Abstract
High expression of Notch-1 and Jagged-1 mRNA correlates with poor prognosis in breast cancer. Elucidating the cross-talk between Notch and other major breast cancer pathways is necessary to determine which patients may benefit from Notch inhibitors, which agents should be combined with them, and which biomarkers indicate Notch activity in vivo. We explored expression of Notch receptors and ligands in clinical specimens, as well as activity, regulation, and effectors of Notch signaling using cell lines and xenografts. Ductal and lobular carcinomas commonly expressed Notch-1, Notch-4, and Jagged-1 at variable levels. However, in breast cancer cell lines, Notch-induced transcriptional activity did not correlate with Notch receptor levels and was highest in estrogen receptor α–negative (ERα–), Her2/Neu nonoverexpressing cells. In ERα+ cells, estradiol inhibited Notch activity and Notch-1IC nuclear levels and affected Notch-1 cellular distribution. Tamoxifen and raloxifene blocked this effect, reactivating Notch. Notch-1 induced Notch-4. Notch-4 expression in clinical specimens correlated with proliferation (Ki67). In MDA-MB231 (ERα–) cells, Notch-1 knockdown or γ-secretase inhibition decreased cyclins A and B1, causing G2 arrest, p53-independent induction of NOXA, and death. In T47D:A18 (ERα+) cells, the same targets were affected, and Notch inhibition potentiated the effects of tamoxifen. In vivo, γ;-secretase inhibitor treatment arrested the growth of MDA-MB231 tumors and, in combination with tamoxifen, caused regression of T47D:A18 tumors. Our data indicate that combinations of antiestrogens and Notch inhibitors may be effective in ERα+ breast cancers and that Notch signaling is a potential therapeutic target in ERα– breast cancers.
Introduction
Notch receptors regulate cell differentiation, proliferation, and apoptosis during intercellular contact, in response to ligands of the Delta and Jagged/Serrate families (1). Mature Notch receptors are heterodimers composed of a transmembrane subunit (N™) associated with an extracellular subunit (NEC). On ligand binding, N™ is cleaved by an ADAM metalloproteinase and γ-secretase. This releases a cytoplasmic subunit (NIC), which activates CSL transcription factors, modulating cell fate decisions (1).
Deregulated expression of wild-type Notch receptors, ligands, and targets has been described in several solid tumors (1–3). Approximately 50% of T-cell lymphoblastic leukemias (T-ALL) carry activating Notch-1 mutations (4). Mutations in ubiquitin ligase FBW7/SEL10/CDC4 also increase Notch activity in T-ALL (5–7). Intracellular forms of Notch receptors have transforming activity in vitro (8) and in vivo (9–11).
Constitutive activation of Notch-1 (12, 13) or Notch-4 (11) causes mammary carcinogenesis in mice. In human breast cancer, high-level expression of Notch-1 and Jagged-1 correlates with poor prognosis (14, 15). Aberrant Notch-1 activation was reported in 20 breast carcinomas of various subtypes (16). These authors showed that Notch-1 NIC transforms MCF-10 cell and protects transformed cells from p53-mediated induction of proapoptotic protein NOXA. Pece and colleagues (17) suggested that Notch negative regulator Numb is lost in ~50% of human breast carcinomas, leading to increased Notch signaling. We showed that activation of Notch-1 maintains the neoplastic phenotype and induces Notch-4 in Ras-transformed human fibroblasts and kidney epithelial cells (18). In the same study, we detected expression of Notch-1 in seven cases of infiltrating ductal carcinoma, which seemed stronger in H-Ras–overexpressing cases (18). We investigated the expression of Notch pathway components in human breast cancers and the regulation of Notch expression and activity by a novel cross-talk mechanism with estrogen. We present data supporting the hypothesis that Notch inhibitors may be effective in estrogen receptor α–negative (ERα–) breast cancers and that combinations including an antiestrogen and a Notch inhibitor may be effective in ERα+ disease.
Materials and Methods
Clinical specimens and immunohistochemistry
Archival formalin-fixed, paraffin-embedded blocks from the Breast Pathology Divisions, Department of Pathology, Loyola University Chicago and Brigham and Women's Hospital were studied. No patient identifiers were used. See Supplementary Data for immunohistochemistry details.
Cell lines and constructs
MCF-7 and MDA-MB231 cells were from the American Type Culture Collection. T47D:C42 and T47D:A18 cells were from Dr. Debra A. Tonetti (University of Illinois at Chicago, Chicago, IL). Human mammary epithelial cells (HMEC) were from Clonetics. See Supplementary Data for medium details. For coculture experiments, breast cancer cells were cocultured with mouse OP9 stromal cells overexpressing Notch ligand Delta-1 or green fluorescent protein (GFP)-expressing OP9 negative controls (gifts of Dr. Juan-Carlos Zuniga-Pflucker, University of Toronto, Toronto, Ontario, Canada). Notch constructs and CBF-1 luciferase reporter assays have been described (18). The pKA9-CBF-1 used for stable transfection of T47D:A18 (19) was a gift of Dr. Dmitry Gabrilovich (University of South Florida, Tampa, FL). The full-length Notch-1 tagged at the COOH terminus with Renilla luciferase (20) was a gift of Dr. Raphael Kopan (Washington University, St. Louis, MO).
Drugs and chemicals
γ-Secretase inhibitor cbz-Leu-Leu-Nle-CHO (GSI; Calbiochem) was dissolved in DMSO, aliquoted, and stored at –80°C. 17β-Estradiol, 4-hydroxytamoxifen (4-OH-TAM; both from Sigma-Aldrich), fulvestrant (ICI182,780; Tocris Bioscience), and raloxifene (kindly donated by Dr. Judy Bolton, University of Illinois at Chicago) were dissolved in ethanol and stored in aliquots at –80°C.
RNA interference
Double-stranded synthetic 21-mer RNA oligonucleo-tides (siRNA) were from Dharmacon and Santa Cruz Biotechnology. siRNA effectiveness was validated by Western blotting and real-time reverse transcription-PCR (RT-PCR; data not shown). siRNA from Dharmacon was synthesized based on sequences we identified. The most effective sequences selected in pilot experiments were the following: Notch-1, 5¶-AAGTGTCTGAGGCCAGCAAGA-3¶; Notch-4, 5¶-AACCCTGTGCCAATGGAGGCA-3¶. A control siRNA that does not match any known mammalian Genbank sequences (Dharmacon) was used in all experiments: 5¶-AACAGTCGCGTTTGCGACTGG-3¶. See Supplementary Data for additional details.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer containing protease inhibitors. See Supplementary Data for additional details. Antibodies used were the following: Notch-1 (C-20), Notch-4 (L-16), cyclin B (H-433), and c-Myc (9E10; Santa Cruz Biotechnology); cyclin A (BF68; Cell Signaling); phospho-histone 3 (Ser10; Cell Signaling); and NOXA (Calbiochem).
Transfection and luciferase assays
Cells were transfected using either FuGene (Roche Diagnostic) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For cells grown in charcoal-stripped medium, electroporation was used to ensure high efficiency of transfection. Luciferase assay was performed using the Dual Luciferase Assay by Promega using pRL-TK (Renilla luciferase) as internal control.
Proliferation, growth inhibition, and chemoinvasion assays
Growth of attached cells was estimated by a standard assay used for cancer drug screening by either sulforhodamine (21) or crystal violet (22) staining of fixed cells. BioCoat Matrigel invasion chambers (Becton Dickinson) were used to assay matrix invasion following the manufacturer's instructions. See Supplementary Data for details.
