstream of this signaling are highly expressed in the hepatocytes located in this zone (9). Expression of Glutamine Synthetase (GS), cytochrome p450 2e1, and Cyp1a2 are all under the control of β-catenin as shown in preclinical and clinical scenarios (40, 75, 118, 142). It was also shown that β-catenin activity in other zones was maintained at low levels because of increased expression of adenomatous polyposis coli gene product, which is responsible for β-catenin degradation. On the other hand, HNF4α was shown to play an important role as a transcriptional activator while acting as a repressor for pericentral genes such as GS in periportal cells (26). It was shown that both HNF4α and TCF family members interact with β-catenin and regulate expression of GS in pericentral hepatocytes, and when HNF4α was bound alone, GS expression was repressed (134). The mechanism of HNF4α action as a repressor needs additional validation. Simultaneously, it is still unclear how β-catenin activation is maintained in the pericentral hepatocytes and what are the upstream effectors and their source, which enable limited activation of this protein to drive expression of certain but not other Wnt targets.
Cholangiocyte proliferation, maturation, and ductal development
Once hepatoblasts differentiate into cholangiocytes, key transcriptional factors are turned on that modulate the process of cholangicoyte differentiation and maturation. Sox9 is the first transcription factor that controls the timing of the cholangiocyte differentiation and appears to be under the control of Notch and TGFβ signaling (4). Notch signaling also regulates the expression of Hes1, which is required later for biliary tubulogenesis (61). It remains to be identified that what are the specific transcriptional targets of biliary transcription factors that are further responsible for ductal morphogenesis although role of Notch and HNF6 in regulating ductal morphogenesis is pivotal (109, 147, 171). FGF, BMP, Wnt, and HGF signaling may be contributing to cholangiocyte differentiation and maturation through incompletely understood mechanisms (Fig. 5). Intriguingly, FoxA1 and FoxA2 loss in hepatoblasts resulted in enhanced differentiation and expansion of cholangiocytes that appeared to be secondary to increased IL-6, which is a known biliary mitogen (74). These observations demonstrate an important balance of biliary and hepatocyte transcription factors that exists during normal hepatobiliary differentiation of hepatoblasts.
While hepatoblasts adjacent to portal mesenchyme differentiate into cholangiocytes, a second phase of ductular morphogenesis was only recently defined. It was reported that adjacent to the single layer of primary cholangiocytes derived from hepatoblasts, a second layer of cholangiocytes appears later in hepatic development but only at specific locations. This layer is derived from undifferentiated hepatoblasts that exist towards the hepatocyte parenchyma in developing livers. There exists a luminal space between the two layers and as these appose to form asymmetric ducts with lumen, the hepatoblasts undergo rapid differentiation to cholangiocytes. It is suggested that Hes1 and thus Notch signaling may be playing an important role in this process of ductal morphogenesis (61).
Lastly, cholangiocyte homeostasis is important postnatally, since these cells may be periodically exposed to harmful microbes or bacterial products. Cholangiocytes have the capability to undergo reactive proliferation as a means to restore and repair the function of bile collection and flow after an injury. Factors such as IL-6 may be critical in modulating such process (100). In addition, since the function of mature cholangiocytes is to biliary flow, these cells have primary cilia, which serve multiple functions as sensors for osmolarsity, chemical composition of bile and mechanical forces. Since cilia are truly the feelers for sensing any changes in bile microenvironment and that cholangiocytes are both absorptive and secretory cells, the implications of cholangiocyte cilia can be of essence in regulating biliary homeostasis through integration to various signaling cascades, the most prominent one being through altered concentrations of intracellular Ca2+ and cyclic adenosine monophosphate (cAMP) [reviewed in reference (83)]. Suffice to say that mutations in ciliary associated proteins do lead to cholangiociliopathies include cystic and/or fibrotic liver diseases such as Autosomal Dominant Polycystic Kidney Disease (caused by mutations in either PKD1 or PKD2 genes), Autosomal Recessive PKD (caused by mutations in PKHD1 gene) and others. Defects in ciliary structure or their sensory and transducing functions induce cAMP to causing cholangiocyte hyperproliferation. In addition, aberrations are evident in cell-matrix interactions, fluid secretion, or absorption and eventually in cyst formation.
Non-epithelial compartment
Hepatic sinusoidal endothelial cells
Hepatic vascular development commences at the inception of the liver primordium. Endothelial cells that exist around the liver diverticulum have been shown to invade the hepatic bud during its growth (85, 98). These cells in may be originating from the omphalomesentric veins or common or posterior cardinal veins. During hepatic morphogenesis at E12.5, liver contains a rather well-developed vascular system including primitive sinusoidal structures (98). Sinusoids are a complex capillary network in the liver that is playing a role in transfer of various molecules to and from blood and hepsatocytes. The endothelial cells of sinusoids are distinct from traditional endothelia in function as well as markers and resemble lymphatic endothelial cells in expressing lymphatic vascular endothelial hyaluronan receptor and low expression of PECAM or CD31 (98). The primitive sinusoids have lumen and are contiguous with one other, and with portal and central veins. In fact, endothelial cells of primitive sinusoids were surrounded by an extracellular matrix and also by hepatic stellate cells. Endothelial cells of primitive sinusoids expressed high levels of Flk-1 (85). Since vascular endothelial growth factor (VEGF) is a known ligand for Flk-1 and is produced by developing hepatoblasts and hematopoietic cells, it is likely driving the proliferation of immature sinusoidal endothelial cells. At later stages, VEGF in addition stimulate branching morphogenesis in the primitive c as angiopoietin or pigment epithelium-derived factor may also be playing a role in the sinusoidal endothelial morphogenesis (114, 115). In addition, role of Wnt signaling in sinusoidal growth may be relevant. VEGF is a known target of β-catenin signaling. Also, Wnt2 was shown to be expressed in rat hepatic sinusoidal endothelial cells, which correlated with expression of VEGFR2 supporting cooperative roles of Wnt2 and VEGF in endothelial cell growth (60). In chick livers during development, Wnt9a is secreted by the hepatic sinusoidal endothelial cells, which induced β-catenin activation in hepatoblasts to stimulate their proliferation and glycogen accumulation (84). Such a paradigm was also recently uncovered during liver regeneration after partial hepatectomy, where sinusoidal endothelial cells were shown to release Wnt2 to then induce β-catenin activation, which is critical for hepatocyte proliferation and restoration of hepatic mass (31). Thus, the endothelial cells of hepatic sinusoids not only play a role as a conduit for transfer and exchange but also act as an instructive niche to modulate hepatoblast proliferation during development and hepctocyte proliferation during liver regeneration.
