Abstract
Cytokines exert a vast array of immunoregulatory actions critical to human biology and disease. However, the desired immunotherapeutic effects of native cytokines are often mitigated by toxicity or lack of efficacy, either of which results from cytokine receptor pleiotropy and/or undesired activation of off-target cells. As our understanding of the structural principles of cytokine–receptor interactions has advanced, mechanism-based manipulation of cytokine signaling through protein engineering has become an increasingly feasible and powerful approach. Modified cytokines, both agonists and antagonists, have been engineered with narrowed target cell specificities, and they have also yielded important mechanistic insights into cytokine biology and signaling. Here we review the theory and practice of cytokine engineering and rationalize the mechanisms of several engineered cytokines in the context of structure. We discuss specific examples of how structure-based cytokine engineering has opened new opportunities for cytokines as drugs, with a focus on the immunotherapeutic cytokines interferon, interleukin-2, and interleukin-4.
Keywords: protein design, ligand-receptor interaction, immune signaling, immunotherapeutics, interferon, interleukin
INTRODUCTION
This review focuses on the extracellular interactions of helical cytokines with their receptors (Figure 1) (1–7) and how such interactions have been manipulated using protein engineering to modulate affinity, potency, and target cell specificity. Helical cytokines regulate many important facets of immune function and numerous other aspects of mammalian physiology (Figure 1) (8–11). In the canonical cytokine signaling pathway, assembly of the cytokine–receptor complex activates intracellularly associated Janus kinases (JAKs) (Figure 2) (12). JAKs, in turn, phosphorylate and activate signal transducer and activator of transcription (STAT) transcription factors (13) to modulate gene expression and, ultimately, determine cell fate (11, 14). In addition to JAK/STAT signaling, some cytokines also activate the Akt and Erk pathways (15) as well as other signaling networks (16–19). The intracellular signaling of cytokines has been extensively reviewed (11, 12, 14), so here we discuss aspects of extracellular domain (ECD) recognition between receptors and engineered cytokines that have been manipulated to alter their biological activity (see Supplemental Table 1; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org). As several comprehensive reviews on cytokine–receptor structure have been published (2, 4, 5, 20, 21), this review focuses instead on the interplay between structure and engineering from several key systems.
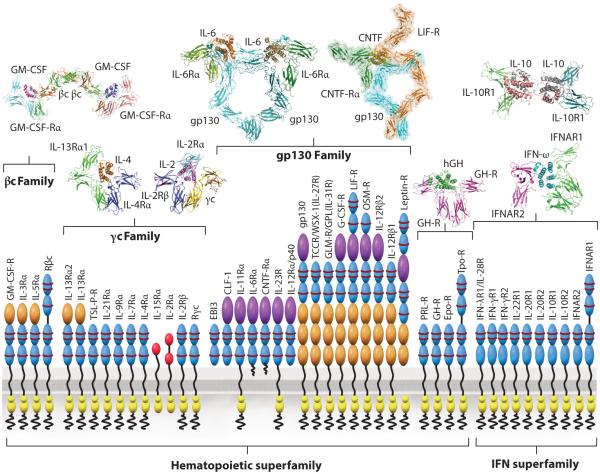
Figure 1.
Structural taxonomy of cytokine receptors and their respective signaling complexes. The receptor classes are subdivided broadly based on the hematopoietic (type I) and interferon (IFN) (type II) cytokine receptors. Type I cytokines are further subdivided based on the three principal shared receptor families (βc, γc, and gp130). The cytokine-binding homology regions (CHR), composed of tandem fibronectin type III (FNIII) domains, are colored blue on the receptor cartoons. The structures demonstrate conservation of the cytokine-CHR complex module across all systems, but also the distinctiveness of the overall complex structures. The structures shown are the receptor complexes for GM-CSF (55); IL-4 and IL-2 (36, 133); IL-6 (45, 194, 195) and CNTF (196, 197), both assembled from a compilation of X-ray diffraction and electron microscopy data; hGH (32); IL-10 (198); and type I IFN (40). (Abbreviations: GM-CSF, granulocyte macrophage colony-stimulating factor; CNTF, ciliary neurotrophic factor; hGH, human growth hormone.)
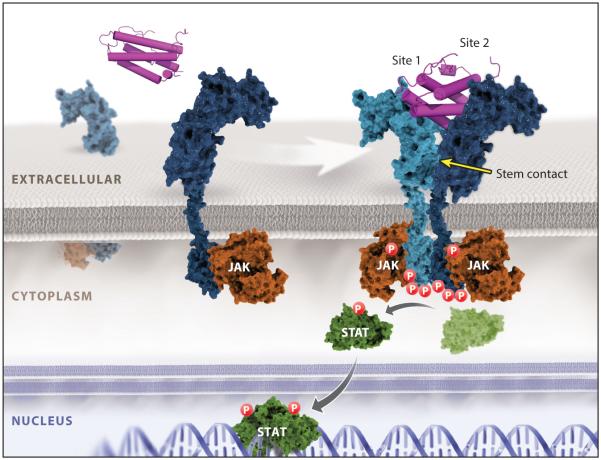
Figure 2.
Cytokine receptor–ligand engagement, dimerization, and signaling. A cytokine interaction with its cognate receptor is shown, based on the structure of the IL-4 receptor signaling complex. This complex is representative of the dimeric unit mediating canonical JAK/STAT cytokine receptor signaling, forming site 1 and site 2 contacts to dimerize the receptors and initiate JAK/STAT signaling.
Structural Taxonomy of Cytokine–Receptor Interactions
Although each cytokine system possesses unique properties, there exist convergent molecular and structural principles to be considered when undertaking a cytokine engineering experiment (2, 6, 22). The paradigm of cytokine receptor signaling is through ligand-mediated dimerization or ligand-mediated reorganization of preformed receptor dimers (23–28). Regardless of whether cytokine receptors exist as preformed dimers in the unliganded state, as some studies suggest (26–28), the signaling complex requires simultaneous binding of two JAK-associated receptor subunits with the cytokine in order to activate signaling (Figures 1 and 2). A dimerization paradigm is also evident for tyrosine kinase receptors (RTKs), and it appears that essentially all single-pass transmembrane receptors require some form of ligand-mediated oligomerization or reorganization to initiate signaling (3, 29). Some cytokines, such as human growth hormone (hGH), erythropoeitin (EPO), and thrombopoietin (TPO), homodimerize two identical copies of their receptors. However, the vast majority of cytokines form heterodimeric receptor complexes containing a shared receptor subunit [e.g., common gamma chain (γc), gp130, common beta chain (βc)] together with a ligand-specific subunit [e.g., interleukin-2 receptor beta (IL-2Rβ), IL-6Rα, IL-3Rα] (Figure 1) (30, 31). Cytokines utilize the sides of their helical faces to engage loops projected from conjoined fibronectin type III (FNIII) domains comprising the cytokine-binding homology regions (CHR) of receptor chains. Early structural studies in the hGH system established this feature as well as the site 1/site 2 binding architecture (25, 32), in which the cytokine binds to a first receptor subunit through site 1, followed by recruitment of the second receptor subunit through site 2. Every cytokine–receptor complex structure exhibits some semblance of this basic architectural building block (Figure 1) (2, 33).
The respective engagements of site 1 and site 2 can be cooperative, as is observed for hGH and γc cytokines (Figure 1), as well as others, wherein an initial high-affinity site 1 interaction results in presentation of a new composite surface that recruits the second receptor subunit to the lower-affinity site 2 (25, 34–37). A significant portion of this cooperativity can be attributed to receptor-receptor stem contact between the membrane-proximal FNIII domains of the CHR, which can bury a large amount of surface area between them, in addition to the smaller site 2 cytokine–receptor contact. γc class cytokines typify the extensive stem contact that underlies the cooperative assembly sequence of heterotrimeric complexes (Figure 2) (2). Conversely, some cytokines engage each receptor chain in more energetically autonomous fashions, such as EPO and type I interferons (IFNs) (23, 38–40). Such cytokine complexes do not exhibit extensive receptor-receptor stem contact. The different relative affinities of site 1 and site 2 can be exploited in protein engineering to manipulate potency and target cell specificity, as well as receptor internalization rates (21, 39, 41–44). Given the immense contribution that receptor stem contact can make to receptor assembly energetics, it is an important consideration in designing engineering experiments.
The current database of cytokine–receptor complex crystal structures reveals that there are many variations on the site 1/site 2 paradigm (Figure 1) (2). A third binding site, site 3, has been found in gp130/IL-6 class cytokines, and it resides at the tip of the α-helical bundle and engages an Ig-like domain that sits atop the CHR of the tall receptor class [e.g., gp130, leukemic inhibitory factor receptor (LIF-R), IL-12Rβ2, granulocyte colony-stimulating factor receptor (GCSF-R), IL-23Rα] (45–48). In the βc family, an intertwined βc receptor dimer results in a site 2 interface that represents a hybrid binding site formed from the conjunction of two head-to-head antiparallel βc chains (49). These structural distinctions endow gp130 and βc class cytokines with the ability to assemble signaling complexes that are of a higher order than the most common 1:2 cytokine:receptor stoichiometry. For example, in tetrameric GCSF/GCSF-R (50) and viral IL-6/gp130 (46) as well as in hexameric IL-6/IL-6Rα/gp130 signaling complexes (45, 51), the basic cytokine-CHR recognition unit is doubled by site 3 interaction. Site 3 has been a major target for engineering antagonists against gp130 class cytokines such as IL-6, IL-23, and LIF (52–54). In the βc system, an unprecedented dodecameric signaling complex of granulocyte macrophage colony-stimulating factor (GM-CSF) bound to GM-CSF-Rα and βc (55) appears to be driven by a broad network of additional interactions that could be specifically targeted in engineering βc cytokines. Several cytokines including IL-5 and IL-13 have an additional site of contact on top of their helical bundle with a top-mounted third Ig domain of their alpha receptors that clearly represents an engineerable site (Figure 1) (36, 56, 57). IL-2 and IL-15 are the only cytokines known to possess a specific third receptor subunit (IL-2Rα and IL-15Rα, respectively) that differs markedly from typical CHR-containing receptors in both structure and mode of cytokine interaction (2, 37, 58). These additional alpha subunits do not possess well-characterized signaling functions and appear to be used primarily for affinity enhancement and modulation of the target cell specificity of their respective cytokines. In type II IFN cytokines (e.g., IFN, IL-22, IL-10), helical bundle structures of the cytokines diverge from the type I four-helix cytokines, in some cases forming intertwined dimers (33, 59–61). Nevertheless, the basic building block of receptor CHR engaging the cytokine α-helical faces is observed in all of these different cytokine classes and remains the central structural blueprint for cytokine engineering studies (5).
Structural Energetic Considerations for Cytokine Engineering
The structural architecture and folding topology of helical cytokines renders them convenient scaffolds for protein engineering (62–64). Cytokines possess a stable hydrophobic core composed of inner facial residues of the α-helical bundle, allowing for presentation of outward-facing amino acids on a structurally stable and relatively flat, exposed surface to contact the receptor (8, 65–67). The cytokine fold is generally tolerant, albeit not entirely so, to substitutions of residues on the helical faces. Pioneering studies by the Wells group, using directed alanine scanning (Ala scanning) (68–72) as well as phage display (44, 68), demonstrated that affinity and specificity of hGH for its receptors could be powerfully manipulated by substitution of helical face residues. In one particularly illustrative example, substitution of 15 hGH residues resulted in a supermutant that exhibited 400-fold higher affinity for the hGH receptor (GH-R) than did the wild-type cytokine (44). Target residues for substitution were initially determined via Ala scanning of hGH (70, 72), and then several cytokine phage libraries were constructed that targeted different quadrants of the receptor binding site, as visualized from the crystal structure of the hGH/GH-R complex (44). Higher affinity for GH-R was attained by separately affinity-maturing different regions of the hGH helical face using phage display and then combining affinity-enhanced mutants from independently sorted libraries.
Even with more current methods today, the overall strategy employed for hGH–GH-R interaction enhancement still provides a sensible framework for affinity maturation of cytokine–receptor complexes. The apparent free energy additivity observed in affinity maturation of hGH is quite unusual for protein-protein interactions (73). A series of studies by Kossiakoff, Sidhu, and colleagues (74) using phage display and shotgun Ala scanning found the underlying energetic basis for hGH supermutant affinity to be more complex than was initially appreciated. The improved binding affinity of hGH site 1 in the supermutant was achieved not by straightforward substitution of residues in the wild-type interface but rather from a large-scale energetic remodeling of the original binding elements. This begs the question of whether it is important for the experimenter to know such mechanistic details. In most cases. it is not necessary to understand the energetic (i.e., thermodynamic, kinetic, structural) basis for the observed behavior of variants. Nevertheless, determining the mechanisms underlying engineered cytokine behavior can be extremely informative for subsequent design efforts, as will become apparent.
