Abstract
Background
Overall, HER2-amplified female breast cancer (FBC) is associated with a high grade, an aggressive phenotype and a poor prognosis. In male breast cancer (MBC) amplification of HER2, located on chromosome 17, occurs at a lower frequency than in FBC, where it is part of complex rearrangements. So far, only few studies have addressed the occurrence of chromosome 17 alterations in small MBC cohorts.
Methods
Multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridization (FISH) were used to detect and characterize copy number changes on chromosome 17 in a cohort of 139 MBC. The results obtained were compared to those in FBC, and were correlated with clinicopathological features and patient outcome data.
Results
We observed a lower frequency of chromosome 17 copy number changes with less complex rearrangement patterns in MBC compared to FBC. Chromosome 17 changes in MBC included gains of 17q and losses of 17p. Whole chromosome 17 polyploidies were not encountered. Two recurrent chromosome 17 amplicons were detected: on 17q12 (encompassing the NEUROD2, HER2, GRB7 and IKZF3 gens) and on 17q23.1 (encompassing the MIR21 and RPS6KB1 genes). Whole arm copy number gains of 17q were associated with decreased 5 year survival rates (p = 0.010). Amplification of HER2 was associated with a high tumor grade, but did not predict patient survival. Although copy number gains of HER2 and NEUROD2 were associated with a high tumor grade, a high mitotic count and a decreased 5 year survival rate (p = 0.015), only tumor size and NEUROD2 copy number gains emerged as independent prognostic factors.
Conclusions
In MBC chromosome 17 shows less complex rearrangements and fewer copy number changes compared to FBC. Frequent gains of 17q, encompassing two distinct amplicons, and losses of 17p were observed, but no whole chromosome 17 polyploidies. Only NEUROD2 gains seem to have an independent prognostic impact. These results suggest different roles of chromosome 17 aberrations in male versus female breast carcinogenesis.
Electronic supplementary material
The online version of this article (doi:10.1007/s13402-015-0227-7) contains supplementary material, which is available to authorized users.
Keywords: Male breast cancer, Oncogenes, Copy number changes, Multiplex ligation-dependent probe amplification
Introduction
Previous studies using multiplex ligation-dependent probe amplification (MLPA) and comparative genomic hybridization (CGH) in male breast cancer (MBC) revealed clear differences in gene copy number changes compared to female breast cancer (FBC), pointing towards differences in carcinogenesis between MBC and FBC [1, 2]. Copy number changes on chromosome 17q have been extensively studied in different cancer types including FBC. This is primarily due to the presence of the ERBB2 oncogene (HER2) on chromosome 17q. Amplification of HER2 is found in about 10–20 % of FBC and usually leads to over-expression of its encoded protein. HER2 amplification/over-expression also correlates with a high grade, a high mitotic index, a worse prognosis and a favorable response to targeted therapy with trastuzumab [3–8]. Next to HER2, several other oncogenes are located on chromosome 17, such as TOP2A and PPM1D [9–11]. To assess the HER2 amplification status by in situ hybridization, correction for polysomy of chromosome 17 is widely applied, although several studies have shown that such polysomy is very rare in FBC, and that the copy number status of the centromere does not reliably represent the number of chromosome 17 copies. Instead, it has been found that chromosome 17 may show very complex rearrangements in FBC [12–15]. It has also been found that in MBC HER2 amplification occurs at a lower frequency than in FBC (2–8 % versus 10–20 %, respectively) [1, 6, 14, 16–18]. So far, only a few (mainly CGH) studies have been published dealing with chromosome 17 alterations in relatively small MBC cohorts [2, 19], and their association with disease outcome has not previously been reported.
In the present study we aimed to characterize copy number changes on chromosome 17 in a large MBC cohort using a dedicated chromosome 17 MLPA kit that has previously been used to study chromosome 17 polysomy in FBC [14]. In addition, we performed HER2 chromogenic in situ hybridization (CISH) and correlated the results with several clinicopathologic features and patient outcome data.
Materials and methods
Patient material and characteristics
Consecutive surgical invasive male breast cancer (MBC) specimens were collected from 1986 to 2011 at four different pathology labs in the Netherlands (i.e., St. Antonius Hospital Nieuwegein, Diakonessenhuis Utrecht, University Medical Center Utrecht, Laboratory for Pathology East Netherlands) as reported in more detail before [1, 20, 21], and at three pathology labs in Germany (i.e., in Paderborn, Cologne, Kassel). Hematoxylin and eosin (HE) stained slides were reviewed by four experienced observers (PJvD, RK, AM, ML) to confirm the diagnosis and to type and grade the tumors according to current WHO standards. Pathology reports were used to retrieve information on age, tumor size and lymph node status. A total of 139 cases, from which the paraffin blocks contained enough tumor cells for DNA isolation, were included in the present study. The clinicopathological features of these cases are listed in Table 1. The average age of the MBC patients was 67 years. The tumor sizes ranged from 0.2 to 7.2 cm. In 114 cases the lymph node status was assessed through axillary lymph node dissection or sentinel node procedures, and in 56 % of these patients lymph node metastases were noted. The majority of the MBC cases was diagnosed (WHO criteria) as invasive ductal carcinoma. According to the modified Bloom and Richardson score [22] most tumors were classified as grade 2 or grade 3. Mitotic activities were assessed as reported before [23], with a mean mitotic index of 12 per 2 mm2. In all cases, the estrogen receptor (ER) and progesterone receptor (PR) expression and HER2 amplification status were re-assessed as described before [20]. Tissue microarray (TMA) slides were used for immunohistochemical detection of ER and PR expression. Chromogenic in situ hybridization (SPoT-Light HER2 CISH kit, Invitrogen) was used for HER2 copy number assessment. The HER2 gene was considered to be amplified when more than 50 % of the tumor cells showed 5–10 single dots or small clusters of dots per nucleus (i.e., low level amplification), or more than 10 single dots or large clusters of dots per nucleus (i.e., high level amplification).