Cell cycle analysis
Cells (106) were washed twice in PBS and then fixed in 80% methanol. Fixed cells were washed twice in PBS and resuspended in 50 μL propidium iodide (50 μg/mL in PBS). After RNase A (100 μg/mL) treatment at 37°C for 1 h, cells were analyzed by flow cytometry using a FACSCalibur instrument.
Real-time RT-PCR
Total RNA was extracted using a commercially available kit (Qiagen) and analyzed by real-time RT-PCR for mRNA for Notch-1 and HEY1. See Supplementary Data for details.
RNase protection assays
Total RNA was extracted using a commercially available kit (Qiagen). The assay was performed using the RiboQuant RPA Starter Package (BD Biosciences) according to the manufacturer's protocol.
Biotinylation experiments
T47D:A18 cell surface proteins were biotinylated in the presence or absence of 17β-estradiol. The amount of Notch-1 on cell membrane was then measured by immunoprecipitation of the biotinylated form of Notch followed by Western blot. See Supplementary Data for additional details.
Confocal microscopy
T47D:A18 and MCF-7 cells were grown on Lab-Tek II chamber slides (Nunc) for 48 h in charcoal-stripped serum plus 1 nmol/L estradiol or 1 μmol/L 4-OH-TAM. Cells were rinsed twice in PBS and fixed in cold methanol. Staining for Notch-1 was performed using 1 μg/mL C-20 antibody (Santa Cruz Biotechnology) and 5 μg/mL of Alexa Fluor 488–labeled anti-rabbit IgG (Invitrogen). Detection was performed with a Zeiss LSM 510 confocal microscope using a C-Apochromat 63×/1.2 W correlation objective.
Statistical analysis
(a) Preclinical studies: For pairwise comparisons, two-tailed unpaired Student's t tests were used with α = 0.05. When more than two samples were compared, one-way ANOVA was used (Student-Newman-Keuls method for multiple comparisons) with α = 0.05. SigmaStat software (Jandel Scientific) was used for preclinical statistical analysis. (b) Clinical studies: Associations involving two-class variables with multilevel variables were performed using a χ2 test and Fisher's exact test statistic. For the analysis of the association between Notch-1 and Notch-4 staining with other variables, we calculated the least square means for each level of Notch staining. Data were parsed into three levels (low, medium, and intense staining). Staining distribution was distinguished into cytoplasmic, nuclear, and membrane. We found a significant association between Ki67 staining and cytoplasmic Notch-4 (P < 0.02). The least square mean values for Ki67 for low, medium, and intense staining were 13.50, 35.00, and 13.20, respectively. The association between Ki67 and Notch-1 staining was only marginal (P = 0.06), with least square mean values for Ki67 breast of 15.83, 32.50, and 33.00 for the low, medium, and intense staining, respectively. Clinical statistical analyses were performed using the Statistical Analysis System software package.
Promoter analysis
The analysis of the cyclin A and cyclin B promoters for nuclear factor-κB (NF-κB) motifs was performed using Motif Search and AliBaba2.1.
Xenograft experiments
Ovariectomized 4- to 6-week-old BALB/c athymic mice (The Jackson Laboratory) were injected s.c. into both axillary mammary fat pads with either 5 × 106 MDA-MB231 cells or 1 × 107 T47D:A18 cells. Tumor size was determined weekly. At the end of treatment, animals were sacrificed, and the tumors were removed and weighed for use in histology.
Results
Notch-1 and Notch-4 expression in breast cancers
We examined normal breast compared with hyperplastic, early, and late neoplastic lesions by immunohistochemistry (see Supplementary Data and Supplementary Table S1 for details). All 41 infiltrating carcinomas (27 ductal and 14 lobular) were positive to some degree for Notch-1 and Notch-4, and at least one Notch ligand. We defined cases as “low” (staining intensity 1–2+) or “high” (3+) expression. Twenty-four of 27 ductal infiltrating carcinomas were Notch-1 high and 3 were Notch-1 low. Twenty-two of 27 were Notch-4 high and 5 were Notch-4 low. Twenty-one were Jagged-1 high. Delta-1 expression showed lower staining intensity, resulting in 21 Delta-1 low and 6 Delta-1 high. Eight of 14 lobular infiltrating carcinomas were Notch-1 high and 6 were Notch-1 low. Thirteen of 14 tumors in this group were Notch-4 high. Nine tumors were Jagged-1 high and 5 were Notch-4 low, whereas for Delta-1 these proportions were reversed.
Breast cancer cell lines have variable levels of CBF-1 transcriptional activity
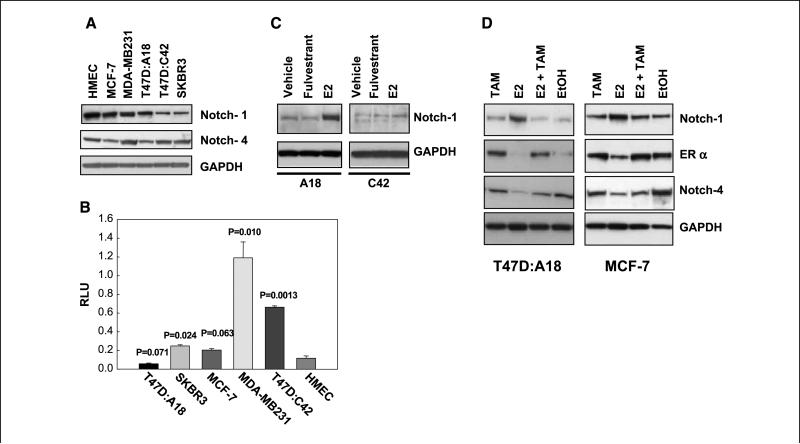
Consistent with the literature (16), Western blot analysis for total Notch-1 with a COOH-terminal antibody showed that several breast cancer cell lines, but also nontransformed HMECs proliferating in culture, had strong Notch-1 expression (Fig. 1A). Notch-4 was also expressed in HMEC and breast cancer cells. Breast cancer cell lines MCF-7, MDA-MB231, T47D ERα– subclone C42 (23), and T47D ERα+ subclone A18 (23) and SKBR3 expressed variable steady-state levels of Notch-1 and Notch-4 proteins. However, when we examined basal CBF-1–dependent reporter activity, there was no obvious correlation between steady-state levels of Notch proteins and Renilla-normalized transcriptional activity, suggesting that factors other than expression levels of Notch-1 and Notch-4 regulate Notch-dependent reporter activity. ERα–, Her2/Neu nonoverexpressing lines MDA-MB231 and T47D:C42 consistently had much higher activity than ERα+ lines MCF-7, T47D:A18, Her2/Neu overexpressing line SKBR3, or HMEC (Fig. 1B). This suggested that there may be an inverse correlation between ERa or Her2/Neu and Notch activity in breast cancer cell lines. Therefore, we examined the role of estrogen in regulating Notch activity. The cross-talk between Her2/Neu and Notch will be described elsewhere.8
Figure 1.