Molecular Engineering Strategies and Platforms
Engineering approaches have been applied to many cytokine systems using a variety of directed mutational strategies (41, 64, 75–78) or combinatorial library-based platforms such as gene shuffling (79) and phage and yeast display (54, 80, 81) (summarized in Supplemental Table 1). Generally, site-directed rational design approaches have been successful when the goal is to weaken a receptor interaction, whereas combinatorial library approaches complemented by structural information are usually required to increase affinity or redirect receptor specificity. This reflects the inherent limitations in predicting free energy changes upon mutation of amino acids within a protein-protein interface. The underlying energetics of cytokine–receptor interactions are unpredictable (82, 83). In most cases, only a subset of the interacting amino acids are energetically important, and these must be determined experimentally through Ala scanning. Thus, directed mutational approaches have a low probability of success for affinity enhancement even if the cytokine–receptor complex structure is known.
Most cytokine engineering efforts to date have focused on receptor interface modulation with the goal of either enhancing affinity, as was implemented for IL-2 (80, 81), IL-4 (84), and IFN (85), or perturbing interaction, as was demonstrated for IL-2 (75, 86, 87), IL-4 (41), IFN (88), and IL-21 (77). Instances of cytokine engineering that have successfully redirected the specificity of a cytokine to alternative receptor subunits or narrowed the specificity of a cross-reactive cytokine (68, 78, 84) are less common. Specificity engineering remains a difficult challenge because in most cases common amino acids are used by a given cytokine (e.g., IL-6, LIF, IL-2, IL-10) to cross-react with shared receptors (e.g., gp130, γc, IL-20R2) (36, 89). In some of the examples shown in Supplemental Table 1, crystal structures were available to guide the engineering of specific subsets of residues known to contact the receptors, but in many cases these structures were not available and yet the experiments were successful. This illustrates that even in the absence of crystal structures, the overall conservation of the cytokine–receptor site 1/site 2 binding paradigm makes it possible to model these interactions well enough, even if crudely, to add significant value to engineering efforts.
Cytokine Engineering Workflow
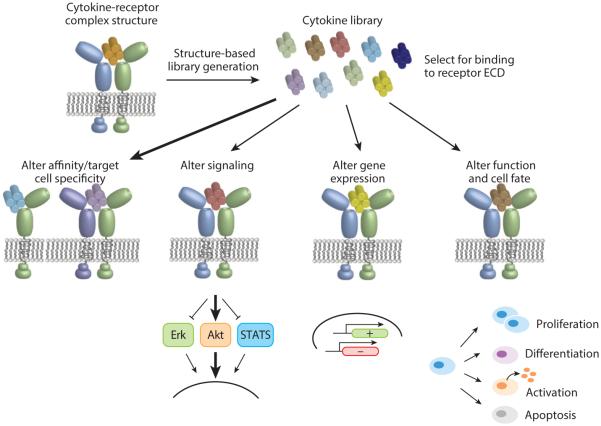
In our laboratory, we have developed a conceptual and experimental cytokine engineering framework that combines traditional approaches with several new strategies (Figure 3). We rely heavily on the existence of a structural template, in the form of cytokine–receptor complex structure coordinates, to guide engineering. A second foundation of our scheme is the use of the yeast surface display platform to select large libraries (>108 clones) in which the receptor binding sites on cytokines have been randomized either completely or in a strategic codon-biased fashion. Yeast surface display has been applied to many systems for protein engineering (90, 91) and offers the advantage of a robust secretory apparatus that enables disulfide bond formation, glycosylation, and secretion of large proteins, all of which pose major challenges for phage display. Further advantages include the ability to carry out nuanced and multivariable selections with differentially labeled receptors using magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). Library design is typically focused on cytokine residues that engage the receptor, and the diversity of these libraries can be controlled by the codon choice at each position that is varied, as has been amply discussed in other forums (92). Selections of yeast-displayed cytokine libraries with recombinant ECDs of their receptors are carried out by MACS or FACS, and evolved cytokine mutants can be characterized for binding affinity and specificity, signaling, gene expression, and induction of phenotypic changes in cells. In our studies on IL-2 using this scheme, we found the convenient property that yeast-displayed cytokines themselves can be used to stimulate responder cells without the need to produce recombinant protein (on-yeast stimulation) (81). This serves as a convenient and rapid initial screen for signaling activity of individual clones. There are many possible variations to the scheme shown in Figure 3 that will suit particular experimental goals. In the following pages of our review, we highlight specific examples of cytokine engineering that have utilized structure, rational mutation, and combinatorial libraries to create variants with novel functional properties.
Figure 3.
Cytokine engineering workflow. A cytokine–receptor complex crystal structure is used as a design template to create receptor binding site libraries of the cytokine that are evolved to isolate clones exhibiting differential receptor interaction properties, as assessed by the metrics shown.
ENGINEERING THE TYPE I INTERFERON SYSTEM
Type I IFNs comprise a family of cytokines with broad immunoregulatory actions and are perhaps best known for their antiviral (AV) and antiproliferative (AP) functions (33, 93, 94). The IFN system represents a fascinating example of naturally engineered variants in the form of 16 IFN subtypes (IFN-α1–12, IFN-β, -ε, -κ, and -ω) (33), as well as synthetically engineered variants produced by directed and combinatorial methods (Supplemental Table 1) (93). The natural IFN subtypes have been used clinically (95, 96), but more widespread use has been impeded by side effects and limited efficacy. These shortcomings could be overcome by engineering IFNs with improved therapeutic properties (95).
Tunability of Type I Interferons
Even though all IFN subtypes signal through a common pair of receptors—the IFN-α receptors IFNAR1 and IFNAR2 (33, 93)—the different subtypes appear to exhibit biased functional activities (40, 97–100). Among many examples that highlight this phenomenon are the functional discrepancies between IFN-α2 and IFN-β. IFN-β is about 50 times more potent in AP activity than IFN-α2, but only approximately 2 times more potent in AV activity (101, 102). The potential of altering the relative AP and AV activities of IFNs has attracted attention to engineering variants with actions that are even more biased than the naturally occurring variants (e.g., versions with enhanced AV but reduced AP activity). With multiple IFN subtypes and a plethora of activities and signaling outcomes, the IFN system appears tantalizingly tunable through ligand engineering.
The signaling properties of IFN receptors have been exhaustively reviewed and are extremely complex (16, 17). IFN receptors principally signal through the receptor-associated TYK2 and JAK1 to initiate multiple STAT phosphorylation cascades, but additional non-STAT pathways have been implicated in the IFN response as well (15, 103). Hundreds of genes associated with AV and AP properties are induced in response to different IFNs (95, 104–108).
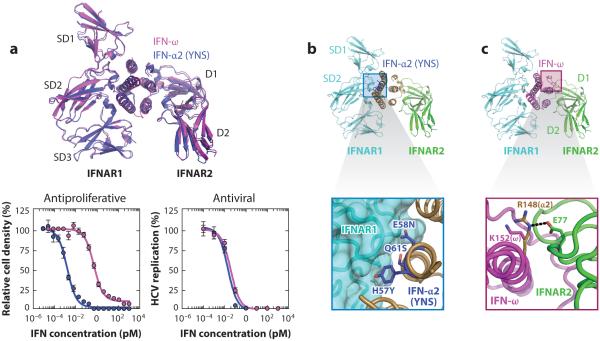
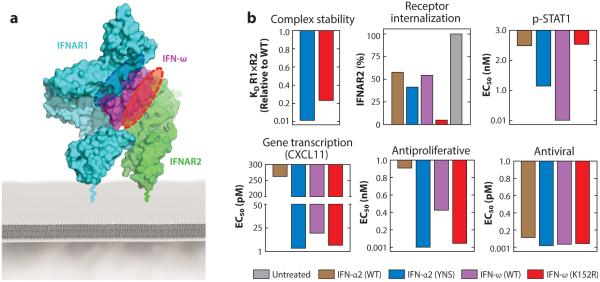
Understanding how different IFNs engage the same pair of receptors, yet are functionally discriminated, was achieved by determination of the crystal structures of two IFN receptor ternary complexes with different IFN ligands, IFN-ω and a mutant of IFN-α2 (termed YNS) (Figure 4a) (39, 40, 85). The two IFNs share nearly identical AV activities but have distinct AP potencies (Figure 4a). The topology of the two different IFN receptor heterodimers was nearly identical (Figure 4a) (40), ruling out large-scale differences in the mode by which different IFNs engage their receptors as an explanation for the differential activities of IFN ligands. With respect to IFN ligand discrimination, the IFN-receptor interfaces contain a mixture of highly conserved and IFN subtype–specific contacts (40). The more conserved contacts, which we termed anchor points, are responsible for broad engagement of all IFN ligands in a conserved docking topology. Interspersed within the anchor points are IFN subtype–specific interactions that endow different IFNs with distinct binding affinities and kinetic properties. The relative binding affinities of IFNs to IFNAR1 and IFNAR2 and the aggregate complex stability are the metrics that appear to dictate biological activity (40, 93, 98, 99, 109). Thus, the interaction chemistry between the outer facial helical residues on the IFNs and the receptors translates into differential biological activity. Conveniently, this chemistry can be manipulated to engineer new cytokine variants.
Figure 4.
Ligand discrimination by type I IFN receptors. (a) IFN-ω and IFN-α2 (YNS) engage IFNAR1 and IFNAR2, forming identical dimer geometries (top panel) that elicit discordant antiproliferative and antiviral potencies (bottom panels) (40). (b) The YNS mutations in IFN-α2 map to the interface with IFNAR1 (top panel) and improve receptor affinity through optimization of van der Waals and electrostatic interactions (bottom panel). (c) Substitution of K152 in IFN-ω with the analogous R148 in IFN-α2 results in strengthening of its interaction free energy with “hot spot” residue E77 in IFNAR2 (bottom panel). The black dashed line indicates a salt bridge. (Abbreviations: HCV, hepatitis C virus; IFNAR, IFN-α receptor.)
Interferon Antiproliferative Activity Is Sensitive to Ternary Complex Stability
In principle, either the IFNAR1 or the IFNAR2 binding interface can be targeted to stabilize the overall ternary complex. However, the various metrics that can be used to characterize stability-altered mutants, including complex stability, receptor internalization, STAT signaling, gene transcription, and AV and AP responses, do not always show concordant changes (Figure 5). The Schreiber group used Ala scanning mutagenesis to determine that simultaneous alanine mutation of three residues (H57, E58, and Q61) in IFN-α2 improved its affinity for IFNAR1 by 30-fold (38, 101). The IFNAR1 affinity was further optimized by screening a phage-displayed library focused on these three positions. The H57Y, E58N, and Q61S (YNS) mutations (Figure 4b) collectively increased the affinity for IFNAR1 by 60-fold relative to the wild-type IFN-α2 (85). AP potency was increased 150-fold, yet AV potency was enhanced only 3.5-fold for this mutant (Figure 5) (39, 85). Crystallization of the IFN-α2 (YNS) ternary complex (40) elucidated the structural basis of YNS affinity enhancement. The YNS mutations on IFN-α2 are distributed across the B helix and engage the IFNAR1 interface in the hinge region between SD1 and SD2 (Figure 4b) (40). These mutations appear to exert their effects through optimization of the electrostatic and van der Waals interactions with the receptor (40).
Figure 5.
Tunability of IFN function through IFN interface engineering. (a) The IFN-ω/IFNAR1/IFNAR2 ternary complex (40) with the SD4 domain of IFNAR1 modeled from the murine IFN-β/IFNAR1 structure (PDB 3WCY). The IFNAR1 (blue dashed oval) or IFNAR2 (red dashed oval) binding interfaces of IFN-ω were engineered to alter the stability of the ternary complex. (b) Characterization of wild-type (WT) versus engineered mutant IFN behavior using various biochemical and functional metrics (40).
On the IFNAR2 side, the crystal structures revealed a potential binding hot spot on the receptor shared between IFN-α2 and IFN-ω that could be targeted to manipulate activity (40). Specifically, R149 in IFN-α2 and the analogous K152 in IFN-ω exhibited structural differences in their interactions with E77 of IFNAR2. In the IFN-α2/IFNAR2 interface, R149 of IFN-α2 and E77 of IFNAR2 form a salt bridge (Figure 4c) (40), which is lacking in the IFN-ω complex. The IFN-ω K152R mutant interaction with IFNAR2 is energetically superior to that of the wild type (Figure 4c) and markedly increases AP activity (Figure 5), with only a modest increase in AV activity (40). Here again, as seen in both the natural and engineered IFNs (40, 85, 101), AP activity is more sensitive to changes in complex stability than is AV activity and therefore is more tunable. The IFN system appears to be incredibly sensitively poised for AV defense even in the face of deleterious mutations to IFN ligands. Importantly, the mechanism by which the K152R mutation exerts its effect does not appear to be a simple extension of the activated complex dwell time on the cell surface. Rather, it appears that higher affinity for IFNAR2 results in its faster receptor internalization and consequently more rapid deactivation of STATs (Figure 5) (40). This emphasizes the multiple layers of regulation of IFN activity that can be exploited by ligand engineering.