Table 1.
Baseline clinicopathological features of 139 male breast cancers
| Characteristics | All cases (n = 139) | Characteristics | All cases (n = 139) |
|---|---|---|---|
| Age, years | Lymph node metastasis | n = 114 | |
| Mean | 67 (range 32–89) | Absent | 50 (44 %) |
| ≤ 50 | 13 (9 %) | Present | 64 (56 %) |
| > 50 | 126 (91 %) | ||
| Immunohistochemistry | |||
| Histological type | ER | ||
| Ductal | 124 (90 %) | (+) | 128 (92 %) |
| Lobular | 3 (2 %) | (−) | 11 (8 %) |
| Invasive cribriform | 3 (2 %) | PR | |
| Mixed (ductal/lobular) | 3 (2 %) | (+) | 93 (67 %) |
| Mucinous | 2 (1 %) | (−) | 46 (33 %) |
| Papillary | 2 (1 %) | AR | |
| Invasive micropapillary | 1 (1 %) | (+) | 112 (81 %) |
| Adenoid cystic | 1 (1 %) | (−) | 27 (19 %) |
| HER2 (CISH) | |||
| Tumor size (mean), cm | 2.3 (n = 135) | (+) | 5 (4 %) |
| T1 | 70 (50 %) | (−) | 134 (96 %) |
| T2 | 61 (45 %) | ||
| T3 | 4 (3 %) | Intrinsic subtypes | |
| Luminal A | 108 (74 %) | ||
| Mitotic activity index/2 mm2 | Luminal B | 27 (19 %) | |
| < 8 mitoses | 54 (39 %) | Her2 driven | 0 (0 %) |
| 8–14 mitoses | 34 (24 %) | Basal like / | 10 (7 %) |
| 15 or > mitoses | 51 (37 %) | unclassified triple negative | |
| Histological grade | |||
| I | 33 (24 %) | ||
| II | 60 (43 %) | ||
| III | 46 (33 %) |
Intrinsic subtype classification
Immunohistochemical staining was used to classify the tumors into five intrinsic subtypes: luminal type A (ER+ and/or PR+, HER2- and Ki-67 low), luminal type B (ER+ and/or PR+, and HER2+ and/or Ki67 high), HER2 driven (HER2+ and ER-/PR-), basal-like (ER-/PR-/HER2-, and CK5/6+ and/or CK14+ and/or EGFR+) and unclassifiable triple negative (negative for all six markers), as described before [20].
DNA extraction and MLPA analysis
Representative tumor areas were identified in HE stained slides and dissected with a scalpel from 8 μm thick paraffin sections as reported before [24]. DNA was extracted by overnight incubation of the samples in proteinase K (10 mg/ml; Roche, Almere, The Netherlands) at 56 °C, boiling for 10 min and subsequent clearance by centrifugation. Five μl of the resulting DNA solution was used for MLPA analysis. MLPA was performed according the manufacturers’ instructions (MRC Holland, Amsterdam, The Netherlands), using a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA, USA). A recently designed kit (P004-C1; MRC Holland), containing five probes for five 17p genes and twenty-six probes for seventeen 17q genes, was used. The kit contained twelve additional reference probes. All tests were performed in duplicate. Negative reference samples (normal breast and blood cells) as well as a positive control sample (tumor sample with high level HER2 amplification as determined by CISH) were included in each MLPA run as reported before [1]. The PCR products were separated by electrophoresis on an ABI 3730 capillary sequencer (Applied Biosystems) and the final gene copy number ratios were calculated using Genescan v4.1 (Applied Biosystems) and Coffalyser v9.4 (MRC-Holland) software packages. For genes represented by more than one probe in the kit, the mean of the copy number ratios in duplicate was calculated. Cut-off values were set as reported before, with > 1.3 to 2.0 for gain, > 2.0 for amplification and < 0.7 for loss. Gene copy number increase (or total gain) was defined by values > 1.3, including gain and amplification. Values between 0.7 and 1.3 were considered normal [6, 25]. Whole chromosome arm loss or gain was defined by copy number loss of more than 75 % of all the probes, as reported before using array-CGH [26]. Partial gain on the long arm of chromosome 17 was defined as any probe showing copy number increase. The MBC chromosome 17 copy number data were also compared to chromosome 17 copy number data of 111 FBC cases reported before, but based on a different MLPA design [14].