Hepatic stellate cells
Closely associated with hepatocytes on one surface and with sinusoidal endothelial cells on the other, stellate cells are an important cell type most known for its role in hepatic fibrosis (15, 36). These cells are the source of activated myofibroblasts that are responsible for collagen deposition. In addition, stellate cells are a source of growth factors and cytokines that may be released in response to appropriate cues in the form of changes in sinusoidal circulation, loss of hepatic mass, hypoxia, and others. Stellate cells are derived from mesothelial and submesothelial cells during hepatic development (5, 6, 76, 104). The mesothelium in turn originates from STM and proepicardium. Recent studies in mice suggests that activated leukocyte cell adhesion molecule-positive cells consisting of both mesothelial and submesothelial cells acquired myofibroblastic morphology in cell culture and also in the presence of retinol and upon embedding in collagen formed intracellular lipid droplets suggesting their ability to differentiated into stellate cells (5, 6). Potential role of Wnt/β-catenin and Pitx2 has been suggested in the differentiation and expansion of these cell types during development (21, 57, 165).
The stellate cells and mesothelial cells during hepatic development have also been shown to play an important role in hepatic morphogenesis. The source of Wnt9a was shown to be not only the endothelial cells but also the hepatic stellate cells during development (84). The mesothelial and submesothelial cells have been also shown to be rich sources of HGF, pleiotrophin, and midkine, which are all mitogens for hepatoblasts and hepatocytes (104). Knockout of Wilm’s tumor 1 homologue (WT1), which is a marker of mesothelial cells, yields smaller livers at E13.5 (52). These livers were hypoplastic due to a decrease in the numbers of hepatoblasts and hepatocytes. The mesothelial cells isolated from WT1 null livers expressed low levels of pleiotrophin and midkine (104). In vitro, hepatocytes from WT1 knockouts showed normal growth that was comparable to control genotypes when cocultured in the presence mesothelial cells from control embryos. Thus, hepatic stellate cells and their precursors play an important role in controlling hepatic morphogenesis.
Hematopoietic elements
Liver is a major site of hematopoiesis during development. As the hepatoblasts begin expanding in the primitive liver bud, the organ acquires the function of hematopoiesis. Recently hepatoblasts have been shown to produce factors such as erythropoietin and stem cell factor, which are responsible for inducing the numbers of erythroid progenitors (138). Other nonparenchymal cells such as fetal-liver-derived stromal cells also enhance hematopoietic progenitors and role of pathways such as Wnt may be of essence in this event. As the hepatoblasts mature to hepatocytes, these lose the capability to support hematopoiesis and this process migrates to the bone marrow. However, exact cellular and molecular basis of cessation of hematopoiesis during late hepatic development remains unknown. It does appear that terminal deoxynucleotidyl transferase dUTP nick end labeling-positive hematopoietic cells and basally enhanced oxidative stress is observed at the time when this process is being terminated in the liver and additional mechanisms will need to be elucidated (141).
The hematopoietic cells also play an important role in hepatic morphogenesis. These cells are a rich source of Oncostatin M, which is a single important factor known to enhance hepatocyte differentiation and maturation through gp130 (55, 56, 91). Gp130 knockout livers were composed of immature hepatocytes that lacked glycogen and showed reduced expression of tyrosine aminotransferase mRNA. Thus, the hematopoiesis and hepatopoiesis is truly a symbiotic relationship at least temporally in development.
Kupffer cells that are the resident macrophages in the liver have been suggested to support hematopoiesis in the developing liver. In adult liver, these cells have been shown to be a source of cytokines such as TNFα and IL-6, which are important in the process of liver regeneration (1, 157). However, it remains less clear if Kupffer cells could be supporting hepatic morphogenesis during liver development.
A pathway with many roles
A major observation that is evident in hepatic development is that some times a single signaling cascade can play seemingly multiple roles and sometimes even seemingly opposite roles. A classical example is the Wnt/β-catenin signaling pathway that has a plethora of roles in hepatic development and sometimes these roles are paradoxical temporally (66). Its role in foregut development requires repression of Wnt/β-catenin, but is required for hepatic specification (73, 86, 101, 124). In addition, β-catenin signaling contributes to hepatic morphogenesis in multiple ways. Its role in both hepatoblast proliferation and survival is visible in both in vitro and in vivo studies (41, 84, 96, 141). It has also been shown to play a role in cholangiocyte differentiation of hepatoblasts in both ex vivo and in vivo studies as well as in both loss of function as well as gain of function of studies (29, 50, 96, 141). Intriguingly, β-catenin is also an important component of the hepatocyte maturation process both in vitro and in vivo and the hepatoblasts that lack β-catenin fail to develop cuboidal morphology and express low levels of certain hepatocyte-enriched transcription factors such as HNF4α and C/EBPα (95, 96, 141). In addition, the role of β-catenin in hepatic zonation is also critical (9). How can β-catenin play so many different roles in various phases of hepatic development? It should be noted that although β-catenin can translocate to the nucleus and regulate target gene expression in response to Wnt, HGF, FGF, and additional factors, but in the nucleus it cannot bind DNA directly. Instead, it binds to factors such as TCF/LEF family of transcription factors and governs their transactivation (80). However, β-catenin has now been shown to also interact with factors such as HIF1α, FOXO3, and others and thus may have a wider role in transcriptional regulation of gene expression (33, 38, 70). Another mode of disparate target gene regulation by β-catenin comes from virtue of its interactions with histone acetyltransferases CBP and P300 and has been shown in embryonic stem (ES) cell renewal and differentiation (79, 90). Very recently, and in the context of liver development, β-catenin was shown to undergo calpain-mediated amino terminal cleavage (65). The observed species during mid-to-late gestational development in the liver localizes to both the membrane and in the nuclei of the hepatocytes. RNA-seq analysis identified several novel targets that include genes encoding for histone H3 and H2, which are responsible for nucleosome structure and epigenetic gene regulation. Thus β-catenin signaling may play diverse roles in liver development. Other signaling pathways may also show such divergent complexity and it is the concerted and highly regulated cellular and molecular interactions that enable normal hepatic development.