Breast cancer cell lines have variable levels of CBF-1 transcriptional activity; estrogen up-regulates Notch-1 but decreases Notch-4 expression. A, Western blots showing expression of Notch-1 and Notch-4 in a panel of breast cancer cell lines as well as HMEC controls. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, CBF-1 luciferase activities of the breast cancer cell lines shown in A and HMEC controls. Cells were transfected simultaneously (n = 3) at comparable levels of confluence and tested side by side. Data are normalized for efficiency of transfection using a Renilla luciferase plasmid. Columns, mean; bars, SD. Data are representative of at least three independent experiments. RLU, relative luciferase unit. C and D, Western blots. C, 10–9 mol/L 17β-estradiol (E2) in steroid-free medium for 72 h up-regulated Notch-1 in T47D:A18 but not T47D:C42 cells. Results represent five independent experiments. D, 10–9 mol/L 17β-estradiol in steroid-free medium for 72 h up-regulated Notch-1 but decreased Notch-4 in either MCF-7 or T47D:A18. 4-OH-TAM (1 μmol/L) antagonized both effects. Results represent three independent experiments. EtOH, ethanol.
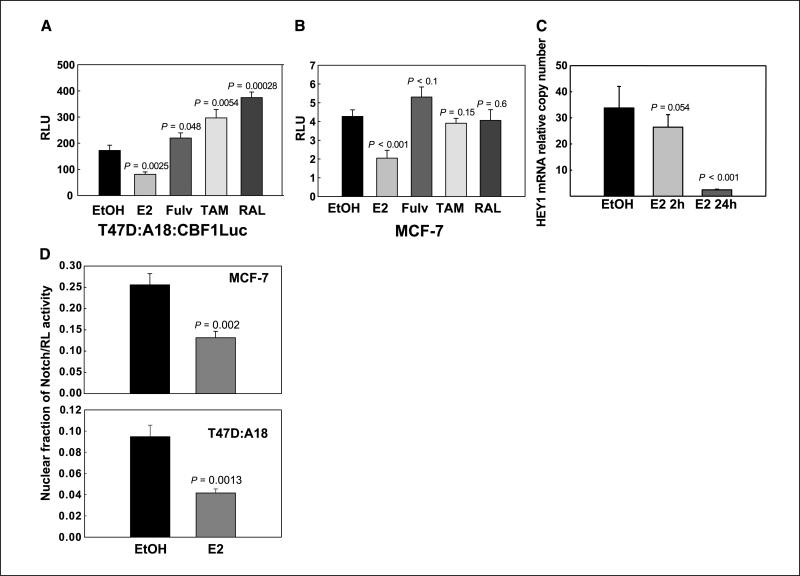
Estradiol inhibits Notch activity and selective ER modulators block its effect
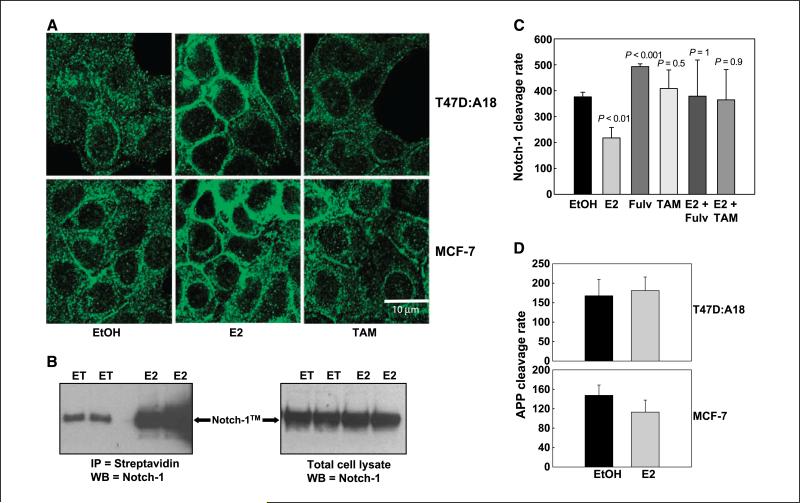
Treatment of ERα+ T47D:A18 cells with 10–9 mol/L estradiol in charcoal-stripped medium strongly up-regulated Notch-1 protein (Fig. 1C) but barely affected it in ERα– T47D:C42 cells. Real-time RT-PCR experiments indicated that these effects are not mediated by transcriptional regulation (data not shown). In two different ERα+ lines, T47D:A18 and MCF-7, treatment with 10–9 mol/L estradiol inhibited Notch-4 expression compared with vehicle (Fig. 1D). These effects were antagonized by 1 μmol/L 4-OH-TAM. Because Notch-4 is a transcriptional target of Notch-1 (18), we hypothesized that estradiol inhibits Notch transcriptional activity while accumulating uncleaved Notch-1 protein. Reporter experiments supported this hypothesis. In T47D:A18 cells stably expressing a CBF-1 reporter, estradiol consistently and significantly (P = 0.0025) inhibited reporter activity, whereas pure antagonist fulvestrant or selective ER modulators (SERM) tamoxifen or raloxifene did not (Fig. 2A). Virtually identical results were obtained in MCF-7 cells transiently transfected with the same reporter (Fig. 2B). We tested Notch activation under physiologic conditions by coculturing MCF-7 cells with mouse OP9 stromal cells expressing Notch ligand Delta-1 or GFP (control). In real-time RT-PCR experiments, Delta-1–expressing stromal cells dramatically increased the expression of Notch target gene HEY1 compared with control stromal cells. Treatment with estradiol for 24 h nearly abolished this effect (P < 0.001; Fig. 2C). Two-hour treatment had a much more modest effect, suggesting that the effect of estradiol on HEY1 expression is indirect. This indicated that estrogen suppresses Notch-1–dependent transcription in ERα+ cells even as it increases the amount of Notch-1 protein. Therefore, we determined nuclear levels of cleaved Notch-1 by using a full-length Notch-1 construct tagged at the COOH terminus with Renilla luciferase (20), which requires a ligand for activation. Figure 2D shows that estradiol significantly decreases the ratio between nuclear and total Notch-1/Renilla luciferase activity in both T47D:A18 (P = 0.01) and MCF-7 cells (P = 0.002). We used confocal immunofluorescence to morphologically define the cellular distribution of Notch-1 in T47D:A18 and MCF-7 cells. Figure 3A shows that in both MCF-7 and T47D:A18 cells under estrogen deprivation conditions in charcoal-stripped medium, Notch-1 could be detected as a punctate signal distributed to cytoplasm and nucleus. Estradiol caused a remarkable redistribution of the signal, which was relatively depleted from the nucleus and showed striking cell membrane accumulation with a “chicken-wire” pattern. 4-OH-TAM treatment produced a pattern similar to control. To further examine the effects of estradiol on Notch-1 cellular distribution, we conducted cell surface biotinylation experiments (Fig. 3B). Cells were treated with vehicle or estradiol for 48 h, a time before significant accumulation of Notch-1 can be detected. Surface proteins were biotinylated with a cell-impermeable reagent, cells were lysed, and equal amounts of total cellular proteins were precipitated with conjugated streptavidin and analyzed by Western blotting for total Notch-1. Under these conditions, estradiol dramatically increased the amount of biotinylated (and thus cell surface associated) Notch-1 compared with vehicle. This difference was much larger than the difference in total amounts of Notch-1 as detected by Western blotting on the same lysates. These data indicated that estrogen-treated cells accumulate inactive Notch at the cell surface.
Figure 2.