Statistical analyses
Statistical calculations were performed using IBM SPSS for Windows version 20.0. Associations between gene copy numbers and clinicopathological features were calculated using the Pearson Chi-square test (or Fisher’s exact test when appropriate) for categorical variables. Grade, tumor size and mitotic count were dichotomized as usual [1, 21]. Unsupervised hierarchical clustering using the statistical program R (www.r-project.org) was performed to identify relevant clusters. We used the maximum distance and Ward’s clustering method, and calculated the stability of the clusters with pvclust, as reported before [1]. Information regarding prognosis and therapy was retrieved from the Integral Cancer registration of the Netherlands (IKNL). Survival data were available from 100 cases with a mean follow-up of 5.6 years. Therefore, we based the survival analyses on 5 year survival rates. For univariate survival estimates, Kaplan-Meier plots were analyzed using the log rank test. Multivariate survival analyses were performed using Cox regression, including the variables that were found to be significant in univariate analyses. Corrections for multiple comparisons were applied according to Holm-Bonferroni.
Results
Identification of chromosome 17 copy number changes in male breast cancers
Multiplex ligation-dependent probe amplification (MLPA) was used to assess the copy number status of chromosome 17 in a cohort of 139 male breast cancers (MBC). The results obtained and its comparison to female breast cancer (FBC) cases are presented in Fig. 1 and supplementary Table A. Overall, we found a lower frequency of copy number changes with less complex patterns in MBC compared to those in FBC. In 51/139 (36.7 %) cases no copy number alterations were seen in MBC in any of the genes included in the MLPA assay. Copy number increases were found to be most frequent on 17q, present in 78/139 (56 %) of the cases, and copy number losses were found to be most frequent on 17p (36/139; 26 %). Six of the 139 MBC cases (4 %) showed a whole 17q arm gain. None of the MBC cases showed a whole chromosome 17 gain. NEUROD2, IKZF3, HER2 and MIR21 were the most commonly amplified genes, and copy number gains were most common for the MIR21 and RPS6KB1 genes. Copy number losses were most frequently observed for the MNT, TP53 and PMP genes, all three located on 17p (Fig. 2).
Fig. 1.
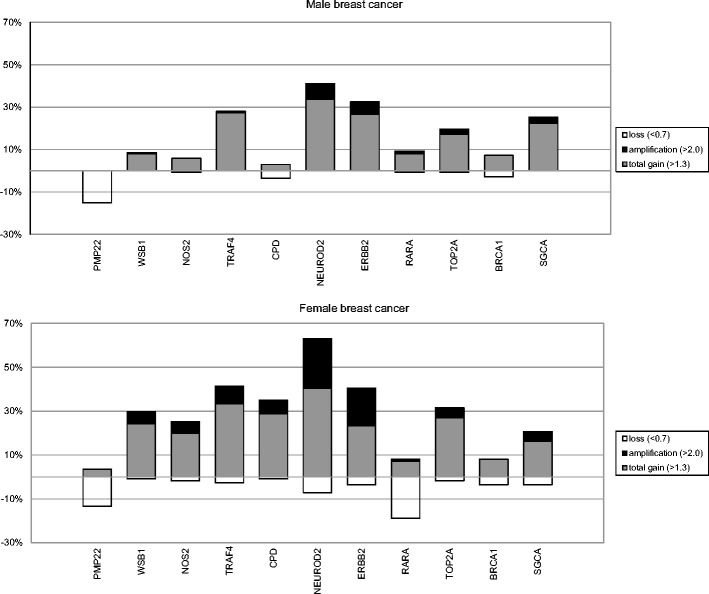
Copy number changes detectecd by MLPA of 11 genes on chromosome 17 in 139 male breast cancers compared to 111 female breast cancers (female data derived from [14])
Fig. 2.
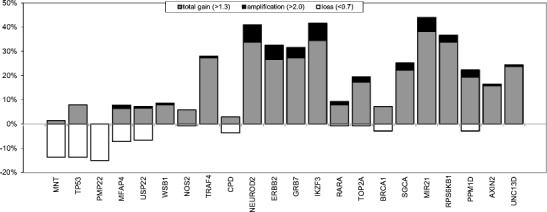
Copy number changes detected by MLPA in 22 genes on chromosome 17 in 139 male breast cancers
Chromosome 17 copy number alterations and associations with clinicopathological features
In 8/139 (5.8 %) of the MBC cases the HER2 status was assessed by MLPA. In four of these eight cases MLPA amplification ratios between 2.0 and 2.5 were observed, whereas the other four showed amplification ratios above 2.5. The cases that exhibited amplification rates between 2.0 and 2.5 by MLPA showed no amplification by CISH, whereas four of the cases with HER2 amplification as detected by CISH showed amplification by MLPA with ratios > 2.5. One case that was interpreted as showing a low level amplification by CISH also showed a gain, but no high-level amplification, by MLPA (Table 2).