Decoupling Interferon Antiviral from Antiproliferative Activity
Despite the apparent ceiling on AV potency of IFNs, attempts have been made to enhance IFN AV activity using DNA family shuffling (79, 110–112). DNA sequences from all IFN-α subtypes were combined to generate a shuffled library that was screened for function based on the metrics of Th1 induction and AP and AV activity. This screening approach identified two shuffled proteins, B9X25 and B9X14, with 20- to 70-fold improvement in AV potencies compared to IFN-α2. Binding experiments on Daudi B cells showed that B9X25 and B9X14 conferred a 9-fold and 100-fold increase in IFNAR complex affinity, respectively, compared to IFN-α2. These mutants were biased toward Th1 induction and AV activities, as the ratios of both AV:AP and Th1:AP potency for the IFN variants were increased approximately 50-fold (79). One caveat in interpreting these results is that the Daudi B cells that were used in the AP assays are exquisitely sensitive to IFN activity, potentially diminishing the AP differences between IFNs and leading to aberrant AV:AP biases compared to other IFN-responsive cell lines (113–115). Nonetheless, this study served as an instructive early example of IFN engineering. Unfortunately, shuffled IFNs did not advance into the clinic, in large part because of immunogenicity resulting from the numerous mutations found in the shuffled IFN products (116). Gene shuffling generates many new potential T cell epitopes compared to more directed mutagenic approaches, which target specific receptor binding residues and thus introduce fewer mutations. Sequence diversity remains an important consideration in selecting a strategy for engineering human cytokine therapeutics.
Further efforts to bias AV:AP ratios have been explored with an IFN-α2 antagonist (117). The D helix of IFN-α2, which engages the upper quadrant of IFNAR1 at the SD1-SD2 hinge region, is rich in positively charged residues (R120, K121, Q124, and R125). A charge reversal mutation on the D helix of IFN-α2, R120E, ablated IFNAR1 binding. IFN-α2 R120E was then fused to a tail region of IFN-α8 that was proposed to form extensive contacts with IFNAR2 and therefore possibly function as a modular affinity-enhancing peptide. Indeed, the IFN-α2 R120E antagonist-IFN-α8 tail region fusion exhibited enhanced affinity for IFNAR2 and acted as a dominant negative antagonist, occupying the IFNAR2 receptor site while not recruiting IFNAR1 efficiently. The antagonist elicited just 1% of the AV activity induced by the wild-type cytokine, but its AP activity was reduced even more dramatically, with the antagonist retaining just 0.1% of wild-type activity. Thus, the respective activities of this mutant IFN were reduced in a biased fashion, favoring the retention of AV activity.
Recent characterization of the IFN-α2 dominant negative antagonist with respect to receptor binding, membrane-proximal signaling, gene transcription, and AV/AP activity elegantly highlighted the tunability of the IFN response (88). A cluster of robust genes was efficiently induced by both the antagonist and an array of IFNs, including IFN-α2, IFN-β, IFN-λ3, and IFN-α2 YNS. Expression of robust genes was regulated by IFN-induced transcription factors directly linked to AV activity (118, 119), again demonstrating the insensitivity of the AV response to variation in binding affinity. A different set of genes, classified as tunable, was induced only by high-affinity IFNs and not by the antagonist. These receptor affinity-sensitive genes mainly correlate with AP activity, contributing to the regulation of cell proliferation and cell death. Thus, the antagonist skews toward AV activity, stimulating the AV-related robust but not the AP-related tunable genes (39, 40, 97, 98). Whether biased IFNs engineered to function as improved AV therapeutics will be clinically useful remains to be determined.
ENGINEERING THE IL-2 SYSTEM
IL-2 is a multifunctional cytokine that plays an instrumental role in the adaptive immune response through its regulation of the homeostasis of T cells and many other immune cell lineages (reviewed in 18, 120, 121). IL-2 signaling controls a balance of immunostimulatory and immunosuppressive responses, rendering it an appealing, yet complicated, target for therapeutic development. The wild-type cytokine has been administered clinically for over 20 years (75, 122–124), but it induces severe toxicity, eliciting side effects such as vascular leak syndrome (125, 126). Additionally, promotion of regulatory T cell (Treg) growth blunts IL-2 efficacy in antitumor applications (127, 128). The ability to decouple the immunostimulatory from the immunosuppressive activities of IL-2 could be therapeutically valuable, so engineering IL-2 to selectively stimulate particular functionalities is highly desirable. Extensive characterization of its structural and molecular properties has enabled the design of IL-2 variants with altered receptor subunit affinity, potency, target cell specificity, and trafficking behavior.
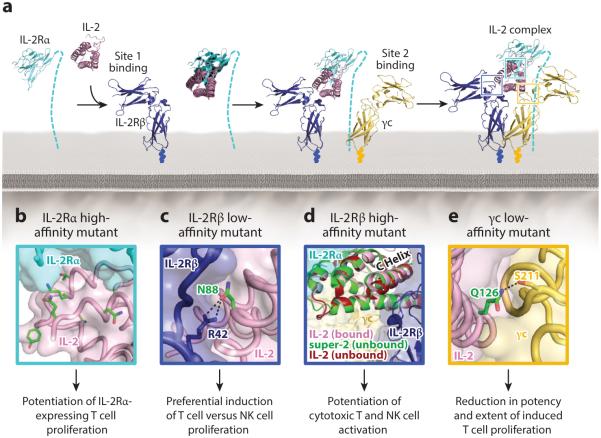
There exist two forms of the IL-2 signaling receptor: the high-affinity (10 pM) heterotrimeric receptor, consisting of the IL-2Rα (also called CD25), IL-2Rβ, and γc chains, and the intermediate-affinity (1 nM) heterodimeric receptor, consisting of only the IL-2Rβ and γc chains (129). Both the high-affinity quaternary and intermediate-affinity ternary IL-2 complexes signal through interaction of the intracellular domains of IL-2Rβ and γc with JAK1 and JAK3, respectively (18, 130). Whereas the IL-2Rα subunit is a private receptor for the IL-2 cytokine, IL-2Rβ is shared with the IL-15 cytokine and γc is shared with five other cytokines (Figure 1) (131). IL-2Rα distinguishes the quaternary from the ternary IL-2 complex, and its expression following TCR stimulation heightens cellular sensitivity to IL-2. The IL-2Rα subunit is constitutively expressed on Tregs but not on natural killer (NK) cells or resting effector CD8+ T cells, resulting in differential IL-2 potency between cell subsets (132). Solution of the IL-2 quaternary complex structure offered extensive insight into the molecular properties of this cytokine system (133, 134). IL-2 employs its helical faces to interact with all three receptor subunits, and there is also extensive stem contact between IL-2Rβ and γc. Assembly of the quaternary complex is thought to occur sequentially, with IL-2 first engaging IL-2Rα, which facilitates binding to IL-2Rβ (via the site 1 interface), and finally recruiting the γc subunit (via the site 2 interface) to lock down the high-affinity complex (Figure 6a).
Figure 6.
IL-2 receptor complex assembly and interface engineering. (a) Assembly of the high-affinity IL-2 quaternary signaling complex is initiated by binding of the cytokine to the IL-2Rα chain, followed by recruitment of the IL-2Rβ chain, and then the γc chain (133). (b–e) Cytokine mutations that have been shown to modulate the IL-2Rα, IL-2Rβ, and γc interfaces are highlighted, with mutated residues indicated in green and hydrogen bonds shown as black dashed lines.
Modulation of IL-2 Receptor Affinity
Many engineering efforts have been focused on altering the affinity of IL-2 for one or more of its receptor chains (Supplemental Table 1). Rao and colleagues (135) created an error-prone IL-2 yeast-displayed library to evolve IL-2 mutants with increased affinity for IL-2Rα. The highest-affinity mutant potentiated growth of IL-2Rα-overexpressing cells (135), and most of its mutations map to the IL-2/IL-2Rα interface (Figure 6b). Liu et al. (136) combined these IL-2Rα affinity-enhancing substitutions with mutations that block the IL-2 interaction with either IL-2Rβ or γc to create antagonists that inhibited signaling and proliferation of an IL-2-dependent cell line. These engineered IL-2 variants could have utility in cancer immunotherapy by binding tightly to and repressing activity of Tregs, which express high levels of the IL-2Rα subunit. Selective Treg inhibition can also be achieved by reducing the affinity of IL-2 for IL-2Rα. Carmenate et al. (137) designed an IL-2 variant that obstructed IL-2Rα binding to inhibit cytokine-mediated promotion of Treg growth without altering its stimulation of CD8+ T cell or NK cell growth. This IL-2 mutant repressed metastasis more effectively than did the wild-type cytokine, while inducing considerably less toxicity (137).
Attention has also been focused on modulating the affinity of IL-2 for the IL-2Rβ and γc receptor subunits. Zurawski and colleagues (86, 138) determined through systematic mutagenesis studies that cytokine variants with mutations at the mouse IL-2 positional analogs of human IL-2 residues D20 and Q126 behave as partial agonists by obstructing IL-2Rβ and γc binding, respectively. The structural basis for the effects of these mutations is now evident (133). IL-2 D20 is engaged in an extensive network of hydrogen bonds to both water molecules and receptor subunit side chains at the IL-2Rβ interface, and Q126 is integral to the γc interaction, forming a hydrogen bond with S211 of the receptor chain (Figure 6e). A directed mutagenesis study by Shanafelt et al. (76)—aimed at crippling the IL-2Rβ interface—identified a single IL-2 point mutant (N88R) that mediated selective growth of T cells over NK cells. In the IL-2 complex crystal structure, N88 is an energetic hot spot for the IL-2/IL-2Rβ interaction, engaging in critical hydrogen bonds with R42 on the receptor chain (Figure 6c). Impairment, but not ablation, of the IL-2/IL-2Rβ interaction affords a growth advantage to cells that highly express IL-2Rα, accounting for the observed preference for T cell versus NK cell proliferation. The authors speculated that the N88R mutant would have a better therapeutic index than wild-type IL-2 in cancer and infectious disease applications by limiting toxicity mediated through NK cell stimulation (76). However, Phase I clinical trials did not show any benefit of N88R compared to wild-type IL-2 treatment in HIV infection, advanced melanoma, or renal cancer (139, 140), as the high doses required for therapeutic effect nullify the selective T cell growth advantage (141). Furthermore, recent findings indicate that vascular leak syndrome is also mediated through IL-2Rα+ endothelial cells in addition to NK cells, and thus inhibition of NK cell growth alone is not sufficient to counteract IL-2 toxicity (121, 142). Another IL-2Rβ binding–impaired IL-2 mutant (D20T) fused to an anti-DNA tumor-targeting antibody shows strong selectivity for promoting T cell over NK cell activation and consequently controls tumor growth with significantly reduced toxicity compared to wild-type IL-2 in early clinical studies (143, 144). In principle, both the N88R and D20T mutants could serve as Treg promoters in autoimmune disease or engraftment applications because the enhanced IL-2 sensitivity of Tregs conferred by IL-2Rα expression may result in a pronounced growth advantage for this cell subset in the context of weakened IL-2Rβ interaction.
Our laboratory recently used the yeast surface display platform to evolve an IL-2 mutant with high affinity for the IL-2Rβ receptor chain (81). Initial error-prone libraries surprisingly directed us toward a single point mutation in the hydrophobic core of IL-2, behind the C helix, rather than outerfacial IL-2 residues that directly contact IL-2Rβ. Based on this founder mutation, a hydrophobic-biased library of core residues surrounding this site in the C helix was designed, with the logic that mutations in this portion of the cytokine could induce a conformation of IL-2 with higher affinity for IL-2Rβ. Indeed, the consensus mutant extracted from our selections (designated super-2) contained mutations that not only reduced the conformational flexibility of IL-2 (as determined by molecular dynamics simulations) but also locked the C helix into a position resembling that seen in the receptor-bound structure of IL-2 (Figure 6d). The 200-fold improvement in IL-2Rβ affinity of super-2 increased the sensitivity of IL-2Rα-deficient cells to signaling, promoting activation of cytotoxic T cells and NK cells, which led to a significant improvement in antitumor activity. Notably, super-2 treatment also resulted in reduced toxicity compared to wild-type IL-2 in mouse tumor models (81). The example of super-2 evolution illustrates how structural subtleties and insights may guide engineering efforts in unexpected ways. By focusing on the core of IL-2 rather than the contact interface between IL-2 and IL-2Rβ, a significant boost in affinity and functionality was achieved.