Estradiol decreases Notch transcriptional activity and Notch nuclear localization in breast cancer cells. A, luciferase assay showing inhibition of CBF-1 transcriptional activity by 48-h treatment with 10–9 mol/L 17β-estradiol in T47D:A18:CBF-1Luc (stably transfected with the firefly luciferase gene under the control of six CBF-1–responsive elements). 4-OH-TAM (10–6 mol/L), raloxifene (10–6 mol/L), or fulvestrant (10–7 mol/L) did not inhibit CBF-1 activity. B, luciferase assay showing inhibition by 24-h treatment with 10–9 mol/L 17β-estradiol of CBF-1 transcriptional activity in MCF-7 transiently transfected with a plasmid containing the firefly luciferase gene under the control of four CBF-1–responsive elements. Renilla luciferase was used to normalize for transfection efficiency. 4-OH-TAM (10–6 mol/L), raloxifene (10–6 mol/L), and fulvestrant (10–7 mol/L) did not inhibit CBF-1 activity. C, real-time RT-PCR showing the relative amount of HEY1 mRNA in MCF-7 cells cocultured for 5 d with OP9 cells expressing Delta-1 with or without 10 nmol/L 17β-estradiol for 2 and 24 h. Data are expressed as fold increase compared with MCF-7 cells cocultured in parallel with control OP9-GFP cells. D, MCF-7 (top) and T47D:A18 (bottom) cells were transfected with a construct coding for full-length Notch-1 fused to Renilla luciferase. Twenty-four hours after transfection, the medium was replaced with hormone-free medium and cells were treated with 10–9 mol/L estradiol for 24 h. Nuclei were isolated and the ratio of Notch-Renilla luciferase in the nuclei versus total cellular Notch-Renilla luciferase was measured. Columns, mean; bar, SD. Data represent three independent experiments.
Figure 3.
Estradiol regulates the cellular localization of Notch-1. A, confocal immunofluorescence showing total cellular Notch-1 staining (green) in MCF-7 and T47D:A18 after treatment with 10–9 mol/L 17β-estradiol, 10–6 mol/L 4-OH-TAM, or vehicle. All pictures were taken at the same magnification. B, left, Western blot showing the amount of biotinylated Notch-1 present on the cell membrane of T47D:A18 with or without 48-h treatment with 10–7 mol/L 17β-estradiol. Similar results were obtained with 10–9 mol/L estradiol and in MCF-7 cells with both treatments. Results of one of two independent experiments are shown. Right, Western blot showing the total amount of Notch-1 in the same cells with or without estradiol treatment. C, T47D:A18 cells were grown in charcoal-stripped medium for 3 d and then transfected by electroporation with GAL4 luciferase reporter and DNA plasmid coding fusion protein GAL4VP16-Notch1ΔE. After transfection, cells were treated for 48 h with hormones. Twenty-four hours before lysing the cells, 25 μmol/L lactacystin was added to inhibit Notch degradation. The experiment was repeated at least thrice. Columns, mean; bars, SD. D, T47D:A18 and MCF-7 cells were grown in charcoal-stripped medium for 3 d and then transfected by electroporation with GAL4 luciferase reporter and DNA plasmid encoding fusion protein GAL4VP16-APPc99 (24). After transfection, cells were treated for 48 h with 10–9 mol/L 17β-estradiol and UAS luciferase activity was measured. The experiment was repeated at least thrice. Columns, mean; bars, SD.
Overall, our data indicate that estrogen inhibits Notch-1 activity at least in part by altering the cellular distribution of Notch-1. Thus, we examined whether estradiol affects the cleavage rate of Notch-1 by γ-Secretase using a quantitative assay (24) in which a Notch-1 N™ construct tagged with GAL4/VP16 is transiently cotransfected with an upstream activation sequence (UAS) reporter plasmid. Activation of the UAS reporter by GAL4/VP16 depends on cleavage of the Notch chimeric protein by γ-Secretase but does not require a ligand. Figure 3C shows that estradiol significantly inhibited Notch activation as measured by this assay (P < 0.01). Fulvestrant modestly stimulated Notch activation, whereas 4-OH-TAM had no significant effects by itself. Both fulvestrant and 4-OH-TAM abrogated estrogen-mediated inhibition. Thus, the activation and nuclear localization of either ligand-dependent or ligand-independent Notch-1 constructs is inhibited by estradiol. These data suggest that estrogen affects a step of Notch activation distal to ligand binding and directly or indirectly inhibits Notch cleavage. This effect is most likely an indirect consequence of the cellular redistribution of Notch caused by estrogen because cleavage of a βh-amyloid substrate under identical assay conditions was not significantly affected by estrogen in either MCF-7 or T47D:A18 cells (Fig. 3D).
Notch-1 and Notch-4 stimulate proliferation and invasion in breast cancer cell lines
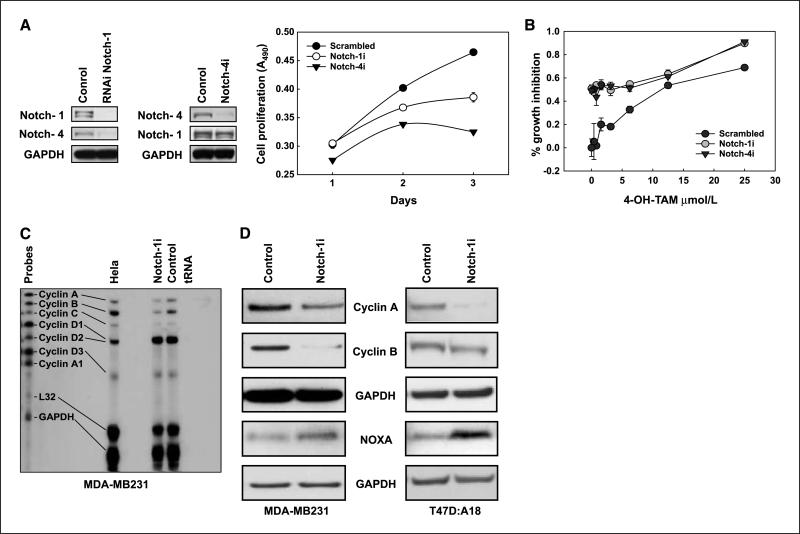
Our data suggested a model in which estrogen inhibits Notch via ERa. ERα– breast cancer cell lines have unfettered Notch activity and are presumably Notch dependent. ERα+ breast cancer cell lines increase Notch activity when estrogen deprived or when estrogen is antagonized by SERMs, and may become more dependent on Notch under these conditions. We tested this hypothesis by siRNA knockdown. In ERα– MDA-MB231 cells, knockdown of Notch-1 or Notch-4 had strong antiproliferative effects (Fig. 4A). Notch-1 knockdown also down-regulated Notch-4, whereas Notch-4 silencing had no effect on Notch-1, indicating that Notch-1 is upstream of Notch-4 in these cells. Additionally, Notch-1 or Notch-4 knockdown significantly inhibited Matrigel invasion (Supplementary Fig. S2A), whereas transfecting Notch-1IC into MCF-7 cells up-regulated Notch-4, increased proliferation, and induced Matrigel invasion (Supplementary Fig. S2B–D). These data suggest that Notch-4 either is necessary for the effects of Notch-1 in these cells or has oncogenic downstream pathways independent of Notch-1. Similarly, in ERα+ T47D:A18 cells, siRNA knockdown of either Notch-1 or Notch-4 caused significant growth inhibition, which was enhanced by addition of 4-OH-TAM (0.37–25 μmol/L; Fig. 4B).
Figure 4.