Table 2.
Her2/Neu status based on immunohistochemistry and CISH in correlation with Her2/Neu amplification status based on MLPA
| Her2/Neu immuno- histochemistry | Chromogenic in situ hybridization (CISH) Her2/Neu | Her2/Neu amplification status | Multiplex ligation-dependent probe amplification (MLPA) |
|---|---|---|---|
| Positive (3+) | high amplification | positive | 3.795 |
| Positive (3+) | low amplification | positive | 4.859 |
| Positive (3+) | high amplification | positive | 4.292 |
| Positive (3+) | high amplification | positive | 2.536 |
| Negative (2+) | low amplification | positive | 1.448* |
| Negative (0) | no amplification | negative | 2.100 |
| Negative (0) | no amplification | negative | 2.003 |
| Negative (1+) | no amplification | negative | 2.023 |
| Negative (2+) | no amplification | negative | 2.269 |
(*only gain by MLPA, no amplification)
Three of the eight cases (37.5 %) showing HER2 amplification by MLPA also exhibited a whole arm gain of 17q, including gain of the WSB1 gene located near the centromere. Two of these cases also showed a partial gain of the short arm, combined with copy number loss of other loci on the short arm. Amplification of the 17q12 region, including the NEUROD2, GRB7 and IKZF3 genes, was observed in 6/8 (75 %) of the cases with HER2 amplification, and two of these cases showed an additional amplification of the RARA/TOP2A gene region on 17q21.2. The NEUROD2, HER2, GRB7 and IKZF3 genes were also frequently found to be gained, whereas copy number loss of this region was observed in none of the cases. Another region of frequent copy number gain was found on 17q23.1, were the MIR21 and RPS6KB1 genes are located (Fig. 2). Taken together, we identified two recurrent amplicons: one on 17q12 (encompassing the NEUROD2, HER2, GRB7 and IKZF3 genes) and one on 17q23.1 (encompassing the MIR21 and RPS6KB1 genes).
As can be deduced from Table 3, copy number increases of several genes on 17q were correlated with unfavorable clinicopathological features, such as a high mitotic count and a high grade (NEUROD2, HER2, GRB7, IKZF3, RPS6KB1, PPM1D, AXIN2 and UNC13D), a high grade and a large size (SGCA), or a high grade alone (BRCA1 and MIR21). Amplification of NEUROD2 was found to be correlated with a high mitotic count and a high grade. Amplification of the HER2 and GRB7 genes was found to be correlated with a high grade, and amplification of the IKZF3 gene with a high mitotic count. After correction for multiple comparisons, the correlation between copy number gains of the NEUROD2 gene and a high mitotic count and a high grade remained significant (Table 3).
Table 3.
Correlations between gene copy number changes and clinicopathological features in 139 male breast cancers
| Gene | Location | Mitotic index | Size | Grade | Lymph node status |
|---|---|---|---|---|---|
| Correlation between gene copy number increases (>1.3 (including amplified cases)) and clinicopathological features | |||||
| USP22 | 17p11.2 | 0.006 | |||
| WSB1 | 17q11.1 | 0.042 | |||
| CPD | 17q11.2 | 0.035 | |||
| NEUROD2 | 17q12 | <0.0001 | <0.0001 | ||
| ERBB2 | 17q12 | 0.046 | 0.017 | ||
| GRB7 | 17q12 | 0.016 | 0.006 | ||
| IKZF3 | 17q12 | 0.001 | 0.028 | ||
| RARA | 17q21.2 | 0.01 | |||
| BRCA1 | 17q21.31 | 0.01 | 0.021 | ||
| SGCA | 17q21.33 | 0.025 | 0.021 | ||
| MIR21 | 17q23.1 | 0.019 | |||
| RPS6KB1 | 17q23.1 | 0.002 | 0.001 | ||
| PPM1D | 17q23.2 | 0.047 | 0.033 | ||
| AXIN2 | 17q24.1 | 0.008 | <0.0001 | ||
| UNC13D | 17q25.1 | <0.0001 | <0.0001 | ||
| Correlation between gene amplification (>2.0) and clinicopathological features | |||||
| NEUROD2 | 17q12 | 0.049 | 0.015 | ||
| ERBB2 | 17q12 | 0.016 | |||
| GRB7 | 17q12 | 0.001 | |||
| IKZF3 | 17q12 | 0.007 | |||
Blank: non-significant results. Bold: after correction for multiple comparisons
Cluster analysis reveals association with luminal breast cancer sub-type
After unsupervised hierarchical cluster analysis, we found that the NEUROD2, HER2, GRB7 and IKZF3 genes clustered together (p < 0.001) (Supplementary Fig. A). Considering the MBC cases, an interesting cluster emerged encompassing 12 cases characterized by chromosome 17 whole arm copy number gains and amplifications of the NEUROD2, HER2, GRB7 and IKZF3 genes with significantly more luminal type B cases than luminal type A cases (p = 0.010). The distribution of other clinicopathological features (age, grade, mitotic index, size and lymph node status) was not found to be significantly different in this cluster compared to the other remaining cases.