Modulation of IL-2 Trafficking
As the IFN variants showed, modification of cytokine trafficking properties is another important mechanism through which engineered variants can impact biological activity in ways that would not be apparent from affinity or signaling measurements alone. IL-2 signaling complexes are rapidly internalized upon assembly. Whereas IL-2Rβ and γc proceed to late endosomes and are degraded, IL-2Rα remains in early endosomes and is efficiently recycled back to the surface (145, 146). Consequently, the trafficking fate of the IL-2 cytokine is governed by its relative affinities for the receptor chains at endosomal pH, presenting an additional layer of tunability for the IL-2 system.
The high-affinity IL-2Rα-binding mutants developed by Rao et al. (135) persisted on the surface of IL-2Rα-expressing cells significantly longer than did wild-type IL-2, presumably due to a combination of slower receptor dissociation and more efficient recycling. Similar surface persistence was observed for an L18M/L19S IL-2 mutant, which was found to be more potent than wild-type IL-2 despite its identical affinity for the receptor (147). It was later established that a higher proportion of L18M/L19S molecules are shunted toward recycling rather than degradation (148). The IL-2 complex structure reveals that the L18M/L19S mutations disrupt the IL-2Rβ interface, rationalizing preferential partitioning toward IL-2Rα-mediated recycling. IL-2 variants that slow internalization, allowing for the cytokine and its receptors to persist on the surface and potentiate signaling, have also been identified. The aforementioned substitution mutants of the mouse Q126 counterpart internalize more slowly than does wild-type IL-2 due to acceleration of the complex dissociation rate (149). Chang et al. (150, 151) also identified an internalization-impaired mutant (T51P) with enhanced signaling potency. The decreased complex stability and concomitant increase in dissociation rate for both internalization mutants are consistent with crystallographic insights, as the Q126 mutation directly perturbs the γc binding interface and T51P introduces a kink at the N-terminal end of the B helix, also disrupting the γc interface.
Modulation of IL-2 Behavior with Small Molecules and Antibodies
Although not strictly examples of cytokine engineering, there are instances of cytokine activity modulation by small molecules and antibodies that complement and inform the engineering approach. IL-2 small molecule antagonists were identified that bind to a surface crevasse on the cytokine, blocking IL-2Rα subunit binding (152). These antagonists engage the same epitope of IL-2 as the receptor chain but recognize a distinct conformation of the cytokine (153). This study highlights that the conformational malleability of IL-2 can play an important role in its receptor interactions and is an important consideration in engineering the cytokine.
Anti-IL-2 antibodies that modify cytokine behavior provide an alternative approach to modulating IL-2 activity without directly changing the biochemical properties of the cytokine. Boyman and colleagues (154) discovered that two mouse IL-2-directed antibodies (designated S4B6 and JES6-1) biased the effects of the cytokine toward specific T cell subsets. In particular, complexes of IL-2 with the S4B6 antibody stimulated proliferation of NK cells and CD8+ T cells to a much greater extent than the cytokine alone, whereas treatment with complexes of IL-2 and the JES6-1 antibody markedly induced proliferation of Tregs (154). Selective stimulation of effector cells by the IL-2/S4B6 complex resulted in potent antitumor activity (155, 156), and, in contrast with IL-2 monotherapy, complex treatment did not induce vascular leak syndrome (142). A chimera consisting of mouse IL-2 fused to the full S4B6 antibody was engineered to prevent cytokine/antibody complex dissociation and maintain stoichiometric amounts of each component, and this fusion construct induced more potent in vivo expansion of effector T cells than did untethered IL-2/S4B6 complexes (157). Boyman & Sprent (121) proposed that the distinct behavior of the S4B6 and JES6-1 antibodies results from obstruction of particular receptor subunit epitopes on the IL-2 cytokine, but the mechanism underlying the behavior of these antibodies remains unclear. Establishing a structural understanding of the effects of these two antibodies could guide engineering of IL-2 mutants that elicit biased functional outcomes.
ENGINEERING THE IL-15 SYSTEM
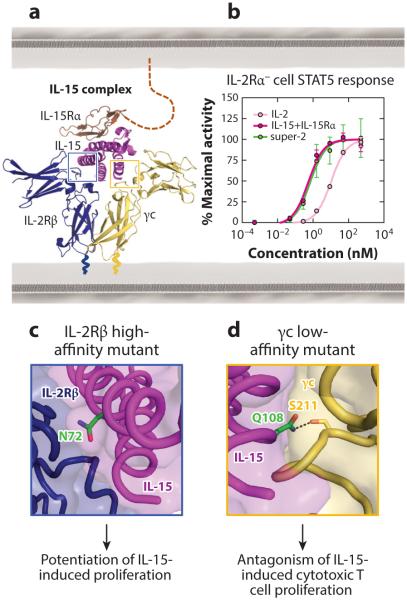
The IL-15 cytokine shares its IL-2Rβ and γc signaling subunits with IL-2, so it can be considered a naturally engineered cytokine variant. Structural studies from our laboratory demonstrated that, despite differences in their binding chemistries, IL-2 and IL-15 dimerize IL-2Rβ and γc with identical topologies in their respective quaternary complexes (Figure 7a) (37). The key distinctions between these two cytokines are (a) IL-15 exhibits an affinity for its private alpha receptor subunit that is 400-fold higher than that of IL-2 (135) and (b) IL-15 primarily signals in trans, with monocytes or dendritic cells (DCs) presenting IL-15Rα to NK cells or T cells rather than the conventional cis activation mechanism favored by IL-2 (158). Both IL-2 and IL-15 induce T cell proliferation, stimulate cytotoxic T lymphocyte and NK cell differentiation, and trigger antibody production by B cells (158–160). However, IL-2 uniquely activates immunosuppressive responses through mediation of activation-induced cell death and support of Treg differentiation and growth (161, 162), whereas IL-15 plays a prominent immunostimulatory role in the response to pathogens by inducing the survival and proliferation of CD8+ memory T cells (163–165). Functional differences between IL-2 and IL-15 do not appear to emanate from divergence in signal activation, but rather from differences in complex stability and tissue expression of the cytokine and receptor subunits (37, 135, 160). In fact, simply by enhancing the affinity of IL-2 for IL-2Rβ in super-2, we recapitulated IL-15 signaling potency in IL-2Rα-deficient cells (Figure 7b).
Figure 7.
Structure-based modulation of IL-15 affinity for shared IL-2 receptor chains. (a) The IL-15 receptor complex architecture (37) and (b) comparison of the STAT5 phosphorylation profiles for IL-2, super-2, and IL-15 in IL-2Rα-deficient NK cells (37). Through a 200-fold increase in its interaction with IL-2Rβ, super-2 matches the signaling potency of IL-15. (c,d) Mutations on the IL-15 cytokine that alter the IL-2Rβ and γc interactions are highlighted, with mutated residues shown in green and a hydrogen bond displayed as a black dashed line.
One focus of IL-15 engineering has been the development of antagonists to counteract its immunostimulatory effects. Pettit and colleagues (166) used molecular modeling to identify a point residue of IL-15 (Q108) that we now know to be critical for γc interaction (Figure 7d) (37), which abrogated cytokine-mediated proliferation. The authors built upon this development to design an IL-15 antagonist (Q101D/Q108D) that prevented graft rejection (167) and inhibited collagen-induced arthritis progression in mice (168) by curbing CD8+ T cell proliferation.
IL-15 has also been engineered to exert agonistic effects. Mortier and colleagues (169) designed a soluble truncated version of the IL-15Rα ECD fused to IL-15 that potently activated IL-15Rα-deficient cells by stabilizing signaling complex formation, analogous to the effect of super-2 on cells that lack IL-2Rα. Accordingly, their IL-15 fusion enhanced the proliferative and antiapoptotic effects of the cytokine (169). Zhu et al. (170) pursued a different approach to developing IL-15 agonists, enhancing the affinity of the cytokine for IL-2Rβ. As the structure of the IL-15 complex was not yet available, they used the IL-2 complex as a model to predict that N72 of IL-15 would play a significant role in IL-2Rβ interaction (170). The recent IL-15 complex structure reveals that N72 is buried within the IL-15 interface with IL-2Rβ, so changes in its chemistry would be predicted to impact interface stability (Figure 7c) (37). By screening multiple residues at this position, the authors determined that an N72D substitution increased the affinity of IL-15 for IL-2Rβ by approximately 10-fold, enhancing the potency of induced cell proliferation (170). In subsequent work, the N72D mutant of IL-15 was copurified in complex with an IL-15Rα Fcfusion to create a superagonist that exhibited potent antitumor activity by stimulating proliferation of CD8+ T cells and converting them into innate-like immune effector cells (171, 172). These examples accentuate that, as was the case for IL-2, differential tissue expression patterns of the alpha receptor subunit for IL-15 enable selective targeting of particular cell subtypes through affinity engineering.
ENGINEERING THE IL-4 SYSTEM
IL-4 is an important immunoregulatory cytokine that contributes to the modulation of both innate and adaptive immunity through activation of macrophages and DCs and induction of T cell differentiation, respectively. As with IL-2, IL-4 has not found broad therapeutic utility, largely due to its complicated receptor usage and target cell specificity (173, 174). For this reason, IL-4 is an ideal candidate for engineering. The wild-type IL-4 cytokine has been used clinically as an agonist to augment the immune response against cancer (175) and as an antagonist to blunt allergic reactions (176). However, its use as an agonist was terminated owing to toxicity resulting from the cytokine’s pleiotropic activities (175). IL-4 acts through the engagement of both type I and type II IL-4 receptors that utilize γc and IL-13Rα1 as second chains, respectively, to form heterotrimeric signaling complexes with the IL-4/IL-4Rα complex (Figure 8a) (2, 36, 177). Early studies using type I and type II IL-4 receptor knockout mice indicated that IL-4-mediated enhancement of DCIL-12 production occurs exclusively via the type I receptor, whereas maturation of DCs occurs mainly via the type II receptor (178).
Figure 8.
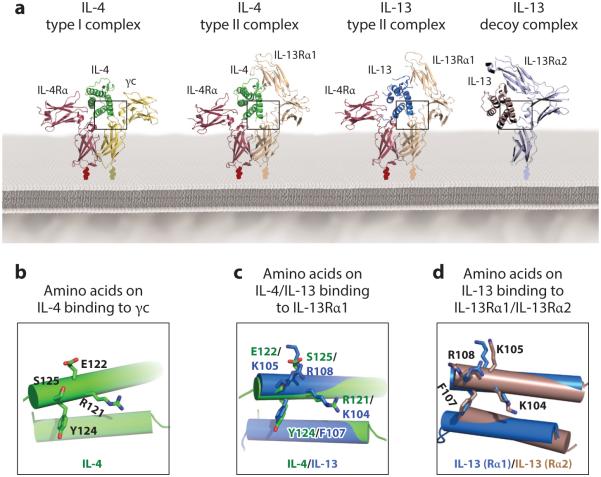
Cross-reactivity of the IL-4 and IL-13 receptor binding interfaces. (a) Crystal structures of the complete set of IL-4 and IL-13 type I and type II receptor complexes as well as the IL-13/IL-13Rα2 receptor complex (36, 57). Site 2 binding interfaces are detailed in the inset boxes below, depicting γc-binding residues on the IL-4 type I complex (b), IL-13Rα1-binding residues on the overlaid IL-4 type II and IL-13 type II complexes (c), and IL-13Rα-1- and IL-13Rα2-binding residues on the overlaid IL-13 type II and IL-13 decoy complexes, respectively (d).
IL-4 variants that induce exclusively type I–dependent or type II–dependent responses could preserve the benefits of IL-4 immunotherapy but with reduced side effects. IL-4 is poised on a razor’s edge, serving as a regulatory cytokine through the type I receptor (mediating Th2 development and class switching) and an effector cytokine through the type II receptor. Deconvoluting these dual reactivities at the structural level has been a major goal of IL-4 engineering. However, the highly cross-reactive nature of the shared binding interface utilized by IL-4 to engage the IL-13Rα1 and γc chains (36) presents a significant technical obstacle to creating type I or type II receptor–selective IL-4 variants. In both instances, IL-4 uses the same amino acids in its D helix (R121, E122, Y124, and S125) to engage the second receptor chains (Figure 8b,c) (36). Decoupling this binding specificity by predictive mutagenesis would be unlikely to succeed. In contrast, this intrinsic difficulty was not an issue for creating IL-4 antagonists to treat allergy and asthma, where simply ablating affinity for the second chain while retaining affinity for IL-4Rα could be easily achieved (41, 176). The IL-4 Y124D mutation disrupted a key amino acid interaction network with Y182 and H159 on γc and R256 and L315 on IL-13Rα1 that reduced binding affinity for both receptor chains, without affecting binding to the IL-4Rα chain (41). In addition to Y124D, another second receptor chain binding disruption mutation (R121D) was introduced to develop an IL-4 antagonist drug known as Pitrakinra (AerovantTM), which is currently undergoing clinical trials in the treatment of asthma (176).