Notch signaling is required for proliferation and survival in breast cancer cell lines and regulates cyclin A, cyclin B1, and NOXA. A, left, Western blots showing that RNAi silencing of Notch-1 in MDA-MB231 cells also down-regulated Notch-4, whereas nearly quantitative silencing of Notch-4 by RNAi had no effects on Notch-1. Control was a siRNA with no homology with known mammalian genes. Right, RNAi silencing of Notch-1 (which also affects Notch-4) or silencing of Notch-4 alone inhibited the proliferation of MDA-MB231 cells. Data are representative of three experiments each conducted in triplicate. SDs where not visible were smaller than data point symbols. B, silencing of either Notch-1 or Notch-4 in T47D:A18 cells inhibits proliferation and potentiates the effects of 4-OH-TAM. T47D:A18 cells were transfected with siRNA to Notch-1 or Notch-4 or scrambled control siRNA. Twenty-four hours after transfection, cells were seeded in a 96-well plate (8,000 per well) and treated with decreasing concentrations of 4-OH-TAM (25–0.38 μmol/L) in 0.5% ethanol for 24 h. At the end of treatment, cytotoxicity was evaluated by crystal violet staining. C, RNase protection assay showing that mRNA levels for cyclins A and B1 were reduced in MDA-MB231 cells transfected with Notch-1 siRNA. L32 (rRNA) and glyceraldehyde-3-phosphate dehydrogenase were internal controls. Additional controls were untreated cells and tRNA (nonspecific protection control). D, Western blots showing that cells transfected with Notch-1 siRNA had dramatically reduced levels of cyclin B1 and cyclin A proteins, as well as increased NOXA expression, compared with controls. Left, MDA-MB231 cells (48 h after transfection); right, T47D:A18 cells (72 h after transfection).
Inhibition of Notch signaling causes G2 growth arrest in breast cancer cells with decreased cyclin A and B levels and induction of NOXA
We investigated the mechanism of the apparent antiproliferative effects of Notch knockdown. In MDAMB231 cells, RNA interference (RNAi) knockdown of Notch-1 or Notch-4 caused accumulation of cells in G2-M and a corresponding decrease of cells in G1 and S (Supplementary Fig. S3A). An increase in the sub-G1 fraction, indicative of cell death, was seen at 48 h. Histone H3 phosphorylation was virtually abolished by Notch-1 silencing, suggesting cell cycle arrest in G2, before mitosis (Supplementary Fig. S3B). S-phase transcription factor E2F-1, which up-regulates its own transcription (25), accumulated after Notch-1 knockdown (Supplementary Fig. S3C), suggesting a deregulated S-G2 transition. With both Notch-1 and Notch-4 RNAi, G2 accumulation was maximal at 24 h, and at 48 h, it was replaced by an increased fraction of cells in sub-G1 (Supplementary Fig. S3D). G2-M accumulation followed by death was described on Notch inhibition in mesothelioma (26), melanoma (27), and Kaposi sarcoma (28). We therefore explored the effects of Notch-1 siRNA on the expression of cyclins, some of which were described as Notch targets. RNase protection data (Fig. 4C) indicated that Notch-1 knockdown in MDA-MB231 cells decreases mRNA expression for cyclins A and B1, but not C, or D. Western blot data confirmed these findings. Figure 4D (left) shows that Notch-1 knockdown in MDA-MB231 caused a striking decrease in steady-state levels of cyclins A and B1. The ratio between nuclear and cytoplasmic levels of cyclin B1 was not affected (data not shown). At the same time, NOXA was markedly up-regulated. Consistent with MDA-MB231 data, in ERα+ T47D:A18 cells, Notch-1 siRNA treatment for 72 h caused strong down-regulation of cyclin A, a more modest decrease of cyclin B1, and NOXA accumulation (Fig. 4D, right). Notch-1IC overexpression inhibits p53-induced NOXA accumulation in MCF-10A cells (16). Our data indicate that endogenous Notch-1 suppresses NOXA expression even in p53-mutant MDA-MB231 cells. Cyclin A/cyclin-dependent kinase 2 (cdk2) phosphorylates and inactivates E2F-1/DP complexes at the end of S phase. Failure of cyclin A/cdk2 to inactivate E2F results in inappropriately persistent E2F activity (29), which induces NOXA (30), causing apoptosis. Thus, loss of cyclin A may contribute to NOXA expression via E2F-1.
Inhibition of γ-secretase has antineoplastic effects on breast cancer cell lines
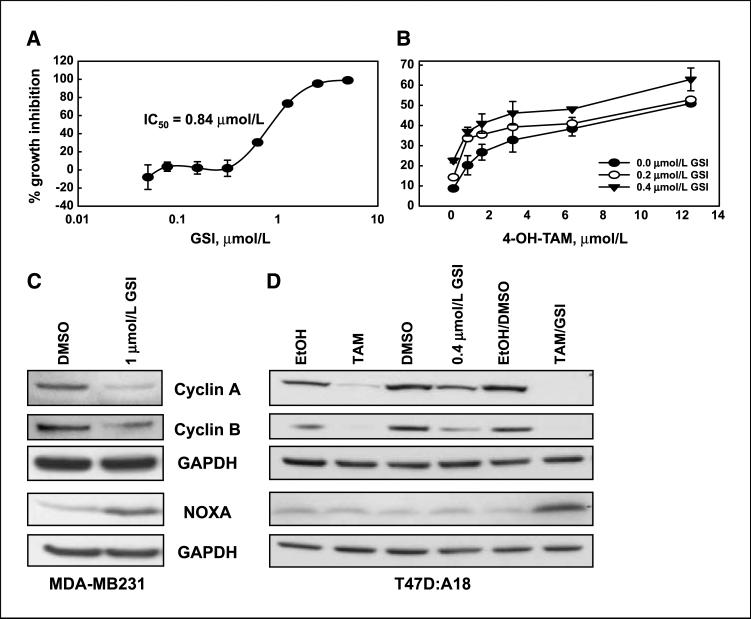
We investigated whether the effects of Notch knockdown can be replicated by nonselective, pharmacologic inhibition of Notch activation with γ-Secretase inhibitors. We used a potent, commercially available compound, GSI, which inhibits Notch cleavage and has antineoplastic activity in melanoma (27) and Kaposi sarcoma cells (28). We validated the activity, specificity, and dose response of this GSI in our cell lines (see Supplementary Data and Supplementary Fig. S4A–D). In T47D:A18 cells grown in complete medium, GSI had strong growth-inhibitory effects, with an IC50 of ~0.84 Amol/L (Fig. 5A). When we combined GSI at sub-IC50 concentrations (0.2 and 0.4 μmol/L) with 4-OH-TAM (0–12.5 μmol/L), the combination caused significantly more growth inhibition after 48-h treatment than either drug alone, particularly at very low (0.8–1.5 μmol/L) 4-OH-TAM concentrations (Fig. 5B). Isobologram analysis (data not shown) indicated that the effect was additive. The IC50 of 4-OH-TAM without GSI was 11 μmol/L. Together with RNAi data, these findings strongly support the hypothesis that genetic or pharmacologic inhibition of Notch signaling has antineoplastic effects on some breast cancer cells.
Figure 5.
Inhibition of γ-Secretase has antineoplastic effects on breast cancer cell lines and affects cyclins A and B1 as well as NOXA. A, dose-response curve of growth inhibition by GSI alone in T47D:A18 breast cancer cells. B, dose response of 4-OH-TAM in the same cells in the absence of GSI and in the presence of two sub-IC50 concentrations of GSI. Growth inhibition assays showed that 4-OH-TAM at a concentration as low as 0.8 μmol/L caused significant growth inhibition in T47D:A18 cells in the presence of 0.2 and 0.4 μmol/L of GSI (IC50 of GSI alone, 0.84 μmol/L). The IC50 of 4-OH-TAM alone in this assay is 11 μmol/L. C, Western blots of MDA-MB231 cells treated for 24 h with GSI (1 μmol/L). This treatment decreased cyclins A and B1 and induced NOXA. D, Western blots of T47D:A18 cells treated for 48 h, showing that 0.4 μmol/L GSI alone, 10 μmol/L 4-OH-TAM alone, or a combination inhibited expression of cyclins A and B1. Only the combination caused induction of NOXA at 24 h.