Tumor size and NEUROD2 copy number gain act as independent prognostic factors
Survival data were available from 100 cases with a mean follow up of 5.6 years. Grade 3 (p = 0.026), high mitotic count (>8 mitoses/2 mm2; p = 0.028), large tumor size (>2.0 cm; p = 0.031), luminal type B (p = 0.042), positive HER2 status by CISH (low and high level amplification; p = 0.039), NEUROD2 copy number gain (p = 0.015), HER2 copy number gain (p = 0.015) and whole chromosome 17q arm gain (p = 0.010) were found to be associated with a decreased 5 year survival rate. The above (3.3) described clusters showed no correlation to survival. After multivariate Cox regression only tumor size and NEUROD2 gene copy number gain remained as independent prognostic factors (Fig. 3).
Fig. 3.
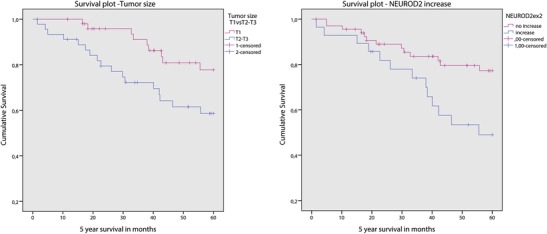
Survival plots of 100 male breast cancers stratified for NEUROD2 copy number status (right) and tumor size (left)
Discussion
The aim of the present study was to detect and characterize copy number changes on chromosome 17 in a large cohort of male breast cancers (MBC) using MLPA. The majority of cases showed chromosome 17 aberrations, mainly copy number gains on 17q (78/139; 56 %) and copy number losses on 17p (36/139; 26 %). Only six of the 139 cases (4 %) showed a whole 17q arm gain. None of the cases showed whole chromosome 17 gains, which is in line with previous female breast cancer (FBC) studies [13–15]. Compared to FBC [12], however, we found a lower frequency of chromosome 17 copy number changes with less complex patterns of genomic rearrangements in MBC.
Previously, chromosome CGH was used to assess copy number gains and losses in MBC [2, 19]. Rudlowski et al. [19] reported gains on 17q in 36 % of the MBC tested. In their study 17q gain was not associated with any of the clinicopathological features studied. In the present study we found that 58 % of the MBC showed gains on 17q and 14 % showed high-level amplifications on 17q. In line with our study, Tommasi et al. [2] found that both losses and gains on chromosome 17 were less prominent in MBC than in FBC.
Whole arm gains of 17q and concomitant gains of the WSB1 gene located near the centromere were frequently seen in association with amplification of the HER2 gene (37.5 %). The 17p arm, however, only showed partial copy number gains and losses, or no alterations at all in these cases, arguing against the occurrence of whole chromosome 17 polysomies in MBC, as has previously been observed in FBC [12]. These findings suggest that FISH correction for polysomy with centromere probes may result in a misleading HER2 gene status assessment, as described before in FBC [12].
Although in itself rare, the co-amplification and co-clustering of genes on the HER2 neighboring segment containing NEUROD2, GRB7 and IKZF3 in 75 % of the MBC cases points towards the existence of a large amplicon on 17q, which is different from what has previously been seen in FBC [12, 27, 28]. This latter amplicon includes both the HER2 and NEUROD2 genes, and is in line with a similar prognostic value of both NEUROD2 and HER2 copy number gains in the present study.
We found that the HER2 gene exhibited copy number gains in 21 % of the MBC cases, but a true amplification (by MLPA) was only seen in 5.8 % of the cases, which is in line with previous HER2 expression studies [16, 18], but lower than that reported by Tommasi et al. [2], who found HER2 to be amplified in 30 % of MBC cases by CGH. This discrepancy may be explained by the different technique and smaller sample size used in the latter study. We found that HER2 was amplified at a lower frequency in MBC compared to previous MLPA studies in FBC (5.8 % versus 20 %) [6]. In concordance with our previous studies in FBC and MBC, HER2 amplification detected by MLPA correlated strongly with HER2 amplification detected by CISH [1, 14].