Differentiation of Stem Cells to Hepatocytes
Liver transplantation is the only effective treatment for severe liver diseases, but the shortage of donor livers makes this therapy extremely limited. Cell-based transplantation of hepatocytes is considered as an excellent alternative to liver transplantation. Hepatocytes can be differentiated from ES or induced pluripotent stem (iPS) cells or directly reprogrammed from adult fibroblasts in vitro, providing a potential source of autologous cells for transplantation. Understanding of the developmental processes of liver formation has significantly contributed to the development of such differentiation and reprogramming protocols (Fig. 7). Currently mature hepatocytes are being differentiated from mesenchymal stem cells, ES cells, and iPS cells among other types of stem cells (127, 129, 136). However, the current protocols need to be further improved in order to make mature, fully functional hepatocytes appropriate for cell-transplantation therapy. A better understanding of liver developmental processes will allow one to improve the current protocols for the transplantation therapy.
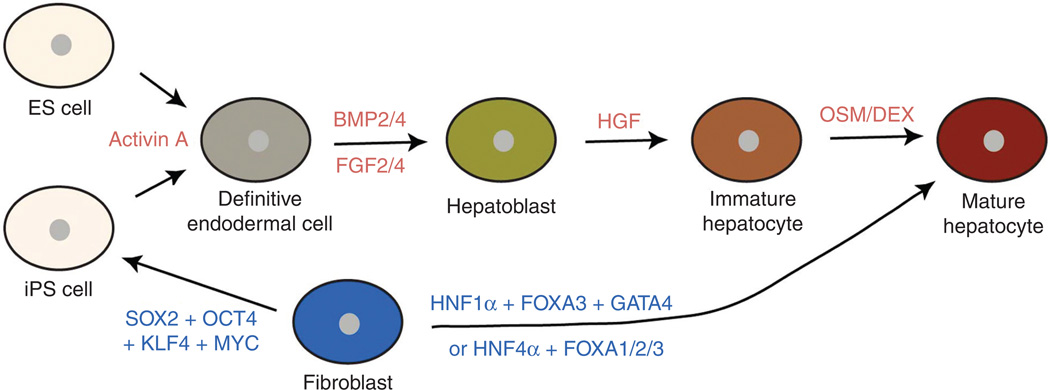
Figure 7.
Diagrams depicting hepatocyte differentiation from embryonic stem (ES) or induced pluripotent stem (iPS) cells and the direct reprogramming of fibroblasts. Exogenous factors that induce differentiation are written in red; transcription factors that are required for fibroblast reprogramming are written in blue.
ES or iPS cells can be differentiated into hepatocytes
Hepatocytes can be differentiated from ES cells in vitro by mimicking the developmental processes of liver formation. Since the liver is derived from the definitive endoderm and Nodal, a member of the TGFβ superfamily, signaling is essential for endoderm specification, ES cells were first differentiated into definitive endodermal cells with the treatment of Activin A, which binds the same receptor as Nodal and elicits similar intracellular signaling events (28). During development, the ventral foregut endoderm receives BMP and FGF from its surrounding tissues and gives rise to hepatoblasts. Therefore, Activin A treatment was followed by BMP2/4 and FGF2/4 treatment (17, 42). To differentiate hepatoblasts into immature hepatocytes and for further maturation, HGF, oncostatin M, dexamethasone, and/or EGF were added into culture media [reviewed in reference (8)]. Although there are some differences in treatment conditions among hepatocyte differentiation protocols, stepwise differentiation (ES cells → the definitive endoderm → hepatoblasts → hepatocytes) adopted from liver developmental processes was applied to these protocols (Fig. 7).
The efficiency of hepatocyte differentiation from ES cells is quite variable probably due to the use of serum and other undefined culture medium components. By eliminating serum and using well-defined culture components, Dr. Duncan’s group has recently established a hepatocyte differentiation protocol that can elicit the efficient and reproducible generation of hepatocytes from human ES and iPS cells (127). This protocol allowed one to obtain more than 80% definitive endodermal cells, subsequently more than 80% hepatoblasts, and finally about 80% Alb-expressing hepatocytes, indicating 80% efficiency of hepatocyte differentiation from human ES and iPS cells.
Hepatocytes differentiated from a patient’s iPS cells have a great potential for understanding disease mechanisms and for autologous hepatocyte transplantation. iPS cells derived from patients with inherited metabolic disorders of the liver were differentiated into hepatocytes, and these differentiated hepatocytes recapitulated key pathological features of the diseases (108), indicating the potential of iPS cells to model liver diseases. The differentiation protocol again was based on normal liver developmental programs elucidated over last couple of decades (Fig. 7). Furthermore, hepatocytes differentiated from human iPS cells were transplanted into immunodeficient Alb-urokinase transgenic mice (110) whose hepatocytes are functionally compromised, resulting in the distribution of the iPS-derived hepatocytes throughout the liver and their integration into the host parenchyma (158). In this study, a mutation in the α1-antitrypsin gene that is responsible for α1-antitrypsin deficiency was corrected in patient-derived iPS cells by genome modification, and the corrected iPS cell-derived hepatocytes were transplanted (158), providing the potential of human iPS cells for autologous cell-based therapies.
Fibroblasts can be reprogrammed into hepatocyte-like cells
Two groups have recently reported that the overexpression of a few transcription factors that play a crucial role in liver development sufficiently reprograms mouse fibroblasts into induced hepatocyte-like (iHep) cells. One group identified that three specific combinations of two transcription factors, HNF4α plus FOXA1, FOXA2, or FOXA3, from 12 candidate transcription factors are sufficient for iHep cell generation (119); the other group identified three transcription factors, GATA4, HNF1α, and FOXA3, from 14 transcription factors in the p19Arf mutant background (48). It is not too surprising that these transcription factors are identified because of their roles in liver development. In case of the reprogramming of fibroblasts into cardiomyocytes, several transcription factors, GATA4, MEF2C, and TBX5, which play crucial roles in heart development, also elicit such reprogramming (51). Since FOXA and GATA transcription factors are implicated in hepatic competence (13, 23, 24), these pioneer factors may open compact chromatin regions of liver-specific genes in fibroblasts during reprogramming. Since HNF4α (71, 130) and HNF1α (106) are required for hepatocyte differentiation, and regulate each other (64, 103), they may induce and stabilize the expression of hepatocyte-specific genes during reprogramming.
iHep cells have many of the morphological and functional characteristics associated with hepatocytes. Moreover, unlike primary hepatocytes, they can be maintained in culture due to their proliferation, allowing for the sufficient supply of hepatocytes for cell-based therapies. The transplantation of iHep cells into fumarylacetoacetate hydrolase (Fah)-deficient mice, a model of human liver disease tyrosinemia, increased the survival rate of Fah mice by restoring liver functions, although less efficient than that of primary hepatocytes did (48, 119). iHep cells together with hepatocytes differentiated from iPS cells have a great potential for liver research and cell-based therapies.