We then investigated whether GSI affects the same targets we identified using Notch-1 siRNA. GSI treatment of MDA-MB231 cells at 1 μmol/L (IC50, 1.23 μmol/L; Supplementary Fig. S4C) had identical effects as Notch-1 siRNA on cyclin A and B1 levels and caused NOXA induction (Fig. 5C). We used T47D:A18 cells to determine if these putative Notch targets are modulated by a combined treatment with a SERM and GSI. In T47D:A18 cells, either 4-OH-TAM or a subtoxic concentration of GSI alone (0.4 μmol/L; IC50, 0.84 μmol/L) reduced cyclins A and B1 (Fig. 5D). The effects of 4-OH-TAM are most likely mediated by Notch-independent mechanisms because 4-OH-TAM does not inhibit Notch activity. 4-OH-TAM suppresses cyclin A transcription via p38 (31). Combined treatment of these cells with 4-OH-TAM and GSI completely abolished expression of cyclins A and B. Interestingly, only the combination caused induction of NOXA. Dose-dependent, additive, or synergistic effects on cyclin A and B expression were observed at 1, 2, and 5 μmol/L of 4-OH-TAM (Supplementary Fig. S5). However, NOXA induction was only observed at 4-OH-TAM concentrations >5 μmol/L. Unlike Notch-1 siRNA, GSI alone did not induce NOXA. It should be noted that for these experiments we used a subtoxic concentration of GSI for 48 h to avoid masking any synergistic effects with 4-OH-TAM, whereas siRNA treatment was continued for 72 h. This suggests that near-complete and/or prolonged inhibition of Notch signaling is necessary for NOXA induction in these cells. The combination used in Fig. 5D (0.4 μmol/L GSI and 10 μmol/L 4-OH-TAM) caused ~50% growth inhibition. These data also suggest that loss of cyclin A is not sufficient to cause NOXA induction in T47D:A18 cells. This may be due to the fact that these cells proliferate slowly due to the cytostatic effects of 4-OH-TAM or to additional cyclin A–independent mechanisms of NOXA suppression by Notch in these cells.
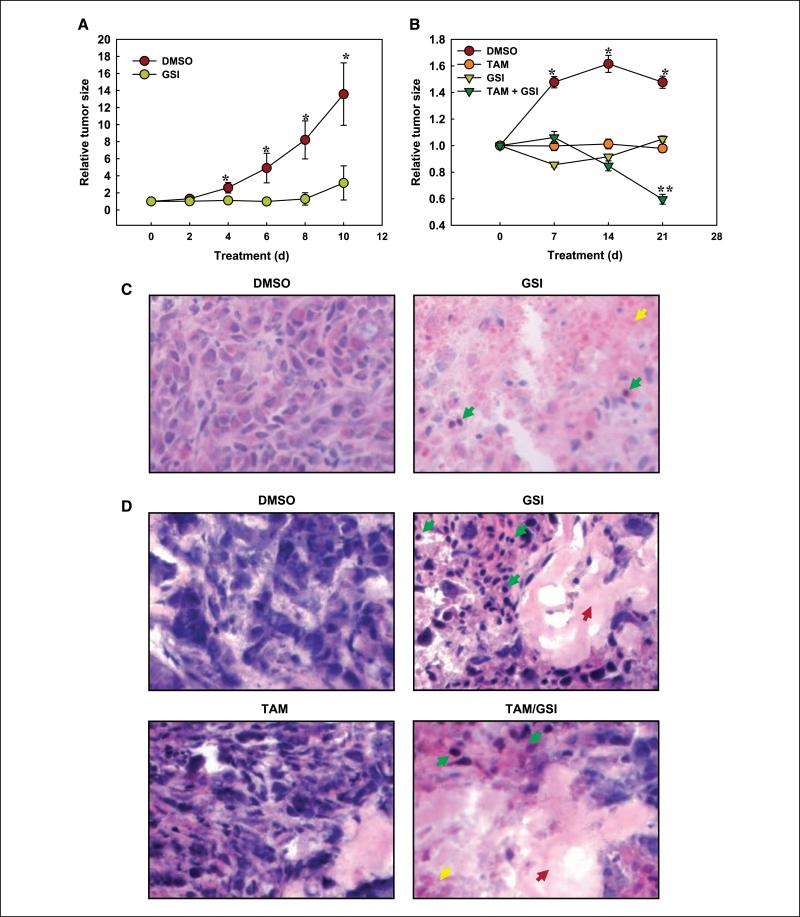
In vivo treatment with a γ-secretase inhibitor blocks the growth of MDA-MB231 xenografts and causes regression of T47D:A18 xenografts in combination with tamoxifen
Our in vitro data support a model in which ERα– breast cancer cells have high Notch activity and are dependent on Notch signaling for growth, survival, and possibly invasion, whereas ERα+ cells have lower basal Notch activity but are highly sensitive to a combination including a Notch inhibitor and a SERM. We tested this model in vivo in established tumor xenograft models. Figure 6A shows that treatment with GSI at 1.2 mg/kg s.c. near the xenografts every other day caused virtually complete growth arrest in established MDA-MB231 tumors compared with DMSO vehicle (P < 0.002 at each time point after day 4). This dose of GSI has similar effects on melanoma xenografts (27). Given a body weight of 25 g, and assuming 100% bioavailability, free diffusion throughout body fluids, no metabolism, and no excretion, the maximum theoretical extracellular fluid concentration of GSI at this dose would be in the order of 2.5 μmol/L. We did inject the drug s.c. near the tumor area to minimize the effects of degradation and excretion because GSI is a tripeptide that is likely rapidly eliminated. However, we have no reason to believe that the drug remained unabsorbed in the tumor area. Under these conditions, average in vivo concentrations are most likely in the order of what we used in cell culture or lower. No overt toxicity of GSI was observed at these doses for 10 days, and mouse weight was not significantly affected. The nonstatistically significant increase in average volume at this time may have been due to edema and hemorrhage (see below). We obtained virtually identical data with a chemically unrelated GSI, LY411,575.9 In established T47D:A18 xenografts receiving a menopausal level of estradiol, GSI alone (1.2 mg/kg every other day) or tamoxifen alone blocked further tumor progression with similar efficacy. However, when given in combination, the two agents caused significant regression of established tumors (P < 0.05 by ANOVA corrected for multiple comparison for the combined treatment compared with each agent alone and for each agent alone compared with vehicle controls; Fig. 6B). Histologically, MDA-MB231 tumors at 10 days were mostly necrotic and hemorrhagic (Fig. 6C). In established T47D:A18 tumors, GSI either alone or with tamoxifen caused extensive cell death with nuclear pyknosis. Hemorrhage and edema were also visible in GSI-treated T47D:A18 tumors (Fig. 6D), suggesting an effect on endothelial cells, where Notch-1 and Notch-4 are known to play active roles (32–34).
Figure 6.