Interestingly, the R121E mutant of IL-4, similar to the R121D mutant described for Pitrakinra, was reported to bind more tightly to the type I than to the type II receptor (179). The structure of the type II receptor complex reveals that the R121E mutation disrupts a critical salt bridge between R121 on IL-4 and E322 on IL-13Rα1 (36). However, the contacts between IL-4 R121E and γc remain unchanged because γc does not rely on this salt bridge for cytokine interaction (36). This result demonstrated that, in principle, the type I/type II binding specificity of IL-4 could be decoupled, although the R121E mutant still retained significant cross-reactivity for the two IL-4 receptor complexes.
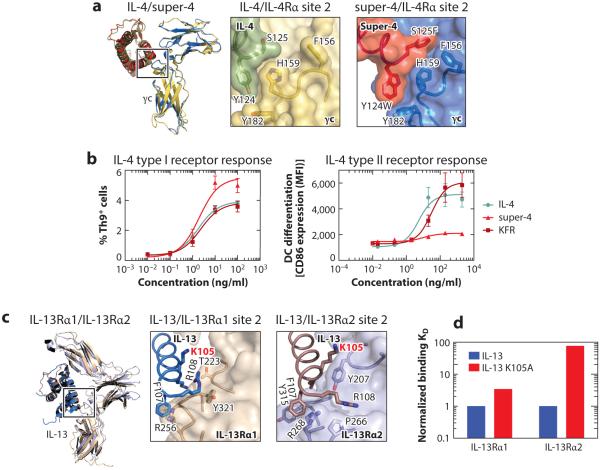
Our laboratory engineered IL-4 mutants with a high degree of selectivity for the IL-4 type I versus type II receptor complexes (84). These mutants are referred to as super-4 (type I receptor specific) and KFR (type II receptor specific) and have proven useful in deconvolving some of the functional redundancies exhibited by IL-4 and IL-13. Different experimental approaches were used to obtain these two IL-4 mutants. Super-4 was engineered through a combinatorial approach involving yeast surface display of IL-4. Based on the interatomic contacts visualized in the IL-4/γc binding interface (36), we generated a focused IL-4 contact library. Through multiple rounds of selection against γc, we isolated an IL-4 mutant with ~20,000-fold higher affinity for the type I compared to the type II receptor. To rationalize how the super-4 mutations (in particular Y124W and S125) manifested higher affinity, we determined the structure of the super-4/γc complex (Figure 9a). F125 on super-4 fills a large hydrophobic pocket on γc that is unoccupied in the wild-type complex, contributing an additional 52.5 Å2 of buried surface, and mutation of Y124 to W created a new hydrogen bond interaction between this residue and a main chain carbonyl on γc (36, 84).
Figure 9.
Structural rationale for engineering IL-4- and IL-13-binding specificity. (a) Superposition of the IL-4/γc and super-4/γc complexes (36, 84) with a close-up view of their respective site 2 binding interfaces. (b) Type I and type II receptor signaling responses activated by IL-4, super-4, and KFR. (c) Superposition of the IL-13/IL-13Rα1 and IL-13/IL-13Rα2 complexes (36, 57) with a close-up view of their respective site 2 interactions. (d) The discordant effect of the K105A mutation on IL-13 binding to the IL-13Rα1 and IL-13Rα2 receptor chains.
To engineer the IL-4 type II receptor–specific mutant KFR, we used a rational structure-based approach. We aligned the structure of IL-4 with IL-13 in the two type II receptor ternary complexes in order to identify important IL-13 receptor–interacting residues that could be grafted into the corresponding positions on IL-4 (Figure 8c) (36, 84). Three centrally located amino acids on IL-4 in the IL-4/IL-13Rα1 binding interface (R121, Y124, and S125) were mutated to their positional equivalents in IL-13 (K104, F107, and R108) to generate the IL-4 type II–specific variant KFR, which binds the type II IL-4 receptor ~500-fold better than the type I receptor (36, 84).
Isolation of IL-4 receptor–selective variants allowed us to independently examine activities emanating from the IL-4 type I and type II receptors. Although super-4 and KFR exhibited differential activation potencies, we did not observe qualitative differences in the signaling pathways that were induced. Both mutants led to an increase in the levels of phosphorylated STAT6 and insulin receptor substrate (IRS)-1, with potencies of induction that paralleled the expression levels of type I and type II receptors on various cell types (84). We did observe differences in the kinetics of signaling activation that could, in principle, perturb gene expression programs and bioactivities induced by the two IL-4 receptor complexes. Indeed, we found that although super-4 was more potent than IL-4 and KFR in inducing the differentiation of CD4+ T cells into a Th9 subset, this mutant was unable to promote the differentiation of monocytes into DCs as seen for IL-4 and KFR (Figure 9b) (84). Because CD4+ T cells express only the type I IL-4 receptor, it appears that Th9 differentiation is a type I receptor–dependent response (180). Monocytes, in contrast, express high levels of both type I and type II receptors (181), making the lack of DC differentiation induction by super-4 unexplainable. These results suggest that although super-4 engages the type I receptor and promotes STAT6 activation in monocytes, this signaling is not sufficient to drive monocyte differentiation into DCs, arguing in favor of signaling by the type II receptor for the induction of this latter activity. The differential activities initiated by super-4 when compared to IL-4 could be exploited for therapeutic development. By activating only type I receptor responses, super-4 could potentially preserve the benefits of an IL-4 therapy while mitigating toxicity associated with the type II receptor responses.
ENGINEERING THE IL-13 SYSTEM
IL-13 and IL-4 share many immunological functions, as both cytokines signal through the type II receptor and activate STAT6 (182, 183). IL-13 uniquely binds to a decoy receptor, IL-13Rα2 (Figure 8a), which lacks a well-characterized signaling function (184) while exhibiting one of the highest affinities reported for a cytokine–receptor interaction (on the order of femtomolar) (57). Thus, IL-13Rα2 is a very efficient IL-13 sink that blunts the agonistic actions of IL-13 in vivo. The restricted IL-13Rα2 expression pattern has made this receptor an attractive target for protein engineering. Several tumor types, including glioblastoma multiforme (GBM), express high levels of this decoy receptor (185, 186), and engineering efforts have focused on designing therapies that would specifically target toxins to the tumor via IL-13Rα2 (186). One approach linked the IL-13 cytokine to Pseudomonas exotoxin (PE) to specifically deliver the toxin to IL-13Rα2-positive GBM cells (187). Unfortunately, this therapy failed in clinical studies, as most patients experienced dose-related toxic side effects (186, 188) resulting from binding of the IL-13-PE construct to IL-13 type II complex receptors expressed in the healthy brain tissue surrounding the tumor (189, 190).
In a second-generation design of the IL-13-PE construct, the authors focused on reducing the toxicity associated with this therapy by introducing mutations that selectively disrupted the IL-13/IL-4Rα interaction to prevent formation of the type II complex (191). An E12K mutation on IL-13 resulted in decreased binding affinity for IL-4Rα and concomitant reduction of IL-13 agonist activity through the type II receptor (191). However, when the IL-13 E12K mutant was linked to PE toxin, it induced similar levels of toxicity to the wild-type cytokine (192), suggesting that binding to IL-13Rα1 is the main driver of toxicity associated with the IL-13-PE therapy. Engineering of IL-13 variants that would specifically bind with high affinity to IL-13Rα2 and with reduced affinity to IL-13Rα1 could potentially reduce the toxicity of this therapy.
The crystal structure of the IL-13/IL-13Rα2 binary complex (57) together with the structures for the IL-4 and IL-13 type I and type II complexes (36) provide a complete set of structures to guide the engineering of IL-13Rα2-specific variants of IL-13 (Figure 8a). Comparison of the IL-13/IL-13Rα1 and IL-13/IL-13Rα2 binding interfaces reveals a highly cross-reactive interface on the cytokine, with IL-13 residues K104, K105, F107, and R108 on the IL-13 D helix mediating most of the contacts with both receptor chains (Figure 8c). This presents a major challenge for the generation of receptor-specific IL-13 mutants (36, 57). In a structure-guided Ala scan, most of the interface mutations introduced on IL-13 led to a parallel decrease in the IL-13 binding affinity for IL-13Rα1 and IL-13Rα2 (57). However, one mutation (K105A) led to a more pronounced decrease in IL-13 affinity for IL-13Rα2 (>70-fold) than for IL-13Rα1 (3-fold) (Figure 9d) (57). The IL-13/IL-13Rα2 crystal structure shows that K105 on IL-13 forms critical contacts with Y207 on IL-13Rα2, but it does not interact with any residue on IL-13Rα1 (Figure 9c) (36, 57). This structural analysis is consistent with previous data demonstrating that mutation of K105 to R on IL-13 increased the binding affinity of IL-13 for IL-13Rα2 (193). These results imply that receptor-specific variants could be engineered by remodeling the IL-13 receptor binding interface, although a more combinatorial approach in tandem with structure-guided library design will be necessary to achieve fine receptor specificity.