References
- 1.Abshagen K, Eipel C, Kalff JC, Menger MD, Vollmar B. Loss of NF-kappaB activation in Kupffer cell-depleted mice impairs liver regeneration after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1570–G1577. doi: 10.1152/ajpgi.00399.2006. [DOI] [PubMed] [Google Scholar]
- 2.Ader T, Norel R, Levoci L, Rogler LE. Transcriptional profiling implicates TGFbeta/BMP and Notch signaling pathways in ductular differentiation of fetal murine hepatoblasts. Mech Dev. 2006;123:177–194. doi: 10.1016/j.mod.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Ang SL, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse - involvement of Hnf3/ forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, Stanger BZ, Jacquemin P, Pierreux CE, Clotman F, Lemaigre FP. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–2333. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asahina K, Tsai SY, Li P, Ishii M, Maxson RE, Jr, Sucov HM, Tsukamoto H. Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development. Hepatology. 2009;49:998–1011. doi: 10.1002/hep.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asahina K, Zhou B, Pu WT, Tsukamoto H. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology. 2011;53:983–995. doi: 10.1002/hep.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 8.Behbahan IS, Duan YY, Lam A, Khoobyari S, Ma XC, Ahuja TP, Zern MA. New approaches in the differentiation of human embryonic stem cells and induced pluripotent stem cells toward hepatocytes. Stem Cell Rev. 2011;7:748–759. doi: 10.1007/s12015-010-9216-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benhamouche S, Decaens T, Godard C, Chambrey R, Rickman DS, Moinard C, Vasseur-Cognet M, Kuo CJ, Kahn A, Perret C, Colnot S. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev Cell. 2006;10:759–770. doi: 10.1016/j.devcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Berg T, Rountree CB, Lee L, Estrada J, Sala FG, Choe A, Veltmaat JM, De Langhe S, Lee R, Tsukamoto H, Crooks GM, Bellusci S, Wang KS. Fibroblast growth factor 10 is critical for liver growth during embryogenesis and controls hepatoblast survival via beta-catenin activation. Hepatology. 2007;46:1187–1197. doi: 10.1002/hep.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnard M, Mirtsos C, Suzuki S, Graham K, Huang J, Ng M, Itie A, Wakeham A, Shahinian A, Henzel WJ, Elia AJ, Shillinglaw W, Mak TW, Cao Z, Yeh WC. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 2000;19:4976–4985. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bort R, Signore M, Tremblay K, Martinez Barbera JP, Zaret KS. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev Biol. 2006;290:44–56. doi: 10.1016/j.ydbio.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- 14.Bossard P, Zaret KS. Repressive and restrictive mesodermal interactions with gut endoderm: possible relation to Meckel’s Diverticulum. Development. 2000;127:4915–4923. doi: 10.1242/dev.127.22.4915. [DOI] [PubMed] [Google Scholar]
- 15.Brenner DA. Molecular pathogenesis of liver fibrosis. Trans Am Clin Climatol Assoc. 2009;120:361–368. [PMC free article] [PubMed] [Google Scholar]
- 16.Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev. 2002;118:147–155. doi: 10.1016/s0925-4773(02)00240-x. [DOI] [PubMed] [Google Scholar]
- 17.Cai J, Zhao Y, Liu YX, Ye F, Song ZH, Qin H, Meng S, Chen YZ, Zhou RD, Song XJ, Guo YS, Ding MX, Deng H. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 18.Calmont A, Wandzioch E, Tremblay KD, Minowada G, Kaestner KH, Martin GR, Zaret KS. An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell. 2006;11:339–348. doi: 10.1016/j.devcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao WL, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Kwong M, Lu R, Ginzinger D, Lee C, Leung L, Chan JY. Nrf1 is critical for redox balance and survival of liver cells during development. Mol Cell Biol. 2003;23:4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng JH, She H, Han YP, Wang J, Xiong S, Asahina K, Tsukamoto H. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294:G39–G49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 22.Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 24.Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. Embo J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colletti M, Cicchini C, Conigliaro A, Santangelo L, Alonzi T, Pasquini E, Tripodi M, Amicone L. Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology. 2009;137:660–672. doi: 10.1053/j.gastro.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 27.Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 28.D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 29.Decaens T, Godard C, de Reynies A, Rickman DS, Tronche F, Couty JP, Perret C, Colnot S. Stabilization of beta-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology. 2008;47:247–258. doi: 10.1002/hep.21952. [DOI] [PubMed] [Google Scholar]
- 30.Dessimoz J, Opoka R, Kordich JJ, Grapin-Botton A, Wells JM. FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mech Dev. 2006;123:42–55. doi: 10.1016/j.mod.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, Sato TN, Rabbany SY, Rafii S. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eferl R, Sibilia M, Hilberg F, Fuchsbichler A, Kufferath I, Guertl B, Zenz R, Wagner EF, Zatloukal K. Functions of c-Jun in liver and heart development. J Cell Biol. 1999;145:1049–1061. doi: 10.1083/jcb.145.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 34.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 35.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 36.Friedman SL. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gebhardt R. Metabolic zonation of the liver: Regulation and implications for liver function. Pharmacol Ther. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 38.Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: The role of Wnt/beta-catenin signaling in liver zonation and beyond. Dev Dyn. 2010;239:45–55. doi: 10.1002/dvdy.22041. [DOI] [PubMed] [Google Scholar]
- 39.Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, Radtke F, Schmid RM, Siveke JT. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607–616. doi: 10.1002/hep.22381. [DOI] [PubMed] [Google Scholar]