GSI has antineoplastic effects in vivo in two breast cancer models. A, MDA-MB231 established xenografts were treated with GSI alone (see Supplementary Materials and Methods). GSI caused statistically significant decrease of tumor volume at all time points, virtually abolishing tumor growth. B, T47D:A18 established xenografts receiving menopausal estrogen levels were treated with GSI, tamoxifen, or a combination thereof. Either drug alone virtually arrested tumor growth. *, statistically significant differences between controls and each individual agent. **, the combination GSI/tamoxifen caused tumor regression and was significantly different not only from controls but also from each individual treatment. C, histologic aspects of MDA-MB231 tumors. Final magnification, ×400. Note nuclear pyknosis (green arrows), necrosis, and hemorrhage (yellow arrow) in GSI-treated tumor. D, histologic aspects of T47D:A18 tumors. Magnification, ×400. Note that GSI, either alone or with tamoxifen, caused widespread cell death with nuclear pyknosis (green arrows), edema (red arrows), and hemorrhage (yellow arrows).
Discussion
High expression of Notch-1 and/or Jagged-1 has negative prognostic significance in breast cancer (14, 15), and Notch-1 can transform HMECs (16). Our data confirm that Notch-1 and Notch-4 are commonly coexpressed in infiltrating breast cancers of ductal and lobular histologies, which also express Notch ligands Jagged-1 and Delta-1. However, our observations on breast cancer cell lines suggest that there may not be a simple correlation between protein levels of Notch receptor and ligands and Notch pathway activity level. Our data suggest that estrogen inhibits Notch signaling through an ERa-dependent effect, which is at least in part mediated by inhibition of Notch cleavage by γ-Secretase. Inhibition of Notch activation by estrogen is observed under physiologic, ligand-induced Notch activation conditions, but it is independent of Notch ligands. Estradiol did not affect expression of Jagged-1, the most abundant Notch ligand in these cells (data not shown). The membrane accumulation of uncleaved Notch-1 in estrogen-treated cells suggests an effect on Notch intracellular trafficking or induction of other γ-Secretase substrates that could compete with Notch-1. In a previous report, estrogen was suggested to activate Notch signaling in MCF-7 cells (35). However, this conclusion was based exclusively on reporter assays, which did not seem to contain transfection efficiency internal controls. Because estrogen increases transfection efficiency in estrogen-dependent cell lines, these results may reflect estrogen-induced increased reporter plasmid transfection. Notch-1 induces Notch-4 expression in breast cancer cell lines, similar to BJ fibroblasts (18). In MDA-MB231 and T47D:A18 cells, Notch-4 seems to contribute to the pro-oncogenic effects of Notch-1. Whether Notch-4 can mediate these effects in the absence of Notch-1 remains to be determined. Notch-4 is a well-known mammary oncogene in mice, but before this report, its expression in human breast cancer had not been carefully analyzed. To test the hypothesis that Notch-4 may be relevant to human breast cancer biology, we studied an additional, prospectively collected group of clinical specimens (n = 31) that did not overlap with our original sample population. We correlated expression of Notch-1 and Notch-4 with known prognostic factors in breast cancer, including node status, tumor size, age, ER status, lymphovascular invasion, Her2/Neu status, and Ki67. Logistic regression analysis revealed that a high proliferation index (Ki67) was significantly associated with high Notch-4 receptor expression (P < 0.02; Supplementary Fig. S6). These data support the hypothesis that Notch-4 may have an important biological role in breast cancer and may be a biomarker and/or a candidate therapeutic target.
Our data suggest a model in which in ERα+ breast epithelial cells Notch-1 is a hormone-modulated signal that controls proliferation and survival. In ERα+ neoplastic cells, estrogen deprivation, loss of ERa, or treatment with antiestrogens reactivates Notch-1 and induces Notch-4, stimulating proliferation, survival, and invasion. In normal human breasts, ERα+ cells are nonproliferative, unlike ERα– cells (36). Our findings are consistent with this model. Clarke and colleagues (37) propose that side population (SP) cells from normal human breast are enriched in ERa and express Notch positive regulator Msi-1, suggesting active Notch signaling in an ERα+ background. We do not know how our findings in transformed cells relate to normal SP cells. However, the effects of Notch-1 siRNA in ERα+ T47D:A18 cells suggest that even low levels of Notch signaling may have important physiologic consequences.
Notch-1IC expression in murine T cells (38) or mammary epithelial cells (39) induces c-Myc. In coculture assays, Delta-1 strongly induced c-Myc in T47D:A18 cells (data not shown). However, Notch-1 silencing by siRNA in these cells only modestly decreased c-Myc, and GSI only decreased it at 2 μmol/L (>2× IC50; Supplementary Fig. S7). In MDA-MB231 cells, even at 2 μmol/L, we could not observe inhibition of c-Myc expression (Supplementary Fig. S7). This suggests that although c-Myc is a transcriptional target of Notch-1 in human breast cancer cell lines, Notch is only one of the factors that regulate its expression. Another putative Notch-1 target is cyclin D1 (40). We did not observe cyclin D1 down-regulation on Notch-1 knockdown in our cells. This does not necessarily mean that Notch-1 does not regulate the cyclin D1 promoter, as we did observe cyclin D1 mRNA accumulation in cells transfected with Notch-1IC (data not shown). The effects of Notch-1 silencing in our cells may be masked by redundant pathways that regulate cyclin D1 expression.
Our study reveals that Notch signaling controls expression of cyclins A and B1 in breast cancer cells. Cyclin A overexpression has been described as a negative prognostic marker in breast cancer (41). Drosophila Notch is required for cyclin A expression in the developing eye (42). Analysis of the human cyclin A promoter revealed two putative CBF-1–responsive elements in the first intron (nucleotides 82637 and 82655 of minus strand, locus AC079341) as well as two putative NF-κB–responsive elements at –2113 nucleotides from the transcriptional start (nucleotide 85677 of minus strand, locus AC079341) and in the first intron (nucleotide 82632 of minus strand, locus AC079341). Up-regulation of cyclin B1 by Notch had not been described to our knowledge. The mechanism may be direct or indirect, but we have identified putative CBF-1–binding elements (TGGGAA) at –616 and –19 nucleotides from the transcriptional start (nucleotides 496 and 1093 of plus strand, locus AY338491) and in the first intron (nucleotide 1570 of plus strand, locus AY338491) of the human cyclin B1 promoter.
Our data support a role for NOXA suppression in Notch-mediated survival effects. Inhibition of γ-Secretase in melanoma cells causes apoptosis by induction of NOXA (27). Overexpression of Notch-1 in MCF-10A cells suppresses NOXA induction by chemotherapy agents (16). Our data indicate that both RNAi knockdown of Notch-1 and γ-Secretase inhibition induce NOXA in breast cancer cells. NOXA up-regulation in response to Notch inhibition may be mediated by p53 (16, 43). However, here we describe NOXA induction on Notch inhibition in two cell lines with mutant p53. Additional mechanisms that may mediate NOXA up-regulation include uncontrolled E2F-1 activation (44), possibly consequence of cyclin A loss, or activation of c-Jun NH2-terminal kinase (45, 46), which is inhibited by Notch-1 via JNP-1 (47).
Taken together, our clinical, in vitro, and in vivo data support the notion that inhibition of Notch signaling may be an attractive therapeutic strategy in breast cancer. Our data also indicate that the optimal use of Notch inhibitors depends on the ER status of the tumor. GSIs may be useful, possibly in combination with chemo-therapy, in the poor-prognosis “triple-negative” cancers that are ERa/progesterone receptor negative and do not overexpress Her2/Neu. Additionally, our data indicate that inhibition of Notch signaling in combination with an antiestrogen may be a promising therapeutic strategy for ERα+ breast cancers.