CONCLUSIONS AND FUTURE PERSPECTIVES
Engineered examples of IFN, IL-2, and IL-4 cytokines could have therapeutic utility in many facets of immune regulation. It remains to be seen how engineered cytokines will fare in the treatment of immune-related diseases, as significant questions remain about potential immunogenicity and pharmacokinetics. Nevertheless, our ability to tune these molecules to evoke biased or novel functional outcomes has deepened our understanding of immune signaling and, coupled with continued new structural insights into these systems, further empowers us to design highly specific immunotherapeutics.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to acknowledge Eric Smith for his assistance with graphic design in Figures 1 and 2. This work was supported by NIH-R01-AI51321 (to K.C.G.), the Mathers Fund, and the Ludwig Foundation. K.C.G. is an investigator of the Howard Hughes Medical Institute. J.B.S. is the recipient of a Leukemia & Lymphoma Society Career Development Program fellowship. J.L.M. is supported by NIH-K01CA175127.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Watowich SS, Hilton DJ, Lodish HF. Activation and inhibition of erythropoietin receptor function: role of receptor dimerization. Mol. Cell. Biol. 1994;14:3535–49. doi: 10.1128/mcb.14.6.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu. Rev. Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stroud RM, Wells JA. Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci. STKE. 20042004:re7. doi: 10.1126/stke.2312004re7. [DOI] [PubMed] [Google Scholar]
- 4.Kossiakoff AA, De Vos AM. Structural basis for cytokine hormone-receptor recognition and receptor activation. Adv. Protein Chem. 1998;52:67–108. doi: 10.1016/s0065-3233(08)60433-7. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber G, Walter MR. Cytokine-receptor interactions as drug targets. Curr. Opin. Chem. Biol. 2010;14:511–19. doi: 10.1016/j.cbpa.2010.06.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broughton SE, Hercus TR, Lopez AF, Parker MW. Cytokine receptor activation at the cell surface. Curr. Opin. Struct. Biol. 2012;22:350–59. doi: 10.1016/j.sbi.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Boulanger MJ, Garcia KC. Shared cytokine signaling receptors: structural insights from the gp130 system. Adv. Protein Chem. 2004;68:107–46. doi: 10.1016/S0065-3233(04)68004-1. [DOI] [PubMed] [Google Scholar]
- 8.Bazan JF. Haemopoietic receptors and helical cytokines. Immunol. Today. 1990;11:350–54. doi: 10.1016/0167-5699(90)90139-z. [DOI] [PubMed] [Google Scholar]
- 9.Bazan JF. A novel family of growth factor receptors: a common binding domain in the growth hormone, prolactin, erythropoietin and IL-6 receptors, and the p75 IL-2 receptor β-chain. Biochem. Biophys. Res. Commun. 1989;164:788–95. doi: 10.1016/0006-291x(89)91528-3. [DOI] [PubMed] [Google Scholar]
- 10.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by γc family cytokines. Nat. Rev. Immunol. 2009;9:480–90. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–50. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ihle JN, Witthuhn BA, Quelle FW, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu. Rev. Immunol. 1995;13:369–98. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 13.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 14.Murray PJ. The JAK-STAT signaling pathway: input and output integration. J. Immunol. 2007;178:2623–29. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 15.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 16.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–72. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282:20059–63. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 18.Malek TR. The biology of interleukin-2. Annu. Rev. Immunol. 2008;26:453–79. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 19.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broughton SE, Dhagat U, Hercus TR, Nero TL, Grimbaldeston MA, et al. The GM-CSF/IL-3/IL-5 cytokine receptor family: from ligand recognition to initiation of signaling. Immunol. Rev. 2012;250:277–302. doi: 10.1111/j.1600-065X.2012.01164.x. [DOI] [PubMed] [Google Scholar]
- 21.Kallen KJ, Grotzinger J, Rose-John S. New perspectives on the design of cytokines and growth factors. Trends Biotechnol. 2000;18:455–61. doi: 10.1016/s0167-7799(00)01492-x. [DOI] [PubMed] [Google Scholar]
- 22.Wells JA, de Vos AM. Hematopoietic receptor complexes. Annu. Rev. Biochem. 1996;65:609–34. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- 23.Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, et al. Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature. 1998;395:511–16. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 24.Watowich SS, Wu H, Socolovsky M, Klingmuller U, Constantinescu SN, Lodish HF. Cytokine receptor signal transduction and the control of hematopoietic cell development. Annu. Rev. Cell Dev. Biol. 1996;12:91–128. doi: 10.1146/annurev.cellbio.12.1.91. [DOI] [PubMed] [Google Scholar]
- 25.Cunningham BC, Ultsch M, De Vos AM, Mulkerrin MG, Clauser KR, Wells JA. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science. 1991;254:821–25. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 26.Constantinescu SN, Keren T, Socolovsky M, Nam H, Henis YI, Lodish HF. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. PNAS. 2001;98:4379–84. doi: 10.1073/pnas.081069198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–90. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- 28.Brooks AJ, Dai W, O’Mara ML, Abankwa D, Chhabra Y, et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014;344:1249783. doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- 29.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozaki K, Leonard WJ. Cytokine and cytokine receptor pleiotropy and redundancy. J. Biol. Chem. 2002;277:29355–58. doi: 10.1074/jbc.R200003200. [DOI] [PubMed] [Google Scholar]
- 31.Hercus TR, Dhagat U, Kan WL, Broughton SE, Nero TL, et al. Signalling by the βc family of cytokines. Cytokine Growth Factor Rev. 2013;24:189–201. doi: 10.1016/j.cytogfr.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 32.de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–12. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 33.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 34.Rickert M, Boulanger MJ, Goriatcheva N, Garcia KC. Compensatory energetic mechanisms mediating the assembly of signaling complexes between interleukin-2 and its α, β, and γc receptors. J. Mol. Biol. 2004;339:1115–28. doi: 10.1016/j.jmb.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 35.Letzelter F, Wang Y, Sebald W. The interleukin-4 site-2 epitope determining binding of the common receptor gamma chain. Eur. J. Biochem. 1998;257:11–20. doi: 10.1046/j.1432-1327.1998.2570011.x. [DOI] [PubMed] [Google Scholar]
- 36.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–72. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ring AM, Lin JX, Feng D, Mitra S, Rickert M, et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat. Immunol. 2012;13:1187–95. doi: 10.1038/ni.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roisman LC, Jaitin DA, Baker DP, Schreiber G. Mutational analysis of the IFNAR1 binding site on IFNα2 reveals the architecture of a weak ligand-receptor binding-site. J. Mol. Biol. 2005;353:271–81. doi: 10.1016/j.jmb.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 39.Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J. Biol. Chem. 2008;283:32925–36. doi: 10.1074/jbc.M806019200. [DOI] [PubMed] [Google Scholar]
- 40.Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146:621–32. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruse N, Tony HP, Sebald W. Conversion of human interleukin-4 into a high affinity antagonist by a single amino acid replacement. EMBO J. 1992;11:3237–44. doi: 10.1002/j.1460-2075.1992.tb05401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraich M, Klein M, Patino E, Harrer H, Nickel J, et al. A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor. BMC Biol. 2006;4:13. doi: 10.1186/1741-7007-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuh G, Cunningham BC, Fukunaga R, Nagata S, Goeddel DV, Wells JA. Rational design of potent antagonists to the human growth hormone receptor. Science. 1992;256:1677–80. doi: 10.1126/science.256.5064.1677. [DOI] [PubMed] [Google Scholar]
- 44.Lowman HB, Wells JA. Affinity maturation of human growth hormone by monovalent phage display. J. Mol. Biol. 1993;234:564–78. doi: 10.1006/jmbi.1993.1612. [DOI] [PubMed] [Google Scholar]
- 45.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 α-receptor/gp130 complex. Science. 2003;300:2101–4. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 46.Chow D, He X, Snow AL, Rose-John S, Garcia KC. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–55. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]
- 47.Bravo J, Heath JK. Receptor recognition by gp130 cytokines. EMBO J. 2000;19:2399–411. doi: 10.1093/emboj/19.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matadeen R, Hon WC, Heath JK, Jones EY, Fuller S. The dynamics of signal triggering in a gp130-receptor complex. Structure. 2007;15:441–48. doi: 10.1016/j.str.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carr PD, Gustin SE, Church AP, Murphy JM, Ford SC, et al. Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell. 2001;104:291–300. doi: 10.1016/s0092-8674(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 50.Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, et al. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. PNAS. 2006;103:3135–40. doi: 10.1073/pnas.0511264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boulanger MJ, Chow D, Brevnova E, Martick M, Sandford G, et al. Molecular mechanisms for viral mimicry of a human cytokine: activation of gp130 by HHV-8 interleukin-6. J. Mol. Biol. 2004;335:641–54. doi: 10.1016/j.jmb.2003.10.070. [DOI] [PubMed] [Google Scholar]
- 52.Beyer BM, Ingram R, Ramanathan L, Reichert P, Le HV, et al. Crystal structures of the proinflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody. J. Mol. Biol. 2008;382:942–55. doi: 10.1016/j.jmb.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 53.Lupardus PJ, Garcia KC. The structure of interleukin-23 reveals the molecular basis of p40 subunit sharing with interleukin-12. J. Mol. Biol. 2008;382:931–41. doi: 10.1016/j.jmb.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fairlie WD, Uboldi AD, McCoubrie JE, Wang CC, Lee EF, et al. Affinity maturation of leukemia inhibitory factor and conversion to potent antagonists of signaling. J. Biol. Chem. 2003;279:2125–34. doi: 10.1074/jbc.M310103200. [DOI] [PubMed] [Google Scholar]
- 55.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134:496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 56.Patino E, Kotzsch A, Saremba S, Nickel J, Schmitz W, et al. Structure analysis of the IL-5 ligand-receptor complex reveals a wrench-like architecture for IL-5Rα. Structure. 2011;19:1864–75. doi: 10.1016/j.str.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 57.Lupardus PJ, Birnbaum ME, Garcia KC. Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Rα2. Structure. 2010;18:332–42. doi: 10.1016/j.str.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chirifu M, Hayashi C, Nakamura T, Toma S, Shuto T, et al. Crystal structure of the IL-15-IL-15Rα complex, a cytokine-receptor unit presented in trans. Nat. Immunol. 2007;8:1001–7. doi: 10.1038/ni1492. [DOI] [PubMed] [Google Scholar]
- 59.Jones BC, Logsdon NJ, Walter MR. Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure. 2008;16:1333–44. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- 61.Walter MR, Windsor WT, Nagabhushan TL, Lundell DJ, Lunn CA, et al. Crystal structure of a complex between interferon-γ and its soluble high-affinity receptor. Nature. 1995;376:230–35. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 62.Nicola NA, Hilton DJ. General classes and functions of four-helix bundle cytokines. Adv. Protein Chem. 1998;52:1–65. doi: 10.1016/s0065-3233(08)60432-5. [DOI] [PubMed] [Google Scholar]
- 63.Domingues H, Cregut D, Sebald W, Oschkinat H, Serrano L. Rational design of a GCN4-derived mimetic of interleukin-4. Nat. Struct. Biol. 1999;6:652–56. doi: 10.1038/10706. [DOI] [PubMed] [Google Scholar]
- 64.Laporte SL, Forsyth CM, Cunningham BC, Miercke LJ, Akhavan D, Stroud RM. De novo design of an IL-4 antagonist and its structure at 1.9 Å. PNAS. 2005;102:1889–94. doi: 10.1073/pnas.0408890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sprang SR, Bazan JF. Cytokine structural taxonomy and mechanisms of receptor engagement. Curr. Opin. Struct. Biol. 1993;3:815–27. [Google Scholar]
- 66.Bazan JF. Emerging families of cytokines and receptors. Curr. Biol. 1993;3:603–6. doi: 10.1016/0960-9822(93)90009-d. [DOI] [PubMed] [Google Scholar]
- 67.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. PNAS. 1990;87:6934–38. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cunningham BC, Henner DJ, Wells JA. Engineering human prolactin to bind to the human growth hormone receptor. Science. 1990;247:1461–65. doi: 10.1126/science.247.4949.1461. [DOI] [PubMed] [Google Scholar]
- 69.Cunningham BC, Bass S, Fuh G, Wells JA. Zinc mediation of the binding of human growth hormone to the human prolactin receptor. Science. 1990;250:1709–12. doi: 10.1126/science.2270485. [DOI] [PubMed] [Google Scholar]
- 70.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–85. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 71.Cunningham BC, Jhurani P, Ng P, Wells JA. Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis. Science. 1989;243:1330–36. doi: 10.1126/science.2466339. [DOI] [PubMed] [Google Scholar]