- 40.Giera S, Braeuning A, Kohle C, Bursch W, Metzger U, Buchmann A, Schwarz M. Wnt/beta-catenin signaling activates and determines hepatic zonal expression of glutathione S-transferases in mouse liver. Toxicol Sci. 2010;115:22–33. doi: 10.1093/toxsci/kfq033. [DOI] [PubMed] [Google Scholar]
- 41.Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, Strijbosch R, Haramis AP, Puder M, Clevers H, Moon RT, Zon LI. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 42.Gouon-Evans V, Boussemart L, Gadue P, Nierhoff D, Koehler CI, Kubo A, Shafritz DA, Keller G. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- 43.Gualdi R, Bossard P, Zheng MH, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 44.Gunes C, Heuchel R, Georgiev O, Muller KH, Lichtlen P, Bluthmann H, Marino S, Aguzzi A, Schaffner W. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J. 1998;17:2846–2854. doi: 10.1093/emboj/17.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 46.Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- 47.Houssaint E. Differentiation of the mouse hepatic primordium. I. An analysis of tissue interactions in hepatocyte differentiation. Cell Differ. 1980;9:269–279. doi: 10.1016/0045-6039(80)90026-3. [DOI] [PubMed] [Google Scholar]
- 48.Huang PY, He ZY, Ji SY, Sun HW, Xiang D, Liu CC, Hu YP, Wang X, Hui LJ. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475 doi: 10.1038/nature10116. 386-U142. [DOI] [PubMed] [Google Scholar]
- 49.Hunter MP, Wilson CM, Jiang X, Cong R, Vasavada H, Kaestner KH, Bogue CW. The homeobox gene Hhex is essential for proper hepatoblast differentiation and bile duct morphogenesis. Dev Biol. 2007;308:355–367. doi: 10.1016/j.ydbio.2007.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain SZ, Sneddon T, Tan XP, Micsenyi A, Michalopoulos GK, Monga SPS. Wnt impacts growth and differentiation in ex vivo liver development. Exp Cell Res. 2004;292:157–169. doi: 10.1016/j.yexcr.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 51.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ijpenberg A, Perez-Pomares JM, Guadix JA, Carmona R, Portillo-Sanchez V, Macias D, Hohenstein P, Miles CM, Hastie ND, Munoz-Chapuli R. Wt1 and retinoic acid signaling are essential for stellate cell development and liver morphogenesis. Dev Biol. 2007;312:157–170. doi: 10.1016/j.ydbio.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 53.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 54.Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu Rev Nutr. 1996;16:179–203. doi: 10.1146/annurev.nu.16.070196.001143. [DOI] [PubMed] [Google Scholar]
- 55.Kamiya A, Kinoshita T, Ito Y, Matsui T, Morikawa Y, Senba E, Nakashima K, Taga T, Yoshida K, Kishimoto T, Miyajima A. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J. 1999;18:2127–2136. doi: 10.1093/emboj/18.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamiya A, Kinoshita T, Miyajima A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett. 2001;492:90–94. doi: 10.1016/s0014-5793(01)02140-8. [DOI] [PubMed] [Google Scholar]
- 57.Kieusseian A, Chagraoui J, Kerdudo C, Mangeot PE, Gage PJ, Navarro N, Izac B, Uzan G, Forget BG, Dubart-Kupperschmitt A. Expression of Pitx2 in stromal cells is required for normal hematopoiesis. Blood. 2006;107:492–500. doi: 10.1182/blood-2005-02-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura J, Deutsch GH. Key mechanisms of early lung development. Pediatr Dev Pathol. 2007;10:335–347. doi: 10.2350/07-06-0290.1. [DOI] [PubMed] [Google Scholar]
- 59.Kimura T, Christoffels VM, Chowdhury S, Iwase K, Matsuzaki H, Mori M, Lamers WH, Darlington GJ, Takiguchi M. Hypoglycemia-associated hyperammonemia caused by impaired expression of ornithine cycle enzyme genes in C/EBPalpha knockout mice. J Biol Chem. 1998;273:27505–27510. doi: 10.1074/jbc.273.42.27505. [DOI] [PubMed] [Google Scholar]
- 60.Klein D, Demory A, Peyre F, Kroll J, Augustin HG, Helfrich W, Kzhyshkowska J, Schledzewski K, Arnold B, Goerdt S. Wnt2 acts as a cell type-specific, autocrine growth factor in rat hepatic sinusoidal endothelial cells cross-stimulating the VEGF pathway. Hepatology. 2008;47:1018–1031. doi: 10.1002/hep.22084. [DOI] [PubMed] [Google Scholar]
- 61.Kodama Y, Hijikata M, Kageyama R, Shimotohno K, Chiba T. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology. 2004;127:1775–1786. doi: 10.1053/j.gastro.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–122. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- 63.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 64.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lade A, Ranganathan S, Luo J, Monga SP. Calpain induces N-terminal truncation of beta-catenin in normal murine liver development: Diagnostic implications in hepatoblastomas. J Biol Chem. 2012 doi: 10.1074/jbc.M112.378224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lade AG, Monga SPS. Beta-catenin signaling in hepatic development and progenitors: Which way does the WNT blow? Dev Dyn. 2011;240:486–500. doi: 10.1002/dvdy.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laverriere AC, Macneill C, Mueller C, Poelmann RE, Burch JBE, Evans T. Gata-4/5/6, a subfamily of 3 transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 68.Le Douarin NM. Experimental analysis of liver development. Med Biol. 1975;53:427–455. [PubMed] [Google Scholar]
- 69.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 70.Lehwald N, Tao GZ, Jang KY, Sorkin M, Knoefel WT, Sylvester KG. Wnt-beta-catenin signaling protects against hepatic ischemia and reperfusion injury in mice. Gastroenterology. 2011;141:707–718. e701–e705. doi: 10.1053/j.gastro.2011.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li JX, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4 alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 73.Li Y, Rankin SA, Sinner D, Kenny AP, Krieg PA, Zorn AM. Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev. 2008;22:3050–3063. doi: 10.1101/gad.1687308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Z, White P, Tuteja G, Rubins N, Sackett S, Kaestner KH. Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest. 2009;119:1537–1545. doi: 10.1172/JCI38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loeppen S, Koehle C, Buchmann A, Schwarz M. A beta-catenin-dependent pathway regulates expression of cytochrome P450 isoforms in mouse liver tumors. Carcinogenesis. 2005;26:239–248. doi: 10.1093/carcin/bgh298. [DOI] [PubMed] [Google Scholar]