Supplementary Material
Acknowledgments
Grant support: NIH grants R01 CA84065 and P01 AG2553101 and Department of Defense IDEA grant W81XWH-04-1-0478.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank Drs. Barbara Osborne, Todd Golde, and Antonio Pannuti for helpful discussions; Dr. Raphael Kopan for the gift of Renilla-tagged Notch construct; Dr. Dmitry Gabrilovich for the CBF-1 reporter; and Dr. Judy Bolton for the gift of raloxifene.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
G. Antico contributed to this work before his current full-time employment at Bristol-Myers Squibb Company.
Osipo C, Patel P, Rizzo P, Clementz AG, Hao L, Golde TE, Miele L. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene. In press, 2008.
J. Mascarenhas et al., in preparation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Miele L, Golde T, Osborne B. Notch signaling in cancer. Curr Mol Med. 2006;6:905–18. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- 2.Miele L. Notch signaling. Clin Cancer Res. 2006;12:1074–9. doi: 10.1158/1078-0432.CCR-05-2570. [DOI] [PubMed] [Google Scholar]
- 3.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–9. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 5.Malyukova A, Dohda T, von der LN, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67:5611–6. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 6.O'neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to g-secretase inhibitors. J Exp Med. 2007;204:1813–24. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson BJ, Buonamici S, Sulis ML, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–35. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–73. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pear WS, Aster JC, Scott ML, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–91. [Google Scholar]
- 10.Bellavia D, Campese AF, Alesse E, et al. Constitutive activation of NF-nB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–48. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callahan R, Raafat A. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:23–36. doi: 10.1023/a:1009512414430. [DOI] [PubMed] [Google Scholar]
- 12.Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;18:5973–81. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- 13.Kiaris H, Politi K, Grimm LM, et al. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;165:695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reedijk M, Odorcic S, Chang L, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 15.Dickson BC, Mulligan AM, Zhang H, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–93. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 16.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–25. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 17.Pece S, Serresi M, Santolini E, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–21. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weijzen S, Rizzo P, Braid M, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 19.Cheng P, Zlobin A, Volgina V, et al. Notch-1 regulates NF-nB activity in hemopoietic progenitor cells. J Immunol. 2001;167:4458–67. doi: 10.4049/jimmunol.167.8.4458. [DOI] [PubMed] [Google Scholar]
- 20.Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R. Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J Biol Chem. 2004;279:50864–73. doi: 10.1074/jbc.M409430200. [DOI] [PubMed] [Google Scholar]
- 21.Likhitwitayawuid K, Angerhofer CK, Cordell GA, Pezzuto JM, Ruangrungsi N. Cytotoxic and antimalarial bisbenzylisoquinoline alkaloids from Stephania erecta. J Nat Prod. 1993;56:30–8. doi: 10.1021/np50091a005. [DOI] [PubMed] [Google Scholar]
- 22.Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 23.Pink JJ, Bilimoria MM, Assikis J, Jordan VC. Irreversible loss of the oestrogen receptor in T47D breast cancer cells following prolonged oestrogen deprivation. Br J Cancer. 1996;74:1227–36. doi: 10.1038/bjc.1996.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlstrom H, Bergman A, Lendahl U, Naslund J, Lundkvist J. A sensitive and quantitative assay for measuring cleavage of presenilin substrates. J Biol Chem. 2002;277:6763–6. doi: 10.1074/jbc.C100649200. [DOI] [PubMed] [Google Scholar]
- 25.Lavia P, Jansen-Durr P. E2F target genes and cell-cycle checkpoint control. Bioessays. 1999;21:221–30. doi: 10.1002/(SICI)1521-1878(199903)21:3<221::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 26.Bocchetta M, Miele L, Pass HI, Carbone M. Notch-1 induction, a novel activity of SV40 required for growth of SV40-transformed human mesothelial cells. Onco-gene. 2003;22:81–9. doi: 10.1038/sj.onc.1206097. [DOI] [PubMed] [Google Scholar]
- 27.Qin JZ, Ziffra J, Stennett L, et al. Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer Res. 2005;65:6282–93. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- 28.Curry CL, Reed LL, Golde TE, et al. g Secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi's sarcoma tumor cells. Oncogene. 2005;24:6333–44. doi: 10.1038/sj.onc.1208783. [DOI] [PubMed] [Google Scholar]
- 29.Nevins JR, Chellappan SP, Mudryj M, et al. E2F transcription factor is a target for the RB protein and the cyclin A protein. Cold Spring Harb Symp Quant Biol. 1991;56:157–62. doi: 10.1101/sqb.1991.056.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–34. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- 31.Buck MB, Pfizenmaier K, Knabbe C. Antiestrogens induce growth inhibition by sequential activation of p38 mitogen-activated protein kinase and transforming growth factor-h pathways in human breast cancer cells. Mol Endocrinol. 2004;18:1643–57. doi: 10.1210/me.2003-0278. [DOI] [PubMed] [Google Scholar]
- 32.Liu ZJ, Shirakawa T, Li Y, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003;23:14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uyttendaele H, Marazzi G, Wu G, et al. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–9. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 34.Zeng Q, Li S, Chepeha DB, et al. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell. 2005;8:13–23. doi: 10.1016/j.ccr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Soares R, Balogh G, Guo S, et al. Evidence for the notch signaling pathway on the role of estrogen in angiogenesis. Mol Endocrinol. 2004;18:2333–43. doi: 10.1210/me.2003-0362. [DOI] [PubMed] [Google Scholar]
- 36.Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 1997;57:4987–91. [PubMed] [Google Scholar]
- 37.Clarke RB, Spence K, Anderson E, et al. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol. 2005;277:443–56. doi: 10.1016/j.ydbio.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 38.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klinakis A, Szabolcs M, Politi K, et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci U S A. 2006;103:9262–7. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol Cell Biol. 2001;21:5925–34. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michalides R, van Tinteren H, Balkenende A, et al. Cyclin A is a prognostic indicator in early stage breast cancer with and without tamoxifen treatment. Br J Cancer. 2002;86:402–8. doi: 10.1038/sj.bjc.6600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baonza A, Freeman M. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev Cell. 2005;8:529–39. doi: 10.1016/j.devcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Beverly LJ, Felsher DW, Capobianco AJ. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 2005;65:7159–68. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- 44.Flinterman M, Guelen L, Ezzati-Nik S, et al. E1A activates transcription of p73 and Noxa to induce apoptosis. J Biol Chem. 2005;280:5945–59. doi: 10.1074/jbc.M406661200. [DOI] [PubMed] [Google Scholar]
- 45.Wong HK, Fricker M, Wyttenbach A, et al. Mutually exclusive subsets of BH3-only proteins are activated by the p53 and c-Jun N-terminal kinase/c-Jun signaling pathways during cortical neuron apoptosis induced by arsenite. Mol Cell Biol. 2005;25:8732–47. doi: 10.1128/MCB.25.19.8732-8747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang WH, Gregori G, Hullinger RL, Andrisani OM. Sustained activation of p38 mitogen-activated protein kinase and c-Jun N-terminal kinase pathways by hepatitis B virus X protein mediates apoptosis via induction of Fas/FasL and tumor necrosis factor (TNF) receptor 1/TNF-α expression. Mol Cell Biol. 2004;24:10352–65. doi: 10.1128/MCB.24.23.10352-10365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JW, Kim MJ, Kim KJ, et al. Notch interferes with the scaffold function of JNK-interacting protein 1 to inhibit the JNK signaling pathway. Proc Natl Acad Sci U S A. 2005;102:14308–13. doi: 10.1073/pnas.0501600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.