- 72.Cunningham BC, Wells JA. Rational design of receptor-specific variants of human growth hormone. PNAS. 1991;88:3407–11. doi: 10.1073/pnas.88.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frisch C, Schreiber G, Johnson CM, Fersht AR. Thermodynamics of the interaction of barnase and barstar: changes in free energy versus changes in enthalpy on mutation. J. Mol. Biol. 1997;267:696–706. doi: 10.1006/jmbi.1997.0892. [DOI] [PubMed] [Google Scholar]
- 74.Pal G, Ultsch MH, Clark KP, Currell B, Kossiakoff AA, Sidhu SS. Intramolecular cooperativity in a protein binding site assessed by combinatorial shotgun scanning mutagenesis. J. Mol. Biol. 2005;347:489–94. doi: 10.1016/j.jmb.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 75.Cassell DJ, Choudhri S, Humphrey R, Martell RE, Reynolds T, Shanafelt AB. Therapeutic enhancement of IL-2 through molecular design. Curr. Pharm. Des. 2002;8:2171–83. doi: 10.2174/1381612023393260. [DOI] [PubMed] [Google Scholar]
- 76.Shanafelt AB, Lin Y, Shanafelt MC, Forte CP, Dubois-Stringfellow N, et al. A T-cell-selective interleukin 2 mutein exhibits potent antitumor activity and is well tolerated in vivo. Nat. Biotechnol. 2000;18:1197–202. doi: 10.1038/81199. [DOI] [PubMed] [Google Scholar]
- 77.Kang L, Bondensgaard K, Li T, Hartmann R, Hjorth SA. Rational design of interleukin-21 antagonist through selective elimination of the γC binding epitope. J. Biol. Chem. 2010;285:12223–31. doi: 10.1074/jbc.M110.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kallen KJ, Grotzinger J, Lelievre E, Vollmer P, Aasland D, et al. Receptor recognition sites of cytokines are organized as exchangeable modules. Transfer of the leukemia inhibitory factor receptor-binding site from ciliary neurotrophic factor to interleukin-6. J. Biol. Chem. 1999;274:11859–67. doi: 10.1074/jbc.274.17.11859. [DOI] [PubMed] [Google Scholar]
- 79.Brideau-Andersen AD, Huang X, Sun SC, Chen TT, Stark D, et al. Directed evolution of gene-shuffled IFN-α molecules with activity profiles tailored for treatment of chronic viral diseases. PNAS. 2007;104:8269–74. doi: 10.1073/pnas.0609001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rao BM, Girvin AT, Ciardelli T, Lauffenburger DA, Wittrup KD. Interleukin-2 mutants with enhanced α-receptor subunit binding affinity. Protein Eng. 2003;16:1081–87. doi: 10.1093/protein/gzg111. [DOI] [PubMed] [Google Scholar]
- 81.Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, et al. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine.’. Nature. 2012;484:529–33. doi: 10.1038/nature10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–86. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 83.Zhang JL, Simeonowa I, Wang Y, Sebald W. The high-affinity interaction of human IL-4 and the receptor α chain is constituted by two independent binding clusters. J. Mol. Biol. 2002;315:399–407. doi: 10.1006/jmbi.2001.5243. [DOI] [PubMed] [Google Scholar]
- 84.Junttila IS, Creusot RJ, Moraga I, Bates DL, Wong MT, et al. Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines. Nat. Chem. Biol. 2012;8:990–98. doi: 10.1038/nchembio.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kalie E, Jaitin DA, Abramovich R, Schreiber G. An interferon α2 mutant optimized by phage display for IFNAR1 binding confers specifically enhanced antitumor activities. J. Biol. Chem. 2007;282:11602–11. doi: 10.1074/jbc.M610115200. [DOI] [PubMed] [Google Scholar]
- 86.Zurawski SM, Imler JL, Zurawski G. Partial agonist/antagonist mouse interleukin-2 proteins indicate that a third component of the receptor complex functions in signal transduction. EMBO J. 1990;9:3899–905. doi: 10.1002/j.1460-2075.1990.tb07610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zurawski SM, Vega F, Jr, Huyghe B, Zurawski G. Receptors for interleukin-13 and interleukin-4 are complex and share a novel component that functions in signal transduction. EMBO J. 1993;12:2663–70. doi: 10.1002/j.1460-2075.1993.tb05927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levin D, Schneider WM, Hoffmann HH, Yarden G, Busetto AG, et al. Multifaceted activities of type I interferon are revealed by a receptor antagonist. Sci. Signal. 2014;7:ra50. doi: 10.1126/scisignal.2004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol. Cell. 2003;12:577–89. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 90.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–68. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 91.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–57. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 92.Levin AM, Weiss GA. Optimizing the affinity and specificity of proteins with molecular display. Mol. Biosyst. 2006;2:49–57. doi: 10.1039/b511782h. [DOI] [PubMed] [Google Scholar]
- 93.Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 2012;250:317–34. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012;12:125–35. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang BX, Rahbar R, Fish EN. Interferon: current status and future prospects in cancer therapy. J. Interferon Cytokine Res. 2011;31:545–52. doi: 10.1089/jir.2010.0158. [DOI] [PubMed] [Google Scholar]
- 97.Moraga I, Harari D, Schreiber G, Uze G, Pellegrini S. Receptor density is key to the α2/β interferon differential activities. Mol. Cell. Biol. 2009;29:4778–87. doi: 10.1128/MCB.01808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, et al. Binding and activity of all human α interferon subtypes. Cytokine. 2011;56:282–89. doi: 10.1016/j.cyto.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 99.Jaks E, Gavutis M, Uze G, Martal J, Piehler J. Differential receptor subunit affinities of type I interferons govern differential signal activation. J. Mol. Biol. 2007;366:525–39. doi: 10.1016/j.jmb.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 100.Subramaniam PS, Khan SA, Pontzer CH, Johnson HM. Differential recognition of the type I interferon receptor by interferons τ and α is responsible for their disparate cytotoxicities. PNAS. 1995;92:12270–74. doi: 10.1073/pnas.92.26.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, et al. Inquiring into the differential action of interferons (IFNs): an IFN-α2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-β. Mol. Cell. Biol. 2006;26:1888–97. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schiller JH, Willson JK, Bittner G, Wolberg WH, Hawkins MJ, Borden EC. Antiproliferative effects of interferons on human melanoma cells in the human tumor colony-forming assay. J. Interferon Res. 1986;6:615–25. doi: 10.1089/jir.1986.6.615. [DOI] [PubMed] [Google Scholar]
- 103.de Weerd NA, Vivian JP, Nguyen TK, Mangan NE, Gould JA, et al. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 2013;14:901–7. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- 104.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–85. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khabar KS, Al-Haj L, Al-Zoghaibi F, Marie M, Dhalla M, et al. Expressed gene clusters associated with cellular sensitivity and resistance towards anti-viral and anti-proliferative actions of interferon. J. Mol. Biol. 2004;342:833–46. doi: 10.1016/j.jmb.2004.07.065. [DOI] [PubMed] [Google Scholar]
- 106.Bolen CR, Ding S, Robek MD, Kleinstein SH. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–72. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.da Silva AJ, Brickelmaier M, Majeau GR, Lukashin AV, Peyman J, et al. Comparison of gene expression patterns induced by treatment of human umbilical vein endothelial cells with IFN-α2b versus IFN-β1a: understanding the functional relationship between distinct type I interferons that act through a common receptor. J. Interferon Cytokine Res. 2002;22:173–88. doi: 10.1089/107999002753536149. [DOI] [PubMed] [Google Scholar]
- 109.Moraga I, Spangler J, Mendoza JL, Garcia KC. Multifarious determinants of cytokine receptor signaling specificity. Adv. Immunol. 2014;121:1–39. doi: 10.1016/B978-0-12-800100-4.00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. PNAS. 1994;91:10747–51. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–91. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 112.Chang CC, Chen TT, Cox BW, Dawes GN, Stemmer WP, et al. Evolution of a cytokine using DNA family shuffling. Nat. Biotechnol. 1999;17:793–97. doi: 10.1038/11737. [DOI] [PubMed] [Google Scholar]
- 113.Bekisz J, Baron S, Balinsky C, Morrow A, Zoon KC. Antiproliferative properties of type I and type II interferon. Pharmaceuticals. 2010;3:994–1015. doi: 10.3390/ph3040994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grimley PM, Fang H, Rui H, Petricoin EF, 3rd, Ray S, et al. Prolonged STAT1 activation related to the growth arrest of malignant lymphoma cells by interferon-α. Blood. 1998;91:3017–27. [PubMed] [Google Scholar]
- 115.Silverman RH, Watling D, Balkwill FR, Trowsdale J, Kerr IM. The ppp(A2′p)nA and protein kinase systems in wild-type and interferon-resistant Daudi cells. Eur. J. Biochem. 1982;126:333–41. doi: 10.1111/j.1432-1033.1982.tb06783.x. [DOI] [PubMed] [Google Scholar]
- 116.Pereira AA, Jacobson IM. New and experimental therapies for HCV. Nat. Rev. Gastroenterol. Hepatol. 2009;6:403–11. doi: 10.1038/nrgastro.2009.92. [DOI] [PubMed] [Google Scholar]
- 117.Pan M, Kalie E, Scaglione BJ, Raveche ES, Schreiber G, Langer JA. Mutation of the IFNAR-1 receptor binding site of human IFN-α2 generates type I IFN competitive antagonists. Biochemistry. 2008;47:12018–27. doi: 10.1021/bi801588g. [DOI] [PubMed] [Google Scholar]
- 118.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 119.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012;12:180–90. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 122.Klapper JA, Downey SG, Smith FO, Yang JC, Hughes MS, et al. High-dose interleukin-2 for the treatment of metastatic renal cell carcinoma: a retrospective analysis of response and survival in patients treated in the surgery branch at the National Cancer Institute between 1986 and 2006. Cancer. 2008;113:293–301. doi: 10.1002/cncr.23552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smith FO, Downey SG, Klapper JA, Yang JC, Sherry RM, et al. Treatment of metastatic melanoma using interleukin-2 alone or in conjunction with vaccines. Clin. Cancer Res. 2008;14:5610–18. doi: 10.1158/1078-0432.CCR-08-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pellegrini M, Mak TW, Ohashi PS. Fighting cancers from within: augmenting tumor immunity with cytokine therapy. Trends Pharmacol. Sci. 2010;31:356–63. doi: 10.1016/j.tips.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 125.Boyman O, Surh CD, Sprent J. Potential use of IL-2/anti-IL-2 antibody immune complexes for the treatment of cancer and autoimmune disease. Expert Opin. Biol. Ther. 2006;6:1323–31. doi: 10.1517/14712598.6.12.1323. [DOI] [PubMed] [Google Scholar]
- 126.McDermott DF, Atkins MB. Application of IL-2 and other cytokines in renal cancer. Expert Opin. Biol. Ther. 2004;4:455–68. doi: 10.1517/14712598.4.4.455. [DOI] [PubMed] [Google Scholar]
- 127.Rosenberg SA, Yang JC, White DE, Steinberg SM. Durability of complete responses in patients with metastatic cancer treated with high-dose interleukin-2: identification of the antigens mediating response. Ann. Surg. 1998;228:307–19. doi: 10.1097/00000658-199809000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rosenberg SA. Raising the bar: the curative potential of human cancer immunotherapy. Sci. Transl. Med. 2012;4:127. doi: 10.1126/scitranslmed.3003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349–66. doi: 10.1016/j.cytogfr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 130.Taniguchi T, Minami Y. The IL-2/IL-2 receptor system: a current overview. Cell. 1993;73:5–8. doi: 10.1016/0092-8674(93)90152-g. [DOI] [PubMed] [Google Scholar]
- 131.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nat. Rev. Immunol. 2004;4:665–74. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 133.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its α, β, and γc receptors. Science. 2005;310:1159–63. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 134.Stauber DJ, Debler EW, Horton PA, Smith KA, Wilson IA. Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. PNAS. 2006;103:2788–93. doi: 10.1073/pnas.0511161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rao BM, Driver I, Lauffenburger DA, Wittrup KD. High-affinity CD25-binding IL-2 mutants potently stimulate persistent T cell growth. Biochemistry. 2005;44:10696–701. doi: 10.1021/bi050436x. [DOI] [PubMed] [Google Scholar]
- 136.Liu DV, Maier LM, Hafler DA, Wittrup KD. Engineered interleukin-2 antagonists for the inhibition of regulatory T cells. J. Immunother. 2009;32:887–94. doi: 10.1097/CJI.0b013e3181b528da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carmenate T, Pacios A, Enamorado M, Moreno E, Garcia-Martinez K, et al. Human IL-2 mutein with higher antitumor efficacy than wild type IL-2. J. Immunol. 2013;190:6230–38. doi: 10.4049/jimmunol.1201895. [DOI] [PubMed] [Google Scholar]
- 138.Zurawski SM, Zurawski G. Receptor antagonist and selective agonist derivatives of mouse interleukin-2. EMBO J. 1992;11:3905–10. doi: 10.1002/j.1460-2075.1992.tb05483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Margolin K, Atkins MB, Dutcher JP, Ernstoff MS, Smith JW, 2nd, et al. Phase I trial of BAY 50-4798, an interleukin-2-specific agonist in advanced melanoma and renal cancer. Clin. Cancer Res. 2007;13:3312–19. doi: 10.1158/1078-0432.CCR-06-1341. [DOI] [PubMed] [Google Scholar]
- 140.Davey RT, Pertel PE, Benson A, Cassell DJ, Gazzard BG, et al. Safety, tolerability, pharmacokinetics, and efficacy of an interleukin-2 agonist among HIV-infected patients receiving highly active antiretroviral therapy. J. Interferon Cytokine Res. 2008;28:89–100. doi: 10.1089/jir.2007.0064. [DOI] [PubMed] [Google Scholar]