We found that a positive HER2 status, defined by both CISH (p = 0.039) and MLPA (p = 0.015), was correlated with a decreased 5 year survival rate. HER2 amplification was relatively rare (5.8 %) and, by itself, not found to serve as a predictor of survival in univariate survival analyses. This notion may be due to the relatively small number of cases showing HER2 amplification. Copy number gain of NEUROD2 was also found to be correlated with a decreased 5 year survival (p = 0.015), as was whole 17q arm gain (p = 0.010). In multivariate Cox regression analyses, NEUROD2 copy number gains exhibited an independent prognostic value next to tumor size. NEUROD2 itself, coding for a protein that plays a role in neuronal differentiation and neuronal cell fate, is unlikely to be involved in breast cancer prognosis. Therefore, other neighbouring genes are more likely to act as drivers of this amplicon. Some candidate genes such as MED1, MED24 and DARPP-32 are located near the NEUROD2 gene. MED1 encodes a subunit of the master transcriptional co-regulator Mediator/TRAP co-activator complex [29] and is a key ERα co-activator [30, 31]. The MED1/MED24 complex was previously found to be frequently and simultaneously over-expressed in FBC and to play an important role in the growth of breast cancer cells via the RAS-mitogen-activated protein kinase (MAPK) pathway [32]. Several studies have indicated that the MED1 gene may be located within the HER2 amplicon [27, 28] and that it may play a key role in HER2-mediated tamoxifen resistance [33]. The neighboring DARPP-32 gene codes for a Dopamine and cAMP-regulated phosphoprotein, and its over-expression has been implicated in resistance to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptor targeted therapy in cancer [34]. It would be interesting to further assess their copy number status and prognostic value in MBC.
The second amplicon encompasses RPS6KB1, a protein coding gene reported to be amplified and over-expressed in 10-30 % of FBC. We observed RPS6KB1 copy number gains in 30 % of the MBC cases. Its encoded protein, a ribosomal protein S6 kinase, is positioned downstream of the PI3K and mTOR pathways and is involved in protein synthesis, cellular growth and proliferation, which makes it an interesting target for therapy. Further research is, however, needed to clarify the exact role of RPS6KB1 copy number changes in MBC.
The TOP2A gene encodes topoisomerase 2 alpha, a nuclear protein which plays an important role in DNA replication and mitosis. It is the main target of adjuvant anthracycline-based chemotherapy. TOP2A has previously been reported to be frequently amplified in FBC. The prognostic value of TOP2A gene amplification in FBC is as yet, however, controversial [9]. We found a lower percentage of TOP2A copy number gain in MBC compared to FBC (15 % versus 27 %) [14]. Gain or amplification of TOP2A does not seem to be of prognostic value for MBC.
The PPM1D gene, previously reported to be frequently amplified in FBC (25 %), was found to be amplified in a low percentage (2.9 %) of MBC, and to be gained in 16.5 % of MBC in our current study. Gain of PPM1D showed a trend towards a correlation with high grade and high mitotic count, but did not appear to be a predictor of survival, as has been reported before in FBC [11].
In conclusion, we found that MBC is characterized by copy number gains on 17q, with two distinct amplicons, and copy number losses on 17p. Like in FBC, no whole chromosome 17 polysomy was found. MBC shows a similar, but less complex pattern of chromosome 17 rearrangements and fewer copy number changes than FBC. These results suggest a different role of chromosome 17 in male and female breast cancer development. Whole arm copy number gain of 17q was associated with HER2 copy number gain. HER2 and NEUROD2 copy number gains were found to be associated with a high tumor grade, a high mitotic count and a decreased 5 year survival rate. NEUROD2 copy number gain seems to serve as an independent prognostic factor, but is unlikely to be a driver of the associated amplicon. Further research is needed to assess copy number changes in neighboring genes, including their putative prognostic/therapeutic role in MBC.
Electronic supplementary material
(DOCX 335 kb)
(DOCX 19 kb)
Acknowledgments
We thank Dr. Bernd Hinrichs, Institute of Pathology and Cytological Diagnostics, Cologne for providing patient material. We also thank Erwin van der Biezen, Remco Radersma, Marja van Blokland and Roel de Weger for their technical support and IKNL for providing survival data.
Ethical Standards
Use of anonymous or coded left-over material for scientific purposes is part of the standard treatment contract with patients and, therefore, informed consent was not required according to our institutional review board and Dutch legislation.
Conflict of interest
The authors declare no conflict of interest.