- 76.Loo CK, Wu XJ. Origin of stellate cells from submesothelial cells in a developing human liver. Liver Int. 2008;28:1437–1445. doi: 10.1111/j.1478-3231.2008.01788.x. [DOI] [PubMed] [Google Scholar]
- 77.Lupien M, Eeckhoute J, Meyer CA, Wang QB, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lyons KM, Pelton RW, Hogan BLM. Patterns of expression of murine Vgr-1 and Bmp-2a Rna suggest that transforming growth factor-beta-like genes coordinately regulate aspects of embryonic-development. Genes Dev. 1989;3:1657–1668. doi: 10.1101/gad.3.11.1657. [DOI] [PubMed] [Google Scholar]
- 79.Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 80.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Margagliotti S, Clotman F, Pierreux CE, Beaudry JB, Jacquemin P, Rousseau GG, Lemaigre FP. The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration. Dev Biol. 2007;311:579–589. doi: 10.1016/j.ydbio.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 82.Margagliotti S, Clotman F, Pierreux CE, Lemoine P, Rousseau GG, Henriet P, Lemaigre FP. Role of metalloproteinases at the onset of liver development. Dev Growth Differ. 2008;50:331–338. doi: 10.1111/j.1440-169X.2008.01031.x. [DOI] [PubMed] [Google Scholar]
- 83.Masyuk AI, Masyuk TV, LaRusso NF. Cholangiocyte primary cilia in liver health and disease. Dev Dyn. 2008;237:2007–2012. doi: 10.1002/dvdy.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matsumoto K, Miki R, Nakayama M, Tatsumi N, Yokouchi Y. Wnt9a secreted from the walls of hepatic sinusoids is essential for morphogenesis, proliferation, and glycogen accumulation of chick hepatic epithelium. Dev Biol. 2008;319:234–247. doi: 10.1016/j.ydbio.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 85.Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- 86.Mclin VA, Rankin SA, Zorn AM. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- 87.Michalopoulos GK, Bowen WC, Mule K, Luo J. HGF-, EGF-, and dexamethasone-induced gene expression patterns during formation of tissue in hepatic organoid cultures. Gene Expr. 2003;11:55–75. doi: 10.3727/000000003108748964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Micsenyi A, Tan X, Sneddon T, Luo JH, Michalopoulos GK, Monga SP. Beta-catenin is temporally regulated during normal liver development. Gastroenterology. 2004;126:1134–1146. doi: 10.1053/j.gastro.2003.12.047. [DOI] [PubMed] [Google Scholar]
- 89.Minguet S, Cortegano I, Gonzalo P, Martinez-Marin JA, de Andres B, Salas C, Melero D, Gaspar ML, Marcos MA. A population of c-Kit(low)(CD45/TER119)- hepatic cell progenitors of 11-day postcoitus mouse embryo liver reconstitutes cell-depleted liver organoids. J Clin Invest. 2003;112:1152–1163. doi: 10.1172/JCI17409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, Kahn M. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2007;104:5668–5673. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miyajima A, Kinoshita T, Tanaka M, Kamiya A, Mukouyama Y, Hara T. Role of Oncostatin M in hematopoiesis and liver development. Cytokine Growth Factor Rev. 2000;11:177–183. doi: 10.1016/s1359-6101(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 92.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 93.Monaghan AP, Kaestner KH, Grau E, Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/Hnf-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- 94.Monga SP, Hout MS, Baun MJ, Micsenyi A, Muller P, Tummalapalli L, Ranade AR, Luo JH, Strom SC, Gerlach JC. Mouse fetal liver cells in artificial capillary beds in three-dimensional four-compartment bioreactors. Am J Pathol. 2005;167:1279–1292. doi: 10.1016/S0002-9440(10)61215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monga SP, Micsenyi A, Germinaro M, Apte U, Bell A. beta-Catenin regulation during matrigel-induced rat hepatocyte differentiation. Cell Tissue Res. 2006;323:71–79. doi: 10.1007/s00441-005-0045-8. [DOI] [PubMed] [Google Scholar]
- 96.Monga SP, Monga HK, Tan X, Mule K, Pediaditakis P, Michalopoulos GK. Beta-catenin antisense studies in embryonic liver cultures: Role in proliferation, apoptosis, and lineage specification. Gastroenterology. 2003;124:202–216. doi: 10.1053/gast.2003.50000. [DOI] [PubMed] [Google Scholar]
- 97.Nejak-Bowen K, Monga SP. Wnt/beta-catenin signaling in hepatic organogenesis. Organogenesis. 2008;4:92–99. doi: 10.4161/org.4.2.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nonaka H, Tanaka M, Suzuki K, Miyajima A. Development of murine hepatic sinusoidal endothelial cells characterized by the expression of hyaluronan receptors. Dev Dyn. 2007;236:2258–2267. doi: 10.1002/dvdy.21227. [DOI] [PubMed] [Google Scholar]
- 99.North TE, Goessling W. Endoderm specification, liver development, and regeneration. Methods Cell Biol. 2011;101:205–223. doi: 10.1016/B978-0-12-387036-0.00010-4. [DOI] [PubMed] [Google Scholar]
- 100.O’Hara SP, Splinter PL, Trussoni CE, Gajdos GB, Lineswala PN, LaRusso NF. Cholangiocyte N-Ras protein mediates lipopolysaccharide-induced interleukin 6 secretion and proliferation. J Biol Chem. 2011;286:30352–30360. doi: 10.1074/jbc.M111.269464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 102.Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 103.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Onitsuka I, Tanaka M, Miyajima A. Characterization and functional analyses of hepatic mesothelial cells in mouse liver development. Gastroenterology. 2010;138:1525–1535. 1535, e1521–e1526. doi: 10.1053/j.gastro.2009.12.059. [DOI] [PubMed] [Google Scholar]