- 141.Matthews L, Chapman S, Ramchandani MS, Lane HC, Davey RT, Jr, Sereti I. BAY 50-4798, a novel, high-affinity receptor-specific recombinant interleukin-2 analog, induces dose-dependent increases in CD25 expression and proliferation among unstimulated, human peripheral blood mononuclear cells in vitro. Clin. Immunol. 2004;113:248–55. doi: 10.1016/j.clim.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 142.Krieg C, Letourneau S, Pantaleo G, Boyman O. Improved IL-2 immunotherapy by selective stimulation of IL-2 receptors on lymphocytes and endothelial cells. PNAS. 2010;107:11906–11. doi: 10.1073/pnas.1002569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gillessen S, Gnad-Vogt US, Gallerani E, Beck J, Sessa C, et al. A phase I dose-escalation study of the immunocytokine EMD 521873 (Selectikine) in patients with advanced solid tumours. Eur. J. Cancer. 2013;49:35–44. doi: 10.1016/j.ejca.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 144.Laurent J, Touvrey C, Gillessen S, Joffraud M, Vicari M, et al. T-cell activation by treatment of cancer patients with EMD 521873 (Selectikine), an IL-2/anti-DNA fusion protein. J. Transl. Med. 2013;11:5. doi: 10.1186/1479-5876-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hemar A, Subtil A, Lieb M, Morelon E, Hellio R, Dautry-Varsat A. Endocytosis of interleukin 2 receptors in human T lymphocytes: distinct intracellular localization and fate of the receptor α, β, and γ chains. J. Cell Biol. 1995;129:55–64. doi: 10.1083/jcb.129.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gesbert F, Sauvonnet N, Dautry-Varsat A. Clathrin-independent endocytosis and signalling of interleukin 2 receptors IL-2R endocytosis and signalling. Curr. Top. Microbiol. Immunol. 2004;286:119–48. [PubMed] [Google Scholar]
- 147.Berndt WG, Chang DZ, Smith KA, Ciardelli TL. Mutagenic analysis of a receptor contact site on interleukin-2: preparation of an IL-2 analog with increased potency. Biochemistry. 1994;33:6571–77. doi: 10.1021/bi00187a026. [DOI] [PubMed] [Google Scholar]
- 148.Fallon EM, Liparoto SF, Lee KJ, Ciardelli TL, Lauffenburger DA. Increased endosomal sorting of ligand to recycling enhances potency of an interleukin-2 analog. J. Biol. Chem. 2000;275:6790–97. doi: 10.1074/jbc.275.10.6790. [DOI] [PubMed] [Google Scholar]
- 149.Imler JL, Zurawski G. Receptor binding and internalization of mouse interleukin-2 derivatives that are partial agonists. J. Biol. Chem. 1992;267:13185–90. [PubMed] [Google Scholar]
- 150.Chang DZ, Wu Z, Ciardelli TL. A point mutation in interleukin-2 that alters ligand internalization. J. Biol. Chem. 1996;271:13349–55. doi: 10.1074/jbc.271.23.13349. [DOI] [PubMed] [Google Scholar]
- 151.Chang DZ, Tasayco ML, Ciardelli TL. Structural analogs of interleukin-2: a point mutation that facilitates biological response. Mol. Pharmacol. 1995;47:206–11. [PubMed] [Google Scholar]
- 152.Braisted AC, Oslob JD, Delano WL, Hyde J, McDowell RS, et al. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. J. Am. Chem. Soc. 2003;125:3714–15. doi: 10.1021/ja034247i. [DOI] [PubMed] [Google Scholar]
- 153.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. PNAS. 2006;103:15422–27. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–27. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 155.Verdeil G, Marquardt K, Surh CD, Sherman LA. Adjuvants targeting innate and adaptive immunity synergize to enhance tumor immunotherapy. PNAS. 2008;105:16683–88. doi: 10.1073/pnas.0805054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jin GH, Hirano T, Murakami M. Combination treatment with IL-2 and anti-IL-2 mAbs reduces tumor metastasis via NK cell activation. Int. Immunol. 2008;20:783–89. doi: 10.1093/intimm/dxn036. [DOI] [PubMed] [Google Scholar]
- 157.Tomala J, Kovarova J, Kabesova M, Votavova P, Chmelova H, et al. Chimera of IL-2 linked to light chain of anti-IL-2 mAb mimics IL-2/anti-IL-2 mAb complexes both structurally and functionally. ACS Chem. Biol. 2013;8:871–76. doi: 10.1021/cb3007242. [DOI] [PubMed] [Google Scholar]
- 158.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 159.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97:14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 160.Castro I, Yu A, Dee MJ, Malek TR. The basis of distinctive IL-2- and IL-15-dependent signaling: weak CD122-dependent signaling favors CD8+ T central-memory cell survival but not T effector-memory cell development. J. Immunol. 2011;187:5170–82. doi: 10.4049/jimmunol.1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 2005;6:1142–51. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 162.Maloy KJ, Powrie F. Fueling regulation: IL-2 keeps CD4+ Treg cells fit. Nat. Immunol. 2005;6:1071–72. doi: 10.1038/ni1105-1071. [DOI] [PubMed] [Google Scholar]
- 163.Zhang X, Sun S, Hwang I, Tough DF, Sprent J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8:591–99. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 164.Schluns KS, Klonowski KD, Lefrancois L. Transregulation of memory CD8 T-cell proliferation by IL-15Rα+ bone marrow-derived cells. Blood. 2004;103:988–94. doi: 10.1182/blood-2003-08-2814. [DOI] [PubMed] [Google Scholar]
- 165.Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, et al. Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J. Exp. Med. 2002;195:1541–48. doi: 10.1084/jem.20020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pettit DK, Bonnert TP, Eisenman J, Srinivasan S, Paxton R, et al. Structure-function studies of interleukin 15 using site-specific mutagenesis, polyethylene glycol conjugation, and homology modeling. J. Biol. Chem. 1997;272:2312–18. doi: 10.1074/jbc.272.4.2312. [DOI] [PubMed] [Google Scholar]
- 167.Ferrari-Lacraz S, Zheng XX, Kim YS, Li Y, Maslinski W, et al. An antagonist IL-15/Fc protein prevents costimulation blockade-resistant rejection. J. Immunol. 2001;167:3478–85. doi: 10.4049/jimmunol.167.6.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ferrari-Lacraz S, Zanelli E, Neuberg M, Donskoy E, Kim YS, et al. Targeting IL-15 receptor-bearing cells with an antagonist mutant IL-15/Fc protein prevents disease development and progression in murine collagen-induced arthritis. J. Immunol. 2004;173:5818–26. doi: 10.4049/jimmunol.173.9.5818. [DOI] [PubMed] [Google Scholar]
- 169.Mortier E, Quemener A, Vusio P, Lorenzen I, Boublik Y, et al. Soluble interleukin-15 receptor α (IL-15Rα)-sushi as a selective and potent agonist of IL-15 action through IL-15Rβ/γ: hyperagonist IL-15· IL-15Rα fusion proteins. J. Biol. Chem. 2006;281:1612–19. doi: 10.1074/jbc.M508624200. [DOI] [PubMed] [Google Scholar]
- 170.Zhu X, Marcus WD, Xu W, Lee HI, Han K, et al. Novel human interleukin-15 agonists. J. Immunol. 2009;183:3598–607. doi: 10.4049/jimmunol.0901244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Han KP, Zhu X, Liu B, Jeng E, Kong L, et al. IL-15:IL-15 receptor α superagonist complex: high-level co-expression in recombinant mammalian cells, purification and characterization. Cytokine. 2011;56:804–10. doi: 10.1016/j.cyto.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wong HC, Jeng EK, Rhode PR. The IL-15-based superagonist ALT-803 promotes the antigen-independent conversion of memory CD8 T cells into innate-like effector cells with antitumor activity. Oncoimmunology. 2013;2:e26442. doi: 10.4161/onci.26442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Junttila IS, Mizukami K, Dickensheets H, Meier-Schellersheim M, Yamane H, et al. Tuning sensitivity to IL-4 and IL-13: differential expression of IL-4Rα, IL-13Rα1, and γc regulates relative cytokine sensitivity. J. Exp. Med. 2008;205:2595–608. doi: 10.1084/jem.20080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Andrews AL, Holloway JW, Holgate ST, Davies DE. IL-4 receptor α is an important modulator of IL-4 and IL-13 receptor binding: implications for the development of therapeutic targets. J. Immunol. 2006;176:7456–61. doi: 10.4049/jimmunol.176.12.7456. [DOI] [PubMed] [Google Scholar]
- 175.Sosman JA, Fisher SG, Kefer C, Fisher RI, Ellis TM. A phase I trial of continuous infusion interleukin-4 (IL-4) alone and following interleukin-2 (IL-2) in cancer patients. Ann. Oncol. 1994;5:447–52. doi: 10.1093/oxfordjournals.annonc.a058878. [DOI] [PubMed] [Google Scholar]
- 176.Wenzel S, Wilbraham D, Fuller R, Getz EB, Longphre M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet. 2007;370:1422–31. doi: 10.1016/S0140-6736(07)61600-6. [DOI] [PubMed] [Google Scholar]
- 177.Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300:1527–28. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 178.Lutz MB, Schnare M, Menges M, Rossner S, Rollinghoff M, et al. Differential functions of IL-4 receptor types I and II for dendritic cell maturation and IL-12 production and their dependency on GM-CSF. J. Immunol. 2002;169:3574–80. doi: 10.4049/jimmunol.169.7.3574. [DOI] [PubMed] [Google Scholar]
- 179.Shanafelt AB, Forte CP, Kasper JJ, Sanchez-Pescador L, Wetzel M, et al. An immune cell-selective interleukin 4 agonist. PNAS. 1998;95:9454–58. doi: 10.1073/pnas.95.16.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wong MT, Ye JJ, Alonso MN, Landrigan A, Cheung RK, et al. Regulation of human Th9 differentiation by type I interferons and IL-21. Immunol. Cell Biol. 2010;88:624–31. doi: 10.1038/icb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dauer M, Obermaier B, Herten J, Haerle C, Pohl K, et al. Mature dendritic cells derived from human monocytes within 48 hours: a novel strategy for dendritic cell differentiation from blood precursors. J. Immunol. 2003;170:4069–76. doi: 10.4049/jimmunol.170.8.4069. [DOI] [PubMed] [Google Scholar]
- 182.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Curr. Allergy Asthma Rep. 2004;4:123–31. doi: 10.1007/s11882-004-0057-6. [DOI] [PubMed] [Google Scholar]
- 183.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 2010;28:445–89. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Caput D, Laurent P, Kaghad M, Lelias JM, Lefort S, et al. Cloning and characterization of a specific interleukin (IL)-13 binding protein structurally related to the IL-5 receptor α chain. J. Biol. Chem. 1996;271:16921–26. doi: 10.1074/jbc.271.28.16921. [DOI] [PubMed] [Google Scholar]
- 185.Debinski W, Miner R, Leland P, Obiri NI, Puri RK. Receptor for interleukin (IL) 13 does not interact with IL4 but receptor for IL4 interacts with IL13 on human glioma cells. J. Biol. Chem. 1996;271:22428–33. doi: 10.1074/jbc.271.37.22428. [DOI] [PubMed] [Google Scholar]
- 186.Candolfi M, Kroeger KM, Xiong W, Liu C, Puntel M, et al. Targeted toxins for glioblastoma multiforme: pre-clinical studies and clinical implementation. Anticancer Agents Med. Chem. 2011;11:729–38. doi: 10.2174/187152011797378689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Debinski W, Obiri NI, Powers SK, Pastan I, Puri RK. Human glioma cells overexpress receptors for interleukin 13 and are extremely sensitive to a novel chimeric protein composed of interleukin 13 and pseudomonas exotoxin. Clin. Cancer Res. 1995;1:1253–58. [PubMed] [Google Scholar]
- 188.Kunwar S, Prados MD, Chang SM, Berger MS, Lang FF, et al. Direct intracerebral delivery of cintredekin besudotox (IL13-PE38QQR) in recurrent malignant glioma: a report by the Cintredekin Besudotox Intraparenchymal Study Group. J. Clin. Oncol. 2007;25:837–44. doi: 10.1200/JCO.2006.08.1117. [DOI] [PubMed] [Google Scholar]
- 189.Liu H, Jacobs BS, Liu J, Prayson RA, Estes ML, et al. Interleukin-13 sensitivity and receptor phenotypes of human glial cell lines: non-neoplastic glia and low-grade astrocytoma differ from malignant glioma. Cancer Immunol. Immunother. 2000;49:319–24. doi: 10.1007/s002620000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu H, Prayson RA, Estes ML, Drazba JA, Barnett GH, et al. In vivo expression of the interleukin 4 receptor α by astrocytes in epilepsy cerebral cortex. Cytokine. 2000;12:1656–61. doi: 10.1006/cyto.2000.0773. [DOI] [PubMed] [Google Scholar]
- 191.Debinski W, Gibo DM, Obiri NI, Kealiher A, Puri RK. Novel anti-brain tumor cytotoxins specific for cancer cells. Nat. Biotechnol. 1998;16:449–53. doi: 10.1038/nbt0598-449. [DOI] [PubMed] [Google Scholar]
- 192.Candolfi M, Xiong W, Yagiz K, Liu C, Muhammad AK, et al. Gene therapy-mediated delivery of targeted cytotoxins for glioma therapeutics. PNAS. 2010;107:20021–26. doi: 10.1073/pnas.1008261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Madhankumar AB, Mintz A, Debinski W. Interleukin 13 mutants of enhanced avidity toward the glioma-associated receptor, IL13Rα2. Neoplasia. 2004;6:15–22. doi: 10.1016/s1476-5586(04)80049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Skiniotis G, Boulanger MJ, Garcia KC, Walz T. Signaling conformations of the tall cytokine receptor gp130 when in complex with IL-6 and IL-6 receptor. Nat. Struct. Mol. Biol. 2005;12:545–51. doi: 10.1038/nsmb941. [DOI] [PubMed] [Google Scholar]
- 195.Xu Y, Kershaw NJ, Luo CS, Soo P, Pocock MJ, et al. Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J. Biol. Chem. 2010;285:21214–18. doi: 10.1074/jbc.C110.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Huyton T, Zhang JG, Luo CS, Lou MZ, Hilton DJ, et al. An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. PNAS. 2007;104:12737–42. doi: 10.1073/pnas.0705577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol. Cell. 2008;31:737–48. doi: 10.1016/j.molcel.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Walter MR. Structural analysis of IL-10 and type I interferon family members and their complexes with receptor. Adv. Protein Chem. 2004;68:171–223. doi: 10.1016/S0065-3233(04)68006-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.