Author’s contribution
PD conceived of the study and drafted the manuscript. ML and RK carried out the molecular genetic studies. ML performed the statistical analysis and drafted the manuscript. CM participated in the design and coordination of the study and drafted the manuscript. CP, AW, EW, JR and HB participated in the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Declaration of interest and Funding
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
References
- 1.Kornegoor R, Moelans CB, Verschuur-Maes AH, Hogenes MC, de Bruin PC, Oudejans JJ, Marchionni L, van Diest PJ. Oncogene amplification in male breast cancer: analysis by multiplex ligation-dependent probe amplification. Breast Cancer Res. Treat. 2012;135:49–58. doi: 10.1007/s10549-012-2051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.S. Tommasi, A. Mangia, G. Iannelli, P. Chiarappa, E. Rossi, L. Ottini, M. Mottolese, W. Zoli, O. Zuffardi, A. Paradiso, Gene copy number variation in male breast cancer by aCGH. Anal. Cell. Pathol. 33, 113–119 (2010) [DOI] [PMC free article] [PubMed]
- 3.S. Tabarestani, S.M. Ghaderian, H. Rezvani, R. Mirfakhraie, A. Ebrahimi, H. Attarian, J. Rafat, M. Ghadyani, H.A. Alavi, N. Kamalian, A. Rakhsha, E. Azargashb, Prognostic and predictive value of copy number alterations in invasive breast cancer as determined by multiplex ligation-dependent probe amplification. Cell Oncol. 37, 107–118 (2014) [DOI] [PubMed]
- 4.A.H. Verschuur-Maes, C.B. Moelans, P.C. de Bruin, P.J. van Diest, Analysis of gene copy number alterations by multiplex ligation-dependent probe amplification in columnar cell lesions of the breast. Cell Oncol. 37, 147–154 (2014) [DOI] [PubMed]
- 5.A. Halon, P. Donizy, P. Surowiak, R. Matkowski, ERM/Rho protein expression in ductal breast cancer: a 15 year follow-up. Cell Oncol. 36, 181–190 (2013) [DOI] [PMC free article] [PubMed]
- 6.Moelans CB, de Weger RA, Monsuur HN, Vijzelaar R, van Diest PJ. Molecular profiling of invasive breast cancer by multiplex ligation-dependent probe amplification-based copy number analysis of tumor suppressor and oncogenes. Mod. Pathol. 2010;23:1029–1039. doi: 10.1038/modpathol.2010.84. [DOI] [PubMed] [Google Scholar]
- 7.J.P. Baak, D. Chin, P.J. van Diest, R. Ortiz, P. Matze-Cok, S.S. Bacus, Comparative long-term prognostic value of quantitative HER-2/neu protein expression, DNA ploidy, and morphometric and clinical features in paraffin-embedded invasive breast cancer. Lab. Invest. 64, 215–223 (1991) [PubMed]
- 8.Hudis CA. Trastuzumab–mechanism of action and use in clinical practice. N. Engl. J. Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 9.Jacot W, Fiche M, Zaman K, Wolfer A, Lamy PJ. The HER2 amplicon in breast cancer: Topoisomerase IIA and beyond. Biochim. Biophys. Acta. 2013;1836:146–157. doi: 10.1016/j.bbcan.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Zaczek A, Markiewicz A, Supernat A, Bednarz-Knoll N, Brandt B, Seroczynska B, Skokowski J, Szade J, Czapiewski P, Biernat W, Welnicka-Jaskiewicz M, Jassem J. Prognostic value of TOP2A gene amplification and chromosome 17 polysomy in early breast cancer. Pathol. Oncol. Res. 2012;18:885–894. doi: 10.1007/s12253-012-9518-8. [DOI] [PubMed] [Google Scholar]
- 11.Lambros MB, Natrajan R, Geyer FC, Lopez-Garcia MA, Dedes KJ, Savage K, Lacroix-Triki M, Jones RL, Lord CJ, Linardopoulos S, Ashworth A, Reis-Filho JS. PPM1D gene amplification and overexpression in breast cancer: a qRT-PCR and chromogenic in situ hybridization study. Mod. Pathol. 2010;23:1334–1345. doi: 10.1038/modpathol.2010.121. [DOI] [PubMed] [Google Scholar]
- 12.Moelans CB, Reis-Filho JS, van Diest PJ. Implications of rarity of chromosome 17 polysomy in breast cancer. Lancet Oncol. 2011;12:1087–1089. doi: 10.1016/S1470-2045(11)70234-0. [DOI] [PubMed] [Google Scholar]
- 13.Marchio C, Lambros MB, Gugliotta P, Di Cantogno LV, Botta C, Pasini B, Tan DS, Mackay A, Fenwick K, Tamber N, Bussolati G, Ashworth A, Reis-Filho JS, Sapino A. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. J. Pathol. 2009;219:16–24. doi: 10.1002/path.2574. [DOI] [PubMed] [Google Scholar]
- 14.Moelans CB, de Weger RA, van Diest PJ. Absence of chromosome 17 polysomy in breast cancer: analysis by CEP17 chromogenic in situ hybridization and multiplex ligation-dependent probe amplification. Breast Cancer Res. Treat. 2010;120:1–7. doi: 10.1007/s10549-009-0539-2. [DOI] [PubMed] [Google Scholar]
- 15.Yeh IT, Martin MA, Robetorye RS, Bolla AR, McCaskill C, Shah RK, Gorre ME, Mohammed MS, Gunn SR. Clinical validation of an array CGH test for HER2 status in breast cancer reveals that polysomy 17 is a rare event. Mod. Pathol. 2009;22:1169–1175. doi: 10.1038/modpathol.2009.78. [DOI] [PubMed] [Google Scholar]