- 105.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 106.Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach JP, Babinet C, Yaniv M. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 107.Poulain M, Ober EA. Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregut-derived organ development. Development. 2011;138:3557–3568. doi: 10.1242/dev.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raynaud P, Carpentier R, Antoniou A, Lemaigre FP. Biliary differentiation and bile duct morphogenesis in development and disease. Int J Biochem Cell Biol. 2011;43:245–256. doi: 10.1016/j.biocel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 110.Rhim JA, Sandgren EP, Degen JL, Palmiter RD, Brinster RL. Replacement of diseased mouse-liver by hepatic cell transplantation. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 111.Rogler LE. Selective bipotential differentiation of mouse embryonic hepatoblasts in vitro. Am J Pathol. 1997;150:591–602. [PMC free article] [PubMed] [Google Scholar]
- 112.Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Santisteban P, Recacha P, Metzger DE, Zaret KS. Dynamic expression of groucho-related genes Grg1 and Grg3 in foregut endoderm and antagonism of differentiation. Dev Dyn. 2010;239(3):980–986. doi: 10.1002/dvdy.22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sato T, El-Assal ON, Ono T, Yamanoi A, Dhar DK, Nagasue N. Sinusoidal endothelial cell proliferation and expression of angiopoietin/Tie family in regenerating rat liver. J Hepatol. 2001;34:690–698. doi: 10.1016/s0168-8278(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 115.Sawant S, Aparicio S, Tink AR, Lara N, Barnstable CJ, Tombran-Tink J. Regulation of factors controlling angiogenesis in liver development: A role for PEDF in the formation and maintenance of normal vasculature. Biochem Biophys Res Commun. 2004;325:408–413. doi: 10.1016/j.bbrc.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 116.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth-factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 117.Sekhon SS, Tan X, Micsenyi A, Bowen WC, Monga SP. Fibroblast growth factor enriches the embryonic liver cultures for hepatic progenitors. Am J Pathol. 2004;164:2229–2240. doi: 10.1016/S0002-9440(10)63779-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sekine S, Lan BYA, Bedolli M, Feng S, Hebrok M. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome P450 expression in mice. Hepatology. 2006;43:817–825. doi: 10.1002/hep.21131. [DOI] [PubMed] [Google Scholar]
- 119.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 120.Sekiya T, Adachi S, Kohu K, Yamada T, Higuchi O, Furukawa Y, Nakamura Y, Nakamura T, Tashiro K, Kuhara S, Ohwada S, Akiyama T. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. J Biol Chem. 2004;279:6840–6846. doi: 10.1074/jbc.M310876200. [DOI] [PubMed] [Google Scholar]
- 121.Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development. 2005;132:35–47. doi: 10.1242/dev.01570. [DOI] [PubMed] [Google Scholar]
- 122.Shin D, Lee Y, Poss KD, Stainier DYR. Restriction of hepatic competence by Fgf signaling. Development. 2011;138:1339–1348. doi: 10.1242/dev.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 124.Shin D, Weidinger G, Moon RT, Stainier DY. Intrinsic and extrinsic modifiers of the regulative capacity of the developing liver. Mech Dev. 2012;128:525–535. doi: 10.1016/j.mod.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shiojiri N, Takeshita K, Yamasaki H, Iwata T. Suppression of C/EBP alpha expression in biliary cell differentiation from hepatoblasts during mouse liver development. J Hepatol. 2004;41:790–798. doi: 10.1016/j.jhep.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 126.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 127.Si-Tayeb K, Noto FK, Nagaoka M, Li JX, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- 129.Soto-Gutierrez A, Navarro-Alvarez N, Zhao D, Rivas-Carrillo JD, Lebkowski J, Tanaka N, Fox IJ, Kobayashi N. Differentiation of mouse embryonic stem cells to hepatocyte-like cells by co-culture with human liver nonparenchymal cell lines. Nat Protoc. 2007;2:347–356. doi: 10.1038/nprot.2007.18. [DOI] [PubMed] [Google Scholar]
- 130.Spath GF, Weiss MC. Hepatocyte nuclear factor 4 provokes expression of epithelial marker genes, acting as a morphogen in dedifferentiated hepatoma cells. J Cell Biol. 1998;140:935–946. doi: 10.1083/jcb.140.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Spear BT, Jin L, Ramasamy S, Dobierzewska A. Transcriptional control in the mammalian liver: liver development, perinatal repression, and zonal gene regulation. Cell Mol Life Sci. 2006;63:2922–2938. doi: 10.1007/s00018-006-6258-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stafford D, Hornbruch A, Mueller PR, Prince VE. A conserved role for retinoid signaling in vertebrate pancreas development. Dev Genes Evol. 2004;214:432–441. doi: 10.1007/s00427-004-0420-6. [DOI] [PubMed] [Google Scholar]
- 133.Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- 134.Stanulovic VS, Kyrmizi I, Kruithof-de Julio M, Hoogenkamp M, Vermeulen JL, Ruijter JM, Talianidis I, Hakvoort TB, Lamers WH. Hepatic HNF4alpha deficiency induces periportal expression of glutamine synthetase and other pericentral enzymes. Hepatology. 2007;45:433–444. doi: 10.1002/hep.21456. [DOI] [PubMed] [Google Scholar]
- 135.Stark KL, Mcmahon JA, Mcmahon AP. Fgfr-4, a new member of the fibroblast growth-factor receptor family, expressed in the definitive endoderm and skeletal-muscle lineages of the mouse. Development. 1991;113:641–651. doi: 10.1242/dev.113.2.641. [DOI] [PubMed] [Google Scholar]
- 136.Stock P, Bruckner S, Ebensing S, Hempel M, Dollinger MM, Christ B. The generation of hepatocytes from mesenchymal stem cells and engraftment into murine liver. Nat Protoc. 2010;5:617–627. doi: 10.1038/nprot.2010.7. [DOI] [PubMed] [Google Scholar]
- 137.Sugi Y, Sasse J, Barron M, Lough J. Developmental expression of fibroblast growth-factor receptor-1 (Cek-1-Flg) during heart development. Dev Dyn. 1995;202:115–125. doi: 10.1002/aja.1002020203. [DOI] [PubMed] [Google Scholar]
- 138.Sugiyama D, Kulkeaw K, Mizuochi C, Horio Y, Okayama S. Hepatoblasts comprise a niche for fetal liver erythropoiesis through cytokine production. Biochem Biophys Res Commun. 2011;410:301–306. doi: 10.1016/j.bbrc.2011.05.137. [DOI] [PubMed] [Google Scholar]