- 16.Muir D, Kanthan R, Kanthan SC. Male versus female breast cancers. A population-based comparative immunohistochemical analysis. Arch. Pathol. Lab. Med. 2003;127:36–41. doi: 10.5858/2003-127-36-MVFB. [DOI] [PubMed] [Google Scholar]
- 17.Fonseca RR, Tomas AR, Andre S, Soares J. Evaluation of ERBB2 gene status and chromosome 17 anomalies in male breast cancer. Am. J. Surg. Pathol. 2006;30:1292–1298. doi: 10.1097/01.pas.0000213354.72638.bd. [DOI] [PubMed] [Google Scholar]
- 18.Bloom KJ, Govil H, Gattuso P, Reddy V, Francescatti D. Status of HER-2 in male and female breast carcinoma. Am. J. Surg. 2001;182:389–392. doi: 10.1016/S0002-9610(01)00733-4. [DOI] [PubMed] [Google Scholar]
- 19.Rudlowski C, Schulten HJ, Golas MM, Sander B, Barwing R, Palandt JE, Schlehe B, Lindenfelser R, Moll R, Liersch T, Schumpelick V, Gunawan B, Fuzesi L. Comparative genomic hybridization analysis on male breast cancer. Int. J. Cancer. 2006;118:2455–2460. doi: 10.1002/ijc.21646. [DOI] [PubMed] [Google Scholar]
- 20.Kornegoor R, Verschuur-Maes AH, Buerger H, Hogenes MC, de Bruin PC, Oudejans JJ, van der Groep P, Hinrichs B, van Diest PJ. Molecular subtyping of male breast cancer by immunohistochemistry. Mod. Pathol. 2012;25:398–404. doi: 10.1038/modpathol.2011.174. [DOI] [PubMed] [Google Scholar]
- 21.Kornegoor R, Moelans CB, Verschuur-Maes AH, Hogenes MC, de Bruin PC, Oudejans JJ, van Diest PJ. Promoter hypermethylation in male breast cancer: analysis by multiplex ligation-dependent probe amplification. Breast Cancer Res. 2012;14:R101. doi: 10.1186/bcr3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 23.van Diest PJ, Baak JP, Matze-Cok P, Wisse-Brekelmans EC, van Galen CM, Kurver PH, Bellot SM, Fijnheer J, van Gorp LH, Kwee WS, et al. Reproducibility of mitosis counting in 2,469 breast cancer specimens: results from the Multicenter Morphometric Mammary Carcinoma Project. Hum. Pathol. 1992;23:603–607. doi: 10.1016/0046-8177(92)90313-R. [DOI] [PubMed] [Google Scholar]
- 24.Moelans CB, de Weger RA, Ezendam C, van Diest PJ. HER-2/neu amplification testing in breast cancer by Multiplex Ligation-dependent Probe Amplification: influence of manual- and laser microdissection. BMC Cancer. 2009;9:4. doi: 10.1186/1471-2407-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunyan DJ, Eccles DM, Sillibourne J, Wilkins E, Thomas NS, Shea-Simonds J, Duncan PJ, Curtis CE, Robinson DO, Harvey JF, Cross NC. Dosage analysis of cancer predisposition genes by multiplex ligation-dependent probe amplification. Br. J. Cancer. 2004;91:1155–1159. doi: 10.1038/sj.bjc.6602121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hungermann D, Schmidt H, Natrajan R, Tidow N, Poos K, Reis-Filho JS, Brandt B, Buerger H, Korsching E. Influence of whole arm loss of chromosome 16q on gene expression patterns in oestrogen receptor-positive, invasive breast cancer. J. Pathol. 2011;224:517–528. doi: 10.1002/path.2938. [DOI] [PubMed] [Google Scholar]
- 27.Luoh SW. Amplification and expression of genes from the 17q11 approximately q12 amplicon in breast cancer cells. Cancer Genet. Cytogenet. 2002;136:43–47. doi: 10.1016/S0165-4608(01)00657-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Qi C, Jain S, Le Beau MM, Espinosa R, 3rd, Atkins GB, Lazar MA, Yeldandi AV, Rao MS, Reddy JK. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10848–10853. doi: 10.1073/pnas.96.19.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Warnmark A, Almlof T, Leers J, Gustafsson JA, Treuter E. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERalpha and ERbeta. J. Biol. Chem. 2001;276:23397–23404. doi: 10.1074/jbc.M011651200. [DOI] [PubMed] [Google Scholar]
- 31.Kang YK, Guermah M, Yuan CX, Roeder RG. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasegawa N, Sumitomo A, Fujita A, Aritome N, Mizuta S, Matsui K, Ishino R, Inoue K, Urahama N, Nose J, Mukohara T, Kamoshida S, Roeder RG, Ito M. Mediator subunits MED1 and MED24 cooperatively contribute to pubertal mammary gland development and growth of breast carcinoma cells. Mol. Cell. Biol. 2012;32:1483–1495. doi: 10.1128/MCB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui J, Germer K, Wu T, Wang J, Luo J, Wang SC, Wang Q, Zhang X. Cross-talk between HER2 and MED1 regulates tamoxifen resistance of human breast cancer cells. Cancer Res. 2012;72:5625–5634. doi: 10.1158/0008-5472.CAN-12-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belkhiri A, Zhu S, Chen Z, Soutto M, El-Rifai W. Resistance to TRAIL is mediated by DARPP-32 in gastric cancer. Clin. Cancer Res. 2012;18:3889–3900. doi: 10.1158/1078-0432.CCR-11-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 335 kb)
(DOCX 19 kb)