- 139.Suksaweang S, Lin CM, Jiang TX, Hughes MW, Widelitz RB, Chuong CM. Morphogenesis of chicken liver: Identification of localized growth zones and the role of beta-catenin/Wnt in size regulation. Dev Biol. 2004;266:109–122. doi: 10.1016/j.ydbio.2003.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suzuki A, Zheng Y, Kondo R, Kusakabe M, Takada Y, Fukao K, Nakauchi H, Taniguchi H. Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology. 2000;32:1230–1239. doi: 10.1053/jhep.2000.20349. [DOI] [PubMed] [Google Scholar]
- 141.Tan X, Yuan Y, Zeng G, Apte U, Thompson MD, Cieply B, Stolz DB, Michalopoulos GK, Kaestner KH, Monga SP. Beta-catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology. 2008;47:1667–1679. doi: 10.1002/hep.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tan XP, Behari J, Cieply B, Michalopoulos GK, Monga SPS. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 143.Tanaka M, Itoh T, Tanimizu N, Miyajima A. Liver stem/progenitor cells: Their characteristics and regulatory mechanisms. J Biochem. 2011;149:231–239. doi: 10.1093/jb/mvr001. [DOI] [PubMed] [Google Scholar]
- 144.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299:574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 145.Tatsumi N, Miki R, Katsu K, Yokouchi Y. Neurturin-GFRalpha2 signaling controls liver bud migration along the ductus venosus in the chick embryo. Dev Biol. 2007;307:14–28. doi: 10.1016/j.ydbio.2007.03.519. [DOI] [PubMed] [Google Scholar]
- 146.Tiso N, Filippi A, Pauls S, Bortolussi M, Argenton F. BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mech Dev. 2002;118:29–37. doi: 10.1016/s0925-4773(02)00252-6. [DOI] [PubMed] [Google Scholar]
- 147.Vanderpool C, Sparks EE, Huppert KA, Gannon M, Means AL, Huppert SS. Genetic interactions between hepatocyte nuclear factor-6 and Notch signaling regulate mouse intrahepatic bile duct development in vivo. Hepatology. 2012;55:233–243. doi: 10.1002/hep.24631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science. 2009;324:1707–1710. doi: 10.1126/science.1174497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 150.Watt AJ, Zhao R, Li JX, Duncan SA. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol. 2007;7:37. doi: 10.1186/1471-213X-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weinstein M, Monga SP, Liu Y, Brodie SG, Tang Y, Li C, Mishra L, Deng CX. Smad proteins and hepatocyte growth factor control parallel regulatory pathways that converge on beta1-integrin to promote normal liver development. Mol Cell Biol. 2001;21:5122–5131. doi: 10.1128/MCB.21.15.5122-5131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–1572. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- 153.Wills A, Dickinson K, Khokha M, Baker JC. Bmp signaling is necessary and sufficient for ventrolateral endoderm specification in Xenopus. Dev Dyn. 2008;237:2177–2186. doi: 10.1002/dvdy.21631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yamasaki H, Sada A, Iwata T, Niwa T, Tomizawa M, Xanthopoulos KG, Koike T, Shiojiri N. Suppression of C/EBPalpha expression in periportal hepatoblasts may stimulate biliary cell differentiation through increased Hnf6 and Hnf1b expression. Development. 2006;133:4233–4243. doi: 10.1242/dev.02591. [DOI] [PubMed] [Google Scholar]
- 156.Yanai M, Tatsumi N, Hasunuma N, Katsu K, Endo F, Yokouchi Y. FGF signaling segregates biliary cell-lineage from chick hepatoblasts cooperatively with BMP4 and ECM components in vitro. Dev Dyn. 2008;237:1268–1283. doi: 10.1002/dvdy.21520. [DOI] [PubMed] [Google Scholar]
- 157.Yang L, Magness ST, Bataller R, Rippe RA, Brenner DA. NF-kappaB activation in Kupffer cells after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2005;289:G530–G538. doi: 10.1152/ajpgi.00526.2004. [DOI] [PubMed] [Google Scholar]
- 158.Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordonez A, Hannan NRF, Rouhani FJ, Darche S, Alexander G, Marciniak SJ, Fusaki N, Hasegawa M, Holmes MC, Di Santo JP, Lomas DA, Bradley A, Vallier L. Targeted gene correction of alpha(1)-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zaret KS. Liver specification and early morphogenesis. Mech Dev. 2000;92:83–88. doi: 10.1016/s0925-4773(99)00326-3. [DOI] [PubMed] [Google Scholar]
- 160.Zaret KS. Hepatocyte differentiation: From the endoderm and beyond. Curr Opin Genet Dev. 2001;11:568–574. doi: 10.1016/s0959-437x(00)00234-3. [DOI] [PubMed] [Google Scholar]
- 161.Zaret KS. Regulatory phases of early liver development: Paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 162.Zaret KS, Carroll JS. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zaret KS, Watts J, Xu E, Wandzioch E, Smale ST, Sekiya T. Pioneer factors, genetic competence, and inductive signaling: Programming liver and pancreas progenitors from the endoderm. Cold Spring Harb Symp Quant Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zeng G, Awan F, Otruba W, Muller P, Apte U, Tan X, Gandhi C, Demetris AJ, Monga SP. Wnt’er in liver: Expression of Wnt and frizzled genes in mouse. Hepatology. 2007;45:195–204. doi: 10.1002/hep.21473. [DOI] [PubMed] [Google Scholar]
- 166.Zhang W, Yatskievych TA, Baker RK, Antin PB. Regulation of Hex gene expression and initial stages of avian hepatogenesis by Bmp and Fgf signaling. Dev Biol. 2004;268:312–326. doi: 10.1016/j.ydbio.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 167.Zhao R, Duncan SA. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- 168.Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhu XL, Sasse J, McAllister D, Lough J. Evidence that fibroblast growth factors 1 and 4 participate in regulation of cardiogenesis. Dev Dyn. 1996;207:429–438. doi: 10.1002/(SICI)1097-0177(199612)207:4<429::AID-AJA7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 170.Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol. 2011;43:257–264. doi: 10.1016/j.biocel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zorn AM, Butler K, Gurdon JB. Anterior endomesoderm specification in Xenopus by Wnt/beta-catenin and TGF-beta signalling pathways. Dev Biol. 1999;209:282–297. doi: 10.1006/dbio.1999.9257. [DOI] [PubMed] [Google Scholar]